Abstract
Mcm2–7 is recruited to eukaryotic origins of DNA replication by origin recognition complex, Cdc6 and Cdt1 thereby licensing the origins. Cdc6 is essential for origin licensing during DNA replication and is readily destabilized from chromatin after Mcm2–7 loading. Here, we show that after origin licensing, deregulation of Cdc6 suppresses DNA replication in Xenopus egg extracts without the involvement of ATM/ATR-dependent checkpoint pathways. DNA replication is arrested specifically after chromatin binding of Cdc7, but before Cdk2-dependent pathways and deregulating Cdc6 after this step does not impair activation of origin firing or elongation. Detailed analyses revealed that Cdc6 deregulation leads to strong suppression of Cdc7-mediated hyperphosphorylation of Mcm4 and subsequent chromatin loading of Cdc45, Sld5 and DNA polymerase α. Mcm2 phosphorylation is also repressed although to a lesser extent. Remarkably, Cdc6 itself does not directly inhibit Cdc7 kinase activity towards Mcm2–4–6–7 in purified systems, rather modulates Mcm2–7 phosphorylation on chromatin context. Taken together, we propose that Cdc6 on chromatin acts as a modulator of Cdc7-mediated phosphorylation of Mcm2–7, and thus destabilization of Cdc6 from chromatin after licensing is a key event ensuring proper transition to the initiation of DNA replication.
INTRODUCTION
To ensure stable maintenance of the genome, initiation of DNA replication needs to be tightly controlled by the strict regulation of origin licensing, which is executed by sequential binding of the origin recognition complex (ORC), Cdc6, Cdt1 and Mcm2–7 to form the pre-replicative complex (pre-RC) onto origins of DNA replication (1–3). Cdc6 belongs to the AAA+ ATPase family and functions in chromatin loading of Mcm2–7, a putative helicase for DNA replication (4). While individual subunits of Mcm2–7 are differentially phosphorylated during the cell cycle, phosphorylation of Mcm4 is most pronounced (5). Cdc7-dependent phosphorylation of Mcm4 facilitates interaction of Mcm2–7 complex with Cdc45 (6). It has also been demonstrated that phosphorylation of Mcm2 during DNA replication depends on Cdc7 and Mcm2 in an Mcm2–7 complex is efficiently phosphorylated by Cdc7 in vitro (7–9).
Cdc7, also known as Dbf4- and Drf1-dependent kinase (DDK), is a serine/threonine kinase conserved from yeasts to human, which is required for the initiation of DNA replication (10,11). The kinase activity peaks at the G1/S transition with the association of the regulatory subunit, Dbf4, in yeast (12,13). In embryonic cell cycle in Xenopus, Drf1 has a more important role than Dbf4 (14). Following Cdc7-mediated phosphorylation of Mcm subunits, Sld5–Psf1–Psf2–Psf3 (Go-Ichi-Ni-San, i.e. 5-1-2-3 in Japanese; GINS) complex, Cdc45 and DNA polymerase α (pol α) are loaded onto chromatin in a Cdk2-dependent manner, thereby starting DNA synthesis.
Regulation of Cdc6 activity during conversion from pre-RC to pre-initiation complex (pre-IC) is crucial but still elusive. Cdc6 mostly dissociates from chromatin after licensing (15,16) and rebinds to chromatin in S phase as a consequence of progression of replication forks (17). This dynamic loading–dissociation–reloading behavior of Cdc6 creates a window of time when Cdc6 is almost absent or loosely bound to chromatin, and the question arises why Cdc6 is destabilized even though it is recruited again in a later stage. Most of the previous studies suggested that inactivation of Cdc6 prevents re-replication. It has been shown that over-expression of Cdc18, the Cdc6 ortholog in Schizosaccharomyces pombe, causes multiple rounds of DNA replication while over-expression of Cdc6 alone is not sufficient to induce re-replication in Saccharomyces cerevisiae and higher eukaryotes (18,19). Thus, the significance of regulating Cdc6 after licensing and the exact consequence of deregulating Cdc6 protein on DNA replication still remain to be investigated.
In this study, we aimed to elucidate why Cdc6 needs to be destabilized from chromatin after licensing using Xenopus egg extracts. We found that deregulation of Cdc6 suppresses the initiation of DNA replication and Cdc7-dependent phosphorylation of Mcm4 on chromatin. Our data provide novel molecular insights into the function of Cdc6 beyond origin licensing as well as its communication with other replication factors in regulating the initiation of DNA replication.
MATERIALS AND METHODS
Preparation of Xenopus egg extracts
Metaphase-arrested Xenopus eggs were treated with 0.2 µg/ml of the calcium ionophore A23187 (Roche Diagnostics, Basel) to release them into interphase, and extracts were prepared as described previously (3,20). Xenopus egg extracts were supplemented with 250 µg/ml cycloheximide, 25 mM phosphocreatine and 15 µg/ml creatine phosphokinase before use. Xenopus sperm nuclei were prepared after demembranation with lysolecithin as described previously (20).
Antibodies and recombinant proteins
Complementary DNA encoding wild-type Xenopus laevis Cdc6 was amplified by PCR using primers attached with a recognition site of EcoRI or NotI 2 and subcloned into pGEX-4T3 expression vector (GE Healthcare). Glutathione-S-transferase (GST) fusion protein was expressed in BL21-Codon Plus (DE3)-RIL (Stratagene) and purified based on a protocol described previously (21) with some modifications. After cells transfected with the expression plasmid were grown at 37°C and treated with 0.1 mM isopropyl β-D-thiogalactside for 1 h at 16°C to induce expression of the recombinant protein, the cells were harvested by centrifugation, and resuspended in lysis buffer [10 mM sodium phosphate, pH 7.2, 0.5 M NaCl, 1 mM EGTA, 1 mM dithiothreitol (DTT) and 0.25% Tween-20] containing 0.2 mM phenylmethylsulfonyl fluoride (PMSF). To purifiy the recombinant GST–Cdc6, the lysate was sonicated gently three times and centrifuged at 9100 g for 20 min. The supernatant was mixed with glutathione-sepharose (GE Healthcare) pre-washed with lysis buffer, and incubated with gentle agitation at 4°C for 1 h to facilitate binding of the recombinant protein to the beads. After the sepharose resin was packed into a column and washed with lysis buffer, the adsorbed proteins were eluted with lysis buffer containing reduced glutathione (10 mM). The eluted protein was collected in 200 μl fractions and the fractions containing GST–Cdc6 were pooled, concentrated and dialyzed against 10 mM 4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)–KOH, pH 7.4, 150 mM NaCl, 0.1 mM DTT and 0.2 mM PMSF. The purified fraction was frozen at −80°C in 10 μl aliquots.
N-terminal hexahistidine-tagged fusion protein of geminin from a cDNA encoding Xenopus geminin H lacking a destruction box and His6-tagged p21 were expressed and purified as described previously (3). Human Cdc7/Ask1 and mouse Mcm2–4–6–7 complex was expressed and purified from baculovirus system as described previously (22).
