Abstract
Activation of the MAPK pathway plays a major role in neoplastic cell transformation. Using a proteomics approach we identified α tubulin and β tubulin as proteins that interact with activated MEK1, a central MAPK regulatory kinase. Confocal analysis revealed spatio-temporal control of MEK1-tubulin co-localization that was most prominent in the mitotic spindle apparatus in variant HT1080 human fibrosarcoma cells. Peptide arrays identified the critical role of positively charged amino acids R108, R113, R160 and K157 on the surface of MEK1 for tubulin interaction. Overexpression of activated MEK1 caused defects in spindle arrangement, chromosome segregation and ploidy. In contrast, chromosome polyploidy was reduced in the presence of an activated MEK1 mutant (R108A, R113A) that disrupted interactions with tubulin. Our findings indicate the importance of signaling by activated MEK1-tubulin in spindle organization and chromosomal instability.
Keywords: MEK1, tubulin, mitotic stability, fibrosarcoma
Introduction
The mitogen-actived protein kinase (MAPK) signal transduction pathway plays a pivotal role in cell cycle progression, cell differentiation, and division (1-4). Deregulated MAPK signaling is a common event in tumors and constitutive MAPK signaling transforms mammalian cells in culture (5). The canonical MAPK pathway is initiated by extracellular proliferative signals detected by membrane-tyrosine kinases and the signal is then transduced by a cascade of phosphorylation events in the cytoplasm through Ras, Raf, mitogen-activated/extracellular signal-regulated kinase kinase (MEK) (6), and extracellular signal-regulated kinase (ERK) (7).
The prominent role of the dual-specificity protein kinases MEK1/2 in mitosis has been extensively documented. MEK1 activity is required for normal mitotic progression and activated MEK1 was detected in extracts from meiotic Xenopus eggs (8-10). MEK1 is also active during mitosis in mammalian somatic cells (11-14). Intriguingly, during normal G2/M in HeLa cells, MEK1 becomes activated without the corresponding activation of its substrate ERK (15). Most recently, it has been reported that MEK1/2 regulates microtubule organization, spindle pole tethering, and asymmetric division during meiotic maturation of mouse oocytes (16). MEK1 also promotes the G2/M transition by reorganizing the Golgi apparatus in vivo (10). Furthermore, inhibition of MEK1 signaling via expression of dominant-negative mutants, use of chemical inhibitors, or RNA interference significantly delays meiotic entry (13,17). MEK1 inhibition causes defects in both spindle formation (18-19) and spindle checkpoint control (18,20), as well as disorganized spindle poles, leading to misaligned chromosomes (16).
We have previously studied which of the multiple signaling pathways, downstream of the mutant constitutively active N-Ras, are required for triggering the full spectrum of phenotypic traits associated with in vitro transformation in cell culture and in vivo tumorigenicity in human fibrosarcoma cell lines. The parental HT1080 cells, harboring an endogenous mutated allele of the N-ras gene, exhibit typical features of a neoplastic transformed cell, including poor adherence, anchorage-independent growth, absence of organized actin fibres, and aggressive tumor formation in vivo. The MCH603 variants, lacking the mutant N-ras allele, have more normal growth characteristics, including a flat adherent morphology, anchorage-dependent growth, well-organized actin microfilaments, and only a weak tumorigenic potential in vivo (21). Of the various components of the MAPK pathway, only stable overexpression of a constitutively active MEK1 S218E, S222D mutant (MEK1act) (5) elicited changes resulting in an aggressive tumorigenic phenotype in MCH603 cells (21).
MEK1/2 are believed to be highly specific for their downstream substrate, ERK (7). The potent biological effects of mutant MEKs, especially the constitutively active form, were explained in terms of enhanced signaling in the canonical MAPK pathway through ERK (5). It has become increasingly clear, however, that this simple linear pathway represents only a minor component of a very complex signaling circuitry (22). Our observations with MCH603MEK1act cells provided further support for the more complex role of MEK1 in tumorigenesis than had been previously envisioned. This led to the postulation that MEK1act may activate as yet unidentified target(s) outside the canonical MAPK pathway and that this activation contributes to the aggressive tumorigenic phenotype (21). Increased polyploidy and multiple spindle pole defects observed in MEK1act transfected cells would be compatible with the notion that MEK1-mediated signaling may have novel mitosis-related target(s).
In an attempt to decipher the mechanism of the apparently non-canonical transforming activity of MEK1act, we set out to identify MEK1act interacting partners. In this report we provide evidence that MEK1act is a tubulin-interacting protein. Interactions of MEK1act with tubulin were studied experimentally and by molecular modeling. R108A and R113A double mutation in the tubulin-binding domain negatively affected interaction of MEK1act with tubulin and significantly compromised its effect on normal spindle function and chromosome ploidy in MCH603 cells. Our data reveal the biological relevance of the MEK1act-tubulin interaction and highlight its role in chromosomal stability.
Materials and Methods
Cell culture and transfections
The human fibrosarcoma cell line, HT1080 (containing an endogenous mutated allele of the N-ras gene), and its derivatives, MCH603 (lacking the mutated N-ras gene) and MCH603MEK1act (MCH603 stably transfected with the constitutively active MEK1act) (21), and 293 HEK cells were grown in Dulbecco’s minimal essential medium (DMEM-Life Technologies), supplemented with 10% fetal calf serum (FCS-Life Technologies), 1.102 U/ml penicillin (Sigma), 1.102 μg/ml streptomycin (ICN), and 125 ng/ml amphotericin B (Sigma), and maintained in 5% CO2 at 37°C. Cells were transfected with Effectene Transfection Reagent (QIAGEN) according to the manufacturer’s recommendations. MCH603 cells stably expressing FLAG-MEK1act and FLAG-MEK1act-D were selected in the presence of G418 (Invitrogen, 800 μg/ml). The plasmid constructs were generated as described in the Supplementary Materials and Methods. Individual cell clones were tested for FLAG expression by western blotting. 293 HEK cells were transiently transfected with the FLAG constructs for 48 h.
