Abstract
C-Glycosylflavones are ubiquitous in the plant kingdom, and many of them have beneficial effects on human health. They are a special group of flavonoid glycosides in which the sugars are C-linked to the flavone skeleton. It has been long presumed that C-glycosylflavones have a different biosynthetic origin from O-glycosylflavonoids. In rice (Oryza sativa), a C-glucosyltransferase (OsCGT) that accepts 2-hydroxyflavanone substrates and a dehydratase activity that selectively converts C-glucosyl-2-hydroxyflavanones to 6C-glucosylflavones were recently described. In this study, we provide in vitro and in planta evidence that the rice P450 CYP93G2 protein encoded by Os06g01250 is a functional flavanone 2-hydroxylase. CYP93G2 is related to the CYP93B subfamily, which consists of dicot flavone synthase II enzymes. In the presence of NADPH, recombinant CYP93G2 converts naringenin and eriodictyol to the corresponding 2-hydroxyflavanones. In addition, CYP93G2 generates 2-hydroxyflavanones, which are modified by O-glycosylation in transgenic Arabidopsis (Arabidopsis thaliana). Coexpression of CYP93G2 and OsCGT in Arabidopsis resulted in the production of C-glucosyl-2-hydroxyflavanones in the dibenzoylmethane tautomeric form. The same structure was reported previously for the in vitro OsCGT reaction products. Thus, CYP93G2 generates 2-hydroxyflavanone substrates from flavanones for C-glucosylation by OsCGT in planta. Furthermore, knocking down Os06g01250 in rice (O. sativa subsp. japonica 'Zhonghua 11') preferentially depleted the accumulation of C-glycosylapigenin, C-glycosylluteolin, and C-glycosylchrysoeriol but did not affect the levels of tricin, which is frequently present as O-glycosides in cereals. Taken together, our work conclusively assigned CYP93G2 as the first enzyme that channels flavanones to C-glycosylflavone biosynthesis in rice.
Flavonoids are common and widespread secondary metabolites in plants. They are frequently present as C- or O-glycosides. The sugars of C-glycosides are attached directly to the flavonoid skeleton by a C-C bond that is resistant to acid hydrolysis. On the other hand, sugars O-linked at the phenolic hydroxyl groups of flavonoids are acid hydrolyzable. Most flavonoid C-glycosides are flavones that have been found in bryophytes, ferns, gymnosperms, and angiosperms (Harborne, 1993). In cereals such as rice (Oryza sativa), wheat (Triticum aestivum), and maize (Zea mays), C-glycosylflavones are the major class of flavonoids, accumulating with a diverse range of physiological functions. For example, several C-glycosylflavones isolated from rice were found to function as probing stimulants of planthoppers (Besson et al., 1985). In addition, accumulation of C-glycosylflavones was enhanced by UV-B light in a UV-tolerant rice cultivar but absent in a susceptible cultivar, suggesting their possible UV light protective roles (Markham et al., 1998). One of the UV-B light-induced rice flavones, an isovitexin (6C-glucosylapigenin) derivative, was shown to reduce the number of fertile eggs in African bollworms dramatically when included in their artificial diets (Cassi-Lit et al., 2007). In maize, maysin is an O-glycosylated C-glycosylluteolin well known to confer natural resistance in silk tissues toward different lepidopteran larval pests (McMullen et al., 2004). In contrast to the above flavones, tricin (3′,5′-dimethoxylated flavone) is usually detected as O-glycosides in cereals (Cavaliere et al., 2005; Brazier-Hicks et al., 2009). Tricin and its O-glycosides have been shown to function as allelopathic compounds in rice, interfering with weeds and harmful microorganisms in paddy soil (Kong et al., 2004, 2007).
The perceived health benefits of flavonoids have made them attractive targets for metabolic engineering of rice (Shin et al., 2006). Both isovitexin and tricin derivatives have been widely reported as healthy phytochemical constituents in rice hull and bran. For example, isovitexin extracted from rice bran was shown to confer strong antioxidant activities and to prevent reactive oxygen species damage in an in vitro system (Lin et al., 2002) and mouse microphages (Lin et al., 2005). On the other hand, tricin has in vitro antiproliferative activities on breast and colon cancer cell lines at submicromolar concentrations (Hudson et al., 2000). It effectively inhibits cyclooxygenase enzymes and interferes with murine gastrointestinal carcinogenesis (Cai et al., 2005). However, the preferred consumed form, white rice, consists exclusively of endosperm tissue, which is poor in phytochemicals. Understanding the molecular biology of the flavone biosynthesis pathway in rice will provide insights for metabolic engineering of edible tissues that normally do not accumulate these health-beneficial phytochemicals.
Flavonoid biosynthesis is initiated by chalcone synthase, followed by the activity of chalcone isomerase. The resulting flavanones are precursors for different classes of flavonoids, including flavones, flavonols, proanthocyanidins, and anthocyanins. Two distinct flavone synthase (FNS) systems specified by either FNSI or FNSII have been described in a number of dicot species. FNSI is a soluble Fe2+/2-oxoglutarate-dependent dioxygenase (DOX) restricted to members of Apiaceae (Martens and Mithöfer, 2005). FNSI shows high sequence identity to flavanone 3β-hydroxylase, another DOX enzyme that uses the same flavanone substrates. Results from site-directed mutagenesis strongly suggested that the parsley (Petroselinum crispum) FNSI was evolved from functional diversification after flavanone 3β-hydroxylase gene duplication (Gebhardt et al., 2007). On the other hand, FNSII is a P450 enzyme found in other flavone-accumulating dicot families including Leguminosae, Asteraceae, Plantaginaceae, and Lamiaceae (Martens and Mithöfer, 2005). All the dicot FNSII enzymes belong to the CYP93B subfamily. When expressed in heterologous systems, FNSI and most FNSII enzymes converted flavanones directly to flavones through C2,C3-cis-desaturation (Martens and Mithöfer, 2005). Interestingly, CYP93B members from the legumes licorice (Glycyrrhiza echinata) and Medicago truncatula showed flavanone 2-hydroxylation activities in recombinant enzyme assays, and flavones were formed from the reaction products after acid treatments (Akashi et al., 1998; Zhang et al., 2007). The biosynthesis of flavones was largely unknown in monocots until recent years. A recombinant rice DOX protein, designated as OsFNS1-1, was found to convert naringenin directly to apigenin, and it represented the first FNSI reported outside Apiaceae (Lee et al., 2007). On the other hand, we recently described a sorghum (Sorghum bicolor) pathogen-inducible P450 gene that resulted in the accumulation of 2-hydroxyflavanone O-hexosides in transgenic Arabidopsis (Arabidopsis thaliana; Du et al., 2010). The encoded protein is a new member (CYP93G3) belonging to the CYP93G subfamily, which was first annotated in the rice genome (Nelson et al., 2004).
