SUMMARY
In most eukaryotic cells, subsets of microtubules are adapted for specific functions by post-translational modifications (PTMs) of tubulin subunits. Acetylation of the ε-amino group of K40 on α-tubulin is a conserved PTM on the luminal side of microtubules1 that was discovered in the flagella of Chlamydomonas reinhardtii2,3. Studies on the significance of microtubule acetylation have been limited by the undefined status of the α-tubulin acetyltransferase. Here, we show that MEC-17, a protein related to the Gcn5 histone acetyltransferases4 and required for the function of touch receptor neurons in C. elegans5,6, acts as a K40-specific acetyltransferase for α-tubulin. In vitro, MEC-17 exclusively acetylates K40 of α-tubulin. Disruption of the Tetrahymena MEC-17 gene phenocopies the K40R α-tubulin mutation and makes microtubules more labile. Depletion of MEC-17 in zebrafish produces phenotypes consistent with neuromuscular defects. In C. elegans, MEC-17 and its paralog W06B11.1 are redundantly required for acetylation of MEC-12 α-tubulin, and contribute to the function of touch receptor neurons partly via MEC-12 acetylation and partly via another function, possibly by acetylating another protein. In summary, we identify MEC-17 as an enzyme that acetylates the K40 residue of α-tubulin, the only PTM known to occur on the luminal surface of microtubules.
RESULTS AND DISCUSSION
Acetyl-K40 marks are enriched on a subset of microtubules that turnover slowly (reviewed in7). The K40 residue of α-tubulin is not required for survival in protists, such as Tetrahymena8, or Chlamydomonas9 but appears to be important in vertebrates. In neurons, axonal microtubules have higher levels of K40 acetylation than dendritic microtubules10. Neurons that overexpress a K40A mutant α-tubulin show altered motor-based trafficking and cell differentiation11,12. Kinesin-1, a motor that is preferentially targeted to the axon13, has higher affinity for acetylated as compared to non-acetylated microtubules11,14,15. Two histone deacetylase-related enzymes, HDAC6 and SIRT2, deacetylate α-tubulin16,17. The α-tubulin acetyltransferase (αTAT) has been partially purified18 but the identity of the catalytic subunit remains unknown. Recently Steczkiewicz and colleagues reported that the conserved protein domain DUF738 has weak amino acid sequence homology to the catalytic domain of the Gcn5 histone acetyltransferases4. Among the DUF738 proteins is MEC-17, whose activity is required for the maintenance of touch receptor neurons (TRNs) in C. elegans5,6. Intriguingly, in C. elegans, acetylated α-tubulin (MEC-12) is enriched in the TRNs19. These observations opened the possibility that MEC-17 is involved in K40 acetylation on α-tubulin.
MEC-17 homologs are present in most eukaryotes with exception of fungi and plants (Supplementary Fig. 1). We used DNA homologous recombination to disrupt the gene encoding MEC-17, MEC17, in the ciliate Tetrahymena thermophila (Supplementary Fig. 2). Immunofluorescence with 6–11 B-1, a monoclonal antibody (mAb) that is specific for acetyl-K40 on α-tubulin20 showed a marked loss of acetyl-K40 in Tetrahymena cells lacking MEC17 (MEC17-KO) (Fig. 1a–c). Western blots with 6–11 B-1 mAb showed a nearly complete loss of acetyl-K40 α-tubulin in MEC17-KO cells, comparable to cells carrying a K40R substitution in α-tubulin (Fig. 1g,h). Consistently, 2D SDS-PAGE showed that MEC-17-KO α-tubulin isoforms are more basic than wild-type isoforms (Supplementary Fig. 3). On a western blot with pan-acetyl-K antibodies bands corresponding to α-tubulin and its proteolytic fragments were missing in the MEC-17-KO and K40R cell extracts, while a few non-tubulin bands (including histones) were present (Fig. 1g,h). In wild-type cells analyzed by immunofluorescence, the pan acetyl-K antibodies strongly labeled microtubules and nuclei (Fig. 1d). In the MEC-17-KO and K40R cells, acetyl-K was not detected on microtubules, but nuclei remained labeled (Fig. 1e,f). We conclude that in Tetrahymena, α-tubulin is the major if not the only substrate of MEC-17-dependent K acetylation.
Figure 1. MEC-17 is required for acetylation of K40 on α-tubulin in Tetrahymena.
a–c, Wild-type (prefed with ink) and MEC17-KO (arrow) cells labeled with anti-acetyl-K40 mAb (6–11 B-1) and anti-tubulin antibodies. d–f, Wild-type (d), MEC17-KO (e) and K40R (f) Tetrahymena labeled with pan anti-acetyl-K antibodies. g–h, Western blots of cells (g) or cytoskeletons (h) probed with 6–11 B-1 mAb, pan anti-acetyl-K, anti-α-tubulin (12G10 mAb) and anti-histone hv1 antibodies. Stars mark non-tubulin proteins. Arrows mark acetylated histones. i, Growth curves of Tetrahymena. j–l, Wild-type (left) and GFP-Mec17p overproducing (right) Tetrahymena cells analyzed for GFP (j) or 6–11 B-1 mAb immunofluorescence (k).
The MEC-17-KO Tetrahymena cells had a normal growth rate (results not shown). However, the MEC-17-KO cells grew more slowly than wild type on medium with the microtubule depolymerizing compound oryzalin. In MEC-17-KO cells treated with oryzalin, most axonemes depolymerized or were shorter than similarly-treated wild-type cells (Supplementary Fig. 4). Conversely, the MEC-17 KO cells grew faster than wild-type cells in medium with paclitaxel, a microtubule-stabilizing drug (Fig. 1i). This drug phenotype is consistent with an increase in dynamics of microtubules in MEC17-KO cells21. Tetrahymena cells with K40R α-tubulin had a similar drug phenotype (Fig. 1i, Supplementary Fig. 4). These observations indicate that in Tetrahymena, MEC-17 regulates the dynamics of microtubules by acetylation of K40 on α-tubulin.