Immunofluorescence assay and confocal microscopy
Cells were incubated on chamber slides (Nalge Nunc Int.) for two days, fixed with 2% paraformaldehyde/PBS, and treated with 0.1% Triton X-100/PBS (each 10 min at room temperature). Anti-β-tubulin mouse monoclonal antibody (Santa Cruz Biotechnology) and anti-MEK rabbit antibody (Cell Signaling Technology) were used at 1:50 dilution. Either Texas Red-X conjugated goat anti-mouse IgG (Invitrogen) or swine anti-rabbit-FITC (DAKO) secondary antibody was used as required at 1:200 dilution. Nuclei were stained with 4, 6-diamidino-2-phenylindole (DAPI, Sigma). All incubations were performed at room temperature, followed by three or four washes. Slides were mounted in ProLong antifade reagent (Invitrogen-Molecular Probes) and viewed in a Zeiss LSM 510 confocal microscope equipped with a 63× 1.4 numerical aperture. All analyses were performed as described in the supplementary Materials and methods and Manders et al. (23).
Co-immunoprecipitation of MEK1/2 and tubulin
MCH603MEK1act cells were washed twice with ice-cold PBS and lysed in 10 mM Hepes, pH 7.2, 100 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 0.02% Triton X-100, 2 mM phenymethylsulfonyl fluoride, 10 μg/ml leupeptin, 10 μg/ml pepstatin A, and 10 μg/ml aprotinin on ice (5 min). After preclearing, the lysate (500 μg) was incubated with 30 μl of a 50% protein G-Sepharose slurry (Amersham Biosciences) for 1 h at 4°C. The supernatant was incubated with 30 μl of anti-MEK1/2 or anti-β-tubulin rabbit antibodies (Santa Cruz Biotechnology) at 4°C overnight, then 30 μl of 50% slurry of protein A/G-Sepharose mixture (Amersham Biosciences) was added and incubated for 1 h at 4°C. Immunocomplexes were recovered as described in the supplementary Materials and methods.
Peptide array binding experiments
A custom nitrocellulose-bound peptide array providing a scan of the amino acid sequence of MEK1act was obtained from Jerini Bio Tools GmbH (Berlin, Germany). Spots on the array contain 13-residue peptides overlapping by 11 residues, attached to a cellulose membrane via a C-terminal β-(Ala)2 spacer. All procedures were carried out at room temperature. The membrane was treated with methanol, washed 3× 10 min with TBS buffer (100 mM KCl and 30 mM Tris-HCl, pH 7.6), and blocked with 5% non-fat milk in TBST buffer (0.05% (v/v) Tween 20 in TBS buffer) overnight. The membrane was incubated with tubulin (25 μg/ml, Cytoskeleton Inc.) in blocking buffer with gentle shaking overnight and nonspecific binding was removed by washing with TBST buffer. Bound protein was transferred to a polyvinylidene difluoride membrane by electroblotting and immunodetected with anti-tubulin mouse monoclonal antibody (Santa Cruz Biotechnology, 1:200 dilution). Alanine replacement scans of MEK1 peptides 34 (HKPSGLVMARKLI), 41 (LEIKPAIRNQIIR) and 65-69 (GSLDQVLKKAGRILGK) were carried out using similar procedures.
Flow cytometry
Trypsinized cells (5 × 105) were washed with PBS and incubated in ice-cold 70% ethanol/30% PBS at 4°C for 1 h. Cells were then washed once with PBS, resuspended in 0.5 ml PBS plus 0.5 ml DNA extraction buffer (0.2 M Na2HPO4, 0.4 μM citric acid, pH 7.8), incubated for 5 min at room temperature, resuspended in 1 ml of staining solution [20 μg/ml RNase A (Qiagen), 20 μg/ml propidium iodide (Sigma) in PBS], and incubated for 1 h in the dark at room temperature. Ten thousand cells were acquired in FASCS Calibur and analyzed with Cell Quest software (BD Biosciences).
Actin cytoskeleton staining
Actin stress fibers were visualized by staining with fluorescein-conjugated phalloidin (Molecular Probes). Cells were plated on chamber slides, fixed in 3.7% paraformaldehyde two days later, treated with 0.1% Triton X-100, and then stained with phalloidin (0.005 U/μl) for 20 min at room temperature. Slides were mounted in ProLong fade-antifade, viewed in a Zeiss axioskop epifluorescense microscope, and images were processed by the Smart Capture II, version 2 software (Digital Scientific Ltd.)
Statistical analysis
Statistical analysis was done by paired t-test with two-tail p value. Values of p < 0.05 were considered significant.
Results
Identification of MEK1act as a tubulin-binding protein by mass spectrometry
In an effort to identify novel MEK1act-interacting proteins, we performed affinity purification of a crude MCH603MEK1act lysate on a GST-MEK1act/glutathione 4B Sepharose column. Bound proteins were eluted in two steps, first with 0.3 M glycine (pH 3.0), followed by 0.1 M NaHCO3 (pH 9.0). The majority of proteins were eluted in the latter fraction, identified as five distinct bands on the Coomassie-stained gel, migrating at ~110, 55, 45, 34 and 20 kDa (data not shown). Mass spectrometry of trypsin-digested proteins eluted from the 55 kDa band revealed that thirteen peptides matched to β-tubulin (159 of 444 amino acids, 36%) and six peptides matched to α-tubulin (82 of 416 amino acid, 20%).