It has long been suggested that C-glycosylation takes place as an early reaction during flavonoid biosynthesis, while O-glycosylation usually occurs at terminal stages. For example, early radioisotope tracer experiments in a few Lemnaceae plants indicated that flavone aglycones can be O-glycosylated but not C-glycosylated (Wallace et al., 1969). By contrast, 14C-labeled flavanones were incorporated into C-glycosylflavones in the same species (Wallace and Grisebach, 1973). More evidence for isovitexin and vitexin formation was later obtained with buckwheat (Fagopyrum esculentum) enzyme preparations. 2-Hydroxyflavanones, not flavanones or flavones, were identified to be substrates for C-glucosylation (Kerscher and Franz, 1987, 1988). Consistent with this, the rice C-glucosyltransferase OsCGT was recently demonstrated to catalyze the UDP-Glc-dependent C-glucosylation of 2-hydroxyflavanone substrates (Brazier-Hicks et al., 2009). In addition, dehydratase activities that preferentially converted 2-hydroxyflavanone C-glucosides to 6C-glucosylflavones were demonstrated in both rice and wheat extracts (Brazier-Hicks et al., 2009). Apparently, enzyme activities that generate 2-hydroxyflavanones are necessary for channeling flavanones to C-glycosylflavone production in rice. In this study, we have filled the remaining gap in this biosynthetic route with the characterization of the rice P450 CYP93G2 protein. Flavanone 2-hydroxylation activities of the rice enzyme were established by recombinant enzyme assays and transgenic Arabidopsis analysis. In addition, Arabidopsis plants overexpressing both CYP93G2 and OsCGT were demonstrated to accumulate C-glucosyl-2-hydroxyflavanones. Furthermore, metabolite profiling experiments revealed the depletion of C-glycosylflavone accumulation in the CYP93G2 T-DNA insertion rice mutant.
RESULTS
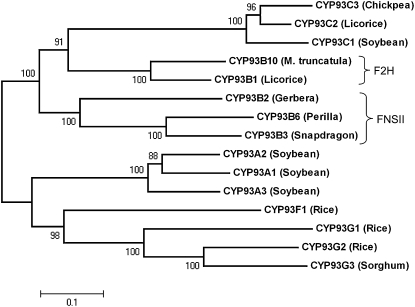
Rice CYP93G2 Is a Close Homolog of Sorghum CYP93G3
The P450 CYP93G subfamily was first described in rice with two sequences (http://drnelson.uthsc.edu/rice.html), and a sorghum protein was subsequently assigned as a new member (Du et al., 2010). Phylogenetic analysis revealed that CYP93G2 is closely related to CYP93G3 (Fig. 1), and the two proteins share 69.7% sequence identity. The cereal proteins are clustered with the soybean (Glycine max) CYP93A sequences but not the dicot FNSII and legume flavanone 2-hydroxylase (F2H) sequences. CYP93G2, consisting of 518 amino acids with a calculated pI of 8.177, is encoded by Os06g01250, which contains two exons separated by a single intron. Multiple sequence alignment analysis of CYP93G2 revealed that the protein harbors the diagnostic sequence signatures of a Pro hinge region, an oxygen-binding pocket, and a heme-binding motif of P450 enzymes (data not shown).
Figure 1.
Sequence analysis of CYP93G2 and other CYP93 family proteins. An unrooted phylogenetic tree of CYP93 proteins was constructed by MEGA4.0. Numbers at nodes represent bootstrap values from 1,000 replications.
Recombinant CYP93G2 Converts Flavanones to 2-Hydroxyflavanones
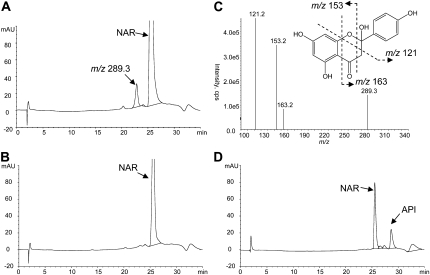
To examine the catalytic functions of CYP93G2, the open reading frame (ORF) was subcloned into the pYES2.1 vector and the recombinant protein was expressed in the Saccharomyces cerevisiae strain INVSc1. Microsomes were isolated from Gal-induced cultures and assayed for enzyme activities toward different flavonoid substrates. After incubation in the presence of NADPH, the reaction mixtures were extracted into ethyl acetate, vacuum dried, and then resuspended in methanol for HPLC-tandem mass spectrometry (MS/MS) analysis. As shown in Figure 2A, a single product that was eluted earlier than the naringenin (molecular weight = 272) substrate was detected in the CYP93G2 reaction, and MS analysis revealed a major [M + H]+ ion at m/z 289. By contrast, the same peak was not observed in reactions performed with microsomes extracted from yeast cultures expressing the empty pYES2.1 vector (Fig. 2B). The 16-D mass increase and the shorter retention time suggested that the reaction product is a hydroxylated derivative of naringenin. MS/MS of the mass-to-charge ratio (m/z) 289 ion generated the characteristic fragmentation pattern (Fig. 2C), which was reported previously for a [2-hydroxynaringenin + H]+ ion (Zhang et al., 2007; Du et al., 2010). Upon acid treatment of the reaction mixture, 2-hydroxynaringenin was dehydrated to apigenin (Fig. 2D), as described previously for the in vitro F2H reaction products (Akashi et al., 1998; Zhang et al., 2007). CYP93G2-expressing microsomes were also found to convert eriodictyol to 2-hydroxyeridictyol (data not shown). On the other hand, they have no detectable catalytic activities on other common flavonoid substrates, including chalcones, dihydroflavonols, and flavonols (Supplemental Fig. S1A). The 2-hydroxyflavanone products were formed in a time-dependent manner (Supplemental Fig. S1B) in the enzyme assays, and the recombinant CYP93G2 enzyme activity was maximal around pH 7.0 and 30°C.
Figure 2.
Recombinant CYP93G2 enzyme activity assay using naringenin (NAR) as a substrate. A and B, HPLC chromatogram of reaction mixtures containing yeast microsomes expressing CYP93G2 (A) or an empty vector (B). C, The single peak detected in the CYP93G2 reaction generated a major [M + H]+ ion at m/z 289 on MS analysis. The MS/MS spectrum revealed the fragmentation pattern consistent with that reported for 2-hydroxynarigenin (Zhang et al., 2007; Du et al., 2010). D, Upon acid treatment, the reaction product was converted to apigenin (API), which was confirmed by an authentic standard. cps, Counts per second; mAU, milli-absorbance unit.