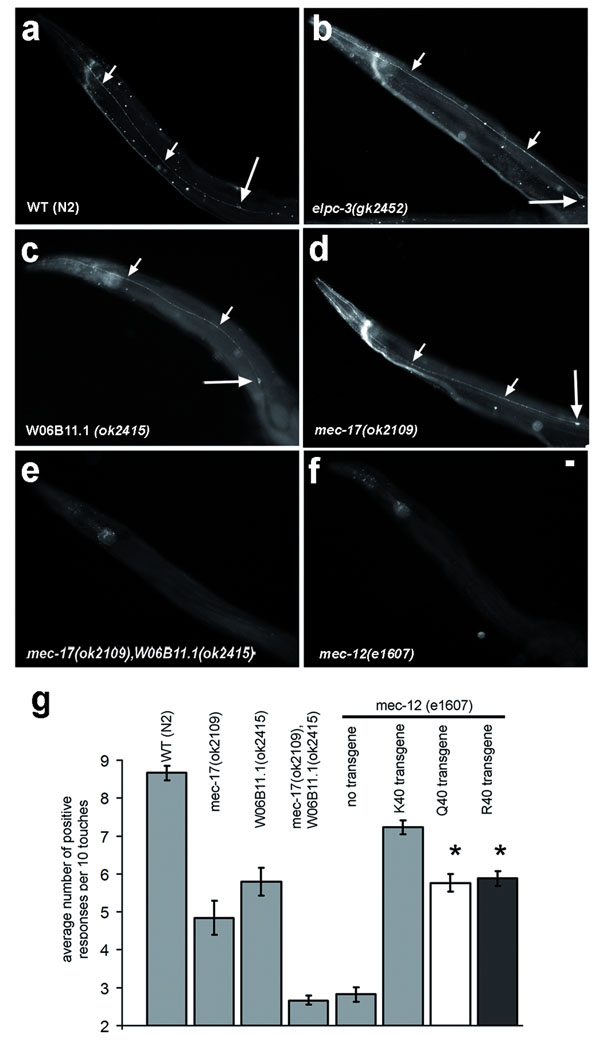
MEC-17 is required for the maintenance of TRNs in C. elegans5,6. The W06B11.1 gene encodes a protein closely related to MEC-175,6. Using 6–11 B-1 mAb, we confirmed that C. elegans wild-type adults have a strong signal for acetylated α-tubulin in the six TRNs19 (Fig. 2a). Single mec-17 or W06B11.1 mutants retained normal levels of acetylated microtubules in the TRNs (Fig. 2c,d). However, double mec-17 and W06B11.1 mutants lacked an acetyl-K40 signal in the TRNs similar to mec-12 α-tubulin mutants (Fig. 2e,f). Thus, MEC-17 and W06B11.1 are redundantly required for acetylation of K40 on MEC-12 α-tubulin. W06B11.1 or mec-17 single deletion mutants had reduced touch responsiveness, and a loss of both genes reduced the touch responsiveness further (Fig. 2g). Next, we investigated the role of MEC-17-dependent acetylatable K40 of α-tubulin. MEC-12 is the only α-tubulin with K40, and mec-12(e1607) (probable null allele22) worms have greatly reduced touch responses. Using Mos1 transposon excision repair23, we integrated single transgenes encoding MEC-12 with either wild-type K40 or K40R or K40Q substitutions into the mec-12(e1607) mutant. The MEC-12-K40 transgene restored the levels of touch response to ~ 80% that of wild type (Fig. 2g), while animals with either MEC-12-K40R or MEC-12-K40Q showed reduced touch response. With the limitation that the wildtype MEC-12 transgene does not fully restore touch sensation, and taking into account that mec12(e1607) mutants have a basal level of touch response, we calculate that a non-acetylatable MEC-12 is 30–33% less efficient than wild-type MEC-12. Nevertheless, animals with K40 substitutions on MEC-12, do respond to touch more frequently than animals lacking MEC-17 and W06B11.1. Thus we surmise that MEC-17 and W06B11.1 contribute to touch sensation partly by acetylating α-tubulin on K40, and through a second mechanism, likely by acetylation of a non-tubulin substrate(s).
Figure 2. MEC-17 and W06B11.1 are required for acetylation of K40 and contribute to touch sensation in C. elegans.
a–f, Wild-type and mutant adult hermaphrodites were labeled using 6–11 B-1 mAb. Small and large arrows mark axons and cell bodies of TRNs, respectively. Scale bar 10 µm. g, Histogram quantifying touch responses. The error bars represent SEM. Asterisk marks significant difference when compared to K40 transgene mec-12(e1607) (p<0.0001). The following numbers of animals were tested: wild type, 69; mec-12(e1607), 49; mec-17(ok2109), 44; W06B11.1(ok2415), 33; mec-17(ok2109) W06B11.1(ok2415), 140; K40 transgene mec-12(e1607), 84; Q40 transgene mec-12(e1607) 78; R40 transgene mec12(e1607) 75.
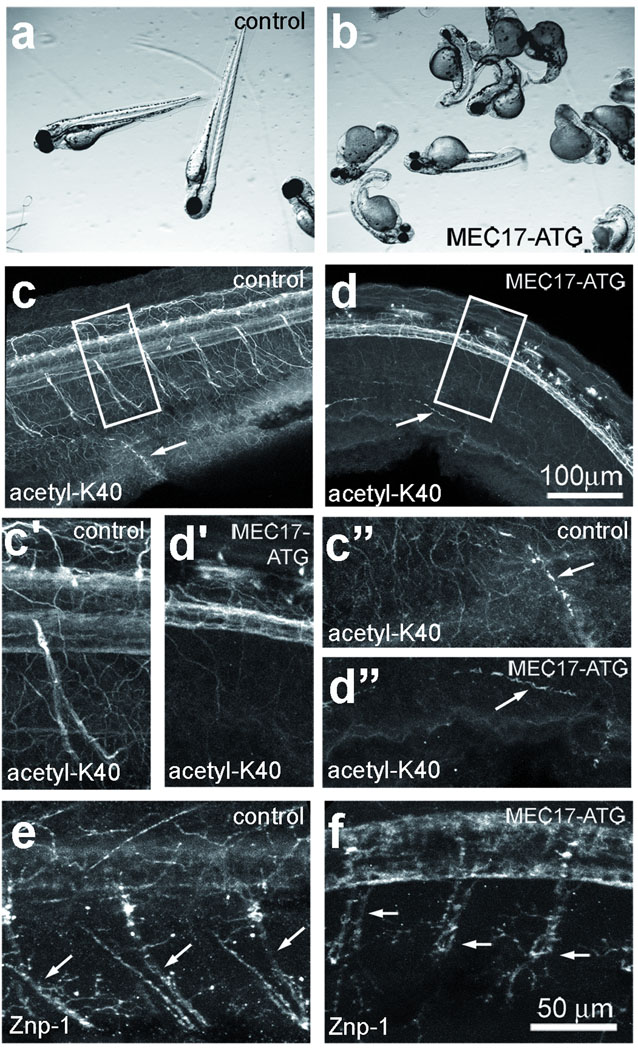
We used zebrafish to test whether MEC-17 is required for α-tubulin acetylation in vertebrates. Acetyl-K40 α-tubulin is enriched in cilia24 and axons of neurons in zebrafish25. Zebrafish has a single MEC-17 ortholog, zgc:65893 (mec17). We injected wild-type zebrafish embryos with morpholinos (MOs) that target either the translation initiation region or a predicted splice junction of mec17. The splice junction MO caused a severe reduction in the levels of mec17 mRNA, possibly by nonsense-mediated mRNA decay (Supplementary Fig. 5). Both MOs produced similar developmental defects, including curved body shape, short body axis, hydrocephalus, small head and small eyes (Fig. 3a,b and results not shown). The vast majority of control embryos injected with random sequence MOs or 5 bp mismatched MOs appeared normal (Supplementary Table 1,2). The mec17 morphants often did not respond or had slow startle response when probed with a needle, consistent with neuromuscular defects (Table 1,2, Supplementary videos S1–3). Immunofluorescence of wild-type embryos with 6–11 B-1 showed that acetyl-K40 carrying microtubules are abundant in the nervous system, including the brain, optical nerves, spinal cord, and axons of peripheral nerves (Fig. 3c–c’’) and in cilia (Fig. 3c”). Strikingly, mec17 morphants showed a nearly complete loss of 6–11 B-1 signal in neurons (Fig. 3d–d’), but not in cilia (Fig. 3d”). The axons of primary motor neurons in the trunk were strongly labeled by the 6–11 B-1 mAb in controls but not in morphants (Fig. 3c’, d’); synaptotagmin 1 localization at synaptic termini26 indicates that the morphants do contain axons (Fig. 3e,f). Depletion of human MEC-17 (C6orf134) in HeLa cells using siRNAs reduced the levels of acetyl-K40 α-tubulin (Fig. 4i), indicating that MEC-17 is also required for α-tubulin acetylation in mammals.
