Abstract
The West Nile virus RNA helicase uses the energy derived from the hydrolysis of nucleotides to separate complementary strands of RNA. Although this enzyme has a preference for ATP, the bias towards this purine nucleotide cannot be explained on the basis of specific protein–ATP interactions. Moreover, the enzyme does not harbor the characteristic Q-motif found in other helicases that regulates binding to ATP. In the present study, we used structural homology modeling to generate a model of the West Nile virus RNA helicase active site that provides instructive findings on the interaction between specific amino acids and the ATP substrate. In addition, we evaluated both the phosphohydrolysis and the inhibitory potential of a collection of 30 synthetic purine analogs. A structure-guided alanine scan of 16 different amino acids was also performed to clarify the contacts that are made between the enzyme and ATP. Our study provides a molecular rationale for the bias of the enzyme for ATP by highlighting the specific functional groups on ATP that are important for binding. Moreover, we identified three new essential amino acids (Arg-185, Arg-202 and Asn-417) that are critical for phosphohydrolysis. Finally, we provide evidence that a region located upstream of motif I, which we termed the nucleotide specificity region, plays a functional role in nucleotide selection which is reminiscent to the role exerted by the Q-motif found in other helicases.
INTRODUCTION
Helicases are widely distributed in nature and are involved in every biological processes involving nucleic acids (1,2). They are responsible for the separation of complementary strands of duplex DNA or RNA molecules that appear during replication, recombination, transcription, splicing, editing and translation (3,4). Helicases use the energy derived from the hydrolysis of nucleoside 5′-triphosphates (NTPs) to separate complementary strands, and many different models have been proposed to explain how the NTPase and unwinding activities are coupled (5–9). Helicases have been divided into distinct subfamilies based upon sequence comparisons and conserved motifs. Extensive sequence analyses of helicases from various organisms have revealed a series of short, conserved amino acids motifs (10). Although these motifs are not common to all helicases, they harbor the amino acid residues that are essential for ATP hydrolysis (motifs I, II, VI and Q), nucleic-acid binding (motifs Ia, Ib, IV and V), and for coupling ATP hydrolysis to strand separation (motif III). Crystallographic studies of various helicases have shown that the conserved motifs are clustered together in the 3D structure of helicases to form the NTP-binding pocket (11–15).
Helicases have now been found in many viral families (16,17). Their importance for the replication of viruses has been highlighted by numerous studies, thereby underscoring their importance for the development of anti-viral drugs (18,19). The West Nile virus (WNV) RNA helicase activity is catalyzed by the non-structural 3 (NS3) protein (19–23), which also harbors the catalytic site for the serine protease activity involved during the proteolytic processing of the viral polyprotein precursor. WNV is a member of the Flaviviridae family which consists of three genera (hepacivirus, pestivirus and flavivirus) encompassing important human pathogens (24). For instance, the hepatitis C virus (HCV) currently infects more than 170 million people worldwide (25), while flaviviruses are also responsible for some of the most important examples of emerging diseases, exemplified by the increasing prevalence of Dengue virus (DENV) in the tropical and subtropical areas of the world, the emergence of West Nile virus (WNV) in North America, and the spread of Japanese encephalitis virus (JEV) through much of Asia (26).
The NS3 protein of Flaviviridae has been extensively characterized both mechanistically and structurally. Analysis of the crystal structure of the helicase domains from HCV, DENV and JEV revealed that it is composed of three structural domains (14,27,28). Domains 1 and 2 are interacting in the 3D structure to form the NTP hydrolyzing engine capable of providing the energy for unwinding, while domain 3 differs most between the different helicases, and is likely involved in the binding to specific protein partners (27,29,30). The NS3 protein likely operates by switching between different structural conformations, as evidenced from the various conformational states that are observed in crystallographic studies. Analysis of the various crystal structures suggest that a tunnel that runs across the interface between domain III and the tip of domains I and II could accommodate a single-stranded nucleic acid along which the enzyme could translocate, following interdomain motions triggered by the hydrolysis of NTPs (27,29).
Biochemical studies have demonstrated that most RNA helicases have a distinct preference for ATP (31). The presence of a glutamine residue (in the Q-motif), which coordinates the N6 and N7 positions of adenine, as well as the presence of a hydrophobic stacking interaction on the adenine base, is a common theme in many RNA helicases (32,33). Surprisingly, structural analyses of the NS3 protein from Flaviviridae bound to nucleotides indicate that both the base and ribose moieties of the ATP substrate are exposed to the solvent (14,27,28). The triphosphate moiety is the only component of the ATP molecule that interacts strongly with the enzyme, being buried between domains I and II (14,27,28). The phosphates are coordinated by four amino acids, either through direct or water-mediated contacts, while the 3′-OH group of the ribose is hydrogen bonded by two amino acids (28). The importance of the 2′- and 3′-OH groups for hydrolysis has also been demonstrated through the use of modified NTPs harboring modified sugars (34). Close inspection of the Dengue virus NS3 protein bound to a non-hydrolyzable ATP substrate indicates that the ε-amino group of a lysine residue forms a hydrogen-bond with the adenine N7 atom, suggesting that the flaviviral helicase would indiscriminately cleave NTPs (28). However, numerous studies have reported a preference of the NS3 protein for certain NTPs (35–38). Moreover, a recent study demonstrated that an ATP analog harboring a methyl group at the N1 position (N1-methyl ATP) weakly supports the unwinding activity of the HCV helicase (34), thereby raising the likelihood of an interaction between the enzyme and the adenine base moiety.
The NS3 protein is one of the most promising targets for drug development against viruses of the Flaviviridae family since it plays such a crucial role in viral replication. Numerous biochemical and structural studies are currently focusing on the NS3 protein of both HCV and DENV. In contrast, analyses of the WNV NS3 protein have been rather limited despite the growing distribution and epidemic character of the virus. Interestingly, mounting evidences indicate that significant structural and mechanistic differences exist between the corresponding proteins of the Flaviviridae family (39). In the present study, we used a panel of nucleotide analogs to explore both the nature of the nucleotide-binding pocket and the specificity of NTP hydrolysis by the WNV NS3 protein. We also applied structural modeling to understand the binding of ATP to the enzyme, and we generated a series of mutants to identify the amino-acid residues that might contribute to the bias towards ATP. Our study highlights the importance of specific functional groups for the binding of the purine substrate, and reveals the complexity of the active site.
MATERIALS AND METHODS
NS3 expression and purification
A truncated version of the WNV NS3 protein (amino acids 168–618) lacking the N-terminal protease domain was expressed and purified as described before (40).
ATPase assays
The standard ATPase reaction was performed in reaction mixtures (20 µl) containing 50 mM Tris–HCl, pH 7.5, 5 mM DTT, 1 mM MgCl2, 100 nM of the purified WNV NS3 protein, and different concentrations of [γ-32P]ATP. The reactions were incubated for 15 min at 37°C. The reactions were quenched by the addition of 5 µl of 5 M formic acid. Aliquots of the reactions were spotted on polyethyleneimine-cellulose thin-layer chromatography plates. The plates were developed in a solution of 1 M formic acid and 0.5 M LiCl and the released inorganic phosphate was quantitated by scanning the plates with a PhosphorImager (Molecular Dynamics). The average of at least two single independent experiments is presented.
The inhibition of the ATPase reaction by various nucleotide analogs was evaluated by performing the standard ATPase assay in the presence of increasing concentrations of nucleotide analogs. The catalytic activity was assayed a minimum of three times independently in the presence of a range of inhibitor concentrations (ranging from 0 to 12 mM) with 50 μM ATP as substrate. The Ki values were determined using the following equation:
where [S] represents the concentration of ATP in the reaction mixture.
Phosphohydrolysis of GTP and nucleotide analogs
Nucleotide analogs were obtained from Jena Bioscience (Germany) and TriLink BioTechnologies (San Diego, USA). The reaction mixtures (20 µl) containing 50 mM Tris–HCl, pH 7.5, 5 mM DTT, 1 mM MgCl2, 100 nM of the WNV NS3 protein, and various concentrations of nucleotide analogs or GTP. The reactions were incubated for 15 min at 37°C, and quenched by the addition of 400 µl of malachite green reagent (BIOMOL Research Laboratories). Released inorganic phosphate was measured spectrophotometrically by monitoring the A620. The values were extrapolated to a standard curve for phosphate. Background levels of contaminating phosphate were subtracted in all cases. As a control, the ATPase reaction was also quantified spectrophotometrically and yielded similar results as previously observed using radiolabeled ATP as a substrate.