Transgenic Arabidopsis Overexpressing CYP93G2 or Coexpressing CYP93G2 and OsCGT Accumulates 2-Hydroxyflavanone Derivatives
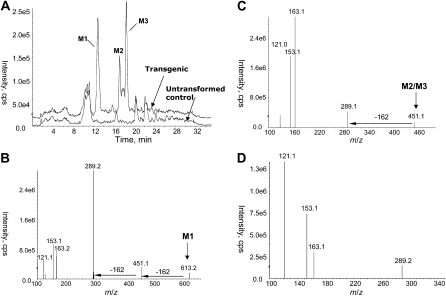
To examine whether CYP93G3 is a functional F2H in planta, the rice ORF was overexpressed in transgenic Arabidopsis under the control of the cauliflower mosaic virus (CaMV) 35S promoter. T1 seeds from at least three independent transgenic lines were germinated on Murashige and Skoog (MS) medium depleted of nitrogen sources to induce the endogenous flavonoid pathway. To preserve the structures of 2-hydroxyflavanone derivatives, plant samples (10-d-old seedlings extracted in methanol) were analyzed directly by HPLC-MS/MS without acid treatments. Since heterologously expressed phenolics are usually O-glycosylated in Arabidopsis (Yu et al., 2005, 2006), we performed precursor ion scan analyses to search for 2-hydroxyflavanone O-glycosides in the nonhydrolyzed extracts. During MS/MS experiments, fragmentations by the loss of sugar moiety are easily induced due to the labile nature of the O-glycosidic linkages. The chromatogram for the precursor ion scan of m/z 289 indicated three distinct peaks (M1, 12.8 min; M2, 17.5 min; M3, 18.5 min) in the transgenic plant extracts that were not detected in the wild-type extracts (Fig. 3A). M1 showed a precursor [M + H]+ ion at m/z 613, while M2 and M3 both gave the precursor [M + H]+ ion at m/z 451. The product ion spectrum of M1 (m/z 613) featured the diagnostic m/z 289 ion and a minor ion at m/z 451 (Fig. 3B). The mass differences between the precursor ion (m/z 613) and the product ions (m/z 451 and 289) indicated the sequential neutral losses of 162 D, which could be attributed to two consecutive losses of hexose moieties. MS3 experiments were also triggered concurrently for the m/z 289 ions for structural confirmation. It is evident that the fragmentation patterns of the m/z 289 ions from M1, M2, and M3 are essentially identical to that of a [2-hydroxynaringenin + H]+ ion (Fig. 3D). Hence, M1 could be annotated as a 2-hydroxynaringenin di-O,O-hexoside and both M2 and M3 as 2-hydroxynarigenin O-hexosides. Similarly, the LC-MS/MS analyses for the precursor ions of m/z 305 revealed the accumulation of one 2-hydroxyeriodictyol di-O,O-hexoside and two 2-hydroxyeriodictyol O-hexosides (Supplemental Table S1).
Figure 3.
Accumulation of 2-hydroxynarigenin O-glycosides in transgenic Arabidopsis overexpressing CYP93G2. A, Precursor ions of m/z 289 were scanned following HPLC separation. Three distinct peaks (M1, M2, and M3) were detected in the transgenic plant extract (top chromatogram) but not the nontransformed sample (bottom chromatogram). B, An MS/MS spectrum of M1 (m/z 613) indicated the two consecutive losses of a hexosyl unit (162 D). C, A representative MS/MS spectrum for M2 and M3 (m/z 451) indicated the loss of a hexosyl unit. D, A representative MS3 spectrum for the m/z 289 daughter ion generated from M1 to M3 revealed the 2-hydroxynarigenin fragmentation pattern. cps, Counts per second.
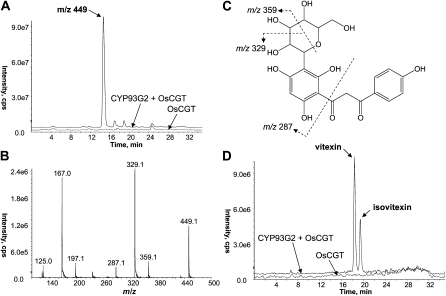
To further investigate whether CYP93G2 and OsCGT function together along the same biosynthetic pathway, the OsCGT ORF driven by the 35S CaMV promoter was transformed into a CYP93G2-expressing Arabidopsis line using a second selection marker (see “Materials and Methods”). The presence of both transgenes in the double transformant plants was confirmed by genomic PCR, and seedlings from three independent lines were prepared for metabolite extractions as described above. As shown in Figure 4A, LC-MS analyses revealed a peak with a parent [M – H]− ion at m/z 449 that was only observed in the double transformant samples. The m/z ratio is consistent with the molecular weight for a [hexosyl-2-hydroxynaringenin – H]− ion. The absence of a dominant 2-hydroxynaringenin aglycone ion in the MS/MS spectrum (Fig. 4B) further indicated that the conjugate was a C-hexoside. In addition, the product ions generated are in accordance with the fragmentation of C-glucosyl-2-hydroxynarigenin existing in the dibenzoylmethane open-chain form (Fig. 4C). In fact, the MS/MS spectrum is consistent with those reported for the OsCGT reaction products formed in vitro from 2-hydroxyflavanones (Brazier-Hicks et al., 2009). When the double transformant extracts were acid treated, the C-glucosyl-2-hydroxynarigenin peak was not detected by LC-MS analysis. Instead, two distinct peaks coeluting with the standards of vitexin (8C-glucosylapigenin) and isovitexin (6C-glucosylapigenin) were observed (Fig. 4D). Furthermore, the C-glucosyl-2-hydroxyeriodictyol [M – H]− ion (m/z 465) was also positively identified in the double transformant lines (Supplemental Fig. S2).
Figure 4.
Accumulation of C-glucosyl-2-hydroxynarigenin in transgenic Arabidopsis coexpressing CYP93G2 and OsCGT. A, Detection of a distinct peak producing a [M − H]− ion at m/z 449 in the double transformant sample (top chromatogram) but not the OsCGT transformant (bottom chromatogram). B and C, MS/MS spectrum (B) of the m/z 449 ion revealed the fragmentation pattern consistent with that reported for C-glucosyl-2-hydroxyflavanone in the open-chain form as shown in (C). D, Upon acid treatment, C-glucosyl-2-hydroxynarigenin was converted to isovitexin and vitexin. cps, Counts per second.
Rice CYP93G2 T-DNA Insertion Mutants Are Depleted in C-Glycosylflavone Accumulation
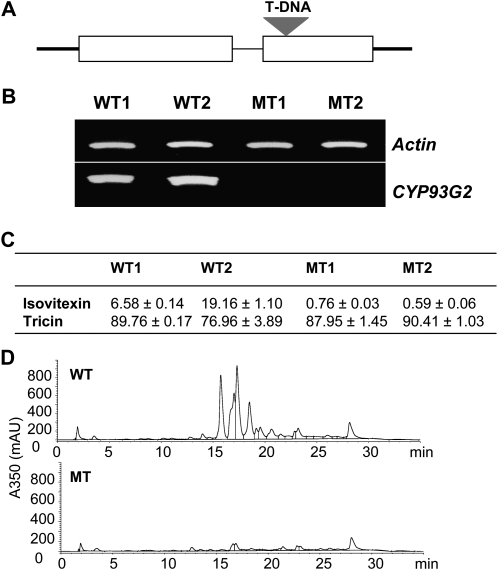
To understand the metabolic functions of CYP93G2 in rice, we analyzed a homozygous T-DNA mutant line with an insertion in the second exon of Os06g01250 (Fig. 5A). Reverse transcription (RT)-PCR analysis confirmed the lack of CYP93G2 transcript accumulation in leaves of 2-month-old mutant plants while the gene was strongly expressed in the wild-type plants (Fig. 5B). The mutant plants did not show observable differences from the wild-type plants with regard to growth, flowering time, or seed yield. We have previously shown that tricin and isovitexin derivatives are the major flavonoids in vegetative tissues of rice (Shih et al., 2008). Both tricin and isovitexin are frequently O-glycosylated (Cavaliere et al., 2005), and the aglycones can be released by acid treatment. In the acid-hydrolyzed methanolic extracts of CYP93G2 mutant leaves, the levels of tricin detected were comparable to those of wild-type samples (Fig. 5C). However, the accumulation of isovitexin was substantially reduced in the mutant plants, suggesting that CYP93G2 is required for C-glycosylflavone production in rice. In nonhydrolyzed leaf extracts, a range of potential flavone metabolites were detected in the wild-type samples following HPLC separation, which was monitored at 350 nm (Fig. 5D). On the other hand, the majority of these UV light-absorbing compounds eluted at 15 to 22 min were markedly reduced in absorbance in the mutant chromatogram.