MEK1 co-localizes with tubulin at the centrosome, spindle and midbody during mitosis and cytokinesis
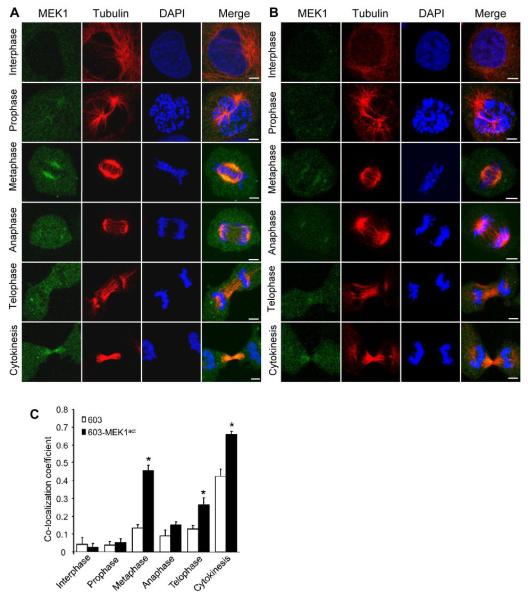
Next we sought to verify the MEK1-tubulin interaction experimentally. Initially, we looked into the extent of MEK1/tubulin co-localization by immunofluorescence and confocal analysis of MEK1 and tubulin in various stages of the cell cycle (Fig. 1A). Parental MCH603 cells were included as a control (Fig. 1B). Analyses of the confocal images of both MCH603MEK1act and MCH603 cells indicated that MEK1 and tubulin do not significantly co-localize during interphase (Fig. 1A, 1B and 1C).The values of co-localization coefficient at this stage appeared to be only 0.03 and 0.04, respectively (Fig. 1C). Additionally, Pearson’s correlation coefficient showed a lack of overlap between the green and red pixels in these cells (Fig. S1A and S1B). A slight increase in the co-localization coefficient was observed in prophase in the MCH603MEK1act population while MCH603 cells remained at 0.04 (Fig. 1C).The pixel distribution at this stage indicated a higher co-localization at the centrosome (Rp = 0.26; Fig. S1A and S1B), compared to other regions of the cell. Interestingly, compared to interphase, the co-localization coefficient in metaphase increased to more than eleven-fold in MCH603MEK1act and to a lesser degree in MCH603 cells (Fig. 1C). In the MCH603MEK1act cells, almost complete co-localization was observed around the centrosome (Rp = 0.7, M1-green = 0.986, M2-red = 0.933) (Fig. S1C). As cells progressed further into anaphase and telophase, the co-localization coefficients decreased and were within the range of 0.1-0.3 in both cell lines (Fig. 1C), representing approximately half of the metaphase signals. As reflected in the whole cell analyses, co-localization signals at the centrosomes were also reduced (Rp = 0.38; Fig. S1D). In cytokinesis, the co-localization coefficients in MCH603MEK1act and MCH603 cells increased again to 0.7 and 0.45, respectively (Fig. 1C). It is worth noting that the extent of co-localization was also high in the midbody (Fig. S1A and S1E, Rp = 0.312, M1-green = 0.98, M2-red = 0.854), compared to the surrounding cellular regions. The difference in the extent of co-localization in both MCH603MEK1act and MCH603 cells was statistically significant in metaphase, telophase and cytokinesis (p < 0.0087), but not in other stages of the cell cycle (Fig. 1C). In summary, the degree of co-localization of MEK1 with tubulin in mitosis increased gradually from prophase, peaking at metaphase, then decreased as cells moved through anaphase and telophase, and finally increased again at cytokinesis. In all stages of the cell cycle, the highest extent of co-localization was observed in the centrosomes in metaphase and to a lesser extent with the spindle fibers, as well as in the midbody during cytokinesis. These data indicate that MEK1-tubulin interaction is temporally and spatially regulated and correlates with spindle organization in the mitotic phase of the cell.
Figure 1.
Co-localization of MEK1 and tubulin in various stages of the cell cycle in MCH603MEK1act and MCH603 cells. (A) MCH603MEK1act cells and B, MCH603 cells were stained for MEK1 (green), tubulin (red), and nuclei were stained with DAPI (blue). Bars, 5 μm. C, Co-localization coefficients in MCH603MEK1act and MCH603 cells. *, p < 0.0087.
We also examined the distribution of activated phospho-ERK (pERK) in these cells. Although we observed association of pERK with the centrosomes during mitosis, we did not see any appreciable association with the mitotic spindle (data not shown). Many proteins localize to the centrosome and we did not further pursue potential interacting protein partners with pERK at this locale.
MEK1 interacts with tubulin via its N-terminal region
We also studied MEK1-tubulin interaction by co-immunoprecipitation and GST pull downs. MEK1 antibody specifically co-immunoprecipitated tubulin and tubulin antibody specifically co-immunoprecipited MEK1 from MCH603MEK1act cells (Fig. 2A). This demonstrates that MEK1 and tubulin associate in vivo. Next, we mapped the tubulin-binding site of MEK1act by testing the tubulin-binding capacities of the N- and C-terminal halves of MEK1act. In GST pull down experiments with purified soluble tubulin dimers, only the full-length MEK1act and the N-terminal half (amino acids 1 to 155) bound tubulin whereas the C-terminal half (amino acids 156 to 373) and GST only did not (Fig. 2B). GST-ERK also failed to pull down tubulin, suggesting that ERK does not directly interact with tubulin (Fig. 2C).
Figure 2.

Co-immunoprecipitation, GST pull down studies and kinase assay of MEK1 and tubulin. A, Total protein lysates from sub-confluent MCH603MEK1act cells were co-immunoprecipitated with MEK1, or tubulin, rabbit antibodies and then tested by immunoblotting with tubulin, and MEK1, mouse monoclonal antibodies. The input proteins were included as controls. B, Mapping of the tubulin-binding region in MEK1act by deletion analysis. C, Characterization of ERK-tubulin interaction using GST pull down studies. D, In vitro kinase assay of GST-MEKact using tubulin and ERK as substrates.