Homology modeling
The crystal structure of the Dengue virus NS3 helicase domain (amino acids 180–618) bound to the non-hydrolyzable ATP analog AMPPNP was used as a template to model the WNV NS3 helicase domain (28). The atomic coordinates were obtained from the Protein Data Bank file 2JLR, representing the crystal structure of the complex between the Dengue virus NS3 helicase domain, manganese and AMPPNP. The predicted 3D structure of the WNV NS3 active site was generated using the Deep View program (41).
RESULTS
Homology modeling
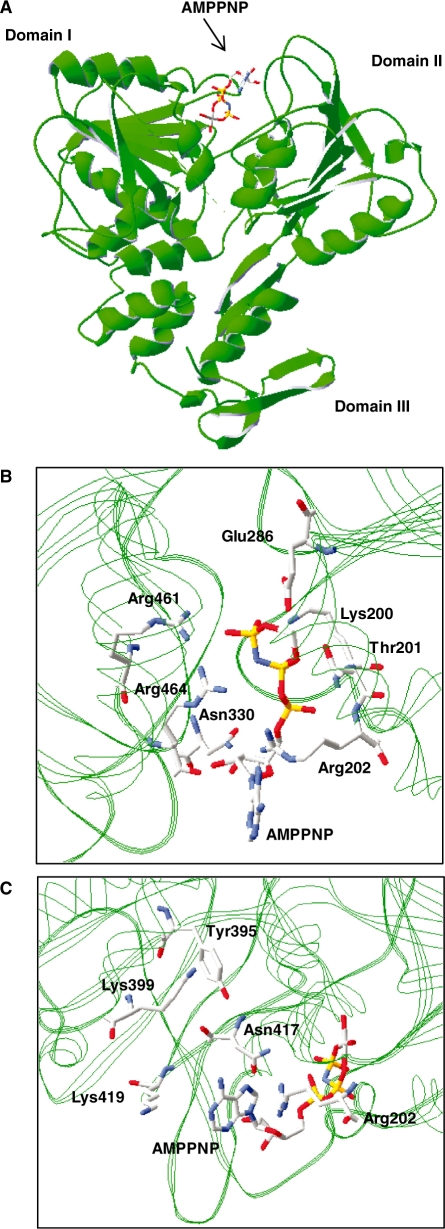
Although important structural information is derived from the currently available crystal structures of the NS3 protein of the Flaviviridae (14,27,28), the specificity of these enzymes for particular NTPs cannot yet be explained on the basis of the interaction between the enzyme and the base moiety of the NTP substrate. Under the standard steady-state conditions used in the current study, the purified WNV NS3 protein displays a ∼2-fold higher phosphohydrolase activity in the presence of ATP than with GTP (Table 1). Since crystal structures of the WNV NS3 protein are not currently available, we set out to generate a 3D model of the protein active site in order to understand the preference of the enzyme for ATP. The closely related NS3 protein from Dengue virus (DENV), which shares 80% sequence homology with the corresponding WNV enzyme, bound to a non-hydrolyzable ATP analog (AMPPNP) was used as a template. The structural model provides instructive findings on the interaction between specific residues and the nucleotide substrate (Figure 1A). As reported for the DENV NS3 crystal (28), a large solvent exposition of the base moiety of the substrate is observed in the structural model of the WNV NS3 protein. The triphosphate moiety of AMPPNP is stabilized by contacts with the side chains of Lys-200, Thr-201, Glu-286, Arg-461 and Arg-464, and with the main chain amide nitrogen of both Lys-200 and Arg-202, while the 3′-OH group of the ribose is hydrogen bonded with the main chain carbonyl oxygen of Arg-464 and the side chain amide group of Asn-330 (Figure 1B). As previously reported for the DENV NS3 protein (28), the interaction of the WNV NS3 protein with the ATP substrate also involves a contact between a basic amino acid (Arg-202) and the N-7 atom of the adenine base (Figure 1B and C). However, closer examination of the homology model reveals the presence of numerous additional amino acids (Tyr-395, Lys-399, Asn-417 and Lys-419) that are in the vicinity of the base moiety of the ATP substrate (Figure 1C). Although most of these residues are probably too far removed to directly interact with the substrate, we hypothesize that they could potentially be important for nucleotide selection by providing a steric gate.
Table 1.
Inhibition of the ATPase activity by nucleotide analogs
| Molecule | IC50 (mM) | Ki (mM) | Phosphohydrolase specific activity (relative to ATP)a |
|---|---|---|---|
| ATP | 0.4 | 0.3 | 1.00 |
| GTP | 11.0 | 8.1 | 0.49 ± 0.11 |
| (P2) 2′,3′ddATP | 0.8 | 0.6 | 0.53 ± 0.10 |
| (P3) 2′-dATP | 0.9 | 0.6 | 0.62 ± 0.12 |
| (P4) 2′F,2′dATP | 0.5 | 0.4 | 0.63 ± 0.12 |
| (P5) ara-ATP | 0.7 | 0.5 | 0.18 ± 0.05 |
| (P6) 3′-dATP | 0.7 | 0.5 | 0.60 ± 0.11 |
| (P7) N1-methyl-ATP | 2.0 | 1.5 | 0.66 ± 0.05 |
| (P8) 2-amino-ATP | 0.3 | 0.2 | 1.03 ± 0.15 |
| (P9) 2-hydroxy-ATP | 3.3 | 2.4 | 0.40 ± 0.11 |
| (P10) N6-methyl-ATP | 0.7 | 0.5 | 0.43 ± 0.07 |
| (P11) ITP | 1.2 | 0.9 | 0.49 ± 0.03 |
| (P12) 6-mercaptopurine-RTP | 1.1 | 0.8 | 1.22 ± 0.23 |
| (P14) 6-methyl-thio-ITP | 0.8 | 0.6 | 0.16 ± 0.09 |
| (P15) 8-bromo-ATP | 0.9 | 0.7 | 1.24 ± 0.19 |
| (P16) 2′,3′ddGTP | 0.5 | 0.3 | 0.28 ± 0.05 |
| (P17) 2′dGTP | >3.5 | 3.4 | 0.39 ± 0.07 |
| (P18) 2′F,2′dGTP | >3.5 | >2.6 | 0.22 ± 0.04 |
| (P19) 2′-O-methyl-GTP | >3.5 | >2.6 | 0.24 ± 0.05 |
| (P20) 2′-deoxy-l-GTP | 1.8 | 1.3 | 0.11 ± 0.02 |
| (P21) 3′dGTP | >3.5 | >2.6 | 0.12 ± 0.02 |
| (P22) 3′-O-methyl-GTP | >3.5 | >2.6 | 0.35 ± 0.07 |
| (P23) N1-methyl-GTP | 3.5 | 2.6 | 0.49 ± 0.04 |
| (P24) XTP | >3.5 | >2.6 | 0.40 ± 0.09 |
| (P25) 2-amino-6-choloro-purine RTP | 2.7 | 2.0 | 0.33 ± 0.09 |
| (P26) 6-thio-GTP | N.D. | N.D. | 0.93 ± 0.17 |
| (P27) 6-methyl-thio-GTP | >3.5 | >2.6 | 0.40 ± 0.08 |
| (P28) O6-methyl-GTP | >3.5 | >2.6 | 0.17 ± 0.03 |
| (P29) m7-GTP | 1.4 | 1.0 | 0.14 ± 0.02 |
| (P31) 8-bromo-GTP | >3.5 | >2.6 | 0.19 ± 0.04 |
| (P32) 8-iodo-GTP | >3.5 | >2.6 | 0.54 ± 0.07 |
| (P33) Ribavirin triphosphate | 3.1 | 2.3 | 0.36 ± 0.05 |
The inhibition and phosphohydrolysis assays were performed a minimum of three times separately. Results are shown as the means of these independent experiments.
aThe phosphohydrolysis specific activities were calculated from the slopes of the titration curves and normalized to the specific activity for the hydrolysis of ATP.
Figure 1.