Figure 5.
Analysis of the rice CYP93G2 T-DNA mutant. A, The T-DNA is inserted into the second exon of Os06g01250. B, RT-PCR analysis of gene expression in leaves of 2-month-old wild-type (WT) and mutant (MT) plants. Two individual plants were analyzed for each sample. C, Detection of isovitexin and tricin in acid-hydrolyzed extracts of WT and MT leaves. Data (μg g−1 fresh weight) are means ± sd of triplicate samples. D, HPLC-UV chromatograms of WT and MT nonhydrolyzed extracts monitored at 350 nm. mAU, Milli-absorbance unit.
To obtain a comprehensive view of the possible metabolic defects in the CYP93G2 mutant plants, nontargeted LC-MS/MS metabolite profiling experiments were performed. A total of 16 peaks (unique m/z retention time pairs) that are depleted in the mutant samples were identified (Table I). The extracted ion chromatograms for the candidate compounds were examined to confirm the differential accumulation levels in the wild-type and CYP93G2 mutant plants (Supplemental Fig. S3). The majority of them were not detectable in the mutant samples, whereas five [M + H]+ ions were substantially lower in abundance when compared with the wild-type samples. As shown in Table I, all of the mutant-depleted ions could be annotated as different derivatives of C-glycosylapigenin, C-glycosylluteolin, and C-glycosylchrysoeriol. A number of these compounds are mono-C-glycosylflavones modified by O-glycosylations, which were inferred from the loss of the labile sugar units following MS/MS fragmentation (i.e. O-hexoside, 162 D; O-p-coumaroylhexoside, 308 D; O-feruloylhexoside, 338 D [Supplemental Fig. S4]). The identities of the C-glycosylflavone aglycones were then confirmed by their product ion spectra with reference to that obtained with an isovitexin standard as well as the published spectra (Cavaliere et al., 2005; Kite et al., 2006; Abad-Garcia et al., 2008). In all cases, the major diagnostic fragment ions [0,1X]+, [0,2X]+, [0,4X – 2H2O]+, and [M + H-nH2O]+ for C-glycosylflavones were detected.
Table I. Accumulation levels of C-glycosylflavones in rice leaves as determined by LC-MS.
| [M + H]+ | m/z | Retention Time | Compound Assignmenta | Peak Area (×108) per Unit Mass Fresh Weightb |
|||
| WT1 | WT2 | MT1 | MT2 | ||||
| min | |||||||
| M4 | 433 | 19.6 | C-Hexosylapigenin | 0.67 ± 0.03 | 1.48 ± 0.07 | N.D. | N.D. |
| M5 | 595 | 18.9 | C-Hexosylapigenin O-hexoside | 0.20 ± 0.03 | 0.24 ± 0.02 | N.D. | N.D. |
| M6 | 595 | 19.8 | C-Hexosylapigenin O-hexoside | 0.69 ± 0.05 | 1.24 ± 0.09 | N.D. | N.D. |
| M7 | 741 | 21.8 | C-Hexosylapigenin O-p-coumaroylhexoside | 1.93 ± 0.04 | 3.86 ± 0.08 | 0.29 ± 0.02 | 0.24 ± 0.03 |
| M8 | 611 | 17.9 | C-Hexosylluteolin O-hexoside | 1.37 ± 0.01 | 2.88 ± 0.08 | N.D. | N.D. |
| M9 | 787 | 19.4 | C-Hexosylluteolin O-feruloylhexoside | 3.01 ± 0.03 | 3.28 ± 0.11 | N.D. | N.D. |
| M10 | 757 | 19.6 | C-Hexosylluteolin O-p-coumaroylhexoside | 6.78 ± 0.04 | 10.38 ± 0.17 | N.D. | N.D. |
| M11 | 625 | 20.1 | C-Hexosylchrysoeriol O-hexoside | 1.11 ± 0.00 | 1.63 ± 0.03 | N.D. | N.D. |
| M12 | 801 | 21.4 | C-Hexosylchrysoeriol O-feruloylhexoside | 3.88 ± 0.09 | 3.92 ± 0.06 | N.D. | N.D. |
| M13 | 771 | 21.6 | C-Hexosylchrysoeriol O-p-coumaroylhexoside | 4.97 ± 0.05 | 5.69 ± 0.05 | 0.35 ± 0.04 | 0.32 ± 0.04 |
| M14 | 565 | 16.6 | C-Hexosyl-C-pentosylapigenin | 2.11 ± 0.04 | 2.47 ± 0.06 | 0.33 ± 0.02 | 0.36 ± 0.03 |
| M15 | 565 | 18.2 | C-Hexosyl-C-pentosylapigenin | 4.18 ± 0.06 | 5.20 ± 0.10 | 1.61 ± 0.08 | 1.23 ± 0.02 |
| M16 | 565 | 19.3 | C-Hexosyl-C-pentosylapigenin | 5.45 ± 0.20 | 6.71 ± 0.20 | 1.02 ± 0.05 | 0.79 ± 0.06 |
| M17 | 581 | 15.1 | C-Hexosyl-C-pentosylluteolin | 3.75 ± 0.09 | 3.96 ± 0.12 | N.D. | N.D. |
| M18 | 581 | 16.8 | C-Hexosyl-C-pentosylluteolin | 3.45 ± 0.03 | 4.57 ± 0.07 | N.D. | N.D. |
| M19 | 581 | 17.7 | C-Hexosyl-C-pentosylluteolin | 5.18 ± 0.06 | 5.89 ± 0.14 | N.D. | N.D. |
Identification was based on MS/MS fragmentations (Supplemental Figs. S4 and S5) with reference to published spectra.
Data are expressed as means ± sd (triplicate samples). MT, Mutant; N.D., not detected; WT, wild type.
In addition, C-hexosyl-C-pentosylapigenin and C-hexosyl-C-pentosylluteolin ions were identified among the mutant-depleted peaks. Their MS/MS spectra (Supplemental Fig. S5) are complex as a consequence of fragmentations in the two different sugars (Abad-Garcia et al., 2008). In addition to the successive losses of three to six water molecules, daughter ions, such as [(0,1Xo)(0,2Xi)]+ and [(0,2Xo)(0,1Xi)]+, deriving from cleavages in both sugars were observed. Three isomers with different retention times have been detected for each of the C-hexosyl-C-pentosylflavones (Table I). They are likely to contain different sugar moieties and glycosylation positions. Taken together, metabolite profiling analysis clearly demonstrated the preferential depletion of C-glycosylflavone derivatives in rice mutant seedlings, further confirming the requirement of CYP93G2 for their biosynthesis.