Anti-Cdc6 and anti-Cdc7 antibodies were obtained as described previously (3,23). Anti-ORC1 antibody was kindly provided by J. Julian Blow (The University of Dundee), anti-Mcm4 and anti-Mcm2 atibodies by Yukio Ishimi (Ibaraki University), anti-Drf1 by Tatsuro Takahashi (Osaka University), anti-Sld5 and anti-Cdc45 by Yumiko Kubota and Haruhiko Takisawa (Osaka University), anti-Smc1 and anti-Smc3 by Keiji Kimura (University of Tsukuba). Anti-histone H3 (ChIP grade) and anti-PSTAIR (CDK) antibodies were purchased from ABcam and Sigma-Aldrich Co. (St Louis, MO, USA), respectively.
Measurement of DNA synthesis in Xenopus egg extracts
For the measurement of DNA replication, [α-32P]dATP was added to the reaction mixture and total DNA synthesized after incubation at 23°C was measured as the radioactivity incorporated into a fraction insoluble in 10% TCA as described previously (20). EcoRI, caffeine and wortmannin were purchased from Takara Bio Inc. (Ohtsu, Japan), Wako Pure Chemical Industries, Ltd. (Osaka, Japan) and Sigma-Aldrich Co. (St Louis), respectively.
Isolation of chromatin fraction
Chromatin fraction was isolated as described previously (3) and the pellet was resuspended in 40 µl of sample buffer for Sodium Dodecyl Sulfate polyacrylamide gel electrophoresis (SDS–PAGE). Samples were then subjected to SDS–PAGE for immunoblotting. When nuclei were isolated to be reincubated in fresh extracts or used in phosphatase treatment, the isolation was performed in a similar way except that Triton X-100 was included at 0.01% and the centrifugation was performed only once at 2100 g for 5 min in a swinging bucket rotor.
Isolation of intact nuclei for Chk1 phosphorylation
After an incubation of sperm nuclei in GST–Cdc6-supplemented extracts for 90 min, intact nuclei were isolated and immunoblotted for phospho-Chk1-S345 (24). For isolation of intact nuclei, extracts containing nuclei were underlayered with 10× volume of intact nuclear isolation buffer (INIB) consisting of 40% sucrose, 50 mM HEPES–KOH, pH 7.5, 100 mM KCl and 2.5 mM MgCl2, and centrifuged at 5800 g in a swinging bucket rotor for 5 min at 4°C. The pellets were resuspended in 1 ml of INIB and recentrifuged under the same conditions. For samples that had been treated with caffeine, 5 mM caffeine was added in INIB to ensure the inhibition of ATM/ATR kinases during the isolation process. The isolated nuclei were subjected to SDS–PAGE and immunoblot analysis to detect phosphorylated Chk1.
Immunodepletion of proteins from Xenopus egg extracts
Immunodepletion was performed as described previously (3). For the depletion of Cdc6, we employed the same protocol using anti-Cdc6 antibody but repeated twice for an efficient removal of Cdc6. The efficiency of the depletion was confirmed by immunoblot analyses or measurement of DNA replication activity.
Phosphatase treatment of chromatin fraction
After incubating sperm nuclei in extracts for 45 min to allow phosphorylation to occur, nuclei were isolated and incubated further with λ-phosphatase (λ-PPase) in the presence or absence of EDTA, by which phosphatase activity was compromised. To detect hyperphosphorylation of Mcm4, half of the chromatin precipitate was subjected to 10% SDS–PAGE and the latter half was subjected to 7.5% SDS–PAGE with Bio-Rad Precision Prestained Marker (Bio-Rad), and were electrophoresed until 25 and 10 kDa marker bands reached bottom of 7.5% and 10% PAGE, respectively. The gels were then subjected to immunoblot analyses as described previously (3).
In vitro kinase assay of Cdc7
Standard in vitro kinase assay for Cdc7 was conducted as described previously (25) using mouse Mcm2–4–6–7 complex as a substrate for human Cdc7–ASK in the absence or presence of GST–Cdc6. Twenty-five µl reactions containing [γ-32P]ATP were incubated at 30°C for 60 min and loaded onto 7.5% SDS–PAGE (59 : 1). The gels were stained with silver, dried and subjected to autoradiography.
Data presentation
All figures in this article indicate representative results obtained from independent experiments to verify their reproducibility.
RESULTS
Deregulated Cdc6 inhibits DNA replication in Xenopus egg extracts independent of ATM/ATR checkpoint pathways
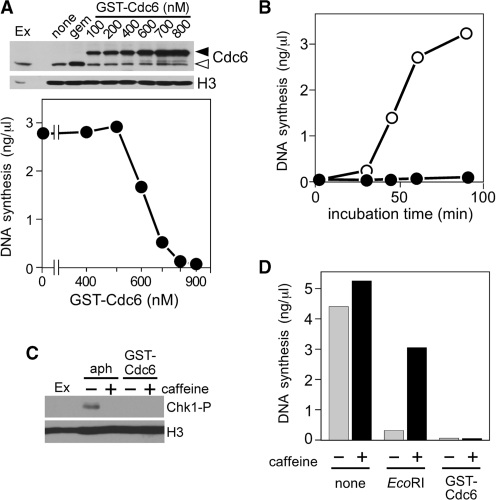
We wanted to directly investigate the consequences of deregulation of Cdc6. To this end, we purified recombinant wild-type GST–Cdc6 from Escherichia coli (Supplementary Figure S1A) and added it to Xenopus egg extracts in order to create a state in which more Cdc6 protein binds on chromatin for longer period even after origins have been licensed. As was expected, Figure 1A (upper panel) shows that the amount of Cdc6 loaded onto chromatin increased with the increase in Cdc6 concentration, and reached plateau at 700 nM of supplemented Cdc6. Under this condition, the amount of chromatin-loaded Cdc6 was higher than the endogenous one ‘after licensing’ but was comparable to that observed ‘before licensing’, which can be physiologically loaded in the presence of geminin, a licensing inhibitor. Then, we investigated the effects of Cdc6 deregulation on DNA replication. To our surprise, as the concentration of Cdc6 increased, the amount of newly synthesized DNA decreased drastically and DNA replication was almost completely inhibited at as low as 700 nM (Figure 1A, lower panel). The dose–response curves varied slightly depending on the preparation of Cdc6 but the overall tendency remained the same. As Cdc6 is a positive regulator of DNA replication, we then asked if the preparation might even be functional and examined the GST–Cdc6 protein for its ability to replace the native Cdc6 after immunodepletion of the egg extract. The result confirmed that the recombinant GST–Cdc6 fraction was fully functional for licensing (Supplementary Figure S1B). The above experiments were repeated with at least three different protein preparations and different egg extracts. For further validation, we have confirmed that DNA replication is also suppressed by endogenous Cdc6 partially purified from Xenopus egg extracts or N-terminal hexahistidine- and FLAG-tagged Cdc6, and that the inhibition is solely caused by the addition of Cdc6, but not GST tag, dialysis buffer or presence of contaminating proteins (Supplementary Figure S1C–E). Therefore, we made use of GST–Cdc6 that sufficiently meets the criteria of endogenous Cdc6. The concentration of endogenous Cdc6 in the extracts used was estimated as ∼200 nM similar to previous reports (21,26) and our results suggest that DNA replication is inhibited by Cdc6 at a concentration <5-fold excess over the endogenous level at a maximal estimation. Hereafter, we used 700 nM of supplemented Cdc6 unless mentioned otherwise.