The evidence that MEK1act interacts with tubulin prompted us to test whether this interaction leads to phosphorylation of tubulin. The GST-MEK1act fusion protein in an in vitro kinase assay failed to phosphorylate tubulin, whereas ERK2 was readily phosphorylated (Fig. 2D). However, we did not investigate whether phosphorylation of tubulin occurred in vivo. In summary, data provided by various methods support the conclusion that MEK1 interacts with tubulin in vitro and in vivo. This interaction is mediated through the N-terminal region of MEK1act.
Identification and characterization of tubulin-binding sites on MEK1act
To identify possible tubulin-binding sites on MEK1act, we used peptide arrays that scanned the MEK1act sequence. As shown in Fig. 3A, several potential tubulin-binding regions were identified: peptides 32-35 share the sequence PSGLVMA (residues 89-95), peptides 41-43 share the sequence PAIRNQIIR (residues 105-113), and peptides 65-69 share the sequence KAGRI (residues 157-161). Many of the residues in the tubulin-binding peptides were non-polar or basic suggesting that hydrophobic and/or electrostatic interactions contribute to the tubulin-MEK1act interaction.
Figure 3.

Peptide array binding analysis of MEK1act to tubulin. A, Tubulin binding to MEK1act peptide arrays. Aligned sequences of peptides 32-35, 41-43, and 65-69 (with MEK1act residue numbers in parentheses) are also shown. B, Binding of tubulin to MEK1act peptides having single alanine replacements. *, parent unsubstituted peptide. Arrows pointed to reduced binding. C, Location of tubulin-binding peptide regions on MEK1. The backbone structure of an N-truncated form of MEK1 [residues 61-382; (41; PDB 1s9j)] is shown as a yellow ribbon with bound ATP (red) and the inhibitor PD184352 (magenta) included as stick models. Side chains of residues common to peptides 32-35, 41-43, and 65-69 are shown as space-filling spheres (white) with arginine and lysine residues colored cyan (carbon) and blue (nitrogen). The view in panel a is similar to that in Figure 4 of (41); the view in panel b is rotated 90° about the Y-axis. The figure was generated using VMD (42). D, Mutation of the key residues in the tubulin-binding site of MEKact results in the loss of tubulin binding. Lanes 1 and 2, the input was 5 and 10 μg of GST-MEKact, respectively. Lanes 3 and 4, the input was 5 and 10 μg of GST-MEK1act-Q, respectively. Lane 5, the input was 10 μg of GST while in lane 6, the input was purified tubulin.
To investigate the possible role of electrostatic interactions in MEK1act-tubulin binding, we carried out “alanine scans”, using peptide arrays of peptides 34 and 41 and a composite peptide corresponding to peptides 65-69. As shown in Fig. 3B, single alanine replacements in peptide 34 had only minor effects on tubulin binding. However, more dramatic effects were observed for peptides 41 and 65-69. Binding was largely lost when R108, R113, K157, or R160 were replaced by alanine suggesting that these residues critically determine interactions with tubulin. Binding was also diminished when K156 or K168 were replaced with alanine, but the lack of requirement for these residues in the arrays shown in Fig. 3A suggests that they are not essential.
The locations of the residues shared by tubulin-binding peptides identified in Fig. 3A and the basic residues identified in Fig. 3B are shown on the crystal structure of MEK1act in Figure 3C. Residues shared in peptides 32-35 and those shared in peptides 41-43 are positioned close to one another on the β-domain on one side of the ATP site, whereas residues shared in peptides 65-69 are located on the α-domain approximately 40Å distant on the other side of the ATP site. Some type of structural rearrangement of MEK1 may be required for both α- and β-domains to interact with tubulin.
The critical role of basic residues of MEK1 for tubulin binding was also confirmed in pull down assays. A mutant protein, designated GST-MEK1act-Q, completely lost tubulin binding activity (Fig. 3D). This finding provides further evidence for the involvement of surface charges in MEK1 in interactions with tubulin.
Effect of GST-MEK1act on microtubule polymerization in vitro
Whether MEK1act affects the assembly of microtubules was investigated in reconstitution experiments. Here, we tested the effect of GST-MEK1act on tubulin polymerization by turbidimetric time course experiments. Microtubule formation appeared to proceed with a lag phase preceding polymerization induced by the GST-MEK1act protein, resulting in an incline shape of increasing absorbance over time (Fig. S2A). Increased concentrations of GST-MEK1act led to an increase in the formation of microtubules, as evidenced by the higher plateau value of absorbance. In addition, transmission electron microscopic imaging has shown that microtubules formed in the presence of GST-MEK1act are structurally similar to those formed in the presence of taxol (Fig. S2B). GST served as a negative control, confirming that the observed effect is specific to MEK1act. Taken together, these results demonstrate that MEK1act affects microtubule polymerization in vitro.
Effect of MEK1act on mitotic spindle formation, cytokinesis, and chromosome ploidy
Having established that MEK1 interacts with tubulin during mitosis, we asked whether the presence of constitutively active MEK1act has any effect on mitotic cells. We noted that MCH603MEK1act cells displayed a disproportionately higher number of cells with aberrant mitotic spindles. Multipolar metaphase spindles were observed in 20 ± 6% of these cells, in contrast to 6 ± 1% of HT1080 and 4 ± 3% of MCH603 cells. Flow cytometry analysis confirmed the strong prevalence of polyploidy in MCH603MEK1act cells (Fig. 4A). Karyotype analysis of these cells detected a number of aberrant chromosome segregations (e.g. anaphase bridges) and sister chromatid separation, which are likely to cause defects in mitosis and/or cytokinesis (Fig. 4B,C). Mitotic indexes indicated that MCH603MEK1act cells are 100% polyploid, whereas HT1080 and MCH603 are mostly diploid (72-80%) (Fig. 4C). It should be noted that that the level of activated MEK in the MCH603MEK1act cells is approximately 3-fold higher than that seen in HT1080 cells (21). Thus, this elevated level of MEK activity may well contribute to the increased chromosomal instability of the MCH603MEK1act cells versus HT1080 cells. These data indicate that constitutive high levels of MEK activity may play a role in defective mitotic spindle formation and cytokinesis that in turn leads to chromosomal aberrations and polyploidy.