DISCUSSION
In flavonoid biosynthesis, the formation of flavones from flavanones is the only chemical conversion known to be specified by two independent enzyme systems (FNSI and FNSII). Added to this complexity is the existence of two catalytic mechanisms for the P450 FNSII enzymes. While most CYP93B proteins convert flavanones to flavones directly, members from licorice (CYP93B1) and M. truncatula (CYP93B10 and CYP93B11) catalyze the 2-hydroxylation of flavanones in vitro (Akashi et al., 1998; Zhang et al., 2007). 2-Hydroxyflavanones are rare secondary metabolites reported in species such as Baccharis bigelovii, Collinsonia canadensis, Galega officinalis, Populus nigra, and Uvaria rufus (Stevens et al., 1999). In legumes, 2-hydroxyflavanones are believed to serve as intermediates in the biosynthesis of secondary metabolites. Silencing of CYP93B10 and CYP93B11 in M. truncatula resulted in flavone-deficient roots, suggesting that flavanone 2-hydroxylation is an important step for flavone formation. In licorice, 2-hydroxyflavanones are also substrates for licodione phytoalexin production (Akashi et al., 1998). Hence, CYP93B1 is believed to provide a common intermediate for the biosynthesis of licodione and flavones.
In this study, we demonstrated that flavanone 2-hydroxylation activities are not restricted to the Leguminosae or the CYP93B subfamily. However, phylogenetic analysis (Fig. 1) suggested an independent origin of F2H in cereals. We provided both in vitro and in planta evidence that the rice P450 enzyme CYP93G2 is a functional F2H. Recombinant enzyme assays demonstrated that the rice protein accepts naringenin and eriodictyol as substrates and converts them to 2-hydroxyflavanones (Fig. 2; Supplemental Fig. S1). Flavones were only detected in the CYP93G2 enzyme reactions after acid treatment. In addition, Arabidopsis plants overexpressing CYP93G2 were shown to produce 2-hydroxyflavanone O-glycosides (Fig. 3). Flavones and their derivatives do not accumulate in many cruciferous plants, including Arabidopsis, whose genome does not harbor any FNSI- or FNSII-encoding sequences (Martens and Mithöfer, 2005). The foreign flavonoid metabolites in transgenic Arabidopsis have been modified by endogenous flavonoid O-glycosyltransferases, which generally show a broad substrate range. Previously, we also detected the presence of 2-hydroxyflavanone O-glycosides in Arabidopsis overexpressing CYP93G3 (Du et al., 2010), which is the closest homolog of CYP93G2.
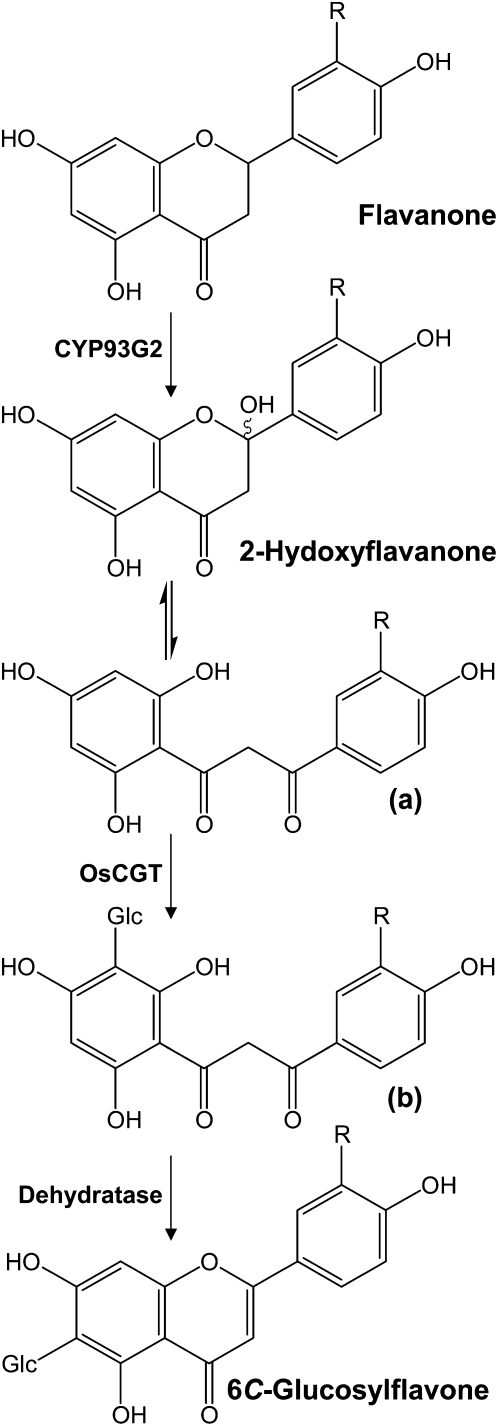
In major cereal crops, flavones occur predominately as C-glycosides that accumulate constitutively in different tissues. The enzymology of flavonoid C-glycosylation had remained elusive until the recent characterization of the rice enzyme OsCGT, which is related to O-glucosyltransferases (Brazier-Hicks et al., 2009). Recombinant OsCGT was found to have a preference for 2-hydroxyflavanones and catalyzed the UDP-Glc-dependent C-glucosyltransferase reaction. Our work provides strong in planta evidence that CYP93G2 and OsCGT participate together in the same biosynthetic pathway leading to C-glucosylflavone formation. Thus, transgenic expression of both rice genes in Arabidopsis resulted in the production of C-hexosyl-2-hydroxyflavanones. MS/MS fragmentations (Fig. 4C; Supplemental Fig. S2C) supported that they exist in the open-chain configuration, which is essentially identical to that of the in vitro reaction products formed from 2-hydroxyflavanones by OsCGT (Brazier-Hicks et al., 2009). Therefore, CYP93G2 evidently provides the in planta 2-hydroxyflavone substrates that are tautomerized to the dibenzoylmethane form for C-glucosylation by OsCGT (Fig. 6). Apparently, enzyme activities that dehydrate C-glucosyl-2-hydroxyflavanones to C-glucosylflavones are absent in Arabidopsis. Nonselective formation of vitexin and isovitexin only occurred following acid treatments of the plant extracts. On the other hand, dehydratase activities that result in the preferential accumulation of 6C-glucosylflavones (Fig. 6) were identified in rice and wheat extracts (Brazier-Hicks et al., 2009).
Figure 6.
Biosynthesis pathway for C-glucosylflavones in rice. Using flavanones as substrates, CYP93G2 generates 2-hydroxyflavanones for C-glucosylation by OsCGT. 2-Hydroxyflavanone exists in equilibrium with the dibenzoylmethane tautomer (a), which serves as the acceptor for C-glucosylation. C-Glucosyl-2-hydroxyflavanones in the open chain form (b) were detected in transgenic Arabidopsis coexpressing CYP93G2 and OsCGT. In rice, dehydratase activities that resulted in the selective formation of 6C-glucosylflavones were demonstrated (Brazier-Hicks et al., 2009).