Figure 1.
DNA replication is inhibited by excess Cdc6 in a manner independent of ATM/ATR-dependent checkpoint pathways. (A) Upper panel: chromatin was isolated after incubation of sperm nuclei (3 ng DNA/µl) for 20 min with extracts containing buffer (none), geminin (100 nM) or various concentrations of GST–Cdc6. Isolated chromatin was subjected to immunoblotting. Open and closed arrowheads represent endogenous Cdc6 and GST–Cdc6, respectively. Egg extracts (Ex; 2 µl) were also applied. Lower panel: DNA synthesis was measured after a 90-min incubation of sperm nuclei (3 ng DNA/µl) with Xenopus egg extracts supplemented with various concentrations of GST–Cdc6. (B) Synthesized DNA at indicated time points was measured after incubation of sperm nuclei (3 ng DNA/µl) with extracts supplemented without (open symbol) or with GST–Cdc6 (700 nM; closed symbol). (C) Nuclei were isolated after sperm nuclei (3 ng DNA/µl) were incubated with egg extracts supplemented with aphidicolin (40 ng/µl) or GST–Cdc6 (700 nM) in the presence or absence of 5 mM caffeine. The isolated fractions were immunoblotted for phospho-Chk1 (Chk1-P) and histone H3 (H3). Egg extracts (Ex; 2 µl) were also applied. (D) DNA synthesis was measured after 90-min incubation of sperm nuclei (3 ng DNA/µl) with extracts supplemented with buffer (none), EcoRI (0.2 U/µl) or GST–Cdc6 (700 nM) in the presence or absence of 5 mM caffeine.
In time-course experiments, we observed a strong suppression of DNA synthesis in Cdc6-supplemented extracts until 90 min, which is sufficient to allow almost complete replication in control extracts (Figure 1B). Interestingly, however, prolonged incubation for 180–240 min rescued the inhibition by Cdc6 (Supplementary Figure S1F) suggesting a kinetic delay of DNA replication in Cdc6-deregulated extracts. It has been reported that Cdc6 is needed to trigger checkpoint activation during replication stress in Xenopus egg extracts (17) and during mitotic exit in yeasts, Cdc6 cooperates with Sic1 to inactivate Cdks, which is probably mediated by activation of Chk1 kinase (27,28). Therefore, we investigated the involvement of checkpoint pathways in Cdc6-induced replication inhibition by monitoring Chk1 phosphorylation. Chk1 is activated by phosphorylation on Ser345 by ATR in response to a variety of genomic insults (29,30). Sperm DNA was incubated with extracts supplemented with aphidicolin or GST–Cdc6 in the presence or absence of caffeine, an inhibitor of ATM/ATR kinases, and the isolated nuclear fraction was subjected to western blotting. Figure 1C shows that aphidicolin induced Chk1 phosphorylation, which was blocked by the addition of caffeine, consistent with activation of ATR checkpoint pathways by aphidicolin treatment. On the other hand, no phospho-Chk1 band was detected in Cdc6-supplemented extracts.
To further assess the involvement of caffeine-sensitive checkpoint pathways, we examined whether Cdc6-mediated inhibition of DNA replication was abrogated by caffeine. Consistent with our previous result (31), DNA replication repressed by EcoRI treatment was restored by co-addition of caffeine (Figure 1D). However, caffeine did not restore DNA replication suppressed by the addition of Cdc6, suggesting that the presence of excess Cdc6 does not activate checkpoint pathways to halt DNA replication. Thus, we speculated that Cdc6 may directly inhibit replication machinery, which likely occurs before checkpoints become able to sense perturbations of DNA replication process.
Deregulated Cdc6 blocks DNA replication after origin licensing, but before Cdk2-dependent steps
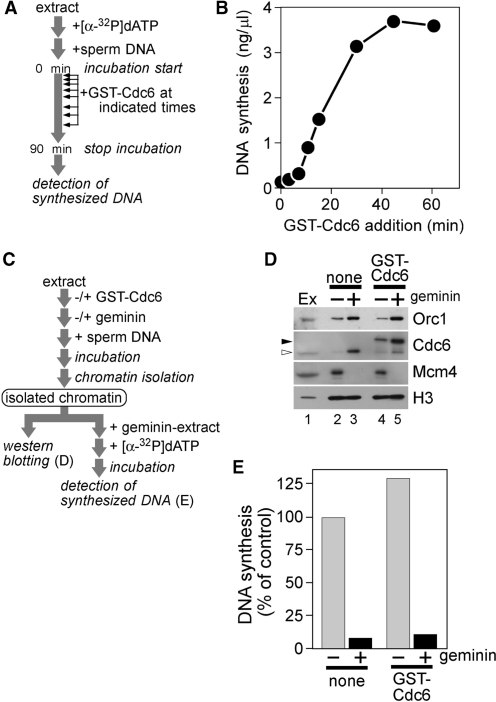
The above results prompted us to determine the step at which DNA replication was arrested by Cdc6. To get an overall idea, we added GST–Cdc6 to egg extracts at various time points after the start of incubation and measured DNA synthesis in a total incubation of 90 min (Figure 2A and B). Whereas the addition of GST–Cdc6 at the start of incubation caused complete inhibition of DNA synthesis, adding Cdc6 between 5 and 15 min caused partial inhibition and that later than 30 min showed little or no suppression of DNA synthesis, suggesting that excess Cdc6 has a target at an earlier stage of DNA replication.
Figure 2.
Cdc6 inhibits DNA replication after origin licensing at an earlier stage of DNA replication. (A and B) GST–Cdc6 (700 nM) was added to the reaction mixture at indicated times after the commencement of incubation of sperm nuclei (3 ng DNA/µl) with egg extracts. Synthesized DNA was measured after 90 min from the start of incubation. (C) A schematic representation of the experimental procedures for D and E. (D) Chromatin was isolated after incubation of sperm nuclei (3 ng DNA/µl) for 20 min with extracts containing buffer (none) or GST–Cdc6 (700 nM) in the presence or absence of geminin (100 nM). Isolated chromatin was subjected to immunoblotting. Open and closed arrowheads represent endogenous Cdc6 and GST–Cdc6, respectively. Egg extracts (Ex; 2 µl) were also applied. (E) Sperm nuclei (15 ng DNA/µl) were incubated with extracts supplemented with buffer (none) or GST–Cdc6 (700 nM) for 20 min in the presence or absence of geminin (100 nM). Chromatin was isolated and incubated for 90 min with fresh geminin-supplemented extracts containing [α-32P]dATP. DNA synthesis in the second incubation was measured and represented as a percentage of the radioactivity incorporated into DNA in the samples of interest to that after incubation with control extracts for 90 min.