Figure 3. MEC-17 is required for K40 acetylation in zebrafish and normal embryonic development.
Control embryos (a,c,c’,c”,e) and embryos injected with MEC17-ATG morpholinos, 48 hr post fertilization (hpf). (b,d,d’,d”,f) were observed live (a,b) or subjected to immunofluorescence 48 hpf using either 6–11 B-1 mAb (c–d”) or Znp1 mAb (e,f), which recognizes synaptotagmin 1. c’ and d’ show higher magnifications of the areas boxed in c and d. c” and d” show higher magnifications of the areas of pronephrons that contain cilia (marked with arrows in c” and d”). In e and f, arrows mark axons of peripheral neurons.
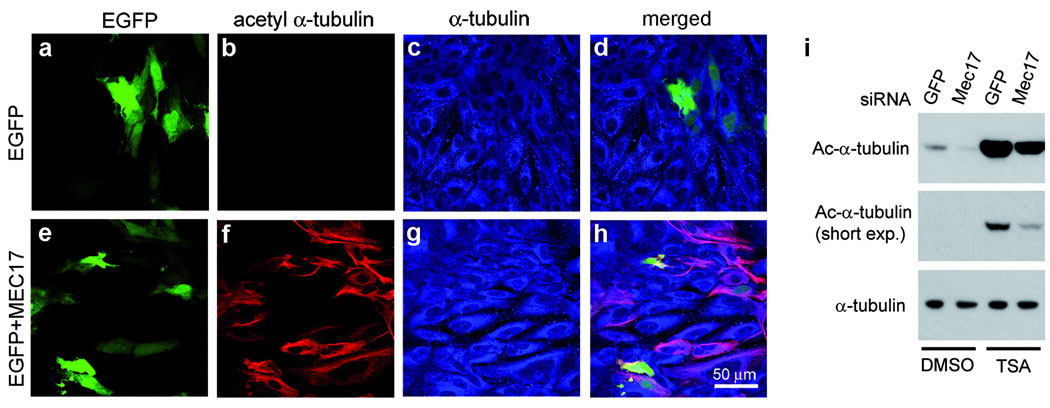
Figure 4. MEC-17 controls the levels of microtubule acetylation in mammalian cells.
a–h, Expression of Mm-MEC-17 in Ptk2 cells increases the levels of acetyl-K40 α-tubulin. Cells expressing either EGFP or EGFP and Mm-MEC17 were stained with 6–11 B-1 mAb and anti-α-tubulin antibodies. i, Depletion of Hs-MEC-17 in HeLa cells reduces the level of acetyl-K40 α-tubulin. Cells were transfected with either GFP or Hs-MEC17 siRNAs and after 50 hr, treated for 7 hr with either 300 nM trichostatin A (TSA, stock solution in DMSO) or DMSO alone. Cell lysates were analyzed by western blot probed with either 6–11 B-1 mAb (top, middle panels) or anti-α-tubulin mAb (bottom panel).
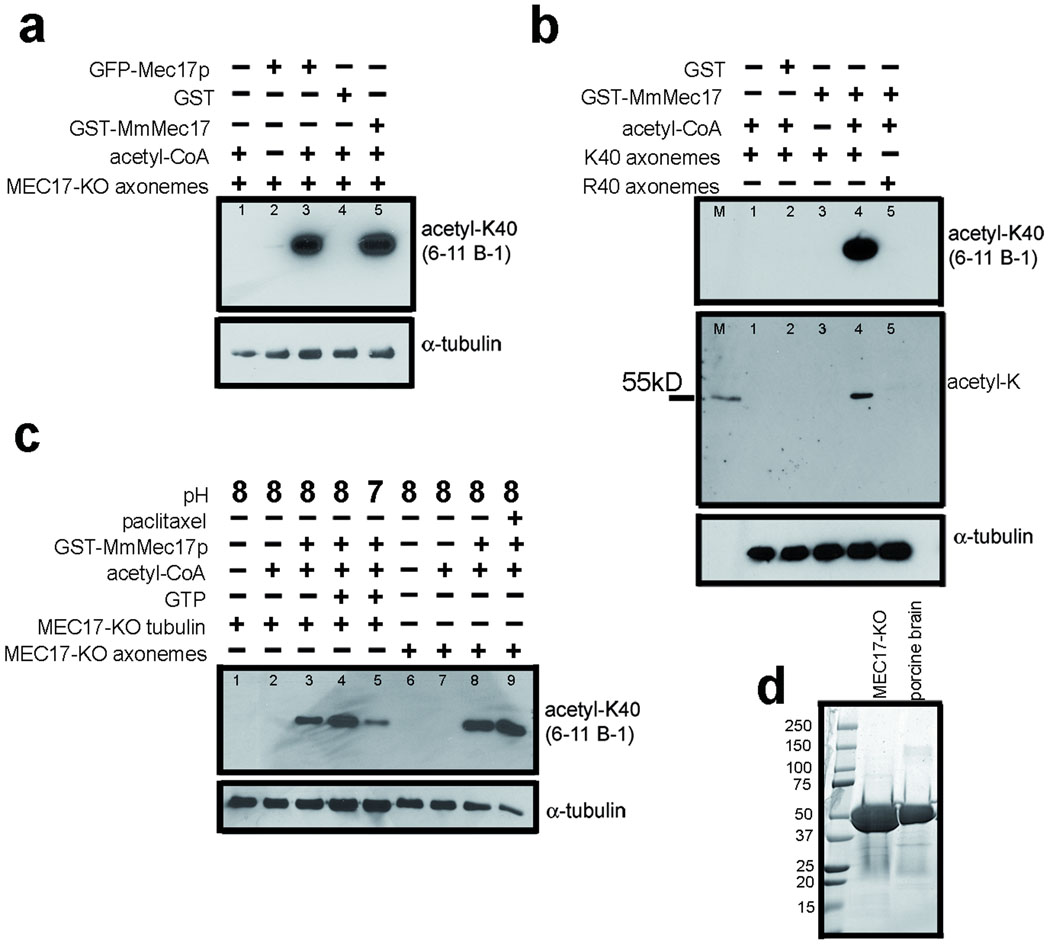
Overexpression of GFP-Mec17p in Tetrahymena greatly increased acetylation of microtubules (Fig. 1j–l). Expression of a murine homolog, MmMEC-17 (Q8K341), in PtK2 cells (which have naturally low acetyl-K40 α-tubulin), induced massive acetylation of cytoplasmic microtubules (Fig. 4a–h). The above observations indicate that either MEC-17 has intrinsic αTAT activity or is an activator of αTAT. To test whether MEC-17 alone can mediate K40 acetyltransferase activity, we established a tubulin acetylation assay using axonemes purified from Tetrahymena MEC17-KO, with acetyl-CoA. A crude GFP-Mec17p-enriched fraction (obtained from transgenic Tetrahymena) had K40 αTAT activity in vitro that was dependent on the presence of acetyl-CoA (results not shown and Fig. 5a, lanes 1–3). Next, we assayed a recombinant MmMEC-17 (expressed in E. coli as a GST fusion, supplementary Fig. 6) on MEC17-KO axonemes. GST-MmMEC-17, but not GST, mediated a robust αTAT activity in vitro (Fig. 5a, lanes 4,5). To test whether the MEC-17 activity is specific to the K40 residue, we assayed GST-MmMEC-17 with axonemes from either a MEC17-KO Tetrahymena (K40 α-tubulin) or from a K40R Tetrahymena mutant (R40 α-tubulin) and used pan acetyl-K antibodies to detect acetyl modification of any K residue. GST-MmMEC-17 modified K40 axonemes (Fig. 5b, lanes 3,4) but failed to acetylate R40 axonemes (Fig. 5b, lane 5). Thus, the activity is specific to K40. Since axonemes are composed of tubulin and MAPs, there is a possibility that MEC-17 activates another protein that is an axoneme-bound αTAT. When MEC-17-KO axonemes were pretreated with 1M salt to remove MAPs, no loss of activity was detected, suggesting that MEC-17 does not require an axoneme-associated cofactor (Supplementary Fig. 7a). To test whether MEC-17 has intrinsic activity, we performed an in vitro acetylation assay with highly purified tubulin obtained from Tetrahymena MEC17-KO cells (Fig. 5d). GST-MmMEC-17 mediated a robust K40 acetylation activity on purified tubulin that was comparable to the level of activity seen with axonemes (Fig. 5c, lanes 4,8). The activity of GST-MmMEC-17 was stimulated when purified tubulin was exposed to GTP to promote tubulin polymerization (Fig. 5c, lanes 3,4). Paclitaxel also stimulated MEC-17 activity, likely by promoting microtubule polymerization (Fig. 5c, lanes 8,9 and Supplementary Fig. 7b). These data indicate that MEC-17 has an intrinsic α-tubulin acetyltransferase activity. The K40 residue of α-tubulin is located on the luminal surface of the microtubule1. When the MEC-17-KO Tetrahymena axonemes were subjected to in vitro acetylation by GST-MmMEC-17, the acetyl-K40 signal was observed near one or both axoneme ends, often as a decreasing gradient from the microtubule end (Supplementary Fig. 8). This supports the model that MEC-17 enters the microtubule lumen from the microtubule end.