Homology model of the WNV RNA helicase active site. (A) Structural model of the WNV RNA helicase bound to a non-hydrolyzable ATP analog AMPPNP. The model was generated with the Deep View program using the crystal structure of the Dengue virus NS3 helicase domain bound to AMPPNP as a template (PDB code: 2JLR). The positions of the three helicase domains are indicated. (B and C) Two close-up views of the WNV NS3 active site are also shown, with emphasis on the residues in the vicinity of the AMPPNP adenine and ribose moieties.
Nucleotide analogs to probe the nucleotide specificity
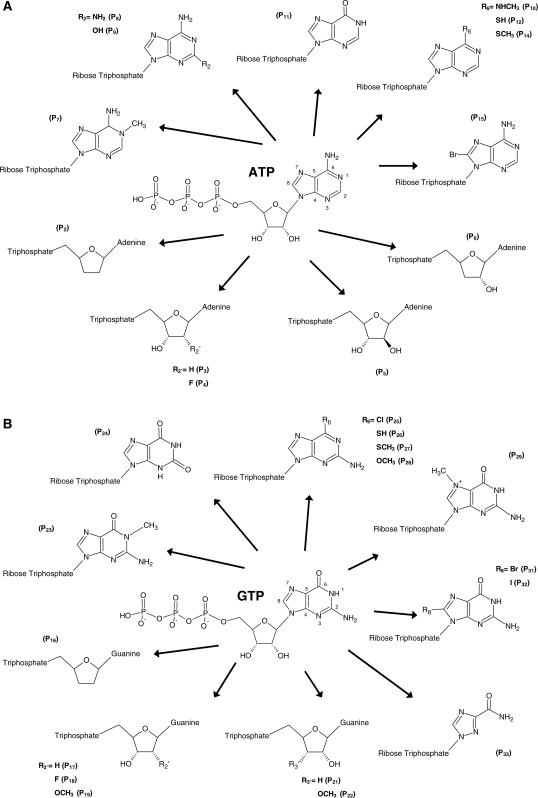
In order to experimentally confirm the homology model and to gain knowledge on how the WNV NS3 protein discriminates between ATP and other nucleotides, we used an extensive collection of nucleotide analogs to monitor their effects on the reaction chemistry. The nucleotide analogs used in the current study displayed numerous modifications both on the ribose and base moieties of the nucleotides (Figure 2A and B). The modifications ranged from relatively minor (modification of a single functional group) to severe (modification of multiple functional groups). Both ATP and GTP analogs were analyzed in order to investigate the specificity of the enzyme for nucleotides. ATP analogs were specifically chosen to investigate the importance of specific functional groups required for efficient hydrolysis, while a variety of GTP analogs were used to monitor if the addition and/or modification of specific functional groups on the molecule could result in an increase of the phosphohydrolase activity.
Figure 2.
Nucleotide analogs used to probe the active site of WNV NS3 helicase. Structure of the nucleotide analogs used in the current study. The ATP (A) and GTP (B) analogs harbor numerous modifications both on the ribose and base moieties of nucleotides.
ATP analogs
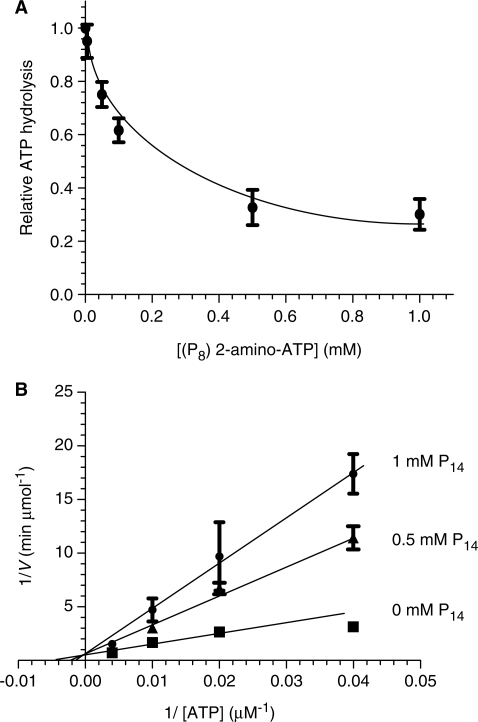
We initially monitored the ability of 13 analogs of ATP to inhibit the ATPase activity of the WNV NS3 protein by evaluating both the IC50 and Ki values for each molecule. An important number of ATP analogs had the ability to inhibit the ATPase reaction, albeit to different extents, as reflected by the apparent IC50 and Ki values (Table 1). All the ATP analogs used in the current study were competitive inhibitors of the ATPase reaction indicating that they bind to the active site of the enzyme. Typical examples using analogs P8 (2-amino-ATP) and P14 (6-methyl-thio-ITP) are shown in Figure 3. In order to gain additional information on the discrimination between ATP and other nucleotides, we also determined the ability of the various ATP analogs to be hydrolyzed by the enzyme. The WNV NS3 protein catalyzed the phosphohydrolysis of all the ATP analogs tested in the current study with specific activity ranging from 0.16 to 1.24 times the hydrolysis of ATP (Table 1), thereby underscoring the high structural flexibility of the active site.
Figure 3.
Inhibition of the phosphohydrolase activity by 6-methyl-thio-ITP. (A) Dose-response inhibition of the ATPase activity by 2-amino-ATP (P8). (B) Competitive inhibition of the ATPase reaction by 6-methyl-thio-ITP (P14). The ATPase activity was evaluated in the absence (filled square) or presence of 0.5 (filled triangle) or 1 mM (filled circle) of P14. Each value represents the average of at least three independent experiments. Error bars represent the S.D. of the mean values.
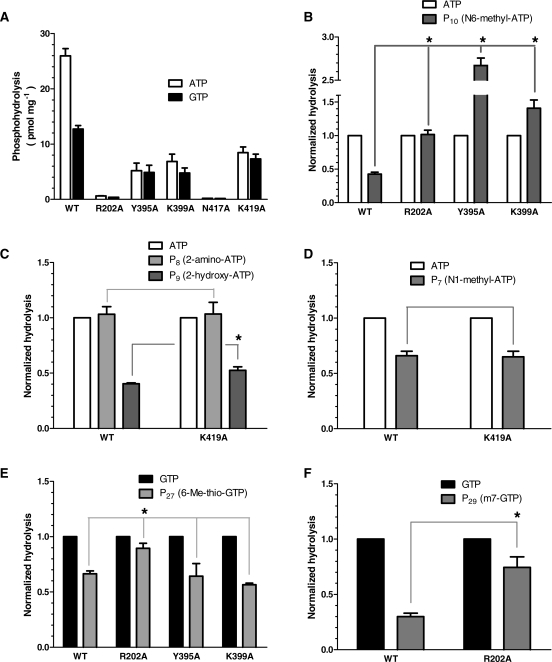
Our study indicates that the ATPase active center can accommodate ATP analogs displaying a number of unusual chemical modifications. The analogs harboring chemical modifications on the adenine base of the ATP molecule (Figure 2A) helped to illuminate the flexibility of the ATPase active site. The 6-amino group of the adenine ring particularly appears important for ATP recognition/alignment/hydrolysis since it possesses both hydrogen bond donor and acceptor capacities. We initially suspected that the ability of this amino group (which is substituted by a 6-oxo group in GTP) to act as a hydrogen bond donor might be a key determinant promoting the catalytic usage of ATP over GTP. The phosphohydrolysis activity of four ATP analogs harboring chemical modifications at this position (P10, P11, P12 and P14) provided clues on the importance of this functional group. We observed that the absence of a hydrogen bond donor at this position considerably decreased the ability of analogs P11 (ITP) and P14 (6-methyl-thio ITP) to be hydrolyzed by the enzyme (Table 1). In contrast, substitution of the 6-amino group of ATP by a thiol group (P12, 6-thio ATP), which is a moderately good hydrogen bond donor (42), resulted in an efficient hydrolysis of the molecule (Table 1). We concluded that the presence of a functional group which can display hydrogen bond donor properties is critical at the C6 position of adenine. However, we observed that the hydrolysis of the P10 analog (N6-methyl ATP), which still displays hydrogen bond donor/acceptor properties, was significantly reduced in comparison to ATP (Table 1). We hypothesized that the addition of the bulkier methyl group at position 6 weakens the ability of the substrate to be hydrolyzed through steric hindrance. To further investigate this possibility, we generated a series of mutants based on amino acids located in the vicinity of the N6 position of ATP. Close inspection of the generated model of the WNV NS3 protein active site suggests that four amino acids are located in proximity to the N6 atom (Figure 1C). These are Arg-202, Tyr-395, Lys-399 and Asn-417, which appear just too far removed to hydrogen-bond with the 6-amino group of ATP, but might potentially interfere with the larger substituent displayed by the P10 analog. The importance of these amino acids was therefore investigated by generating four distinct enzymatic mutants. Arg-202, Tyr-395, Lys-399 and Asn-417 were individually substituted by alanine, and the mutant polypeptides were expressed and purified in parallel with the wild-type enzyme. SDS–polyacrylamide gel electrophoresis analysis showed that the different mutants were expressed at similar levels as the wild-type enzyme (Supplementary Figure S1). It should be noted that the R202A, Y395A, K399A and N417A mutations elicited a significant decrease in the ATPase activity (Figure 4A, Supplementary Figure S2 and Supplementary Table S1), thereby highlighting an important role of these amino acids in the reaction catalyzed by the WNV NS3 protein. The informative finding of the mutagenesis studies is that the replacement of these lateral side chains by alanine resulted in an increase in the relative phosphohydrolysis of the P10 analog which harbors a large NHCH3 substituent at the C6 position (Figure 4B). The relative hydrolysis of the P10 analog (in comparison to ATP) increased from 0.43 for the wild-type enzyme to 1.02, 2.67 and 1.41 for the R202A, Y395A and K399A mutants, respectively (Figure 4B and Supplementary Table S1). The ability of the N417A mutant to hydrolyze the P10 analog could not be investigated because the mutation completely abolished the ATPase activity of the enzyme. Overall, the mutagenesis data are consistent with steric hindrance preventing the phosphohydrolysis of the P10 analog.
