Abstract
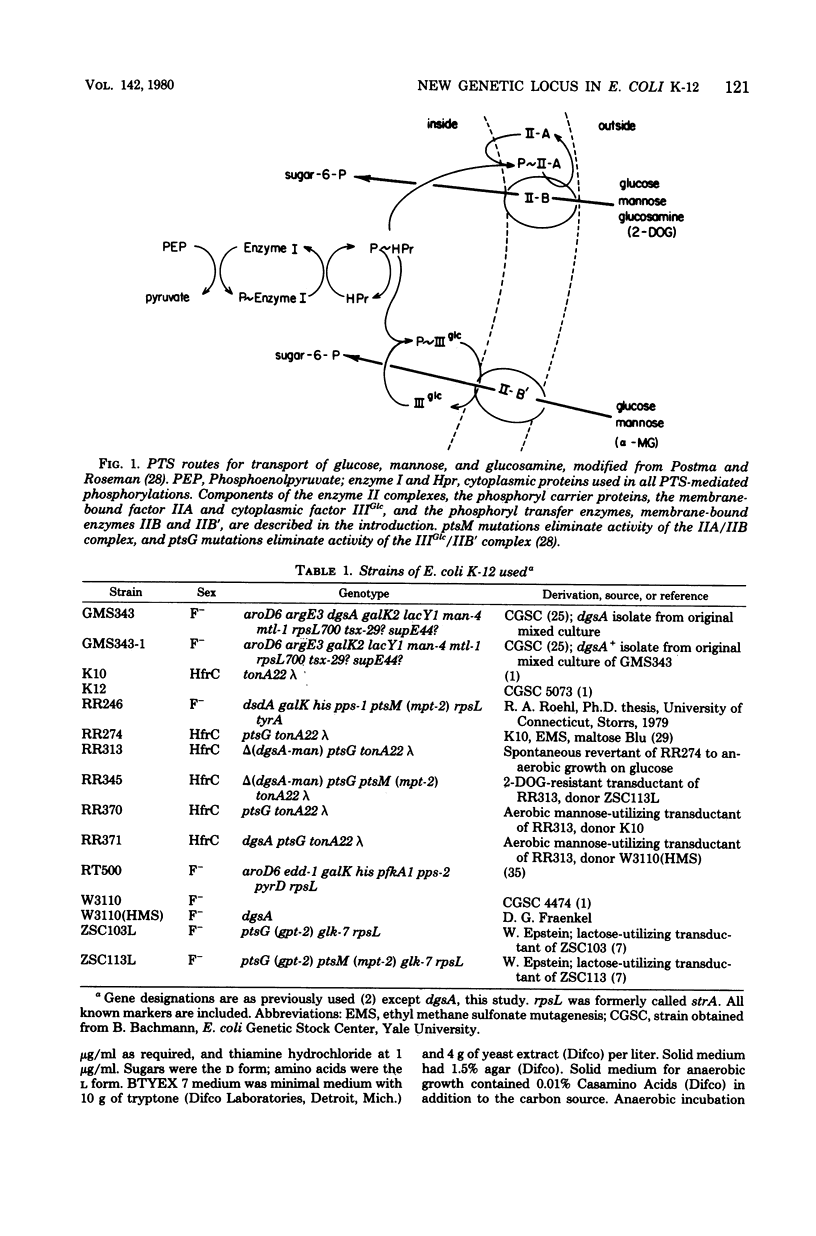
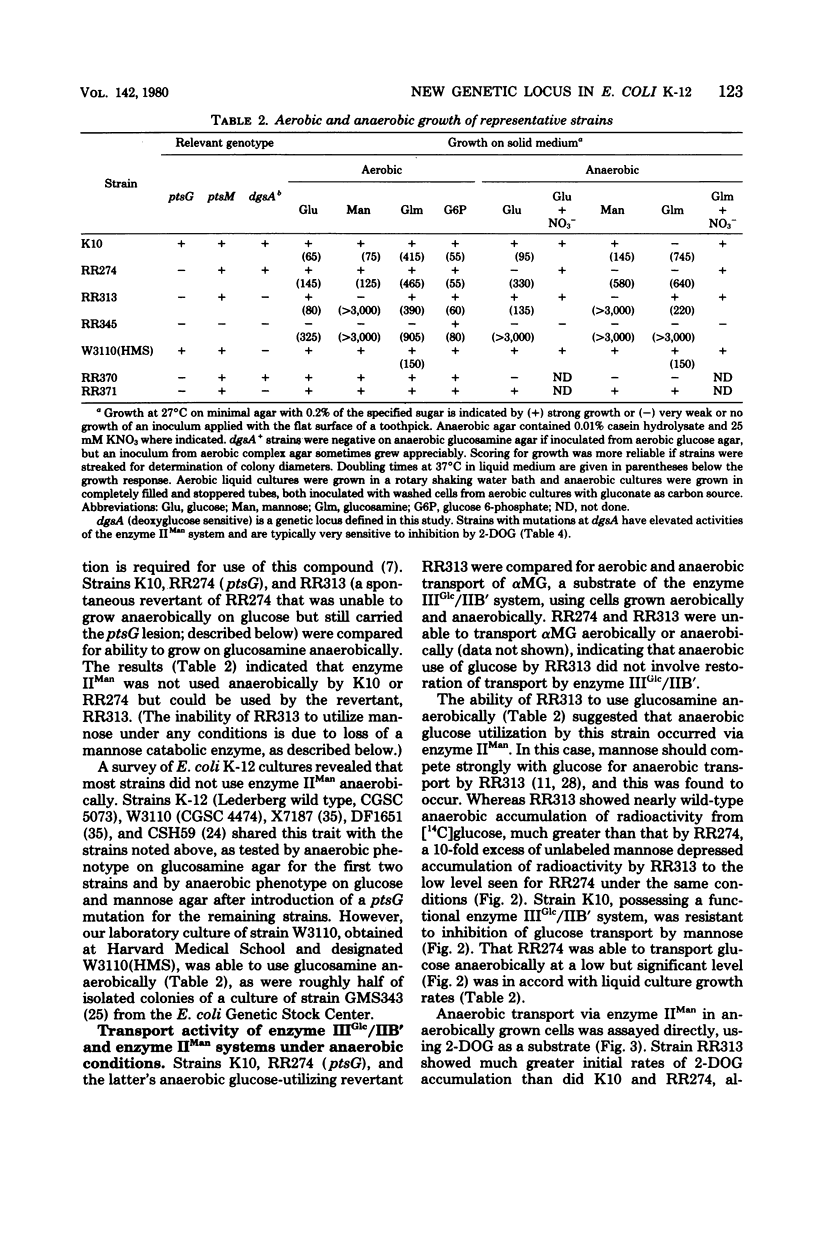
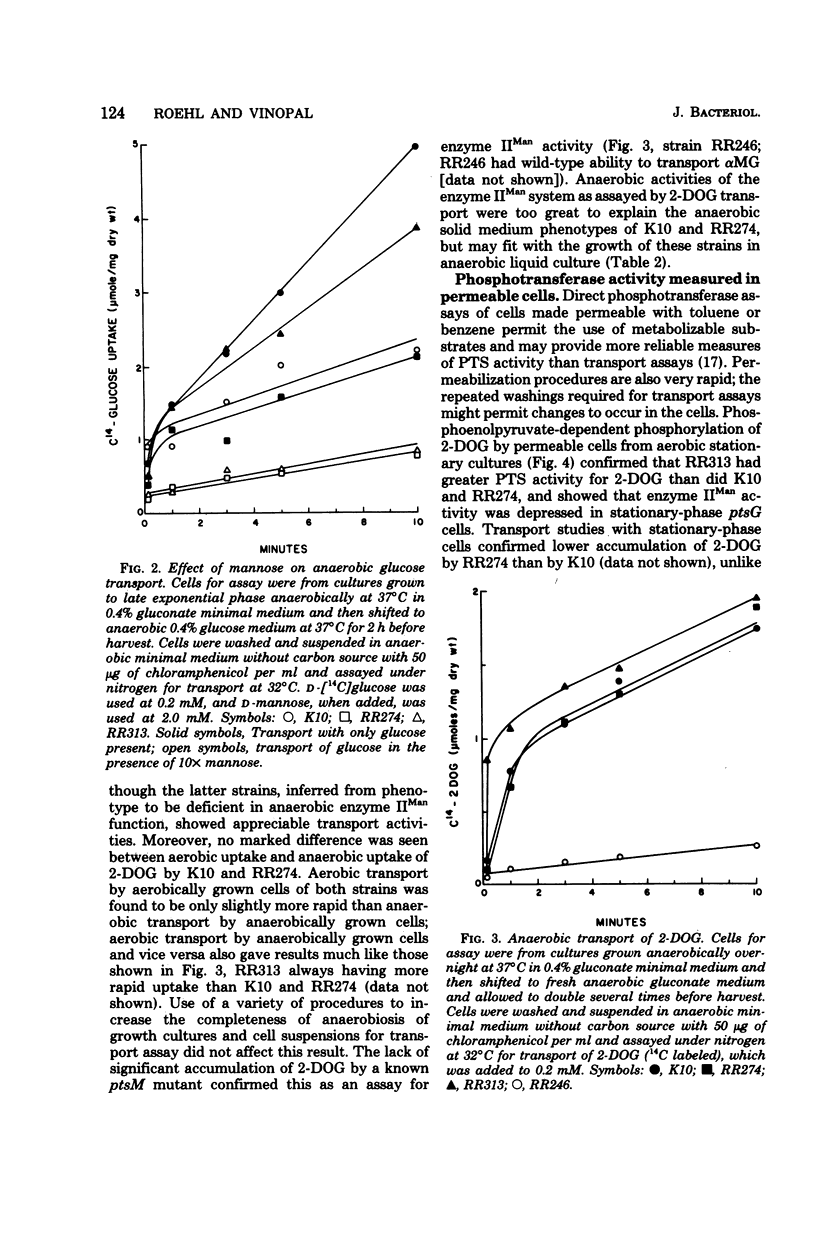
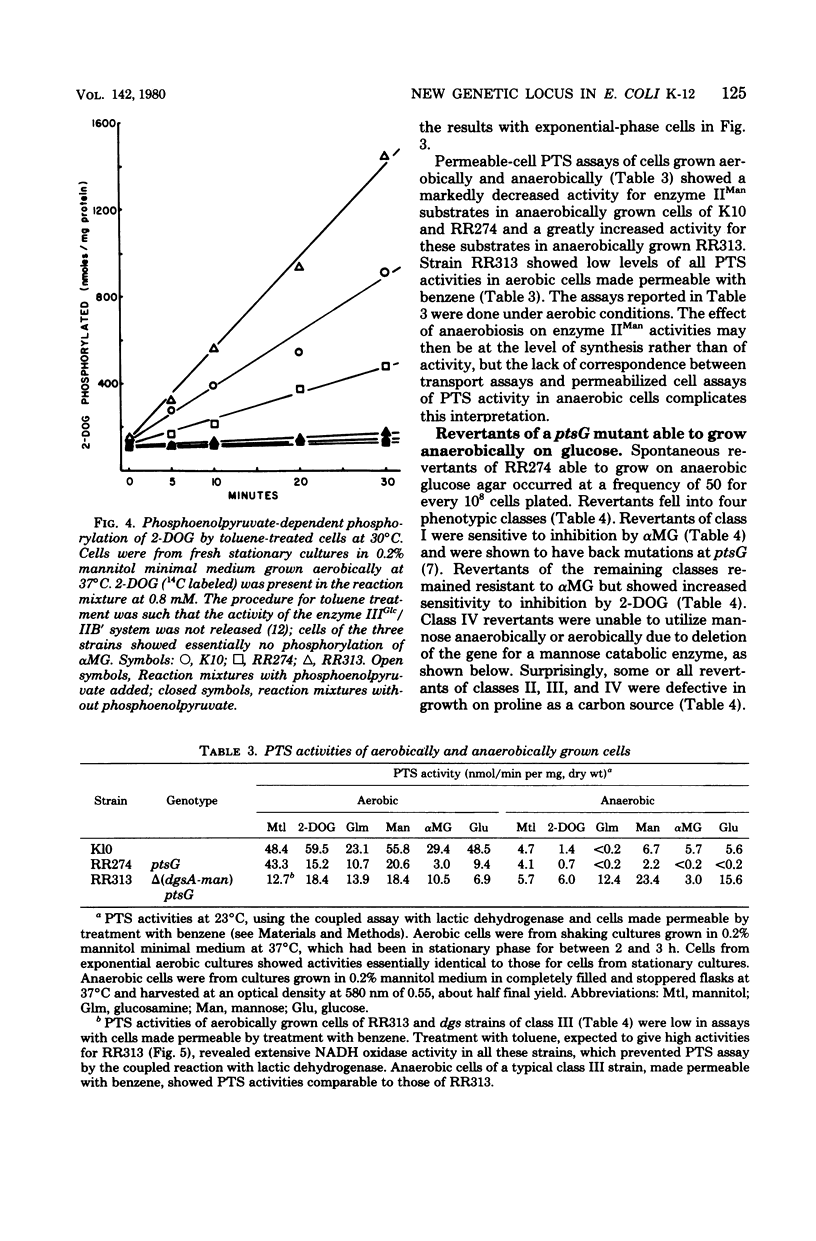
Most strains of Escherichia coli K-12 are unable to use the enzyme IIA/IIB (enzyme IIMan) complex of the phosphoenolpyruvate:sugar phosphotransferase system (PTS) in anaerobic growth and therefore cannot utilize glucosamine anaerobically. Introduction into these strains of a ptsG mutation, which eliminates activity of the enzyme IIIGlc/IIB' complex of the PTS, resulted in inability to grow anaerobically on glucose and mannose. Derivative strains able to grow anaerobically on glucosamine had mutations at a locus close to man, the gene coding for phosphomannose isomerase, and had higher enzyme IIA/IIB activities during anaerobic growth than did the parental strain. These results establish a locus affecting function of enzyme IIA/IIB that maps distant from ptsM, the probable structural gene for enzyme IIB.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissett D. L., Anderson R. L. Genetic evidence for the physiological significance of the D-tagatose 6-phosphate pathway of lactose and D-galactose degradation in staphylococcus aureus. J Bacteriol. 1974 Sep;119(3):698–704. doi: 10.1128/jb.119.3.698-704.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark B., Holms W. H. Control of the sequential utilization of glucose and fructose by Escherichia coli. J Gen Microbiol. 1976 Aug;96(2):191–201. doi: 10.1099/00221287-95-2-191. [DOI] [PubMed] [Google Scholar]