Figure 4.

MEK1act increased mitotic spindle defects and genomic instability in MCH603 cells. A, Flow cytometry analysis of cell DNA content. B, Chromosome ploidy analysis of MCH603 and MCH603MEK1act cells. Bars indicate 2 μM. a, Pseudodiploid M-phase spread of an MCH603 cell; b, Polyploid and metaphase spreads of MCH603MEK1act cells, with one showing an anaphase bridge (arrow head); and c, lack of cytokinesis in MCH603MEK1act cell. C, Ploidy variations in mitotic cells of HT1080 and its variants. Values represent the percentages of diploid and polyploid cells in 400 total cells of each cell line.
Effect of MEK1act-D mutant with disrupted tubulin-binding sites on chromosome ploidy, multiple spindles, and actin fiber organization
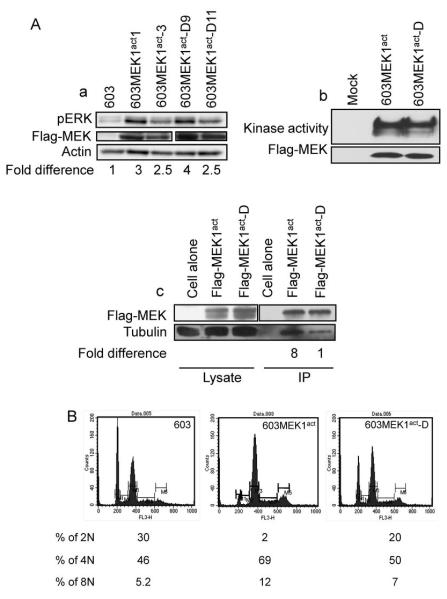
In order to test what role, if any, the MEK1act /tubulin interaction plays in determining the phenotype of cells, we transfected MCH603 cells with FLAG-MEK1act and FLAG-MEK1act-D. Western blotting confirmed that the FLAG-MEK1act-D retained its ability to activate ERK (Fig. 5A, a) and had the same kinase activity as the FLAG-MEK1act (Fig. 5A, b) but exhibited significantly decreased tubulin-binding (Fig. 5A, c). Similar kinase assay and co-immunoprecipitation results were obtained when GFP-MEK constructs were tested (data not shown). Furthermore, co-localization coefficient analyses of MCH603-GFP-MEK1act and MCH603-GFP-MEK1act-D showed that the latter had less colocalization of MEK1act with tubulin (Fig. S3). Therefore, we did comparative flow cytometry analysis, which revealed that MCH603 cells had 30% of 2N, 46% of 4N, and 5% of 8N, whereas MCH603-GFP-MEKact cells had 2% of 2N, 69% of 4N, and 12% of 8N (Fig. 5B). However, MCH603-GFP-MEK1act-D cells had 20% of 2N, 50% of 4 N, and 7% of 8N, a notably lower fraction of > 2N cells. Karyotype analysis further confirmed these results (data not shown). Multiple spindle formation was observed in 5 ± 3% of MCH603-GFP-MEK1act-D cells (similar to MCH603 cells). In contrast, 28 ± 5% of MCH603-GFP-MEK1act mitotic cells had aberrant spindles. Thus, MEK1act/tubulin binding correlates with induction of polyploidy and multiple spindle formation.
Figure 5.
The effect of R108A, R113A double mutation (MEK1act-D) on phospho-ERK levels, kinase activity, interaction with tubulin and ploidy. A,a, Phospho-ERK (pERK) in MCH603-FLAG-MEK1act and FLAG-MEK1act-D cells was assayed by immunoblotting with pERK antibody. Fold difference indicates pERK levels relative to MCH603 cells. b, Total protein lysates from 293 HEK cells transiently transfected with FLAG-MEK1act and FLAG-MEK1act-D were immunoprecipitated with anti-FLAG M2-Affinity gel and their kinase activity was assayed with the MEK Activity Assay Kit. c, Total protein lysates from sub-confluent transfectants were co-immunoprecipitated with anti-FLAG M2 Affinity gel. The immune complexes were tested by immunoblotting with anti-tubulin and anti-MEK1 mouse monoclonal antibodies. Fold difference indicates the level of co-immunoprecipitated tubulin level relative to the FLAG-MEK1act-D in IP sample. B, Ploidy of the indicated cell lines was measured by flow cytometry analysis of DNA content.
Investigation of actin fibers by phalloidin staining also revealed dramatic differences in the cytoskeletal architecture of cells transfected with FLAG-MEK1act and FLAG-MEK1act-D. Actin fibers in MCH603-FLAG-MEK1act cells showed a highly disorganized pattern (Fig. 6), which is typical of the aggressively tumorigenic HT1080 cells (21). MCH603-FLAG-MEK1act-D cells, on the other hand, showed more organized actin fibers, similar to those in the MCH603 cells (Fig. 6).
Figure 6.

Effect of MEK1act and MEK1act-D on actin stress fibers. Actin stress fiber organization in indicated cell lines. Bar indicates 2 μM.