Figure 4.
Phosphohydrolysis activity of WNV RNA helicase mutants. (A) The phosphohydrolase activities of the wild-type enzyme and various mutant NS3 proteins (100 nM) were evaluated in the presence of ATP (100 µM) or GTP (100 µM). (B–D) Normalized phosphohydrolysis of the wild-type enzyme and various NS3 mutants in the presence of different ATP analogs (100 µM). (E–F) Normalized phosphohydrolysis of the wild-type enzyme and various NS3 mutants in the presence of different GTP analogs (100 µM). Each value represents the average of at least three independent experiments. Error bars represent the SD of the mean values. The asterisks indicate that the levels of hydrolysis were significantly different (P < 0.05) than the hydrolysis of the same substrate by the wild-type enzyme. The respective P-values were calculated using Student’s unpaired t-test.
The addition of a halogen substituent with a large atomic radius at the C8 position of the adenine base (P15, 8-bromo ATP) had almost no effect on the phosphohydrolysis activity, most likely reflecting the absence of amino acid of functional importance in the vicinity of the adenine C8 position, and the solvent exposure of the adenine base. Similarly, the addition of an amino group at the C2 position of adenine (P8, 2-amino-ATP) had no significant impact on the catalytic activity of the enzyme. In contrast, the addition of a hydroxyl group at the same position (P9, 2-hydroxy ATP) resulted in a decrease of the phosphohydrolysis activity (Table 1). We speculated that the additional 2-OH group probably makes an unfavorable interaction with an amino acid located in the vicinity of the adenine C2 position. Closer examination of the structural model of the WNV NS3 protein indicates that Lys-419 is the amino acid in closest proximity to the C2 position of adenine (Figure 1C). The importance of the residue was therefore investigated through site-directed mutagenesis. The WNV Lys-419 residue was substituted by alanine, and the mutant polypeptide was expressed and purified in parallel with the wild-type enzyme. Again, the substitution of this residue by alanine elicited a decrease in the ATPase activity of the enzyme, underscoring the importance of the residue in catalysis (Figure 4A, Supplementary Figure S2 and Table S1). The informative finding is that the replacement of the lysine lateral side chain by alanine resulted in a small but significant increase in the relative hydrolysis of the 2-OH ATP analog (Figure 4C). The relative hydrolysis (in comparison to ATP) increased from 0.40 for the wild-type enzyme to 0.53 for the K419A mutant (Figure 4C and Supplementary Table S1). Although the exact nature of the interaction between Lys-419 and the additional 2-OH group remains elusive, our mutagenesis study clearly indicates that the presence of this lysine residue impairs the ability of the 2-OH ATP analog to be hydrolyzed efficiently by the enzyme.
The addition of a methyl group to the N1 position of adenine had a negative effect on the phosphohydrolysis activity. The analog harboring such a modification (P7, N1-methyl ATP) was hydrolyzed to a lesser extent than ATP by the enzyme (Table 1). We reasoned that two factors might be responsible for this reduction in activity. The addition of the methyl group could result in steric hindrance with active site residues, thereby limiting the hydrolysis of the substrate. Alternatively, the alteration of the N1 atom ability to act as hydrogen bond acceptor in the P7 analog might be responsible for the observed decreased hydrolysis. Closer examination of the modelized active site of the WNV NS3 protein (Figure 1C) indicates that Lys-419 is again the amino acid in closest proximity to the N1 position of adenine. In order to illuminate the molecular determinants accountable for the lower catalytic usage of the P7 analog, we once again relied on the K419A mutant which displayed a significant decrease in the ability of the enzyme to hydrolyze ATP (25.9 pmol/mg for the wild-type enzyme versus 8.5 pmol/mg for the K419A mutant) (Figure 4A, Supplementary Figure S2 and Table S1). Our data indicated that the hydrolysis of the N1-methyl ATP analog by the K419A mutant did not result in an increase of the catalytic activity (Figure 4D), thereby revealing that steric hindrance is not a likely factor that contributes to the observed reduction in hydrolysis of the P7 analog. Therefore, our structural model coupled with the mutagenesis data suggest that the Lys-419 residue makes an important interaction with the adenine N1 atom, and that the additional presence of a methyl group at this position significantly impairs the ability of the N1-methyl ATP analog to be hydrolyzed efficiently by the enzyme.
It should be noted that the various mutants that were generated throughout this study are only affecting the hydrolysis of analogs that display chemical modifications at the corresponding positions (Supplementary Figure S3A and B). For instance, the R202A mutation resulted in an increase of the hydrolysis of analogs harboring substitutions at the O6, N7 and C8 positions of the adenine (analogs P10, P29, P31 and P32) but the mutation has no effect on the hydrolysis of analogs harboring modifications at other positions such a the C2 of adenine or the 2′OH of the ribose (analogs P5, P8 and P9) (Supplementary Figure S3A). A similar conclusion is also reached for both the K399A and K419A mutants (Supplementary Figure S3A and B).
GTP analogs
Under the standard steady-state conditions used in our assays, GTP was hydrolyzed approximately two times less efficiently than the ATP substrate (Table 1), highlighting the discriminating ability of the enzyme between various NTPs. We therefore set out to use a collection of GTP analogs (Figure 2B) to evaluate if the addition and/or modification of specific functional groups could increase the capacity of the enzyme to use these molecules as substrates during the phosphohydrolase reaction.
As reported above, one of the most striking features of the ATP-enzyme complex formation is the importance of the hydrogen bond donor properties of the 6-amino group of adenine. Not surprisingly, GTP which lacks a hydrogen bond donor at that position is not hydrolyzed as efficiently by the WNV NS3 protein. In that regard, P26–28 are particularly interesting analogs since they only differ with GTP by the substitution of the 6-oxo moiety by various functional groups (Figure 2B). The importance of the hydrogen bond donor ability at this position is again highlighted by the fact that analog P26 (6-thio GTP) which displays a thiol group with hydrogen bond donor properties is hydrolyzed 1.8 times more efficiently by the enzyme than GTP (Table 1). In contrast, replacement of the 6-oxo group of GTP by SCH3 (P27) or OCH3 (P28), that both lack the capacity to act as hydrogen bond donors, had a negative effect on the hydrolysis of these analogs in comparison to GTP (Table 1). The hydrolysis of the 6-methyl-thio GTP (P27) and O6-methyl GTP analog (P28) were reduced by 18 and 65%, respectively. These results highlighted the importance of the hydrogen bond donor properties at the C6 position. However, the results also raised the possibility that the addition of the bulkier methyl group at position 6 weakens the ability of the substrate to be hydrolyzed by the WNV NS3 protein. To investigate that possibility, we took advantage of the R202A, Y395A and K399A mutants since the side chains of these residues are in the vicinity of the C6 position, and they can potentially interfere with the larger substituent displayed by the P27 analog. The replacement of Arg-202, Tyr-395 and Lys-399 by alanine did not result in an increased ability of these mutants to hydrolyze the P27 analog (Figure 4E), thereby indicating that steric hindrance is not a factor that contributes to the reduced hydrolysis of the P27 analog.