The requirement of CYP93G2 in C-glycosylflavone biosynthesis in rice is further supported by metabolite analyses of the Os06g01250 T-DNA insertion line. As a consequence of gene disruption, the CYP93G2 mutant seedlings showed a substantial reduction in the contents of a range of C-glycosides derived from apigenin, luteolin, and chrysoeriol (Table I). Apparently, 2-hydroxyflavanone formation is important for the formation of both mono- and di-C-glycosylflavone metabolites. By contrast, the accumulation of tricin derivatives was not affected by the absence of Os06g01250 expression in the mutant seedlings (Fig. 5C). In rice and wheat seedlings, tricin derivatives have been detected almost exclusively as O-glycosides (Cavaliere et al., 2005; Brazier-Hicks et al., 2009). Thus, our data strongly indicated that a completely different enzyme system is involved in flavone O-glycoside biosynthesis. A recombinant rice FNSI protein (OsFNS1-1), which converted naringenin to apigenin in vitro, was previously reported (Lee et al., 2007). It remains to be elucidated whether the enzyme shows the same activity in planta. Alternatively, P450 enzymes that desaturate flavanones directly to flavones, similar to the nonleguminous FNSIIs, may participate in the pathway that leads to flavone O-glycoside formation. Possible candidates include CYP93G1 and CYP93F1 (Nelson et al., 2004), which are closely related to CYP93G2 and CYP93G3 (Fig. 1). For the formation of tricin, flavonoid 3′,5′-hydroxylation was proposed to proceed before flavone formation, followed by 3′- and 5′-O-methyltransferase activities (Cummins et al., 2006). Subsequently, O-glycosylation of tricin is expected to occur as a terminal step in flavonoid biosynthesis.
In summary, our work firmly establishes CYP93G2 as the first enzyme for the formation of C-glycosylflavones from flavanones in rice. The concerted activities of F2H, C-glycosyltransferase, and dehydratase may represent a unique pathway to produce flavone metabolites in Poaceae. The existence of separate biosynthetic pathways for flavone glycosides was first proposed by Harborne and Hill (1964), when they reported the regular occurrence of luteolin and apigenin as C-glycosides and tricin as O-glycosides in a number of grass species. The identity of the molecular component responsible for the selective dehydration of 2-hydroxyflavanone C-glycosides remains an intriguing question. On the other hand, a different FNS enzyme is likely to generate flavone aglycones for O-glycosylation. Rice accumulates a variety of secondary metabolites, including flavonoids in different tissues but not in endosperm, which is the major component in polished rice (the preferred consumed form). The successful introduction of the provitamin A pathway into the rice endosperm offers the promise for metabolic engineering of health-beneficial phytochemicals in this important staple food (Ye et al., 2000). Successful elucidation of the biosynthesis pathways for flavone C- and O-glycosides in rice will provide knowledge useful for biotechnological improvements of its nutraceutical values.
MATERIALS AND METHODS
Expression of Rice P450 Protein in Yeast
The ORF of CYP93G2 was amplified from the full-length rice (Oryza sativa subsp. japonica ‘Nipponbare’) cDNA clone AK099468 (National Institute of Agrobiological Sciences of Japan) by the high-fidelity pfx DNA polymerase (Invitrogen). The PCR fragments were then cloned into the yeast vector pYES2.1/V5-His-TOPO (Invitrogen). The expression vector was transformed into Saccharomyces cerevisiae INVSc1 cells (Invitrogen) using the lithium acetate procedure as described in the manufacturer’s manual. Transformed cells were selected on agar plates containing synthetic minimal medium without uracil (SC-U). Microsome extractions were performed as described previously (Urban et al., 1994) with slight modifications. Single colonies harboring the desired expression construct were inoculated overnight at 30°C with shaking in SC-U containing 2% raffinose. The cultures were then diluted to an optical density at 600 nm value of 0.4 in SC-U containing 2% Gal for induction. Yeast cells were harvested by centrifugation after incubation with shaking at 30°C for 20 h. The pellet was resuspended in 50 mm Tris-HCl, pH 7.4, 2 mm EDTA, and 100 mm 2-mercaptoethanol (0.5 g cells mL−1). Following incubation at room temperature for 10 min, the cells were pelleted and washed with the same buffer containing 100 mm KCl. Afterward, the pellet was resuspended in ice-cold buffer (50 mm Tris-HCl, pH 7.4, and 2 mm EDTA) containing 0.6 m sorbitol (0.3 g cells mL−1), followed by the addition of acid-washed glass beads (425–600 nm diameter; Sigma). The cells were vortexed for 5 min at 4°C, incubated on ice for 1 min, and vortexed again for another 5 min at 4°C, and the crude extract was recovered by centrifugation. The glass beads were washed twice in the same buffer, which was then combined with the crude extract. The final extract was centrifuged at 4°C for 3 min at 3,500g followed by 10 min at 10,000g. Subsequently, polyethylene glycol 4000 (Sigma) and NaCl were added to the supernatant at final concentrations of 0.1 g mL−1 and 0.15 m, respectively, followed by ice incubation for 15 min. Microsomes were then pelleted by centrifugation at 10,000g for 10 min at 5°C and resuspended in 50 mm Tris-HCl, pH 7.4, and 1 mm EDTA containing 20% (v/v) glycerol. The final preparations (10–20 mg protein mL−1) were kept at −70°C until further use.
In Vitro CYP93G Enzyme Activity Assays
Protein concentrations of microsome preparations were determined by the Bradford protein assay (Bio-Rad). Enzyme assays were performed for 1 h at 30°C in 0.1 m potassium phosphate buffer (pH 7.0), 5 mm NADPH, 5 mm l-glutathionine, 1 mm substrate, and 500 μg of microsomal protein. The reaction was stopped by extraction with an equal volume of ethyl acetate (twice). The ethyl acetate fraction was evaporated to dryness under vacuum, and the pellet was redissolved in methanol for LC-MS/MS analysis (see below).