Discussion
The critical role of the MAPK cascade in the induction of proliferation and in oncogenic transformation has been firmly established (7). One of the most convincing lines of evidence for the involvement of the MAPK cascade in oncogenic transformation came from the use of MEK1 mutants. Whereas the dominant negative form of MEK1 reversed Ras-mediated transformation (7), the constitutively active form served as an oncogene, suggesting that the deregulation of the MAPK cascade itself is sufficient to induce cell transformation (5).
Until recently, MEK1 was considered to be a highly selective protein, known to interact exclusively with its downstream substrate ERK (7). This led to the proposition that the potent biological effect of MEKact is the consequence of enhanced signaling in the canonical MAPK pathway through ERK (5). However, the paradigm of exclusive specificity of MEKs for ERKs is being challenged at present, e.g. activation of endomembrane-associated MEK1 during normal G2/M in HeLa cells occurs without the corresponding activation of ERK (15). Most recently, it also has been reported that MEK1 regulates microtubule organization, spindle pole tethering, and asymmetric division during meiotic maturation of mouse oocytes (16). Thus, MEK1 might be a more promiscuous protein than previously thought and some of its function could be mediated by signaling outside the canonical MAPK pathway.
In our previous work we observed that among the various components of the MAPK pathway only overexpression of MEK1act generated an aggressive tumorigenic phenotype in MCH603 cells (21). Several factors could contribute to the strong transforming activity of MEK1act, e.g. cross talk to the JNK pathway. However, MCH603 cells transfected with activated Rac, which can also activate JNK and MEK1, did not exhibit the comparable aggressive tumorigenic phenotype (21). We speculated at that time that MEK1act elicits its transforming activity through interaction with an as yet unidentified non-canonical target(s) (21).
To decipher the mechanism of the strong transforming activity of MEK1act, we used a proteomics approach to identify MEK1act-interacting proteins. Affinity purification of MCH603MEK1act cell lysate with GST-MEK1act provided multiple proteins, and mass spectrometry of 55 kDa proteins identified the presence of α and β tubulin. α and β tubulin are abundant structural proteins and their heterodimer is the structural subunit of microtubules (24). Microtubules have a number of essential cellular functions, including chromosome segregation, the maintenance of cell shape, transport, motility, and organelle distribution (25, 26). The notable structure involving microtubules is the mitotic spindle, which is used by eukaryotic cells to segregate their chromosomes precisely during cell division (27, 28). Defects in the organization, constitution, or regulation of the mitotic spindle apparatus are thought to be important causes of chromosome missegregation and aneuploidy in human cancer (29-33). Highly dynamic mitotic-spindle microtubules are among the most successful targets for anticancer therapy (34).
Mapping of the tubulin-binding site by SPOT peptide analysis suggests that three segments of MEK are involved in interactions with tubulin. Alanine scanning mutagenesis of these regions identified four basic residues (R108, R113, R160 and K157) that are critical for interaction with tubulin and a mutant in which these residues were replaced with alanine (MEKact-Q) failed to interact with tubulin in a GST pull-down assay. Examination of the crystal structure revealed that the charged arginine and lysine residues identified in this study are located on the surface of MEK. This could indicate that MEK1-tubulin interaction is dependent to some extent on ionic charges.
Tubulin and microtubules have several features that could contribute to binding positively charged amino acids on MEK1. First, α and β tubulin subunits are slightly acidic with isoelectric points between 5.2 and 5.8. Second, negatively charged amino acids (glutamic and aspartic acid) are abundantly present on the surface of the tertiary structures of α and β tubulin (data not shown). Third, microtubules are intrinsically polar hollow tubes with β tubulin at the faster polymerizing ‘plus’ end and α tubulin at the slower polymerizing ‘minus’ end (35). During mitosis, ‘minus’ ends of astral microtubules associate with spindle poles and are firmly anchored at the centrosome which prevents their depolymerization (35). Several proteins, such as γ tubulin, pericentrin (36), PCM1 (37), and NuMa (38) are known to localize at the centrosome and stabilize microtubules by interacting with their ‘minus’ end (32, 39). In a recent report, phospho-MEK1 was found to associate with spindle poles and cytoplasmic microtubule organizing centers, and co-localized with centrosome proteins γ tubulin and NuMa but not with kinetochore microtubules during mouse oocyte maturation (16). Inhibition of MEK1 led to spindle pole defects, which were characterized by splaying of the ‘minus’ ends of microtubules (16). Spindles in cells lacking either NuMA, dynactin, or cytoplasmic dynein, which are all essential for focusing and stabilizing the spindle pole formation, had a similar morphology (38, 39). Taken together, we have established a number of features of MEK1, including colocalization with tubulin in the centrosome region, positively charged arginine and lysine residues in tubulin binding sites, and polymerization of tubulin in vitro, indicating that MEK1 may play a role in the regulation of microtubule organization through microtubule dynamics and stabilization.