Since Cdc6 is one of the pre-RC components, it was speculated that excess Cdc6 would lead to impaired origin licensing. To address the issue, chromatin was isolated from egg extracts supplemented with or without GST–Cdc6 in the presence or absence of geminin (Figure 2C and D). Previous studies have shown that Cdc6 has high affinity for chromatin before licensing, which drops drastically after Mcm2–7 loading (15–17). Consistent with this, ORC1 and Cdc6 remained at high levels on chromatin in the presence of geminin even in Cdc6-supplemented extracts (Figure 2D). In addition, the chromatin binding of ORC1 and Mcm4 in Cdc6-supplemented extracts did not differ significantly from that in control extracts. This result suggests that excess Cdc6 does not impair licensing-dependent Mcm2–7 loading.
As Cdc6 is directly involved in Mcm2–7 loading, it was possible that excess Cdc6 disturbed normal Mcm2–7 loading, resulting in malfunction of the loaded Mcm2–7 complex without altering the amount of the loaded proteins. If this were the scenario, chromatin licensed under excess Cdc6 should fail to replicate in fresh geminin-treated extracts in which de novo Mcm2–7 loading was inhibited. To address this possibility, sperm DNA was incubated in egg extracts supplemented with buffer or Cdc6 for 20 min to allow sufficient time for Mcm2–7 loading. Chromatin was then isolated and transferred to fresh geminin-treated extracts so that the origins licensed in the first incubation could only support replication in the secondary extracts (Figure 2C). Figure 2E confirmed that the capability of DNA replication was no less for chromatin isolated from Cdc6-supplemented extracts than that from control extracts, indicating that Mcm2–7 complexes loaded in Cdc6-deregulated extracts were fully functional.
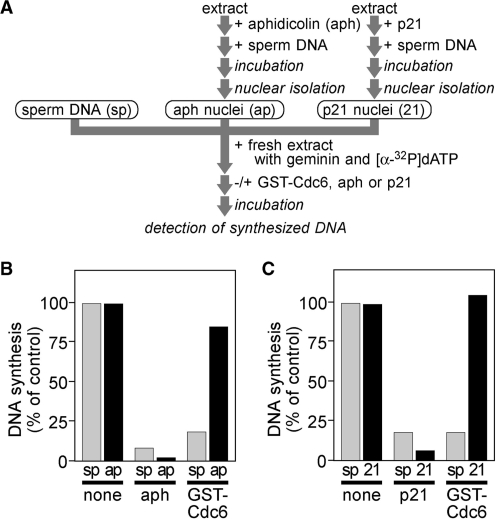
In Xenopus egg extracts, replisome assembly occurs in a stepwise manner with several discrete isolatable intermediates. Then, we investigated the consequences of additive Cdc6 on origin firing or elongation steps separately. First, we examined the effects of Cdc6 on elongation. We incubated sperm chromatin in interphase extracts supplemented with aphidicolin, an inhibitor of DNA polymerases. Under this condition, DNA replication was arrested at the onset of elongation (32). Nuclei were then isolated (aphidicolin nuclei) and individual aliquots were transferred to fresh extracts supplemented with buffer, aphidicolin or Cdc6 (Figure 3A and B). Aphidicolin nuclei fully supported DNA replication even in the presence of additive Cdc6, whereas untreated sperm chromatin failed to replicate, indicating that Cdc6 inhibits DNA replication before elongation steps.
Figure 3.
Cdc6 halts DNA replication before Cdk2-dependent steps. (A) A schematic representation of the experimental procedures. (B) Sperm nuclei (6 ng DNA/µl) were incubated with extracts containing aphidicolin (40 ng/µl) for 60 min and isolated to obtain aphidicolin nuclei. Amount of DNA synthesized was measured during a 90-min incubation after untreated sperm chromatin (sp) or aphidicolin nuclei (ap) were added in extracts containing buffer (none), aphidicolin (aph; 40 ng/µl) or GST–Cdc6 (700 nM). DNA synthesis is represented as a percentage of the radioactivity incorporated into DNA in the samples of interest to that after incubation with control extracts for 90 min. (C) The same experiment as shown in B but using p21 (5 ng/µl) instead of aphidicolin for nuclear preparation to obtain p21 nuclei (21) and for additives of the second incubation (p21).
We exploited similar protocol using Cdk2 inhibitor, p21 (33), instead of aphidicolin in order to determine whether Cdc6 influences Cdk2-dependent processes. Similar to aphidicolin nuclei, p21 nuclei were also fully competent to support DNA replication in Cdc6-supplemented extracts (Figure 3C). Together, these results strongly suggest that Cdc6 deregulation halts DNA replication at a step that lies between pre-RC assembly and Cdk2-dependent events consistent with the idea that Cdc6 deregulation does not affect later stages of DNA replication (Figure 2A).
Cdc6 inhibits Cdc7-dependent hyperphosphorylation of Mcm4
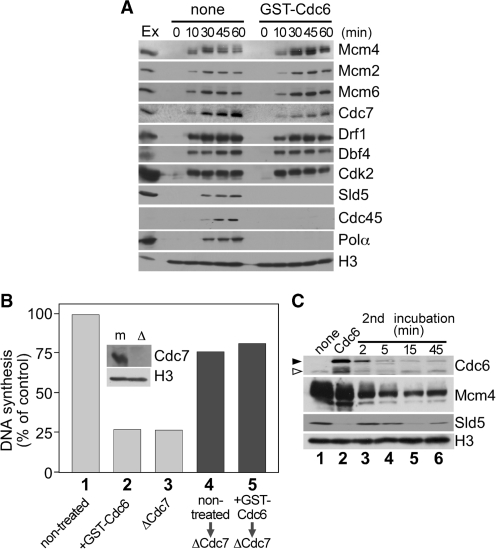
We then focused on the events between pre-RC assembly and Cdk2-dependent origin firing, and investigated chromatin binding of replication-related proteins in Cdc6-supplemented extracts (Figure 4A). In control extracts, chromatin association of Mcm4, Mcm2 and Mcm6 was observed at 10 min and loading of Cdc7 and Drf1 almost overlapped with that of Mcm4. Chromatin binding of Dbf4 was observed from the beginning consistent with a previous report (14). The accumulation of Sld5 and pol α on chromatin peaked around 45 min, concomitant with the peak of nascent strand synthesis (Figure 1B). In Cdc6-supplemented extracts, the chromatin binding of Mcm4, Dbf4 and Cdk2 was not altered compared to that in control extracts although chromatin loading of Cdc7 and Drf1 appeared slightly lower. Then, we assessed whether Cdc6-induced replication block was due to the decrease in Cdc7 on chromatin. However, Figure 4B shows that chromatin isolated from Cdc6-deregulated extracts could replicate well in Cdc7-depleted extracts in which replication of sperm chromatin was severely attenuated, implying that chromatin-loaded Cdc7 in the isolated fraction is sufficient to support replication. This observation suggests that Cdc6 does not essentially inhibit chromatin loading of Cdc7 to suppress DNA replication, rather the inhibition step lies after Cdc7 has been loaded on licensed chromatin. Furthermore, we investigated chromatin-bound Cdc6 and other proteins under these conditions (Figure 4C). The result shows that in control extracts, endogenous Cdc6 bound to chromatin at lower level at 45 min when strong chromatin binding of Cdc6 was still observed in Cdc6-supplemented extracts (first incubation). Chromatin was then isolated and incubated with fresh Cdc7-depleted extracts supplemented with geminin to prevent de novo origin licensing (second incubation). In second incubation, Cdc6 dissociated rapidly from chromatin as soon as 2 min and almost disappeared later than 5 min. Intriguingly, chromatin binding of Sld5, which was suppressed in the first incubation in Cdc6-deregulated extracts, drastically recovered in second incubation paralleling with the dissociation of Cdc6 from chromatin. These results strongly suggest that DNA replication is inhibited after chromatin loading of Cdc7, but before chromatin loading of Sld5 in Cdc6-deregulated extracts.