Figure 5. MEC-17 has intrinsic, K40-specific α-TAT activity.
a, Crude Tetrahymena and recombinant murine MEC-17 were used for in vitro acetylation reactions of MEC17-KO axonemes and analyzed by western using 6–11 B-1 and 12G10 mAb. b, In vitro acetylation assays were performed with GST-MmMEC-17 using axonemes isolated from either the MEC17-KO (K40) strain or a K40R α-tubulin mutant. The marker (M) is acetylated glutamate dehydrogenase (55.6 kD). c, Recombinant GST-MmMEC-17 directly acetylates purified tubulin from the MEC17-KO strain in vitro. d, Coomassie Blue-stained gel with either purified MEC17-KO tubulin (36 ng) or porcine brain tubulin (15 ng, 99% pure, Cytoskeleton Inc).
To conclude, we identified MEC-17 as an α-tubulin K40 acetyltransferase. We show that MEC-17 is important in the nervous system in both vertebrates and invertebrates. Importantly, another αTAT enzyme likely exists. MEC-17 sequences, are absent from Chlamydomonas reinhardtii, an organism that has αTAT activity3,18 and zebrafish embryos depleted in MEC-17 showed a dramatic loss of acetyl-K40 in neurons but not in cilia. A recent study revealed that ELP3, a conserved histone acetyltransferase, is required for normal levels of K40 acetylation and the differentiation of cortical neurons in the mouse12. However, an ELP3 expressed in insect cells and partially purified was associated with only weak αTAT activity in vitro12. Moreover, TRN microtubules remain highly acetylated in C. elegans elpc-3 mutants, which lack the sole ELP3 homolog (Fig. 2b and27,28). NAT1-ARD29 and NAT1030 are also associated with acetylated microtubules, but it is not known whether these proteins have intrinsic αTAT activity. Thus, the identity of the second αTAT remains uncertain.
Tetrahymena cells lacking α-tubulin acetylation are resistant to paclitaxel and sensitive to oryzalin, consistent with an increase in microtubule dynamics21. Based on these studies, MEC-17-mediated K40 acetylation could mildly stabilize microtubules. It remains to be determined whether changes in microtubule dynamics are a direct effect of acetyl-K40 or are mediated by microtubule effector proteins. We show that in C. elegans, MEC-17 contributes to TRN function partly by acetylating K40 on MEC-12 α-tubulin and partly by other means. For example, MEC-17 could acetylate another protein, or act as a MAP, possibly inside the microtubule lumen.
METHODS SUMMARY
To disrupt the MEC-17 gene in Tetrahymena, we used homologous DNA recombination with a fragment carrying the neo4 marker that replaced the coding region. MEC-17 was overexpressed in Tetrahymena using the MTT1 cadmium-dependent promoter. In C. elegans, MEC-12-K40, MEC-12-Q40, and MEC-12-R40 transgenes were introduced into a single site on chromosome II in the EG4322 strain. Animals homozygous for a MEC-12 transgene and homozygous for the mec-12(e1607) allele were obtained by standard crosses. All touch sensation assays in C. elegans were done using blind scoring. To deplete human MEC-17 (C6Orf134) mRNA in Hela cells, we introduced MEC-17-specific siRNAs (ON-TARGETplus pool, Dharmacon) using Oligofectamine (Invitrogen). To knockdown mec17 expression in zebrafish, MOs designed to target the MEC-17 mRNA (Open Biosystems) were injected into early embryos. ATG-MEC17 MO targets the translation initiation site of mec17 mRNA. SP-MEC17 MO targets the exon3/intron3–4 splice junction, and is expected to result in an aberrant splicing isoform of exon2 to exon 4, producing a frameshift mutation and associated protein truncation. As a negative control, we injected MO with a random sequence (oligo-25N, Gene Tools) or a 5bp mismatch to the ATG-MEC17 MO.Live embryos were scored for phenotypes at 48 hpf. To produce a recombinant MEC-17 protein, the cDNA sequence of the murine MEC-17 (BF135007, Open Biosystems) was subcloned into pGEX-3X plasmid (GE Healthcare), expressed in BL21 E. coli cells as a GST fusion and purified using GST-Bind kit (Novagen). The in vitro acetylation assays were performed in 50 mM Tris-HCl pH 8.0, 10 mM glycerol, 0.1 mM EDTA, with purified Tetrahymena MEC-17-KO axonemes or tubulin (purified using DEAE chromatography), recombinant GST-MmMEC-17 enzyme and 10 µM acetyl-CoA. The reaction was detected by western blotting using anti-acetyl–K antibodies.