The expression of MEK1act profoundly affected transfected MCH603 cells and generated a phenotype shift. The most striking feature of MCH603MEK1act cells is that they are 100% polyploid, unlike the non-transfected MCH603 cells. This is consistent with abundant defects in formation of mitotic spindles in MCH603MEK1act cells. Temporal and spatial changes in the association of MEK1 with tubulin during the cell cycle suggest its role in regulation of mitotic spindle formation, mitotic progress, and chromosome segregation. The abundance of MEK1act on the centrosome could impact interaction with tubulin and lead to aberrant spindle formation and chromosome segregation in MCH603MEKact cells. The increased incidence of multipolar spindles in MCH603MEK1act cells might contrast with splaying of the spindle in mouse oocytes in the presence of MEK1 inhibitor. Unlike splaying and unfocused spindle poles caused by the loss of stabilization of microtubule ‘minus’ ends, formation of multipolar spindles (multiple centrosomes) in MCH603MEK1act cells might be the consequence of over-stabilization of the microtubules. Multipolar spindles are induced by stabilizing drugs, such as taxol, but not by destabilizing agents (40). Our data suggest not only a correlation between MEK1act/tubulin interaction and the formation of multipolar spindles and polyploidy in MCH603MEK1act cells, but a possible causal link as well. The MEK1act-D mutant with disrupted tubulin binding was unable to induce polyploidy in MCH603 cells, indicating that MEK1act requires interaction with tubulin during mitosis for aberrant microtubule organization and centrosome formation. Abnormal activity of MEK1act could thus be a major cause of chromosome instability. We propose that polyploid chromosomes in MCH603MEK1act cells are most likely the consequence of mitotic spindle and midbody defects caused by over-stabilization of microtubules in the presence of highly expressed MEK1act that eventually lead to incomplete mitosis and/or cytokinesis. Although MEK1act did not phosphorylate tubulin, it did affect polymerization of tubulin dimers into microtubules in vitro. The mechanism of how MEK1act regulates slower tubulin polymerization in vitro and how MEK1 and MEK1act involve in regulation of the balance between polymerization and depolymerization of microtubules in vivo, as well as in regulation of the centrosome cycle, spindle checkpoint and spindle assembly in metaphase require further study. One possibility is that spindle tubulin acts as a scaffold for MEK and provides a structural base for interaction with other, as yet unidentified, target(s).
Supplementary Material
Acknowledgments
We thank the Mass Spectrometry Core Facility at the Beckman Research Institute, City of Hope Cancer Center for LC/MS analysis and Dr. Ashley Davis (Cytoskeleton Inc.) for his helpful discussion and advice on the microtubule polymerization assay. We thank Ulla Bengtsson for assistance with karyotype analysis. This work was supported in part by NIH grant CA-104214 (EJS), the Avon Foundation-American Association for Cancer Research International Scholar Award in Breast Cancer Research (NS and EJS), and the Malaysian Ministry of Science, Technology and Innovation grants 02-01-04-SF0788, 04-11-08-62FR and 04-01-09-0802RU (NS and EJS).
References
- 1.Giroux S, Tremblay M, Bernard D, et al. Embryonic death of Mek1-deficient mice reveals a role for this kinase in angiogenesis in the labyrinthine region of the placenta. Curr Biol. 1999;9:369–72. doi: 10.1016/s0960-9822(99)80164-x. [DOI] [PubMed] [Google Scholar]
- 2.Roovers K, Assoian RK. Integrating the MAP kinase signal into the G1 phase cell cycle machinery. Bioessays. 2000;22:818–26. doi: 10.1002/1521-1878(200009)22:9<818::AID-BIES7>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 3.Scholl FA, Dumesic PA, Khavari PA. Mek1 alters epidermal growth and differentiation. Cancer Res. 2004;64:6035–40. doi: 10.1158/0008-5472.CAN-04-0017. [DOI] [PubMed] [Google Scholar]
- 4.Scholl FA, Dumesic PA, Khavari PA. Effects of active MEK1 expression in vivo. Cancer Lett. 2005;230:1–5. doi: 10.1016/j.canlet.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 5.Mansour SJ, Matten WT, Hermann AS, et al. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–70. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 6.Schramek H, Feifel E, Healy E, Pollack V. Constitutively active mutant of the mitogen-activated protein kinase kinase MEK1 induces epithelial dedifferentiation and growth inhibition in madin-darby canine kidney-C7 cells. J Biol Chem. 1997;272:11426–33. doi: 10.1074/jbc.272.17.11426. [DOI] [PubMed] [Google Scholar]
- 7.Rubinfeld H, Seger R. The ERK cascade: a prototype of MAPK signaling. Mol Biotechnol. 2005;31:151–74. doi: 10.1385/MB:31:2:151. [DOI] [PubMed] [Google Scholar]
- 8.Walter SA, Guadagno TM, Ferrell JE., Jr Induction of a G2-phase arrest in Xenopus egg extracts by activation of p42 mitogen-activated protein kinase. Mol Biol Cell. 1997;8:2157–69. doi: 10.1091/mbc.8.11.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bitangcol JC, Chau AS, Stadnick E, Lohka MJ, Dicken B, Shibuya EK. Activation of the p42 mitogen-activated protein kinase pathway inhibits Cdc2 activation and entry into M-phase in cycling Xenopus egg extracts. Mol Biol Cell. 1998;9:451–67. doi: 10.1091/mbc.9.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feinstein TN, Linstedt AD. Mitogen-activated protein kinase kinase 1-dependent Golgi unlinking occurs in G2 phase and promotes the G2/M cell cycle transition. Mol Biol Cell. 2007;18:594–604. doi: 10.1091/mbc.E06-06-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shapiro PS, Vaisberg E, Hunt AJ, et al. Activation of the MKK/ERK pathway during somatic cell mitosis: Direct interactions of active ERK with kinetochores and regulation of the mitotic 3F3/2 phosphoantigen. J Cell Biol. 1998;142:1533–45. doi: 10.1083/jcb.142.6.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayne C, Tzivion G, Luo Z. Raf-1/MEK/MAPK pathway is necessary for the G2/M transition induced by nocodazole. J Biol Chem. 2000;275:31876–82. doi: 10.1074/jbc.M002766200. [DOI] [PubMed] [Google Scholar]