Figure 4.
Cdc6 inhibits DNA replication after Cdc7 has been loaded. (A) Sperm nuclei (3 ng DNA/µl) were incubated in extracts supplemented with buffer (none) or GST–Cdc6 (700 nM). Chromatin was isolated at indicated time points and subjected to immunoblotting. Egg extracts (Ex; 2 µl) were also applied. (B) DNA synthesis in sperm nuclei were measured after incubation for 90 min in non-treated extracts (column 1), extracts supplemented with 700 nM of GST–Cdc6 (column 2) or Cdc7-depleted (ΔCdc7) extracts (column 3). Sperm nuclei (15 ng DNA/µl) were incubated in extracts supplemented without (non-treated) or with 700 nM of GST–Cdc6 (+GST–Cdc6) for 45 min, followed by nuclear isolation. DNA synthesis was assayed after the nuclei isolated from non-treated or GST–Cdc6 supplemented extracts were incubated for 90 min with Cdc7-depleted extracts (ΔCdc7) containing 100 nM of geminin (column 4 or 5, respectively). DNA synthesis is represented as a percentage of the radioactivity incorporated into DNA in the samples of interest to that after incubation with control extracts for 90 min. Inset: Mock-treated (m) and Cdc7-depleted (Δ) extracts (Ex; 2 µl) were subjected to immunoblotting. (C) Sperm nuclei (15 ng DNA/µl) were incubated for 45 min in extracts supplemented with buffer (none) or GST–Cdc6 (700 nM) and were isolated (lanes 1 and 2). The isolated nuclei were incubated with Cdc7-depleted extracts supplemented with geminin (100 nM), and the chromatin-bound proteins at indicated time points were detected by immunoblotting (lanes 3–6). Open and closed arrowheads represent endogenous Cdc6 and GST–Cdc6, respectively.
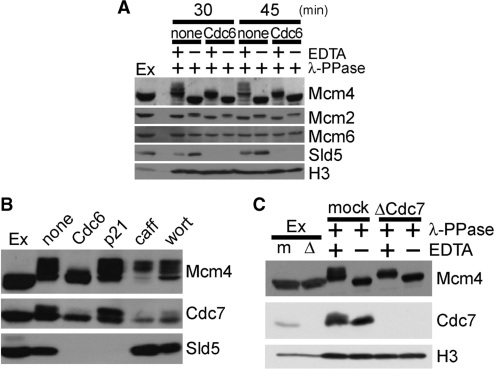
Next, we employed 7.5% SDS–PAGE to address phosphorylation status of Mcm2–7 in the chromatin fraction (Figure 5A and Supplementary Figure S2). While individual subunits of the Mcm2–7 complex are differentially phosphorylated during the cell cycle, the changes in phosphorylation are most pronounced and dynamic for the Mcm4 subunit (5). The phosphatase-treated band is likely to be the non-phosphorylated form (Band 1 in Supplementary Figure S2), and Mcm4 band in extracts migrated more slowly than Band 1 (Band 2, basal phosphorylation), reconfirming that Mcm4 is phosphorylated in interphase prior to DNA replication. During 30–45 min incubation in control extracts, two additional slowly migrating bands appeared (Bands 3 and 4, hyperphosphorylated forms). Figure 5A shows that, although the basal phosphorylation state was not affected in Cdc6-supplemented extracts, hyperphosphorylated forms of Mcm4 were strongly suppressed in the presence of Cdc6.
Figure 5.
Cdc6 inhibits Cdc7-dependent phosphorylation of Mcm4. (A) Chromatin was isolated at 30 and 45 min from extracts supplemented with buffer (none) or GST–Cdc6 (Cdc6; 700 nM) and subjected to λ-PPase treatment in the presence of absence of EDTA, followed by immunoblotting analysis. Egg extracts (Ex; 2 µl) were also applied. (B) Chromatin was isolated after a 45-min incubation of sperm nuclei (3 ng DNA/µl) with extracts supplemented with buffer (none), GST–Cdc6 (Cdc6; 700 nM), p21 (5 ng/µl), caffeine (caff; 5 mM) or wortmannin (wort; 100 µM) and subjected to immunoblotting. Egg extracts (Ex; 2 µl) were also applied. (C) Chromatin was isolated after a 75-min incubation of sperm nuclei (15 ng DNA/µl) with mock-treated (mock) or Cdc7-depleted (ΔCdc7) extracts. Isolated chromatin was treated with λ-PPase in the presence or absence of EDTA, and then subjected to immunoblotting. Mock-treated (m) and Cdc7-depleted (Δ) extracts (Ex; 2 µl) were also applied.
Several lines of studies indicate that subunits of Mcm2–7 complex are phosphorylated by Cdc7, Cdk2 and checkpoint kinases during the course of DNA replication (6,14,22,34). Thus, we attempted to determine the kinase responsible for the hyperphosphorylation observed on Mcm4 that was inhibited by Cdc6. Figure 5B shows that the hyperphosphorylation of Mcm4 was insensitive to the treatment of extracts with p21, caffeine or wortmannin, excluding Cdk2, ATM/ATR or DNA-dependent protein kinase from the kinases potentially affected by the excess Cdc6. On the other hand, immunodepleting Cdc7 from interphase extracts almost completely abolished the hyperphosphorylation of Mcm4 observed in our condition (Figure 5C), suggesting that the presence of excess Cdc6 during DNA replication inhibits Cdc7-dependent hyperphosphorylation of Mcm4. In addition, phosphorylation of chromatin-bound Cdc7 detected in control extracts was also suppressed in the presence of excess Cdc6 (Figure 5B). Importantly, we observed that Cdc6 did not affect the phosphorylation status of Mcm4 that had already been hyperphosphorylated (Supplementary Figure S3) and also, the suppressed phosphorylation of Mcm4 in Cdc6-deregulated extracts recovered paralleling with the dissociation of Cdc6 from chromatin (Figure 4C). Together, these results strongly support the idea that deregulation of Cdc6 blocks DNA replication before Mcm4 has been hyperphosphorylated.