METHODS
Tetrahymena
For disruption of the Tetrahymena MEC17 gene the two targeting fragments (1.4 kb of 5’ UTR and 2.0 kb of the coding region with 3’ UTR) of the MEC17 locus were designed and subcloned on the sides of the neo4 selectable cassette31. The fragments were amplified with the addition of restriction sites with the following pairs of primers: 5’-ATTGTGGGCCCTAGCATTTCTGGAAGATTCATTC-3’ (ApaI), 5’-AATACCCGGGCAATTGAATGTATGTGCTGAT-3’ (SmaI) and 5’-AAATTCTGCAGTTAGTACTTTAGAAGTGATGCT-3’ (PstI), 5’-AAATTGAGCTCTCTAGTTGACTATATTATGCATTC-3’ (SacI). The fragments were designed to remove a small part of the 5’UTR and most of the coding region and insert the neo4 resistance cassette in reverse orientation. CU428 and B2086 mating cells were biolistically transformed as described32. Heterokaryons with a germline disruption of MEC17 and progeny cells homozygous for the disruption in the micronucleus and macronucleus were obtain by a heterokaryon × heterokaryon mating33.
For overexpression of GFP-Mec17p, the coding region of MEC17 was amplified with primers 5’-ATATTACGCGTCATGGAGTTTAACTTCATCATTAATAG-3’ and 5’-ATATTGGATCCTCATTTTTTGTAGTATGTGTAGTGAT-3’ and subcloned between the MluI and BamHI sites of pMTT1-GFP plasmid34 and the MTT1-GFP-MEC17-BTU1 fragment was integrated into the BTU1 locus by biolistic bombardment and paclitaxel selection35. The expression of GFP-Mec17p under the MTT1 promoter was induced with 2 µg/ml CdCl2 for 2 hr.
For immunofluorescence, cells were prepared as described (Wloga et al., 2006) and stained overnight with the following antibodies: anti-acetylated K40 α-tubulin 6–11 B-1 mAb 1:200 dilution36; pan anti-acetyl-K antibodies (ImmuneChem, ICP0380) at 1:150 dilution; anti-α-tubulin 12G10 mAb (37, Developmental Studies Hybridoma Bank at 1:25; and polyclonal anti-tubulin antibodies (SG, 1:600). To compare the levels of tubulin acetylation side-by side, wild type cells were marked by feeding for 10 min with India Ink and mixed with MEC17-KO cells.
For western blotting studies, wild type (CU428), MEC17-KO and K40R mutant cells8 were grown to the mid-log phase. The cytoskeletal fractions were prepared as described38 except that trichostatin A at 1µg/ml was added to concentrated cells prior lysis. Total extracts of 2 ×104 cells, or 5 µg of cytoskeletons per lane were used for western blotting with the following antibodies: 12G10 mAb (1:5000); 6–11 B-1 mAb (1:5000); pan anti-acetyl-K antibodies (1:300); hv1 anti-histone (1:2000).
Zebrafish
To knockdown MEC-17 expression in zebrafish, two morpholinos designed to target the MEC-17 mRNA (Open Biosystems) were injected into embryos (at 3 ng/embryo): ATG-MEC17 (5’CATTCAGGTCGTAAGGGAAATCCAT-3’) and SP-MEC17 (5’-AGAGAAAGCTATTTTACCCGTTCTG-3’). ATG-MEC17 targets the translation initiation site of MEC-17 mRNA. SP-MEC17 MO targets the exon3/intron3–4 splice junction, and is expected to result in an aberrantly spliced isoform in which exon2 is joined to exon 4. The predicted transcript contains a frameshift mutation and encodes a nonsense protein. As a negative control we injected MO with a random sequence (oligo-25N, Gene Tools) or a 5bp mismatch to the ATG-MEC17 MO (5’-CATTgAcGTCcTAAGGcAAATgCAT-3’). Live embryos were scored for phenotypes at 48 hpf, or fixed processed for immunofluorescence as described39. The antibodies were used in the following concentrations: 6–11 B-1 mAb (1: 1000), Znp-1 anti-synaptotagmin 1 mAb (1: 100). After incubation with secondary antibodies (Zymed) overnight at 4°C (1: 500) embryos were mounted in 100 mg/ml of DABCO (Sigma-Aldrich) in PBS and viewed in a Leica TCS SP confocal microscope. Live zebrafish morphants shown in videos S1–S3 were recorded using on the Zeiss Stemi SV11 Apo microscope and a SPOT FLEX camera (Diagnostic Instruments Inc.) at 12 frames per second.
For RT-PCR, mRNA was isolated from 70 embryos 24 hpf using TRIzol reagent (Invitrogen, Carslbad, CA), and total cDNA was synthesized using iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules,CA). The sequences of primers used to amplify the mec17 cDNA were: MEC17EX1F 5'-GGTCGGAAAGCGCATGGGAG-3’ and MEC17EX5R2 5'-GAAGTCGAAGAGCTCTGAGCC-3'. The forward primer binds to exon 1 and the reverse primer binds to exon 5 (the splice blocking morpholino binds to the junction between exon 3 and the intron 3–4). For the control amplification of β-actin cDNA, we used the following primers: 5’-GATTCGCTGGAGATGATG-3’ and 5’-GTCTTTCTGTCCCATACCAA-3’.
C. elegans
C. elegans strains were cultured as described40. The following strains were used: N2, wild type; CB1607, mec-12(e1607) III; RB1696, mec-17(ok2109) IV; RB1869, W06B11.1(ok2415) X; ET389, mec-17(ok2109) IV,W06B11.1(ok2415) X; VC1937, elpc-3(gk2452) X; ET431, mec-12(e1607) III, ekSi1[Pmec-12::MEC-12::3'UTR mec-12] II; ET432, mec-12(e1607) III, ekSi2[Pmec-12::MEC-12(K40Q)::3'UTR mec-12] II; ET433, mec-12(e1607) III, ekSi3[Pmec-12::MEC-12(K40R)::3'UTR mec-12] II; EG4322, ttTi5605; unc-119(ed3).
A MEC-12 plasmid, pMEC-12, was constructed using a MEC-12 cDNA and genomic sequence as described19. Using overlapping PCR, we mutated the K40 codon to either a Q or R codon on pMEC-12 along with silent substitutions creating restriction sites (PvuI site for R40 plasmid and HindIII site for Q40 plasmid) and confirmed the mutagenesis by sequencing the entire plasmids. The K40, R40 or Q40 derivatives of pMEC-12 were used to prepare targeting plasmids for Mos-SCI23 as follows. pCFJ151 is a plasmid vector designed to target fragments into the ttTi5605 locus on chromosome II23. A 1.7Kb fragment pMEC-12 comprising a part of the MEC-12 cDNA sequence and 3’UTR was amplified using the primers 5’-ATTATGTTTAAACCAAGCTCGAGTTCTCCATC-3’ (PmeI site is underlined and XhoI site shown in bold) and 5’ AATTATGATCACAGCA AAG GAT TCA AGG CTC3’ (BclI site is underlined), digested with BclI and BglII and inserted into a modified pCFJ151 lacking its original XhoI site (as a result of earlier XhoI digestion, blunting and religation). The resulting plasmid was digested with PmeI and XhoI and used for insertion of a 5.7 Kb of 5’UTR and a part of cDNA amplified from either pMEC-12-K40, or p-MEC-12Q40 or pMEC12-R40 using primers 5’-ATAATGTTTAAACCGGCGAGAAGAGCTATCAA-3’ (with PmeI site underlined) and 5’-AATTTGGAGAACTCGAGCTTGGCC-3’ (with XhoI underlined). The resulting plasmids: pCFJ151-MEC-12-K40, pCFJ151-MEC-12-Q40 and pCFJ151-MEC-12-R40 were used for introduction of single copy MEC-12 transgenes into a site on chromosome II of the EG4322 strain and integrant animals were identified as described23. Strains homozygous one of the three MEC-12 transgene types and homozygous for the mec-12(e1607) probable null allele22 were obtained by standard crosses. All touch assays were done using blind scoring. To determine the touch response level, 30 L4 larvae were isolated on a 5 cm 1× NGM OP50 seeded plate and adult animals were scored for touch responses after 24 hr. Each animal was touched 10 times by moving an eyebrow hair across the body below the anterior and posterior ends. The level of touch response was calculated as an average number of responses per 10 touches.