- Cordaro C. Genetics of the bacterial phosphoenolpyruvate: glycose phosphotransferase system. Annu Rev Genet. 1976;10:341–359. doi: 10.1146/annurev.ge.10.120176.002013. [DOI] [PubMed] [Google Scholar]
- Curtis S. J., Epstein W. Phosphorylation of D-glucose in Escherichia coli mutants defective in glucosephosphotransferase, mannosephosphotransferase, and glucokinase. J Bacteriol. 1975 Jun;122(3):1189–1199. doi: 10.1128/jb.122.3.1189-1199.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dendinger S., Brill W. J. Regulation of proline degradation in Salmonella typhimurium. J Bacteriol. 1970 Jul;103(1):144–152. doi: 10.1128/jb.103.1.144-152.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall B. G., Hartl D. L. Regulation of newly evolved enzymes. II. The ebg repressor. Genetics. 1975 Nov;81(3):427–435. doi: 10.1093/genetics/81.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter I. S., Kornberg H. L. Glucose transport of Escherichia coli growing in glucose-limited continuous culture. Biochem J. 1979 Jan 15;178(1):97–101. doi: 10.1042/bj1780097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetten A. M. Effects of colicins K and E1 on the glucose phosphotransferase system. Biochim Biophys Acta. 1976 Aug 13;440(2):403–411. doi: 10.1016/0005-2728(76)90074-8. [DOI] [PubMed] [Google Scholar]