Generation of Transgenic Arabidopsis Plants
CYP93G2 ORF was amplified from the full-length cDNA clone mentioned above by the pfx DNA polymerase and subcloned between the CaMV 35S promoter and the nopaline 3′-terminator in the plasmid vector 103c-SK (Yu et al., 2005). The resulting overexpression cassette was cloned into the binary vector pCAMBIA 1300 (CAMBIA), which harbors the hygromycin resistance gene for the selection of plant transformants. Agrobacterium tumefaciens-mediated transformation of the final construct into wild-type Arabidopsis (Arabidopsis thaliana ecotype Columbia) was performed by the floral dip method (Clough and Bent, 1998). Harvested seeds were surface sterilized and germinated on MS (Sigma) agar containing 3% (v/v) Suc and 25 μg mL−1 hygromycin (Sigma). Resistant seedlings were transplanted and placed in a growth chamber (22°C, 16 h of light, 8 h of dark). The OsCGT coding region was amplified with the full-length rice cDNA clone AK071127 (National Institute of Agrobiological Sciences) using the pfx polymerase and then subcloned into the 103c-SK vector. Subsequently, the 35S-OsCGT-nos3′ cassette was inserted into a modified pCAMBIA vector carrying the BASTA resistance gene. The final construct was then transformed into CYP93G2 transgenic plants by Agrobacterium. Harvested seeds were germinated on soil, and the seedlings (six- to eight-leaf stage) were selected by three spray applications of BASTA (120 mg mL−1, 1:1,000 in water; Bayer CropScience). All the transgenic lines were confirmed by genomic PCR. Seeds from at least three independent lines for the CYP93G2 transformant and the double transformant were germinated in MS medium without nitrogen sources for 10 d. Seedlings (0.5 g) were collected and ground to fine powder in liquid nitrogen and extracted in 100% HPLC-grade methanol (500 μL). For acid hydrolysis, an equal volume of 2 n HCl was added to the samples for incubation at 90°C for 1 h.
Characterization of the Rice T-DNA Insertion Mutant
The rice (O. sativa subsp. japonica 'Zhonghua 11') T-DNA insertion mutant line (SHIP_ZSX0568) for Os06g01250 was identified at the RiceGE database (http://signal.salk.edu/cgi-bin/RiceGE). Homozygous seeds were available from the National Key Laboratory of Plant Molecular Genetics, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences. Plants were grown at 26°C to 28°C (12 h of light, 12 h of dark), and leaves of 2-month-old plants were collected for different analyses. RNA extraction and RT-PCR experiments were performed as described previously (Shih et al., 2008), and metabolites were extracted in methanol as described above.
HPLC-MS/MS Analysis of Enzyme Reaction Products and Plant Metabolites
Filtered samples (10 μL) of enzyme reactions and plant (Arabidopsis and rice) extracts were injected onto an Agilent 1100 series HPLC system (Agilent Technologies) connected with a Eclipse XDB-C18 column (5 μm, 2.1 × 150 mm; Agilent Technologies). Separation was performed using a solvent system of 0.5% (v/v) formic acid/water (A) and 0.5% (v/v) formic acid/methanol (B), with a linear gradient of 15% to 60% B over 20 min, then held at 60% B for 5 min, and equilibrated at 15% B for 10 min. The flow rate was maintained at 0.2 mL min−1, and the elution was monitored by a diode-array detector (200–600 nm) in tandem with an API2000-QTRAP mass spectrometer (Applied Biosystems). Enhanced full-scan, product ion scan, precursor ion scan, and MS3 spectra were acquired as described previously (Yu et al., 2006; Shih et al., 2008). Quantification of tricin and isovitexin was achieved by multiple-reaction monitoring using the transition reactions m/z 331.2 → 315.3 and m/z 433.3 → 283.3, respectively. Authentic standards (TransMIT) were used for standard curve construction. Data acquisition, peak integration, and calculations were performed by using the Analyst 1.4.2 software (Applied Biosystems).
For nontargeted metabolite profiling of rice wild-type and mutant extracts, raw LC-MS data were processed by Markerview 1.1 (Applied Biosystems) for mass signal extraction, retention time alignment, and multivariate analysis. Extracted ion chromatograms for the candidate ions (P < 0.01, Student’s t test) were retrieved to confirm their differential accumulation in the wild-type and CYP93G2 mutant plants. Two individual plants each for wild-type and mutant rice plants were analyzed with three extractions per plant.
Primers for vector construction as well as molecular analysis of transgenic Arabidopsis and rice mutant plants are listed in Supplemental Table S2.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AK099468 (OsF2H/CYP93G2) and AK071127 (OsCGT).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Enzyme activities of CYP93G2-expressing yeast microsomes toward different flavonoid substrates.
Supplemental Figure S2. Accumulation of C-glucosyl-2-hydroxyeriodictyol in transgenic Arabidopsis coexpressing CYP93G2 and OsCGT.
Supplemental Figure S3. Representative extracted ion chromatograms (XICs) for the C-hexosylflavone derivatives (M4–M19; Table I) in the rice wild-type (WT) and CYP93G2 mutant (MT) nonhydrolyzed methanol samples.
Supplemental Figure S4. MS/MS spectra for M4 to M13 (C-hexosylflavone O-glycosylated derivatives).
Supplemental Figure S5. MS/MS spectra and fragmentation patterns for M14 to M19 (C-hexosyl-C-pentosyl flavones).
Supplemental Table S1. List of 2-hydroxyflavanone O-glycosides identified in Arabidopsis overexpressing CYP93G2.
Supplemental Table S2. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank the National Key Laboratory of Plant Molecular Genetics (Shanghai, China) for providing the CYP93G2 T-DNA insertion homozygous mutant seeds.
References
- Abad-Garcia B, Garmon-Lobato S, Berrueta LA, Gallo B, Vicente F. (2008) New features on the fragmentation and differentiation of C-glycosidic flavone isomers by positive electrospray ionization and triple quadrupole mass spectrometry. Rapid Commun Mass Spectrom 22: 1834–1842 [DOI] [PubMed] [Google Scholar]
- Akashi T, Aoki T, Ayabe S. (1998) Identification of a cytochrome P450 cDNA encoding (2S)-flavanone 2-hydroxylase of licorice (Glycyrrhiza echinata L.; Fabaceae) which represents licodione synthase and flavone synthase II. FEBS Lett 431: 287–290 [DOI] [PubMed] [Google Scholar]
- Besson E, Dellamonica G, Chopin J, Markham KR, Kim M, Koh HS, Fukami H. (1985) C-Glycosylflavones from Oryza sativa. Phytochemistry 24: 1061–1064 [Google Scholar]
- Brazier-Hicks M, Evans KM, Gershater MC, Puschmann H, Steel PG, Edwards R. (2009) The C-glycosylation of flavonoids in cereals. J Biol Chem 284: 17926–17934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Al-Fayez M, Tunstall RG, Platton S, Greaves P, Steward WP, Gescher AJ. (2005) The rice bran constituent tricin potently inhibits cyclooxygenase enzymes and interferes with intestinal carcinogenesis in ApcMin mice. Mol Cancer Ther 4: 1287–1292 [DOI] [PubMed] [Google Scholar]