For immunofluorescence, animals were made permeable by the ‘freeze-crack’ method, followed by methanol and acetone fixation (10 min at -20°C for each)41, and probed with the primary antibody (6–11 B-1 mAb, 1:500 dilution) and the secondary antibody (anti-mouse rhodamine (Cappel, 1:50). Animals were observed with a Zeiss Axioskop microscope equipped for differential interference contrast (DIC) and fluorescence microscopy. Images were captured with a Hamamatsu ORCA-ER digital camera with Openlab 5.0.2 software (Improvision). Images were processed using Adobe Photoshop CS2. Matched images were taken with the same exposure and processed identically. There was no statistical significance between the acetyl-K40 signal intensities over TRNs (after adjacent background subtraction) in wild type, mec-17, and W06B11.1 single mutant strains (241±117; 294±111; and 252±162 arbitrary pixel intensity units, respectively) (Fig. 2a,c,d). The signal intensity in mec-12 mutants or the double mutant mec-17; W06B11.1 could not be determined because there was no detectable TRN signal to measure (Fig. 2e,f).
Mammalian Cells
PtK2 rat kangaroo kidney epithelial cells were grown in DMEM medium supplemented with 10% fetal calf serum, 2 mM L-glutamine and an antibiotics mix at 37°C with 5% CO2. For transfection, cells were grown in 24-well plates to 80–90% of confluency, and transfected with Lipofectamine 2000 (Invitrogen) according to manufacturer instructions, using either 20 ng of pEGFP-N1 plasmid (Clontech) plasmid alone or 20 ng of pEGFP-N1 and 800 ng of pCMV-SPORT6-mmMEC17 (Open Biosystems, MMM1013-7510854) for 16–20 h. Following transfection, cells were grown for 48 hr, split onto coverslips and grown for another 24 hrs and subjected to immunofluorescence. Coverslips with cells were rinsed with PBS, fixed in 4% paraformaldehyde in PBS for 12 min and permeabilized in 0.5% Triton-X-100 in PBS for 15 min. After permeabilization, coverslips were incubated in 3% BSA in PBS for 10 min and incubated in primary antibodies diluted in 3% BSA in PBS (6–11 B-1 anti-acetyl-K40 at 1:300 1:10 and polyclonal anti-α-tubulin (Sigma-Aldrich) 1:10 for 2 hr. After 3 × 5 min washes with PBS cells were incubated in secondary antibodies: anti-mouse IgG-Cy3 in 3% BSA in PBS at 1:100 for 1 hr, washed 3 times and mounted with 100 mg/ml DABCO (Sigma) in PBS and viewed in a Leica TCS SP confocal microscope.
To deplete human MEC-17 (C6Orf134) mRNA in Hela cells, we used ON-TARGETplus siRNAs from Dharmacon as a pool of four siRNAs; the sequences are as follows: 5’-GUAGCUAGGUCCCGAUAUA-3’ (#1); 5’-GAGUAUAGCUAGAUCCCUU-3’ (#2); 5’-GGGAAACUCACCAGAACGA-3’ (#3); 5’-CUUGUGAGAUUGUCGAGAU-3’ (#4). GFP siRNA, 5’-GCUGACCCUGAAGUUCAUCUGdTdT-3’ (Invitrogen) was used as a negative control. HeLa cells were grown as above and transfected with 100 nM of siRNAs (in the pool, each siRNA was at 25 nM) using Oligofectamine (Invitrogen) transfection reagent in accordance with the manufacturers’ instructions. Transfections were performed three times sequentially, followed by subculturing into the new wells. Fifty hr after the first transfection, 300 nM of Trichostatin A in DMSO or the same volume of DMSO were added to the cell cultures, and cells were grown for another 7 hr. Cells were collected and lysed with boiling Laemmli loading buffer containing 2.5% SDS. Lysates of equal number of cells were analyzed using SDS-PAGE/western blot with mouse antibody against acetylated α-tubulin (6-11B-1, Sigma), 1:1000, and mouse anti-α-tubulin antibody (DM1A, Sigma), 1:10,000.
Substrates for in vitro tubulin acetylation
To prepare MEC17-KO axonemes, Tetrahymena cells were grown to the mid-log phase and deciliated by pH shock42. Cilia were suspended in 1 ml of 1% NP-40 in the axoneme buffer (30 mM HEPES, 20 mM potassium acetate, 5 mM MgSO4, 0.5 mM EDTA, pH 7.6) with Complete protease inhibitors (Roche). After 1–2 min on ice, axonemes were collected by centrifugation (20,000 × g, 15 min, 4°C), suspended in the in vitro acetylation reaction buffer (50 mM Tris HCl, pH 8.0, 10% glycerol, 0.1 mM EDTA, 1mM DTT) with protease inhibitors and stored at -80°C.
Total tubulin was purified from the MEC17-KO strain of Tetrahymena using a protocol modified after Yakovich and colleagues43. Tetrahymena cells (2 × 109) were suspended in 40 ml of PME+P buffer (0.1 M Pipes pH 6.9, 1 mM MgCl2, 1 mM EGTA, 1 mM benzamidine, 0.5 mM phenylmethylsulfonyl fluoride, and 25 µg/ml leupeptin) on ice. Cells were sonicated on ice using a Sonic Dismembrator Model 100 (Fischer Scientific) with ten 30 sec bursts at 25 W with a 2 min cooling interval between each burst. The lysate was incubated on ice for 30 min and centrifuged at 40,000 × g for 30 min at 4°C. The supernatant was filtered through glass wool and loaded into a 10 ml column DEAE-Sepharose Fast Flow Matrix (GE Healthcare, earlier equilibrated with two volumes of PME+P) at a rate of 2.5 ml/min using a peristaltic pump. The column was washed with two column volumes of PME+P and followed by four column volumes PME+P with 0.1 M KCl, 0.25 M glutamate pH 6.9. Tubulin was eluted with two column volumes PME+P and 0.3 M KCl, 0.75 M glutamate pH 6.9. Two and half ml fractions were collected. Fractions 6 through 8 were pooled and supplemented with 10 mM MgCl2, 8% DMSO (v/v), and 2 mM GTP. The tubulin-rich pooled fraction was incubated at 37°C for 60 min to induce microtubule assembly and centrifuged at 50,000 × g at 30°C for 30 min. The pellet consisting of microtubules was rinsed once with warm PME (~37°C), and suspended in ~1.5 ml of ice-cold PME. The pellet was solubilized by sonication (thirty ~5 s bursts at 10 W). The tubulin solution was incubated on ice for 30 min, and centrifuged 50,000 × g at 4°C for 30 min. The supernatant containing highly purified dimeric tubulin was stored at −80°C in 50 µl aliquots.