- Jochimsen B., Nygaard P., Vestergaard T. Location on the chromosome of Escherichia coli of genes governing purine metabolism. Adenosine deaminase (add), guanosine kinase (gsk) and hypoxanthine phosphoribosyltransferase (hpt). Mol Gen Genet. 1975 Dec 30;143(1):85–91. doi: 10.1007/BF00269424. [DOI] [PubMed] [Google Scholar]
- Kang S., Markovitz A. Induction of capsular polysaccharide synthesis by rho-fluorophenylalanine in Escherichia coli wild type and strains with altered phenylalanyl soluble ribonucleic acid synthetase. J Bacteriol. 1967 Feb;93(2):584–591. doi: 10.1128/jb.93.2.584-591.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg H. L., Reeves R. E. Correlation between hexose transport and phosphotransferase activity in Escherichia coli. Biochem J. 1972 Mar;126(5):1241–1243. doi: 10.1042/bj1261241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg H. L., Reeves R. E. Inducible phosphoenolpyruvate-dependent hexose phosphotransferase activities in Escherichia coli. Biochem J. 1972 Aug;128(5):1339–1344. doi: 10.1042/bj1281339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg H. L., Riordan C. Uptake of galactose into Escherichia coli by facilitated diffusion. J Gen Microbiol. 1976 May;94(1):75–89. doi: 10.1099/00221287-94-1-75. [DOI] [PubMed] [Google Scholar]