- Cassi-Lit M, Tanner GJ, Nayudu M, Whitecross MI. (2007) Isovitexin-2′ O-β-[6-O-E-p-coumaroylglucopyranoside] from UV-B irradiated leaves of rice, Oryza sativa L. inhibits fertility of Helocoverpa armigera. Photochem Photobiol 83: 1167–1173 [DOI] [PubMed] [Google Scholar]
- Cavaliere C, Foglia P, Pastorini E, Samperi R, Laganà A. (2005) Identification and mass spectrometric characterization of glycosylated flavonoids in Triticum durum plants by high-performance liquid chromatography coupled with tandem mass spectrometry. Rapid Commun Mass Spectrom 19: 3143–3158 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cummins I, Brazier-Hicks M, Stobiecki M, Franski R, Edwards R. (2006) Selective disruption of wheat secondary metabolism by herbicide safeners. Phytochemistry 67: 1722–1730 [DOI] [PubMed] [Google Scholar]
- Du Y, Chu H, Wang M, Chu IK, Lo C. (2010) Identification of flavone phytoalexins and a pathogen-inducible flavone synthase II gene in sorghum. J Exp Bot 61: 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt YH, Whit S, Steuber H, Matern U, Martens S. (2007) Evolution of flavone synthase I from parsley flavanone 3-β-hydroxylase by site directed mutagenesis. Plant Physiol 144: 1442–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborne JB. (1993) The Flavonoids: Advances in Research Since 1986. Chapman & Hall, London, pp 57–93 [Google Scholar]
- Harborne JB, Hill E. (1964) Plant polyphenols. XII. The occurrence of tricin and of glycoflavones in grasses. Phytochemistry 3: 421–428 [Google Scholar]
- Hudson EA, Dinh EA, Kokubun T, Simmonds MSJ, Gescher A. (2000) Characterization of potentially chemopreventive phenols in extracts of brown rice that inhibit the growth of human breast and colon cancer cells. Cancer Epidemiol Biomarkers Prev 9: 1167–1170 [PubMed] [Google Scholar]
- Kerscher F, Franz G. (1987) Biosynthesis of vitexin and isovitexin: enzymatic synthesis of the C-glucosylflavones vitexin and isovitexin with an enzyme preparation from Fagopyrum esculentum M. seedlings. Z Naturforsch 42c: 519–524 [Google Scholar]
- Kerscher F, Franz G. (1988) Isolation and some properties of an UDP-glucose:2-hydroxyflavanone-6(or 8)-C-glucosyltransferase from Fagopyrum esculentum M. cotyledons. J Plant Physiol 132: 110–115 [Google Scholar]
- Kite GC, Porter EA, Denison FC, Grayer RJ, Veitch NC, Butler I, Simmonds MSJ. (2006) Data-directed scan sequence for the general assignment of C-glycosylflavone O-glycosides in plant extracts by liquid chromatography-ion trap mass spectrometry. J Chromatogr A 1104: 123–131 [DOI] [PubMed] [Google Scholar]
- Kong C, Xu X, Zhou B, Hu F, Zhang C, Zhang M. (2004) Two compounds from allelopathic rice accession and their inhibitory activity on weeds and fungal pathogens. Phytochemistry 65: 1123–1128 [DOI] [PubMed] [Google Scholar]
- Kong CH, Zhao H, Xu XH, Wang P, Gu Y. (2007) Activity and allelopathy of soil of flavone O-glycosides from rice. J Agric Food Chem 55: 6007–6012 [DOI] [PubMed] [Google Scholar]
- Lee YJ, Kim JH, Kim BG, Lim Y, Ahn JH. (2007) Characterization of flavone synthase I from rice. BMB Rep 41: 68–71 [DOI] [PubMed] [Google Scholar]
- Lin CM, Chen CT, Lee HH, Lin JK. (2002) Prevention of cellular ROS damage by isovitexin and related flavonoids. Planta Med 68: 365–367 [DOI] [PubMed] [Google Scholar]
- Lin CM, Huang ST, Liang YC, Lin MS, Shih CM, Chang YC, Chen TY, Chen CT. (2005) Isovitexin suppresses lipopolysaccharide-mediated inducible nitric oxide synthase through inhibition of NF-kappa B in mouse macrophages. Planta Med 71: 748–753 [DOI] [PubMed] [Google Scholar]
- Markham KR, Tanner GJ, Cassi-Lit M, Whitecross MI, Nayudu M, Mitchell KA. (1998) Possible protective role for 3′,4′-dihydroxyflavones induced by enhanced UV-B in a UV-tolerant rice cultivar. Phytochemistry 49: 1913–1919 [Google Scholar]
- Martens S, Mithöfer A. (2005) Flavones and flavone synthases. Phytochemistry 66: 2399–2407 [DOI] [PubMed] [Google Scholar]
- McMullen MD, Kross H, Snook ME, Cortés-Cruz M, Houchins KE, Musket TA, Coe EH. (2004) Salmon silk genes contribute to the elucidation of the flavone pathway in maize (Zea mays L.). J Hered 95: 225–233 [DOI] [PubMed] [Google Scholar]
- Nelson DR, Schuler MA, Paquette SM, Werck-Rechhart D, Bak S. (2004) Comparative genomics of rice and Arabidopsis: analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiol 135: 756–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih CH, Chu H, Tang LK, Sakamoto W, Maekawa M, Chu IK, Wang M, Lo C. (2008) Functional characterization of key structural genes in rice flavonoid biosynthesis. Planta 228: 1043–1054 [DOI] [PubMed] [Google Scholar]
- Shin YM, Park HJ, Yim SD, Baek NI, Lee CH, An G, Woo YM. (2006) Transgenic rice lines expressing maize C1 and R-S regulatory genes produce various flavonoids in the endosperm. Plant Biotechnol J 4: 303–315 [DOI] [PubMed] [Google Scholar]
- Stevens JF, Ivancic Monika I, Deinzer ML, Wollenweber E. (1999) A novel 2-hydroxyflavanone from Collinsonia canadensis. J Nat Prod 62: 392–394 [DOI] [PubMed] [Google Scholar]
- Urban P, Werck-Reichhart D, Teutsch HG, Durst F, Regnier S, Kazmaier M, Pompon D. (1994) Characterization of recombinant plant cinnamate 4-hydroxylase produced in yeast: kinetic and spectral properties of the major plant P450 of the phenylpropanoid pathway. Eur J Biochem 222: 843–850 [DOI] [PubMed] [Google Scholar]
- Wallace JW, Grisebach H. (1973) The in vivo incorporation of flavanone into C-glycosylflavones. Biochim Biophys Acta 304: 837–841 [DOI] [PubMed] [Google Scholar]
- Wallace JW, Mabry TJ, Alston RE. (1969) On the biogenesis of flavone O-glycosides and C-glycosides in the Lemnaceae. Phytochemistry 8: 93–99 [Google Scholar]
- Ye X, Al-Babili S, Kloti A, Zhang J, Lucca P, Beyer P, Potrykus I. (2000) Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287: 303–305 [DOI] [PubMed] [Google Scholar]
- Yu CKY, Lam CNW, Springob K, Schmidt J, Chu IK, Lo C. (2006) Constitutive accumulation of cis-piceid in transgenic Arabidopsis over-expressing a sorghum stilbene synthase gene. Plant Cell Physiol 47: 1017–1021 [DOI] [PubMed] [Google Scholar]
- Yu CKY, Springob K, Schmidt J, Nicholson RL, Chu IK, Yip WK, Lo C. (2005) A stilbene synthase gene is involved in host and non-host defense responses in Sorghum bicolor. Plant Physiol 138: 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Subramanian S, Zhang Y, Yu O. (2007) Flavone synthases from Medicago truncatula are flavanone-2-hydroxylases and are important for nodulation. Plant Physiol 144: 741–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.