To polymerize tubulin, 100 µl of purified Tetrahymena MEC-17-KO tubulin (5.5 mg/ml) in PME buffer (100 mM Pipes, 1 mM MgCl2, 1 mM EGTA, pH=6.9) was combined with 80 µl of BRB80 buffer (80 mM PIPES, 1 mM MgCl2, 1 mM EGTA, pH 6.8), 2 µl of 100 mM GTP and 1µl of 0.2 M DTT and incubated for 1 hr at 37°C. Microtubules were collected by centrifugation (13,000 rpm, 10 min at room temperature) and the pellet was suspended in the acetylation reaction buffer (see above).
Expression and purification of MEC-17 enzymes
Tetrahymena cells with a GFP-Mec17p encoding transgene under MTT1 promoter were grown without paclitaxel to the density 2 × 105 cells/ml (25 ml) and overexpression was induced by incubation with 2.5 µg/ml of CdCl2 for 3 hr. The cells were collected by centrifugation, washed with Tris-HCl buffer, pH7.5, suspended in the cold in vitro acetylation buffer with protease inhibitors and gently homogenized on ice using a Dounce tissue grounder. The homogenate was centrifuged for 20 min at 20,000 × g at 4°C and the supernatant stored in aliquots at −80°C.
To express recombinant MEC-17 proteins, the pCMV-SPORT6 plasmid containing a full cDNA sequence of the murine MEC-17 ortholog (BF135007, Open Biosystems) was used as template for amplification of the coding regions with primers: 5’-AAATTGAGCTCTGGAGTTCCCGTTCGATGTGGAT-3’ and 5’AATAGAATTCCCGCGGACTAAGCTTTGGCCATGGTTACC-3’. The fragment was subcloned into pGEX-3X expression vector (GE Healthcare). The E.coli BL21 cells carrying either pGEX-3X (GST) or pGEX-3X –MmMEC-17 (GST-MmMEC-17) plasmids were grown in 3 ml cultures of LB medium with ampicilin (50 µg/ml concentration) overnight at 37°C with shaking. A 1.5 ml of culture was transferred into 25 ml of LB medium, IPTG was added to 1 mM final concentration and bacteria were grown for 2.5–3 hrs at 37°C with shaking. Bacteria were collected by centrifugation (6000 × g, 10 min), washed with 25 ml of cold washing buffer (20 mM Tris-HCl, pH 8.0, 0.2M NaCl, 10% glycerol, 2 mM EDTA) and centrifuged as above. Bacteria were suspended in 1ml of washing buffer supplied with 50µM β-mercaptoethanol, 0.5mM PMSF, 10 µg/ml leupeptin, 5 µg/ml DNAse I, 10 µg/ml RNAse A, 1 mg/ml lysozyme, subjected to 2–3 rounds of freezing at −80°C (20–30 min each) followed by thawing on ice, followed by 10 passages through a syringe with an 18 gauge needle. The homogenate was centrifuge at 16,000 × g for 20 min at 4°C and GST-tagged recombinant proteins were purified with GST-Bind Kit (Novagen) according to manufacturer instructions. The recombinant proteins were stored aliquoted at −80°C.
In vitro tubulin acetyltransferase assay
The assays were performed in a buffer that was used earlier for histone acetyltransferases containing 50 mM Tris-HCl pH 8.0, 10 mM glycerol, 0.1 mM EDTA, 1 mM DTT 44 in 50 µl volumes that included 5 µl of purified Tetrahymena MEC-17-KO axonemes or tubulin, 10 µl of GFP-Mec-17p supernatant or purified GST-MmMEC-17 enzyme and 0.5 µl of 1mM acetyl-CoA). Samples were incubated for 60–90 min at 28°C. The reaction was stopped by addition of 5X SDS sample buffer and heating for 5 min at 96°C. Proteins from 10 µl of samples were separated on 10% SDS-PAGE gel and transferred onto nitrocellulose and processed with 6–11 B-1 anti-acetylated K40 α-tubulin mAb (1:15,000) or pan anti-acetyl-K antibodies (1:500) or anti-α-tubulin antibodies (12G10, 1:10,000), as described above.
Supplementary Material
Acknowledgments
This work was supported by funds from the National Science Foundation (MBC-033965 to JG), American Cancer Society (RSG DDC-112979 to S.T.D), and National Institutes of Health (R01GM074212 to ETK, R01AI067981 to NSM, R01GM089912 to JG). S.T.D. is a Georgia Cancer Coalition Distinguished Investigator. We are grateful to Martin Chalfie (Columbia University) for the mec-12(e1627) mutant, Joseph Frankel (University of Iowa) for 12G10 mAb (available from the Developmental Studies Hybridoma Bank), Martin Gorovsky (University of Rochester) for SG anti-tubulin antibodies, David Allis (Rockefeller University) for anti-hv1 antibodies, Ben Feldman (NIH) for mismatch morpholinos, and Scott Dougan lab members for advice and assistance with zebrafish experiments.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
Authors contributions: JSA, DW, JK, NGS, SL-A, STD, ETK, and JG designed and performed the experiments. NSM, STD, ETK and JG supervised the work in their respective laboratories. JG integrated data and wrote drafts of the paper that were edited by all co-authors.
Author information: The authors declare no competing financial interests.