Regarding the reduction in chromatin loading of Cdc7 and Drf1 seen in Figure 4A, it is possible that phosphorylation of Mcm4 and/or Cdc7 stabilizes Drf1–Cdc7 on chromatin although the stabilization is not essential for DNA replication. Furthermore, chromatin loading of the cohesin subunits, Smc1 and Smc3, was significantly suppressed in the Cdc6-deregulated condition (Supplementary Figure S4), which reportedly depends on Cdc7 kinase activity (35). Together, these results strongly argue that Cdc6-induced replication inhibition is linked to Cdc7 kinase activity.
Cdc6 inhibits Cdc7-dependent phosphorylation of Mcm2–7 on chromatin
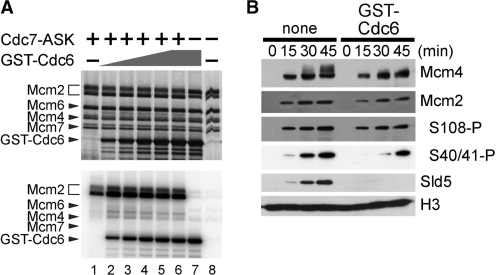
To gain insight into the mechanism of inhibition of Cdc7-dependent phosphorylation events by Cdc6, we examined whether Cdc6 directly modulated the kinase activity of Cdc7. We assayed Cdc7 kinase activity using purified recombinant human Cdc7–ASK and mouse Mcm2–4–6–7 complex as a substrate (22,25). As we mentioned above, Mcm2 is a well-known target of Cdc7 kinase during the course of DNA replication, and Mcm2 in the Mcm2–4–6–7 complex is efficiently phosphorylated by Cdc7–ASK in this assay. Cdc7 also phosphorylates Mcm4 and Mcm6 in vitro, albeit to lesser extent, which can be detected as smeared bands above the lowest Mcm4 band after Cdc7-mediated phosphorylation reaction (see silver staining profile of Figure 6A). Remarkable phosphorylation of Cdc6 was also observed in the presence of Mcm2–4–6–7, but independent of Cdc7. This might be due to a little contamination of Cdk2 in the Mcm fraction, which did not affect phosphorylation status of Mcm proteins by Cdc6 addition. Figure 6A shows that phosphorylation of Mcm2 and Mcm4 by Cdc7–ASK was not suppressed, rather slightly stimulated in the presence of Cdc6 possibly due to the stimulation of Cdc7 kinase by the acidic amino acids on Cdc6 (25). Thus, the above result indicates that Cdc6 itself does not act as a direct inhibitor of Cdc7 kinase.
Figure 6.
Cdc6 inhibits Cdc7-dependent phosphorylation of Mcm2–7 on chromatin during DNA replication. (A) Silver staining (upper) and autoradiography (lower) profiles of the kinase assay using recombinant proteins. Mouse Mcm2–4–6–7 complex was incubated at 30°C for 60 min without (lanes 7 and 8) or with human Cdc7–ASK (lanes 1–6) in the absence (lanes 7 and 8) or presence of GST–Cdc6 (160, 320, 480, 640, 800 and 800 nM for lanes 2, 3, 4, 5, 6 and 7, respectively). (B) Chromatin was isolated after 0-, 15-, 30- or 45-min incubation of sperm nuclei (3 ng DNA/µl) with extracts supplemented with buffer (none) or GST–Cdc6 (700 nM). Isolated chromatin was subjected to immunoblotting.
Finally, we questioned if the suppression by Cdc6 was specific to phosphorylation of Mcm4 subunit. We isolated nuclear fraction at various time points after incubation of sperm nuclei with egg extracts, and detected Mcm4, Mcm2 and phosphorylation of Ser40/41 (S40/41) and Ser108 (S108) in Mcm2 in the fraction (34). As shown in Figure 6B, phosphorylation of Mcm2 on S40/41 and S108 was partially suppressed even under the condition in which hyperphosphorylation of Mcm4 was mostly blocked. These results together suggest an ‘on-chromatin’ regulation of the Cdc7-mediated phosphorylation of selected residues on Mcm2–7 complex, preferentially on Mcm4, rather than a direct inactivation of Cdc7 kinase by Cdc6.
DISCUSSION
In this study, we tried to elucidate the significance of regulating Cdc6 during DNA replication by dissecting the consequences of its deregulation on specific stages of DNA replication and came up with the finding that deregulation of Cdc6 leads to Cdc7-dependent phosphorylation of Mcm2–7 and DNA replication instead of stimulating over-replication in Xenopus egg extracts. These findings extend our current understanding of Cdc6 function beyond origin licensing in two ways: (i) Cdc6 has the capability to halt DNA replication before initiation; and (ii) chromatin-bound Cdc6 acts as a modulator of Cdc7-mediated phosphorylation of Mcm2–7.
Previously, the effects of Cdc6 deregulation have been investigated by several groups in an attempt to study the nuclear transport mechanism mediated by Cdk2. While two of the groups did not find any inhibition by retaining Cdc6 in the nucleus (26,36), two other groups have demonstrated that retention of Cdc6 in the nucleus perturbs DNA replication at an early stage (37,38). The exact reasons behind these controversies remained unclear because of lack of further investigations at molecular level. Furthermore, most of these over-expression studies had been performed using cancer cell lines (36–38) and, therefore, these results do not necessarily reflect those in normal cell cycle. Notably, a previous study using Xenopus egg extracts has shown that retention of excess Cdc6 in the nucleus still support one round of DNA replication (26), which is in apparent contrast to our study. Cells might use this regulation in a reversible manner because we have observed that prolonged incubation drastically rescues the inhibition of DNA replication by excess Cdc6 (Supplementary Figure S1F), and the differences in detailed conditions of experiments might result into contradictory observations. In the present study, we have addressed the effects of Cdc6 on specific stages of DNA replication process rather than simply describing gross effects, and found that deregulated Cdc6 halts DNA replication before Cdc7-dependent phosphorylation of Mcm2–7.
After licensing has been completed, the chromatin association of Cdc6 and ORC is destabilized and Cdc6 is reloaded to chromatin after the commencement of elongation phase. This Cdc6-destabilization period overlaps with the timing of DDK-dependent phosphorylation of Mcm2–7 subunits. Since Cdc6 did not inhibit Cdc7 kinase activity in purified systems, Mcm proteins are supposedly required to be included in pre-RC for Cdc6-induced inhibition of their phosphorylation by Cdc7. Indeed, a recent study has shown that, in the context of pre-RC, DDK preferentially targets a conformationally distinct subpopulation of Mcm2–7 complexes that is tightly linked to the origin DNA (39). A study in yeast showed that the N-terminal DDK-docking domain of Mcm4 interacts with Cdc7 for substrate specification (40). The authors hypothesized that this interaction facilitates ‘autophosphorylation’ of Cdc7 that in turn boosts phosphorylation event of Mcm4. Therefore, it is likely that Cdc6 is topologically situated in close proximity to Mcm4 on chromatin, and thus masks site(s) recognized by Cdc7 thereby suppressing hyperphosphorylation of Mcm4 and autophosphorylation of Cdc7.