One of the most striking effect on the GTPase activity was obtained upon methylation of the N7 atom of the guanine ring (P29, 7-methyl GTP). This modification significantly impaired the ability of the enzyme to use GTP as a substrate which resulted in a decrease of 71% (Table 1) of the hydrolysis activity (relative to GTP). We therefore hypothesized that the nitrogen atom is involved in a critical hydrogen bond interaction with an amino-acid side chain that can act as a hydrogen bond donor. Alternatively, the addition of the methyl group could also result in steric hindrance. Analysis of the structural model of the WNV NS3 protein active site revealed that the N7 atom of the adenine base is in close proximity to the η1-amino group of Arg-202 (Figure 1B and C). However, closer inspection of the model also reveals that the N7 atom appears to make a rather weak interaction with Asn-417. The importance of these two residues in the WNV NS3 protein was again investigated through site-directed mutagenesis. As we reported above for the ATPase activity, substitution of the Arg-202 and Asn-417 residues by alanine also elicited a severe decrease in the GTPase reaction catalyzed by the enzyme, thereby highlighting the importance of these residues for the NTPase activity (Figure 4A and Supplementary Figure S2). In fact, substitution of the Asn-417 residue by alanine completely abolished the GTPase activity of the enzyme. The replacement of the Arg-202 by alanine resulted in a significant increase in the phosphohydrolysis of the 7-methyl GTP (P29) (Figure 4F). The relative hydrolysis (in comparison to GTP) increased from 0.29 for the wild-type enzyme to 0.75 for the R202A mutant (Figure 4F and Supplementary Table S2), thereby underscoring the fact that steric hindrance most likely prevents the binding/catalysis of the P29 analog. The mutagenesis data are consistent with a role of the N7 atom of the purine ring in substrate recognition by the WNV NS3 protein.
Overall, the phosphohydrolysis assays performed with both ATP and GTP analogs allowed us to identify the important structural and functional features for the interaction of the enzyme with the NTP substrate. It should be noted that similar conclusions on the importance of specific functional groups were reached when the phosphohydrolysis assays were performed in the presence of single-stranded RNA, a known stimulator of the NTPase activity of the flavivirus NS3 protein (Supplementary Figure S4).
Effects of mutations on the ATP/GTP specificity
In order to better understand the nucleotide binding activity of the WNV helicase, we set out to identify specific amino-acid residues that might be responsible for this discrimination between ATP and GTP by relying on structural information obtained from other nucleic-acid-dependent NTP phosphohydrolases. For instance, the adenine nucleotide-bound crystal structures of UvrB, UvrD and Hera have shown that the glutamine residue of Q-motif, usually located 21 amino acids upstream of the motif I, makes bidentate hydrogen contacts with the adenine N6 and N7 atoms (33,43–46). It has been suggested that this glutamine residue might enforce specificity for adenine nucleotides (46). In fact, recent structure/function studies of the mycobacterial UvrD1 helicase clearly demonstrated a role for this particular amino-acid residue in dictating substrate specificity (47).
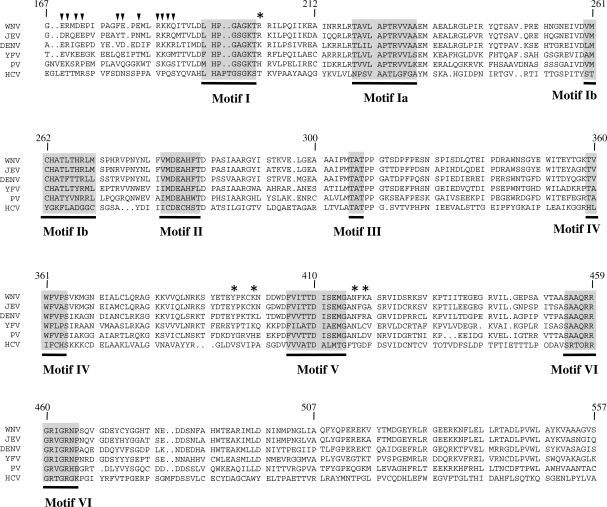
In contrast to many DEAD/H box helicases, the RNA helicases of the Flaviviridae do not harbor the classical Q-motif. In the case of the WNV NS3 protein, a single glutamine residue (Gln-188) is found in this region. Its position (11 amino acids upstream of the glycine of motif I) makes it an unlikely candidate to play an identical role as the conserved glutamine found in the Q-motif. We therefore monitored the effects of single alanine mutations at 11 positions located upstream of motif I (indicated by arrows in Figure 5). The mutant polypeptides were expressed and purified in parallel with the wild-type protein (Supplementary Figure S1), and the NTPase activities of the mutants were assessed and compared with the wild-type enzyme. Table 3 reports the kinetic parameters of both ATP and GTP hydrolysis for each mutant under steady-state conditions. Substitution of the Gln-188 residue by alanine resulted in a 4.6-fold decrease of the affinity of the enzyme for ATP, as evidenced from the higher Km value (Table 3). Similarly, the substitution of Lys-187 by alanine also resulted in a significantly lower affinity of the mutant enzymes for ATP. Surprisingly, the replacement of Arg-185 by alanine completely abrogated the ATPase activity of the enzyme, thereby highlighting the critical importance of this residue for the ATPase reaction. The importance of the amino acids located upstream of motif I is also highlighted by the effects of Ala-substitutions on the other residues of that region. Replacement of Glu-169, Arg-170, Asp-172, Glu-173, Phe-179, Glu-180, Glu-182 and Lys-186 by alanine resulted in a decrease in the rate of the ATP hydrolysis, as evidenced both from the Vmax and kcat values (Table 3). Overall, the entire region located upstream of motif I appears important for the regulation of the catalytic activity as reflected by the decrease in the kcat/KM constants of the mutants. It should be noted that the decreased phosphohydrolytic activities of the mutants are not the result of protein misfolding, as evidenced from circular dichroism assays showing that the mutant polypeptides are correctly folded (Supplementary Figure S5). Moreover, the mutants that are severely affected in their phosphohydrolase activity retain other functions. For instance, we evaluated the ability of three highly deficient mutants (R185A, R202A and N417A) to bind an RNA substrate using fluorescence spectroscopy. Our binding assays demonstrated that both the R185A and N417A mutants bind RNA with a similar affinity than the wild-type enzyme (as judged by the Kd values), while the R202A has a 5-fold decrease in its affinity for the RNA substrate (Supplementary Table S3).
Table 2.
Key interactions between the active site residues of the WNV NS3 protein and ATP as predicted by the structural homology model
| ATP (atom) | Amino acid | Distance from ATP (Å) |
|---|---|---|
| N6 | Arg202 (η1) | 5.9 |
| N7 | Arg202 (η1) | 3.1 |
| C8 | Arg202 (η1) | 2.2 |
| N6 | Tyr395 | 7.4 |
| N6 | Lys399 | 8.0 |
| N6 | Asn417 | 6.0 |
| N7 | Asn417 | 3.9 |
| C8 | Asn417 | 3.3 |
| N1 | Lys419 | 5.2 |
| C2 | Lys419 | 5.3 |
| β-PO4 | Lys200 (main chain amide) | 3.1 |
| β-PO4 | Lys200 (main chain amide) | 2.8 |
| γ-PO4 | Lys200 (main chain amide) | 2.6 |
| β-PO4 | Thr201 | 3.0 |
| α-PO4 | Arg202 (main chain amide) | 2.8 |
| γ-PO4 | Glu286 (O′) | 3.3 |
| γ-PO4 | Glu286 (O′′) | 3.1 |
| 3′OH | Asn330 | 3.4 |
| γ-PO4 | Arg461 | 2.9 |
| 3′OH | Arg464 (main chain carbonyl) | 2.7 |
| γ-PO4 | Arg464 (η1) | 2.8 |
| γ-PO4 | Arg464 (η2) | 3.2 |
Figure 5.