- Kundig W. Molecular interactions in the bacterial phosphoenolpyruvate-phosphotransferase system (PTS). J Supramol Struct. 1974;2(5-6):695–814. doi: 10.1002/jss.400020514. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Lengeler J. Analysis of mutations affecting the dissmilation of galactitol (dulcitol) in Escherichia coli K 12. Mol Gen Genet. 1977 Mar 28;152(1):83–91. doi: 10.1007/BF00264944. [DOI] [PubMed] [Google Scholar]
- Markovitz A., Sydiskis R. J., Lieberman M. M. Genetic and biochemical studies on mannose-negative mutants that are deficient in phosphomannose isomerase in Escherichia coli K-12. J Bacteriol. 1967 Nov;94(5):1492–1496. doi: 10.1128/jb.94.5.1492-1496.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novel G., Novel M. Mutants d'Escherichia coli K 12 affectés pour leur croissance sur méthyl-beta-D-glucuronide: localisation of gène de structure de la beta-D-glucuronidase (uid A. Mol Gen Genet. 1973;120(4):319–335. [PubMed] [Google Scholar]
- Postma P. W. Galactose transport in Salmonella typhimurium. J Bacteriol. 1977 Feb;129(2):630–639. doi: 10.1128/jb.129.2.630-639.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma P. W. Involvement of the phosphotransferase system in galactose transport in Salmonella typhimurium. FEBS Lett. 1976 Jan 1;61(1):49–53. doi: 10.1016/0014-5793(76)80169-x. [DOI] [PubMed] [Google Scholar]
- Postma P. W., Roseman S. The bacterial phosphoenolpyruvate: sugar phosphotransferase system. Biochim Biophys Acta. 1976 Dec 14;457(3-4):213–257. doi: 10.1016/0304-4157(76)90001-0. [DOI] [PubMed] [Google Scholar]