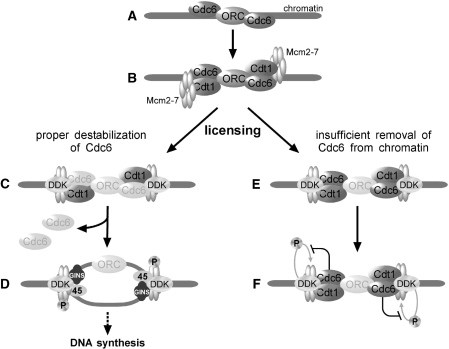
Now, the question arises by what mechanism Cdc6 dissociates from chromatin. A recent report has demonstrated that in human cells, Cdc6 is acetylated by Gcn5, a histone acetyltransferase, which results into chromatin dissociation of human Cdc6 followed by Cdk2–CyclinA-mediated phosphorylation on Ser106 to promote nuclear exclusion (41). Therefore, it is possible that intra-molecular regulatory program by acetylation and/or other post-translational modifications of Cdc6 is linked to subsequent DDK-mediated phosphorylation events of Mcm2–7 complex. Given that phosphorylation of Mcm4 by DDK facilitates its interaction with Cdc45 (6), and that Cdc45, Mcm2–7 and GINS complex probably form the replicative helicase that unwinds double-stranded DNA during the elongation stage (42), it is plausible that regulation of Cdc6 is possibly one of the mechanisms that help insulating pre-RC formation from initiation by modulating establishment of replicative helicases. These ideas are compatible with our observation that Cdc7-catalyzed phosphorylation of Mcm2–7 and subsequent loading of GINS, Cdc45 and pol α remain suppressed in Cdc6-deregulated condition. A summary of our observations is outlined in Figure 7.
Figure 7.
Outline of the observations leading to the inhibition of DNA replication by deregulated Cdc6. (A) Cdc6 is targeted to origin after chromatin binding of ORC. (B) Mcm2–7 is recruited to chromatin by Cdt1. (C) After Mcm2–7 has been loaded onto chromatin (licensing), Cdc6 is destabilized from chromatin. (D) Origin firing occurs by Cdc7 (DDK)-dependent phosphorylation of Mcm2–7 in concert with Cdk2 kinase activity. (E) When Cdc6 is deregulated resulting in insufficient removal from chromatin after licensing, DNA replication is arrested after chromatin loading of DDK. (F) Persisting Cdc6 on chromatin after origin licensing prevents Cdc7-dependent phosphorylation of Mcm2–7 thereby blocking further steps.
Our results do not rule out the possibility that Cdc6 also influences some other unknown pathway(s) that leads to suppression of phosphorylation of Mcm4 paralleling chromatin loading of GINS complex to inhibit DNA replication. Nonetheless, there is no doubt that the inhibitory step lies before DDK-mediated Mcm2–7 phosphorylation events and the suppression of chromatin loading of Sld5 directly reflects the inhibition of DNA replication. Our present work provides insight into the regulation of the activation of replicative helicase by modulating phosphorylation status of one of the subunits, Mcm4, as well as Mcm2, and gives a hint why Cdc6 needs to be regulated as soon as origin has been licensed. In the next stage, it would be interesting to explore how cells overcome this regulation during the progression of carcinogenesis where in many cases Cdc6 is over-expressed.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan; MEXT scholarship from the Ministry of Education, Culture, Sports, Science and Technology of Japan (for undergraduate and graduate studies of L.R.K.); Honjo International Scholarship Foundation (HISF) (for graduate studies of L.R.K.). Funding for open access charge: Grant-in-Aid for Scientific Research from Japan Society for the Promotion of Science.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Yukio Ishimi, Tatsuro Takahashi, Haruhiko Takisawa, Yumiko Kubota, Keiji Kimura and J Julian Blow for kindly providing antibodies. L.R.K. and S.T. designed the work. L.R.K. performed all experiments except in vitro kinase assay. Y.K. supported initial experiments. N.K. performed in vitro kinase assay. Sa.W. constructed plasmids. A.F. and Sh.W. generated Cdc7 and Dbf4 antibodies. L.R.K. and S.T. discussed and wrote the manuscript with M.S., H.M. and T.E.
REFERENCES
- 1.Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- 2.Rowles A, Chong JP, Brown L, Howell M, Evan GI, Blow JJ. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- 3.Tsuyama T, Tada S, Watanabe S, Seki M, Enomoto T. Licensing for DNA replication requires a strict sequential assembly of Cdc6 and Cdt1 onto chromatin in Xenopus egg extracts. Nucleic Acids Res. 2005;33:765–775. doi: 10.1093/nar/gki226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi TS, Wigley DB, Walter JC. Pumps, paradoxes and ploughshares: mechanism of the MCM2–7 DNA helicase. Trends Biochem. Sci. 2005;30:437–444. doi: 10.1016/j.tibs.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Pereverzeva I, Whitmire E, Khan B, Coué M. Distinct phosphoisoforms of the Xenopus Mcm4 protein regulate the function of the Mcm complex. Mol. Cell Biol. 2000;20:3667–3676. doi: 10.1128/mcb.20.10.3667-3676.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masai H, Taniyama C, Ogino K, Matsui E, Kakusho N, Matsumoto S, Kim JM, Ishii A, Tanaka T, Kobayashi T, et al. Phosphorylation of MCM4 by Cdc7 kinase facilitates its interaction with Cdc45 on the chromatin. J. Biol. Chem. 2006;281:39249–39261. doi: 10.1074/jbc.M608935200. [DOI] [PubMed] [Google Scholar]
- 7.Lei M, Kawasaki Y, Young MR, Kihara M, Sugino A, Tye BK. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jares P, Blow JJ. Xenopus Cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes Dev. 2000;14:1528–1540. [PMC free article] [PubMed] [Google Scholar]
- 9.Takeda T, Ogino K, Tatebayashi K, Ikeda H, Arai K, Masai H. Regulation of initiation of S phase, replication checkpoint signaling, and maintenance of mitotic chromosome structures during S phase by Hsk1 kinase in the fission yeast. Mol. Biol. Cell. 2001;12:1257–1274. doi: 10.1091/mbc.12.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masai H, Arai K. Dbf4 motifs: conserved motifs in activation subunits for Cdc7 kinases essential for S-phase. Biochem. Biophys. Res. Commun. 2000;275:228–232. doi: 10.1006/bbrc.2000.3281. [DOI] [PubMed] [Google Scholar]