Sequence alignment of the Flaviviridae RNA helicases. An amino acid alignment of the RNA helicases from WNV, JEV, DENV, YFV, Powassan virus (PV) and HCV is presented. The conserved helicase motifs are shown in grey. Arrows (inverted filled triangle) indicate amino acids that were targeted for mutagenesis in the nucleotide specificity region. Asterisks represent amino acids that are located in the vicinity of the ATP hydrolysis site, and that were targeted for mutagenesis. The numbers above the alignment indicate the amino acid boundaries of the WNV NS3 protein. The conserved sequence motifs found in the flaviviral helicases are also shown under the aligned sequences.
Table 3.
Characterization of the NS3 mutants located in the nucleotide specificity region found upstream of motif I
| Protein | Substrate | KM (µM) | Vmax (pmol min−1) | kcat (min−1) | kcat/KM (µM−1 min−1) | Discrimination (kcat/KM)ATP/(kcat/KM)GTP | Relative ATP/GTP discrimination | ||
|---|---|---|---|---|---|---|---|---|---|
| NS3 WT | ATP | 140 | 700 | 180 | 1.29 | 2.72 | 1.00 | ||
| GTP | 350 | 650 | 165 | 0.47 | |||||
| NS3 E169A | ATP | 220 | 295 | 75 | 0.34 | 1.03 | 0.38 | ||
| GTP | 340 | 440 | 110 | 0.33 | |||||
| NS3 R170A | ATP | 200 | 241 | 60 | 0.31 | 0.84 | 0.31 | ||
| GTP | 270 | 400 | 100 | 0.37 | |||||
| NS3 D172A | ATP | 180 | 133 | 35 | 0.19 | 1.11 | 0.41 | ||
| GTP | 330 | 220 | 55 | 0.17 | |||||
| NS3 E173A | ATP | 200 | 210 | 55 | 0.27 | 0.47 | 0.17 | ||
| GTP | 110 | 250 | 65 | 0.58 | |||||
| NS3 F179A | ATP | 180 | 273 | 70 | 0.39 | 0.53 | 0.19 | ||
| GTP | 90 | 220 | 65 | 0.73 | |||||
| NS3 E180A | ATP | 240 | 257 | 65 | 0.28 | 0.96 | 0.35 | ||
| GTP | 270 | 750 | 80 | 0.29 | |||||
| NS3 E182A | ATP | 270 | 790 | 200 | 0.75 | 0.81 | 0.29 | ||
| GTP | 230 | 840 | 215 | 0.93 | |||||
| NS3 R185A | ATP | Inactive | |||||||
| GTP | |||||||||
| NS3 K186A | ATP | 390 | 630 | 160 | 0.41 | 0.40 | 0.15 | ||
| GTP | 100 | 400 | 100 | 1.02 | |||||
| NS3 K187A | ATP | 940 | 502 | 130 | 0.14 | 0.21 | 0.08 | ||
| GTP | 90 | 240 | 60 | 0.68 | |||||
| NS3 Q188A | ATP | 640 | 790 | 200 | 0.32 | 0.91 | 0.33 | ||
| GTP | 290 | 400 | 100 | 0.35 | |||||
The phosphohydrolysis assays were performed a minimum of three times separately. Results are shown as the means of these independent experiments.
We next sought to investigate whether these residues located upstream of motif I could be responsible for preferential binding of ATP over GTP. The kinetic parameters for the hydrolysis of both ATP and GTP under steady-state conditions are presented in Table 3. The nucleotide discrimination is defined by the ratio (kcat/KM)ATP/(kcat/KM)GTP. Interestingly, the substitutions of the targeted amino acids located upstream of motif I by alanine resulted in a significant loss of discrimination between GTP and ATP. In fact, the E173A, F179A, K186A and K187A mutants hydrolyzed GTP much more efficiently than the ATP substrate, as evidenced from the kcat/KM constants (Table 3). These results provide a clear evidence for a regulatory role for the region located upstream of motif I as a substrate specificity filter.
Since the mutants located upstream of motif I affect the discrimination between ATP and GTP, we monitored the ability of two of these mutants (E173A and K187A) to hydrolyze specific nucleotide analogs. We therefore selected GTP analogs (P11, P23 and P24) harboring substitutions at positions that were shown to affect the binding of nucleotides (N6, C1 and C2) and compared their hydrolysis with the levels obtained in the presence of ATP (Supplementary Figure S6). Our results clearly demonstrate that the substitution of Glu-173 or Lys-187 by alanine results in an increase in the hydrolysis of these GTP analogs (Supplementary Figure S6). This further demonstrates the role for the region located upstream of motif I as a substrate specificity filter.
DISCUSSION
Our study illustrates the important structural and functional features for the interaction of ATP with the WNV NS3 protein. More importantly, it provides a molecular rationale for the preference of the enzyme for the ATP substrate in the following respects: (i) by highlighting the specific functional groups on the nucleotide substrate that are important for binding/catalysis; (ii) by demonstrating the importance of specific amino acids located outside of the conserved helicase domains for the ATPase activity; and (iii) by providing a role for the region located upstream of motif I as a key determinant for the ATP/GTP discrimination.
The combination of nucleotide analogs, site-directed mutagenesis and the use of a structural model exposed the important structural and functional features for the interaction of the flaviviral helicase with ATP. The use of nucleotide analogs highlighted both the complexity and flexibility of the flaviviral ATPase active site. An informative finding was that every ATP analog tested had the ability to inhibit the ATPase reaction and to be used as a substrate, albeit to different extents, thus underscoring the high structural flexibility of the active site. A previous study monitoring the effects of four ATP analogs (with modified bases) on the unwinding activity of the HCV RNA helicase reported that NTPs lacking hydrogen at position N1 of the purine ring supported faster unwinding than NTPs harboring a protonated N1 group (34). The authors concluded that the single greatest factor affecting the rates of unwinding fuelled by NTPs was the hydrogen-bond acceptor/donor properties of the N1 nitrogen (34). In light of the current study, the interaction between the WNV NS3 protein and ATP appears more complex than initially anticipated. The use of a large collection of ATP and GTP analogs allowed us to demonstrate the critical importance of the hydrogen bond donor properties at the C6 position of the nucleotide substrate. The importance of this chemical property is reflected by the fact that the only GTP analog that was hydrolyzed very efficiently by the enzyme was 6-thio-GTP (P26), the single GTP analog displaying hydrogen-bond donor capacities at the C6 position. Interestingly, a previous bioinformatic analysis of the Protein Data Bank performed on enzymes that interact with ATP or GTP (48) revealed that the ligand atoms most commonly involved in recognition are the hydrogen bond donors (N6 in ATP) rather than the acceptors (N1, N3 and N7 in ATP). In accordance with the observations, the importance of the N6 atom of ATP for molecular recognition was clearly demonstrated in the present study of the WNV RNA helicase. The use of nucleotide analogs also allowed us to demonstrate that the presence of a functional group which can display hydrogen bond acceptor properties is important at the N1 position of adenine.
The library of nucleotide analogs used in the current study did not allow us to unambiguously demonstrate the precise molecular contribution of the N7 atom to the binding of the purine analog. This atom has the same hydrogen-bond donor abilities in both ATP and GTP, yet the enzyme has a clear bias towards the ATP substrate. However, the mutagenesis data are undoubtedly consistent with a role of the N7 atom in substrate recognition by the WNV NS3 protein. The fact that the substitution of the two amino acids located in the vicinity of the N7 atom (Arg-202 and Asn-417) elicited a severe decrease in both the ATPase and GTPase activities strongly suggests that the residues are hydrogen-bonding to the N7 atom of both purines. We therefore hypothesize that the presence of a functional group which can display hydrogen-bond acceptor properties at the N7 position of the purine substrate is important for binding.