REFERENCES
- 1.Nogales E, Whittaker M, Milligan RA, Downing KH. High-resolution model of the microtubule. Cell. 1999;96:79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- 2.L'Hernault SW, Rosenbaum JL. Chlamydomonas α-tubulin is posttranslationally modified in the flagella during flagellar assembly. J.Cell Biol. 1983;97:258–263. doi: 10.1083/jcb.97.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LeDizet M, Piperno G. Identification of an acetylation site of Chlamydomonas α-tubulin. Proc.Natl.Acad.Sci.USA. 1987;84:5720–5724. doi: 10.1073/pnas.84.16.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steczkiewicz K, Kinch L, Grishin NV, Rychlewski L, Ginalski K. Eukaryotic domain of unknown function DUF738 belongs to Gcn5-related N-acetyltransferase superfamily. Cell Cycle. 2006;5:2927–2930. doi: 10.4161/cc.5.24.3572. [DOI] [PubMed] [Google Scholar]
- 5.Chalfie M, Au M. Genetic control of differentiation of the Caenorhabditis elegans touch receptor neurons. Science. 1989;243:1027–1033. doi: 10.1126/science.2646709. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, et al. Identification of genes expressed in C elegans touch receptor neurons. Nature. 2002;418:331–335. doi: 10.1038/nature00891. [DOI] [PubMed] [Google Scholar]
- 7.Verhey KJ, Gaertig J. The tubulin code. Cell Cycle. 2007;6:2152–2160. doi: 10.4161/cc.6.17.4633. [DOI] [PubMed] [Google Scholar]
- 8.Gaertig J, et al. Acetylation of lysine 40 in alpha-tubulin is not essential in Tetrahymena thermophila. J.Cell Biol. 1995;129:1301–1310. doi: 10.1083/jcb.129.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozminski KG, Diener DR, Rosenbaum JL. High level expression of nonacetylatable α-tubulin in Chlamydomonas reinhardtii. Cell Motil.Cytoskel. 1993;25:158–170. doi: 10.1002/cm.970250205. [DOI] [PubMed] [Google Scholar]
- 10.Witte H, Neukirchen D, Bradke F. Microtubule stabilization specifies initial neuronal polarization. J Cell Biol. 2008;180:619–632. doi: 10.1083/jcb.200707042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dompierre JP, et al. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington's disease by increasing tubulin acetylation. J Neurosci. 2007;27:3571–3583. doi: 10.1523/JNEUROSCI.0037-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Creppe C, et al. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell. 2009;136:551–564. doi: 10.1016/j.cell.2008.11.043. [DOI] [PubMed] [Google Scholar]
- 13.Nakata T, Hirokawa N. Microtubules provide directional cues for polarized axonal transport through interaction with kinesin motor head. J Cell Biol. 2003;162:1045–1055. doi: 10.1083/jcb.200302175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reed NA, et al. Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biol. 2006;16:2166–2172. doi: 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 15.Konishi Y, Setou M. Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nature neuroscience. 2009;12:559–567. doi: 10.1038/nn.2314. [DOI] [PubMed] [Google Scholar]
- 16.Hubbert C, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 17.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 18.Maruta H, Greer K, Rosenbaum JL. The acetylation of alpha-tubulin and its relationship to the assembly and disassembly of microtubules. J.Cell Biol. 1986;103:571–579. doi: 10.1083/jcb.103.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukushige T, et al. MEC-12, an alpha-tubulin required for touch sensitivity in C.elegans. J. Cell Sci. 1999;112:395–403. doi: 10.1242/jcs.112.3.395. [DOI] [PubMed] [Google Scholar]
- 20.LeDizet M, Piperno G. Detection of acetylated α-tubulin by specific antibodies. Meth.Enzymol. 1991;196:264–274. doi: 10.1016/0076-6879(91)96025-m. [DOI] [PubMed] [Google Scholar]
- 21.Barlow SB, Gonzalez-Garay ML, Cabral F. Paclitaxel-dependent mutants have severely reduced microtubule assembly and reduced tubulin synthesis. J Cell Sci. 2002;115:3469–3478. doi: 10.1242/jcs.115.17.3469. [DOI] [PubMed] [Google Scholar]
- 22.Bounoutas A, O'Hagan R, Chalfie M. The multipurpose 15-protofilament microtubules in C. elegans have specific roles in mechanosensation. Curr Biol. 2009;19:1362–1367. doi: 10.1016/j.cub.2009.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frokjaer-Jensen C, et al. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Z, et al. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 2004;131:4085–4093. doi: 10.1242/dev.01240. [DOI] [PubMed] [Google Scholar]
- 25.Wilson SW, Easter SS., Jr Stereotyped pathway selection by growth cones of early epiphysial neurons in the embryonic zebrafish. Development. 1991;112:723–746. doi: 10.1242/dev.112.3.723. [DOI] [PubMed] [Google Scholar]
- 26.Fox MA, Sanes JR. Synaptotagmin I and II are present in distinct subsets of central synapses. The Journal of comparative neurology. 2007;503:280–296. doi: 10.1002/cne.21381. [DOI] [PubMed] [Google Scholar]
- 27.Solinger JA, et al. The Caenorhabditis elegans Elongator complex regulates neuronal alpha-tubulin acetylation. PLoS Genet. 2010;6:e1000820. doi: 10.1371/journal.pgen.1000820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C, Tuck S, Bystrom AS. Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet. 2009;5:e1000561. doi: 10.1371/journal.pgen.1000561. doi:10.1371/journal.pgen.1000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohkawa N, et al. N-acetyltransferase ARD1-NAT1 regulates neuronal dendritic development. Genes Cells. 2008;13:1171–1183. doi: 10.1111/j.1365-2443.2008.01235.x. [DOI] [PubMed] [Google Scholar]
- 30.Shen Q, et al. NAT10, a nucleolar protein, localizes to the midbody and regulates cytokinesis and acetylation of microtubules. Exp Cell Res. 2009;315:1653–1667. doi: 10.1016/j.yexcr.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 31.Mochizuki K. High efficiency transformation of Tetrahymena using a codon-optimized neomycin resistance gene. Gene. 2008;425:79–83. doi: 10.1016/j.gene.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 32.Cassidy-Hanley D, et al. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics. 1997;146:135–147. doi: 10.1093/genetics/146.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hai B, Gorovsky MA. Germ-line knockout heterokaryons of an essential alpha-tubulin gene enable high-frequency gene replacement and a test of gene transfer from somatic to germ-line nuclei in Tetrahymena thermophila. Proc. Natl. Acad. Sci. U.S.A. 1997;94:1310–1315. doi: 10.1073/pnas.94.4.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wloga D, et al. Members of the Nima-related kinase family prmote disassembly of cilia by multiple mechanisms. Mol. Biol. Cell. 2006;17:2799–2810. doi: 10.1091/mbc.E05-05-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaertig J, Gao Y, Tishgarten T, Clark TG, Dickerson HW. Surface display of a parasite antigen in the ciliate Tetrahymena thermophila. Nat. Biotech. 1999;17:462–465. doi: 10.1038/8638. [DOI] [PubMed] [Google Scholar]
- 36.Piperno G, Fuller MT. Monoclonal antibodies specific for an acetylated form of α-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J.Cell Biol. 1985;101:2085–2094. doi: 10.1083/jcb.101.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jerka-Dziadosz M, Strzyewska-Jowko I, Wojsa-Lugowska U, Krawczynska W, Krzywicka A. The dynamics of filamentous structures in the apical band, oral crescent, fission line and the postoral meridional filament in Tetrahymena thermophila revealed by the monoclonal antibody 12G9. Protist. 2001;152:53–67. doi: 10.1078/1434-4610-00043. [DOI] [PubMed] [Google Scholar]
- 38.Janke C, et al. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science. 2005;308:1758–1762. doi: 10.1126/science.1113010. [DOI] [PubMed] [Google Scholar]
- 39.Wloga D, et al. TTLL3 Is a tubulin glycine ligase that regulates the assembly of cilia. Dev Cell. 2009;16:867–876. doi: 10.1016/j.devcel.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 40.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller DM, Shakes DC. Immunofluorescence microscopy. Method Cell Biol. 1995;48:365–394. [PubMed] [Google Scholar]
- 42.Wloga D, et al. Glutamylation on α-tubulin is not essential but affects the assembly and functions of a subset of microtubules in Tetrahymena. Eukaryot Cell. 2008;7:1362–1372. doi: 10.1128/EC.00084-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yakovich AJ, Ragone FL, Alfonzo JD, Sackett DL, Werbovetz KA. Leishmania tarentolae: purification and characterization of tubulin and its suitability for antileishmanial drug screening. Exp Parasitol. 2006;114:289–296. doi: 10.1016/j.exppara.2006.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuninger D, Lundblad J, Semirale A, Rotwein P. A non-isotopic in vitro assay for histone acetylation. Journal of biotechnology. 2007;131:253–260. doi: 10.1016/j.jbiotec.2007.07.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.