- Roehl R. A., Vinopal R. T. New maltose Blu mutations in Escherichia coli K-12. J Bacteriol. 1979 Aug;139(2):683–685. doi: 10.1128/jb.139.2.683-685.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano A. H., Trifone J. D., Brustolon M. Distribution of the phosphoenolpyruvate:glucose phosphotransferase system in fermentative bacteria. J Bacteriol. 1979 Jul;139(1):93–97. doi: 10.1128/jb.139.1.93-97.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner J. L. Formation, induction, and curing of bacteriophage P1 lysogens. Virology. 1972 Jun;48(3):679–689. doi: 10.1016/0042-6822(72)90152-3. [DOI] [PubMed] [Google Scholar]
- Saier M. H., Jr Bacterial phosphoenolpyruvate: sugar phosphotransferase systems: structural, functional, and evolutionary interrelationships. Bacteriol Rev. 1977 Dec;41(4):856–871. doi: 10.1128/br.41.4.856-871.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinopal R. T., Fraenkel D. G. Phenotypic suppression of phosphofructokinase mutations in Escherichia coli by constitutive expression of the glyoxylate shunt. J Bacteriol. 1974 Jun;118(3):1090–1100. doi: 10.1128/jb.118.3.1090-1100.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. T. A model for three-point analysis of random general transduction. Genetics. 1966 Aug;54(2):405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]