Although the current study largely focused on the importance of specific molecular determinants of the purine rings required for efficient binding/catalysis, some of the analogs contained modifications on the ribose moiety of the nucleotide substrate. Based both on the structural model that we generated and on the available crystal structures of other flaviviral RNA helicases bound to ATP (or ATP analogs), a limited number of interactions appear to occur between the enzymes and the 2′-OH and 3′-OH groups of ATP. Not surprisingly, ATP analogs harboring various modifications/substitutions on the ribose moieties were hydrolyzed by the WNV NS3 protein, albeit with a lower catalytic efficiency (Table 1). The removal of either 2′- or 3′-hydroxyl group or both (P2, P3 and P6) lowered the relative hydrolysis of these substrates by respectively 47, 38 and 40%. Although these data indicate that the ribose moiety of ATP is important for optimal recognition/hydrolysis of the purine nucleotide, the enzyme makes sufficient additional contacts with the ATP substrate to promote efficient catalysis. The interaction between the complete ATP molecule and the enzyme is clearly supported by an intricate network of hydrogen bonds and electrostatic interactions, which explains the ability of the enzyme to hydrolyze numerous analogs that lack key functional groups.
By performing a mutational analysis of 16 different residues of the WNV NS3 protein, we have identified three new amino acids that are essential for the ATPase activity. Interestingly, these three essential residues (Arg-185, Arg-202 and Asn-417) are highly conserved in all the flaviviruses (WNV, yellow fever virus, DENV), but are divergent in hepacivirus (HCV) (Figure 5). Based on the structural model of the WNV RNA helicase, both Arg-202 and Asn-417 appear to be making multiple contacts with the adenine base. However, the precise role of the Arg-185 residue in catalysis remains unsolved at the moment since it is removed from the ATP substrate in the structural model. Similarly, the structural model provides no immediate explanation for the precise role of the nucleotide specificity region (amino acids 169–188) located upstream of motif I since the majority of this region is absent from the DENV NS3 crystal structure that was used to generate the model for the interaction between the WNV NS3 protein and ATP. We speculate that two plausible scenarios could explain how these residues influence the activity of the enzyme: (i) an indirect effect whereby they interact with other amino acids that are involved in catalysis, or (ii) a direct effect by which they interact with the ATP substrate following a conformational change. Because of their close proximity to motif I, the residues responsible for the ATP/GTP discrimination might contact amino acids that directly interact with the ATP substrate. Closer examination of the crystal structures of other helicases suggest that the corresponding region is in close proximity to motif I. For instance, in eIF4A two residues of the Q-motif form hydrogen bonds with the threonine and glycine residues of motif I (33). The second scenario by which residues located upstream of motif I could influence the activity of the enzyme is through a modification of the protein structure during catalysis. In this context, the residues located upstream of motif I could come into contact with ATP following a conformational change. Interestingly, biochemical evidences for a conformational change that occurs upon nucleotide binding have been observed for the related HCV NS3 protein (34). This conformational switch would allow the enzyme to act like a molecular motor on the double-stranded RNA in order to unwind the RNA duplex (34). Clearly future studies will aim at identifying the precise molecular mechanism by which the nucleotide specificity region modulates the phosphohydrolase activity of helicases.
The region located upstream of motif I plays a role not only in regulating the activity of the enzyme, but also in dictating phosphohydrolase substrate specificity. Our study provides clear evidence for a gain-of-function of the E173A, F179A, K186A and K187A mutants which hydrolyzed GTP much more efficiently than the ATP substrate. Thus, the residues of this region clearly play a functional role in nucleotide discrimination. This is reminiscent of the situation observed for the Mycobacterium uvrD1 DNA helicase in which a specific glutamine (Gln-24) of the Q-motif is responsible for the specificity of the enzyme for ATP (47). However, two differences appear to distinguish the WNV NS3 protein from the Mycobacterium enzyme. First, the sequence of nucleotide specificity region of the WNV NS3 protein is divergent from the classical Q-motif. Second, the loss of the glutamine residue in the Mycobacterium enzyme reduces ATP affinity only modestly (47). In the case of the WNV NS3 protein, mutations in the nucleotide specificity region invariably result in a decrease of phosphohydrolysis. A rationale to explain the precise mechanism by which the residues of the nucleotide specificity region exert their effects will ultimately hinge on obtaining crystal structures of the enzyme bound to its nucleotide substrate at discrete functional states along the reaction pathway. In spite of these differences, we propose that the region located upstream of motif I of the WNV NS3 protein is a nucleotide specificity region that plays a functional role in nucleotide discrimination which is reminiscent of the role exerted by the Q-motif found in other helicases.
The current study did not allow us to distinguish between binding and hydrolysis events that are occurring during the phosphohydrolysis reaction. Preliminary assays using various techniques such as fluorescence spectroscopy indicated that metal ions are crucial for the nucleotide-binding activity of the WNV NS3 enzyme (data not shown). The addition of the metal ions to the binding reaction therefore results in the hydrolysis of nucleotides or nucleotide analogs. The use of non-hydrolyzable nucleotide analogs, which are not currently available, could eventually allow us to distinguish between binding and hydrolysis in order to evaluate the molecular determinants involved in both events.
Although flaviviruses are medically relevant, no antiviral therapy is currently available for the clinical treatment of these pathogens. Because of its importance in the replication of the virus, the WNV NS3 protein is an attractive target for the future development of inhibitors targeting the virus. Some of the analogs identified in the current study appear as good starting blocks for the design of more specific inhibitors. For instance, 2′,3′ ddGTP (P16), which bound very tightly to the enzyme active site but was not efficiently hydrolyzed, would be a good candidate to generate novel compounds that could display anti-WNV properties. The potential of nucleotide analogs to inhibit WNV replication is highlighted by previous studies that demonstrated that the broad-spectrum antiviral ribavirin triphosphate, a GTP analog, has inhibitory activity against WNV in cell culture (49). Recent evidences suggest that the antiviral may interfere with the capping of the viral mRNAs, inhibit the viral polymerase and induce transition mutations in the viral genome via ambiguous base-pairing during replication (50). Both structural and functional studies are clearly beginning to reveal new insights that should eventually lead to the design of effective anti-flaviviral drugs to ultimately cure infected patients.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Sciences and Engineering Research Council of Canada and the Canadian Institutes for Health Research (CIHR). The RNA group is supported by a grant from the CIHR and by the Université de Sherbrooke. M.B. is a Chercheur Boursier Junior 2 from the Fonds de Recherche en Santé du Québec and a member of the Infectious Diseases Group of the Centre de Recherche Clinique Étienne-Le Bel. Funding for open access charge: Canadian Institutes of Health Research.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.de la Cruz J, Kressler D, Tollervey D, Linder p. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuller-pace FV. RNA helicases: modulators of RNA structure. Trends Cell Biol. 1994;4:271–274. doi: 10.1016/0962-8924(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 3.Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationship. Curr. Opin. Struct. Biol. 1993;3:419–429. [Google Scholar]
- 4.Hodgman TC. A new superfamily of replicative proteins. Nature. 1988;333:22–23. doi: 10.1038/333022b0. [DOI] [PubMed] [Google Scholar]
- 5.Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, Chu S. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 6.Lohman TM, Bjornson KP. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 7.Maluf NK, Fischer CJ, Lohman TM. A dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J. Mol. Biol. 2003;325:913–935. doi: 10.1016/s0022-2836(02)01277-9. [DOI] [PubMed] [Google Scholar]