- 13.Roberts EC, Shapiro PS, Nahreini TS, Pages G, Pouyssegur J, Ahn NG. Distinct cell cycle timing requirements for extracellular signal-regulated kinase and phosphoinositide 3-kinase signaling pathways in somatic cell mitosis. Mol Cell Biol. 2002;22:7226–41. doi: 10.1128/MCB.22.20.7226-7241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colanzi A, Suetterlin C, Malhotra V. Cell-cycle-specific golgi fragmentation: how and why? Curr Opin Cell Biol. 2003;15:462–7. doi: 10.1016/s0955-0674(03)00067-x. [DOI] [PubMed] [Google Scholar]
- 15.Harding A, Giles N, Burgess A, Hancock JF, Gabrielli BG. Mechanism of mitosis-specific activation of MEK1. J Biol Chem. 2003;278:16747–54. doi: 10.1074/jbc.M301015200. [DOI] [PubMed] [Google Scholar]
- 16.Yu LZ, Xiong B, Gao WX, et al. MEK1/2 regulates microtubule organization, spindle pole tethering and asymmetric division during mouse oocyte meiotic maturation. Cell Cycle. 2007;6:330–8. doi: 10.4161/cc.6.3.3805. [DOI] [PubMed] [Google Scholar]
- 17.Wright JH, Munar E, Jameson DR, et al. Mitogen-activated protein kinase kinase activity is required for the G2/M transition of the cell cycle in mammalian fibroblasts. Proc Natl Acad Sci USA. 1999;96:11335–40. doi: 10.1073/pnas.96.20.11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takenaka K, Gotoh Y, Nishida E. MAP kinase is required for the spindle assembly checkpoint but is dispensable for the normal M phase entry and exit in Xenopus egg cell cycle extracts. J Cell Biol. 1997;136:1091–7. doi: 10.1083/jcb.136.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horne MM, Guadagno TM. A requirement for MAP kinase in the assembly and maintenance of the mitotic spindle. J Cell Biol. 2003;161:1021–8. doi: 10.1083/jcb.200304144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung EN, Chen RH. Phosphorylation of Cdc20 is required for its inhibition by the spindle checkpoint. Nat Cell Biol. 2003;5:748–53. doi: 10.1038/ncb1022. [DOI] [PubMed] [Google Scholar]
- 21.Gupta S, Plattner R, Der CJ, Stanbridge EJ. Dissection of ras-dependent signaling pathways controlling aggressive tumor growth of human fibrosarcoma cells: Evidence for a potential novel pathway. Mol Cell Biol. 2000;20:9294–306. doi: 10.1128/mcb.20.24.9294-9306.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell SL, Khosravi-Far R, Rossman KL, Clark GJ, Der CJ. Increasing complexity of Ras signaling. Oncogene. 1998;17:1395–413. doi: 10.1038/sj.onc.1202174. [DOI] [PubMed] [Google Scholar]
- 23.Manders EMM, Verbeek FJ, Aten JA. Measurement of co-localization of objects in dual-color confocal images. J Microsc. 1993;169:375–82. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 24.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 25.Nogales E. Structural insights into microtubule function. Annu Rev Biochem. 2000;69:277–302. doi: 10.1146/annurev.biochem.69.1.277. [DOI] [PubMed] [Google Scholar]
- 26.Krebs A, Goldie KN, Hoenger A. Structural rearrangements in tubulin following microtubule formation. EMBO Rep. 2005;6:227–32. doi: 10.1038/sj.embor.7400360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inoue S. Cell division and the mitotic spindle. J Cell Biol. 1981;91:131s–47s. doi: 10.1083/jcb.91.3.131s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kline-Smith SL, Walczak CE. Mitotic spindle assembly and chromosome segregation: Refocusing on microtubule dynamics. Mol Cell. 2004;15:317–27. doi: 10.1016/j.molcel.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 29.Duensing S, Lee BH, Dal Cin P, Münger K. Excessive centrosome abnormalities without ongoing numerical chromosome instability in a Burkitt’s lymphoma. Mol Cancer. 2003;2:30. doi: 10.1186/1476-4598-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pihan GA, Wallace J, Zhou Y, Doxsey SJ. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 2003;63:1398–404. [PubMed] [Google Scholar]
- 31.Roh M, Gary B, Song C, et al. Overexpression of the oncogenic kinase Pim-1 leads to genomic instability. Cancer Res. 2003;63:8079–84. [PubMed] [Google Scholar]
- 32.Doxsey S, Zimmerman W, Mikule K. Centrosome control of the cell cycle. Trends Cell Biol. 2005;15:303–11. doi: 10.1016/j.tcb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 33.Sankaran S, Parvin JD. Centrosome function in normal and tumor cells. J Cell Biochem. 2006;99:1240–50. doi: 10.1002/jcb.21003. [DOI] [PubMed] [Google Scholar]
- 34.Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004;4:253–65. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 35.Dammermann A, Desai A, Oegema K. The minus end in sight. Curr Biol. 2003;13:R614–24. doi: 10.1016/s0960-9822(03)00530-x. [DOI] [PubMed] [Google Scholar]
- 36.Zimmerman WC, Sillibourne J, Rosa J, Doxsey SJ. Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol Biol Cell. 2004;15:3642–57. doi: 10.1091/mbc.E03-11-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159:255–66. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levesque AA, Howard L, Gordon MB, Compton DA. A functional relationship between NuMA and kid is involved in both spindle organization and chromosome alignment in vertebrate cells. Mol Biol Cell. 2003;14:3541–52. doi: 10.1091/mbc.E03-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Compton DA. Focusing on spindle poles. J Cell Sci. 1998;111:1477–81. doi: 10.1242/jcs.111.11.1477. [DOI] [PubMed] [Google Scholar]
- 40.Chen JG, Horwitz SB. Differential mitotic responses to microtubule-stabilizing and - destabilizing drugs. Cancer Res. 2002;62:1935–8. [PubMed] [Google Scholar]
- 41.Ohren JF, Chen H, Pavlovsky A, et al. Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat Struct Mol Biol. 2004;11:1192–7. doi: 10.1038/nsmb859. [DOI] [PubMed] [Google Scholar]
- 42.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.