- 11.Jares P, Luciani MG, Blow JJ. A Xenopus Dbf4 homolog is required for Cdc7 chromatin binding and DNA replication. BMC Mol. Biol. 2004;5:5. doi: 10.1186/1471-2199-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson AL, Pahl PM, Harrison K, Rosamond J, Sclafani RA. Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol. Cell Biol. 1993;13:2899–2908. doi: 10.1128/mcb.13.5.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoon HJ, Loo S, Campbell JL. Regulation of Saccharomyces cerevisiae CDC7 function during the cell cycle. Mol. Biol. Cell. 1993;4:195–208. doi: 10.1091/mbc.4.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi TS, Walter JC. Cdc7-Drf1 is a developmentally regulated protein kinase required for the initiation of vertebrate DNA replication. Genes Dev. 2005;19:2295–2300. doi: 10.1101/gad.1339805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hua XH, Newport J. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J. Cell. Biol. 1998;140:271–281. doi: 10.1083/jcb.140.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rowles A, Tada S, Blow JJ. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J. Cell Sci. 1999;112:2011–2018. doi: 10.1242/jcs.112.12.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oehlmann M, Score AJ, Blow JJ. The role of Cdc6 in ensuring complete genome licensing and S phase checkpoint activation. J. Cell Biol. 2004;165:181–190. doi: 10.1083/jcb.200311044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishitani H, Nurse P. p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell. 1995;83:397–405. doi: 10.1016/0092-8674(95)90117-5. [DOI] [PubMed] [Google Scholar]
- 19.Arias EE, Walter JC. Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 2007;21:497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- 20.Chong JP, Thömmes P, Rowles A, Mahbubani HM, Blow JJ. Characterization of the Xenopus replication licensing system. Meth. Enzymol. 1997;283:549–564. doi: 10.1016/s0076-6879(97)83043-1. [DOI] [PubMed] [Google Scholar]
- 21.Frolova NS, Schek N, Tikhmyanova N, Coleman TR. Xenopus Cdc6 performs separate functions in initiating DNA replication. Mol. Biol. Cell. 2002;13:1298–1312. doi: 10.1091/mbc.01-08-0382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masai H, Matsui E, You Z, Ishimi Y, Tamai K, Arai K. Human Cdc7-related kinase complex. In vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a criticial threonine residue of Cdc7 by Cdks. J. Biol. Chem. 2000;275:29042–29052. doi: 10.1074/jbc.M002713200. [DOI] [PubMed] [Google Scholar]
- 23.Furukohri A, Sato N, Masai H, Arai K, Sugino A, Waga S. Identification and characterization of a Xenopus homolog of Dbf4, a regulatory subunit of the Cdc7 protein kinase required for the initiation of DNA replication. J. Biochem. 2003;134:447–457. doi: 10.1093/jb/mvg163. [DOI] [PubMed] [Google Scholar]
- 24.Kumagai A, Guo Z, Emami KH, Wang SX, Dunphy WG. The Xenopus Chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts. J. Cell Biol. 1998;142:1559–1569. doi: 10.1083/jcb.142.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kakusho N, Taniyama C, Masai H. Identification of stimulators and inhibitors of Cdc7 kinase in vitro. J. Biol. Chem. 2008;283:19211–19218. doi: 10.1074/jbc.M803113200. [DOI] [PubMed] [Google Scholar]
- 26.Pelizon C, Madine MA, Romanowski P, Laskey RA. Unphosphorylatable mutants of Cdc6 disrupt its nuclear export but still support DNA replication once per cell cycle. Genes Dev. 2000;14:2526–2533. doi: 10.1101/gad.176300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calzada A, Sacristán M, Sánchez E, Bueno A. Cdc6 cooperates with Sic1 and Hct1 to inactivate mitotic cyclin-dependent kinases. Nature. 2001;412:355–358. doi: 10.1038/35085610. [DOI] [PubMed] [Google Scholar]
- 28.Hermand D, Nurse P. Cdc18 enforces long-term maintenance of the S phase checkpoint by anchoring the Rad3-Rad26 complex to chromatin. Mol. Cell. 2007;26:553–63. doi: 10.1016/j.molcel.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 29.Guo Z, Kumagai A, Wang SX, Dunphy WG. Requirement for ATR in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobayashi T, Tada S, Tsuyama T, Murofushi H, Seki M, Enomoto T. Focus-formation of replication protein A, activation of checkpoint system and DNA repair synthesis induced by DNA double-strand breaks in Xenopus egg extract. J. Cell Sci. 2002;115:3159–3169. doi: 10.1242/jcs.115.15.3159. [DOI] [PubMed] [Google Scholar]
- 32.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strausfeld UP, Howell M, Rempel R, Maller JL, Hunt T, Blow JJ. Cip1 blocks the initiation of DNA replication in Xenopus extracts by inhibition of cyclin-dependent kinases. Curr. Biol. 1994;4:876–883. doi: 10.1016/s0960-9822(00)00196-2. [DOI] [PubMed] [Google Scholar]
- 34.Montagnoli A, Valsasina B, Brotherton D, Troiani S, Rainoldi S, Tenca P, Molinari A, Santocanale C. Identification of Mcm2 phosphorylation sites by S-phase-regulating kinases. J. Biol. Chem. 2006;281:10281–10290. doi: 10.1074/jbc.M512921200. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi TS, Basu A, Bermudez V, Hurwitz J, Walter JC. Cdc7-Drf1 kinase links chromosome cohesion to the initiation of DNA replication in Xenopus egg extracts. Genes Dev. 2008;22:1894–905. doi: 10.1101/gad.1683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petersen BO, Lukas J, Sørensen CS, Bartek J, Helin K. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 1999;18:396–410. doi: 10.1093/emboj/18.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herbig U, Griffith JW, Fanning E. Mutation of cyclin/Cdk phosphorylation sites in HsCdc6 disrupts a late step in initiation of DNA replication in human cells. Mol. Biol. Cell. 2000;11:4117–4130. doi: 10.1091/mbc.11.12.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang W, Wells NJ, Hunter T. Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc. Natl Acad. Sci. USA. 1999;96:6193–6198. doi: 10.1073/pnas.96.11.6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Francis LI, Randell JC, Takara TJ, Uchima L, Bell SP. Incorporation into the prereplicative complex activates the Mcm2-7 helicase for Cdc7-Dbf4 phosphorylation. Genes Dev. 2009;23:643–54. doi: 10.1101/gad.1759609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheu YJ, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol. Cell. 2006;24:101–113. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paolinelli R, Mendoza-Maldonado R, Cereseto A, Giacca M. Acetylation by GCN5 regulates CDC6 phosphorylation in the S phase of the cell cycle. Nat. Struct. Mol. Biol. 2009;16:412–20. doi: 10.1038/nsmb.1583. [DOI] [PubMed] [Google Scholar]
- 42.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2–7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl Acad. Sci. USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.