- 8.Patel SS, Picha KM. Structure and function of hexameric helicases. Annu. Rev. Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 9.Soultanas P, Wigley DB. DNA helicases: “inching forward”. Curr. Opin. Struct. Biol. 2000;10:124–128. doi: 10.1016/s0959-440x(99)00059-7. [DOI] [PubMed] [Google Scholar]
- 10.Hall MC, Matson SW. Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol. 1999;34:867–877. doi: 10.1046/j.1365-2958.1999.01659.x. [DOI] [PubMed] [Google Scholar]
- 11.Subramanya HS, Bird LE, Brannigan JA, Wigley DB. Crystal structure of a DExx box DNA helicase. Nature. 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 12.Koroloev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 13.Yao N, Hesson T, Cable M, Hong Z, Kwong AD, Le HV, Weber pC. Structure of the hepatitis C virus RNA helicase domain. Nat. Struct. Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- 14.Kim JL, Morgenstern KA, Griffith JP, Dwyer MD, Thomson JA, Murcko MA, Lin C, Caron PR. Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding. Structure. 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 15.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 16.Kwong AD, Rao BG, Jeang KT. Viral and cellular RNA helicases as antiviral targets. Nat. Rev. Drug Disc. 2005;4:845–853. doi: 10.1038/nrd1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frick DN, Lam AMI. Understanding helicases as a means of virus control. Curr. Pharm. Des. 2006;12:1315–1338. doi: 10.2174/138161206776361147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grassmann CW, Isken O, Behrens SE. Assignment of the multifunctional NS3 protein of bovine viral diarrhea virus during RNA replication: an in vivo and in vitro study. J. Virol. 1999;73:9196–9205. doi: 10.1128/jvi.73.11.9196-9205.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gwack Y, Kim DW, Han JH, Choe J. Characterization of RNA binding activity and RNA helicase activity of the hepatitis C virus NS3 protein. Biochem. Biophys. Res. Commun. 1996;225:654–659. doi: 10.1006/bbrc.1996.1225. [DOI] [PubMed] [Google Scholar]
- 20.Utama A, Shimizu H, Morikawa S, Hasebe F, Morita K, Igarashi A, Hatsu M, Takamizawa K, Miyamura T. Identification and characterization of the RNA helicase activity of Japanese encephalitis virus NS3 protein. FEBS Lett. 2000;465:74–78. doi: 10.1016/s0014-5793(99)01705-6. [DOI] [PubMed] [Google Scholar]
- 21.Utama A, Shimizu H, Hasebe F, Morita K, Igarashi A, Shoji I, Matsuura Y, Hatsu M, Takamizawa K, Hagiwara A. Role of the DExH motif of the Japanese encephalitis virus and hepatitis C virus NS3 proteins in the ATPase and RNA helicase activities. Virology. 2000;273:316–324. doi: 10.1006/viro.2000.0417. [DOI] [PubMed] [Google Scholar]
- 22.Borowski P, Mueller O, Niebuhr A, Kalitzky M, Hwang LH, Schmitz H, Siwecka MA, Kulikowsk T. ATP-binding domain of NTPase/helicase as a target for hepatitis C antiviral therapy. Acta Biochim. Pol. 2000;47:173–180. [PubMed] [Google Scholar]
- 23.Kim DW, Gwack Y, Han JH, Choe J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem. Biophys. Res. Commun. 1995;215:160–166. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- 24.Mackenzie JS, Gubler DJ, Petersen LR. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 2004;10:S98–S109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- 25.Sarbah S, Younossi Z. Hepatitis C: an update on the silent epidemic. J. Clin. Gastro. 2000;30:125–143. doi: 10.1097/00004836-200003000-00005. [DOI] [PubMed] [Google Scholar]
- 26.Hayes EB, Gubler DJ. West Nile virus: epidemiology and clinical features of an emerging epidemic in the United States. Annu. Rev. Med. 2006;57:181–194. doi: 10.1146/annurev.med.57.121304.131418. [DOI] [PubMed] [Google Scholar]
- 27.Wu J, Bera AK, Kuhn RJ, Smith JL. Structure of the Flavivirus helicase: implications for catalytic activity, protein interactions, and proteolytic processing. J. Virol. 2005;79:10268–10277. doi: 10.1128/JVI.79.16.10268-10277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo D, Xu T, Watson RP, Scherer-Becker D, Sampath A, Jahnke W, Yeong SS, Wang CH, Lim SP, Strongin A. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008;27:3209–3219. doi: 10.1038/emboj.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu T, Sampath A, Chao A, Wen D, Nanao M, Chene P, Vasudevan SG, Lescar J. Structure of the Dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 Å. J. Virol. 2005;79:10278–10288. doi: 10.1128/JVI.79.16.10278-10288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johansson M, Brooks AJ, Jans DA, Vasudevan SG. A small region of the dengue virus-encoded RNA-dependent RNA polymerase, NS5, confers interaction with both the nuclear transport receptor importin-beta and the viral helicase, NS3. J. Gen. Virol. 2001;82:735–745. doi: 10.1099/0022-1317-82-4-735. [DOI] [PubMed] [Google Scholar]
- 31.Tuteja N, Tuteja R. Unraveling DNA helicases. Motif, structure, mechanism and function. Eur. J. Biochem. 2004;271:1849–1863. doi: 10.1111/j.1432-1033.2004.04094.x. [DOI] [PubMed] [Google Scholar]
- 32.Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 33.Tanner NK, Cordin O, Banroques J, Doère M, Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell. 2003;11:127–138. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 34.Belon CA, Frick DN. Fuel specificity of the hepatitis C virus NS3 helicase. J. Mol. Biol. 2009;388:851–864. doi: 10.1016/j.jmb.2009.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzich JA, Tamura JK, Palmer-Hill F, Warrener P, Grakoui A, Rice CM, Feinstone SM, Collett MS. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J. Virol. 1993;67:6152–6158. doi: 10.1128/jvi.67.10.6152-6158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preugschat F, Averett DR, Clarke BE, Porter DJ. A steady-state and pre-steady-state kinetic analysis of the NTPase activity associated with the hepatitis C virus NS3 helicase domain. J. Biol. Chem. 1996;271:24449–24457. doi: 10.1074/jbc.271.40.24449. [DOI] [PubMed] [Google Scholar]
- 37.Wardell AD, Errington W, Ciaramella G, Merson J, McGarvey MJ. Characterization and mutational analysis of the helicase and NTPase activities of hepatitis C virus full-length NS3 protein. J. Gen. Virol. 1999;80:701–709. doi: 10.1099/0022-1317-80-3-701. [DOI] [PubMed] [Google Scholar]
- 38.Lam AM, Keeney D, Eckert PQ, Frick DN. Hepatitis C virus NS3 ATPases/helicases from different genotypes exhibit variations in enzymatic properties. J. Virol. 2003;77:3950–3961. doi: 10.1128/JVI.77.7.3950-3961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bougie I, Bisaillon M. Metal ion-binding studies highlight important differences between flaviviral RNA polymerases. Biochem. Biophys. Acta. 2009;1794:50–60. doi: 10.1016/j.bbapap.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 40.Benzaghou I, Bougie I, Picard-Jean F, Bisaillon M. Energetics of RNA binding by the West Nile virus RNA triphosphatase. FEBS Lett. 2006;580:867–877. doi: 10.1016/j.febslet.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 41.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou P, Tian F, Lv F, Shang Z. Geometric characteristics of hydrogen bonds involving sulfur atoms in proteins. Proteins. 2009;76:151–163. doi: 10.1002/prot.22327. [DOI] [PubMed] [Google Scholar]
- 43.Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Theis K, Chen PJ, Skorvaga M, Van Houten B, Kisker C. Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair. EMBO J. 1999;18:6899–6907. doi: 10.1093/emboj/18.24.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rudolph MG, Heissmann R, Wittmann JG, Klostermeier D. Crystal structure and nucleotide binding of the Thermus thermophilus RNA helicase Hera N-terminal domain. J. Mol. Biol. 2006;361:731–743. doi: 10.1016/j.jmb.2006.06.065. [DOI] [PubMed] [Google Scholar]
- 46.Cordin O, Tanner NK, Moere M, Linder P, Banroques J. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 2004;23:2478–2487. doi: 10.1038/sj.emboj.7600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sinha KM, Glickman MS, Shuman S. Mutational analysis of Mycobacterium UvrD1 identifies functional groups required for ATP hydrolysis, DNA unwinding, and chemomechanical coupling. Biochemistry. 2009;48:4019–4030. doi: 10.1021/bi900103d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nobeli I, Laskowski RA, Valdar WS, Thornton JM. On the molecular discrimination between adenine and guanine by proteins. Nucleic Acids Res. 2001;29:4294–4309. doi: 10.1093/nar/29.21.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Day CW, Smee DF, Julander JG, Yamshchikov VF, Sidwell RW, Morrey JD. Error-prone replication of West Nile virus caused by ribavirin. Antiviral Res. 2005;67:38–45. doi: 10.1016/j.antiviral.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 50.Graci JD, Cameron CE. Therapeutically targeting RNA viruses via lethal mutagenesis. Future Virol. 2008;3:553–566. doi: 10.2217/17460794.3.6.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.