Abstract
While low levels of unesterified long chain fatty acids (LCFAs) are normal metabolic intermediates of dietary and endogenous fat, LCFAs are also potent regulators of key receptors/enzymes, and at high levels become toxic detergents within the cell. Elevated levels of LCFAs are associated with diabetes, obesity, and metabolic syndrome. Consequently, mammals evolved fatty acid binding proteins (FABPs) that bind/sequester these potentially toxic free fatty acids in the cytosol and present them for rapid removal in oxidative (mitochondria, peroxisomes) or storage (endoplasmic reticulum, lipid droplets) organelles. Mammals have a large (15 member) family of FABPs with multiple members occurring within a single cell type. The first described FABP, liver-FABP (L-FABP, or FABP1), is expressed in very high levels (2-5% of cytosolic protein) in liver as well as intestine and kidney. Since L-FABP facilitates uptake and metabolism of LCFAs in vitro and in cultured cells, it was expected that abnormal function or loss of L-FABP would reduce hepatic LCFA uptake/oxidation and thereby increase LCFAs available for oxidation in muscle and/or storage in adipose. This prediction was confirmed in vitro with isolated liver slices and cultured primary hepatocytes from L-FABP gene-ablated mice. Despite unaltered food consumption when fed a control diet ad libitum, the L-FABP null mice exhibited age- and sex-dependent weight gain and increased fat tissue mass. The obese phenotype was exacerbated in L-FABP null mice pair-fed a high fat diet. Taken together with other findings, these data suggest that L-FABP could have an important role in preventing age- or diet-induced obesity.
Keywords: Liver, fatty acid binding protein, fat, oxidation, body weight, obesity
1. Introduction
Although intracellular fatty acid binding proteins (FABPs) were first discovered over 30 years ago [1], physiological functions of this 15 member protein family are not well elucidated. FABPs are the single most abundant proteins in the cytosol of cells most active in long chain fatty acid (LCFA) uptake and metabolism (liver, intestine), oxidation (kidney, heart, skeletal muscle), and storage (adipose) (Fig. 1) (rev. in [2-6]. Several excellent reviews have previously addressed: (i) native and recombinant FABPs with regards to tissue occurrence, intracellular distribution, isolation, ligand specificity, structure, and potential functions in vitro [4, 7-11]; (ii) gene structures, regulation, and functions of FABPs in cultured cells (rev. in [6, 12-16]; (iii) roles in regulation of nuclear receptors [e.g. peroxisome proliferator activated receptors-α (PPARα), hepatocyte nuclear factor-4α (HNF-4α)] (rev. in [17, 18]; and (iv) physiological functions of FABPs in genetically engineered mice (rev. in [19-21]. Of these FABPs, liver fatty acid binding protein (L-FABP, also called FABP1) is the most broadly distributed mammalian FABP, is expressed at very high levels in tissues most active in LCFA metabolism [liver (2-5% of cytosolic protein, 0.1-0.4 mM), intestine, kidney].
Figure 1. Distribution of fatty acid binding proteins (FABPs) in tissues important for long chain fatty acid (LCFA) metabolism.
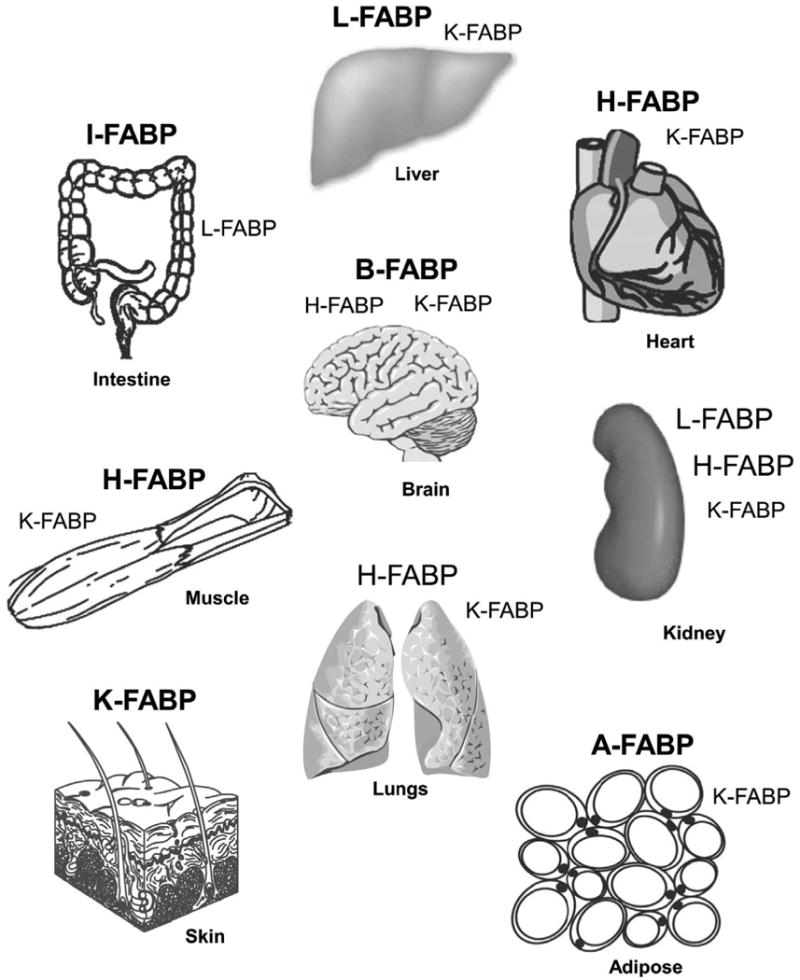
FABPs present in tissues at highest concentration are shown in large bold letters. FABPS present at lower concentration are shown in large unbold letters. Low expression is shown with small bold letters. The nomenclature of the long chain fatty acid binding protein family has been described [21]: L-FABP, liver type fatty acid binding protein (Fabp1 gene); I-FABP, intestinal type fatty acid binding protein (Fabp2 gene); H-FABP, heart type fatty acid binding protein (Fabp3 gene); A-FABP, adipocyte type fatty acid binding protein (Fabp4 gene); K-FABP, keratinocyte type fatty acid binding protein (also called epidermal fatty acid binding protein, E-FABP, Fabp5 gene); B-FABP, brain type fatty acid binding protein (Fabp7 gene). Additional members of the FABP family (not shown) that bind other types of ligands include: M-FABP, myelin (peripheral) type fatty acid binding protein (Fabp8 gene); T-FABP, testis type fatty acid binding protein; ILBP, ileal bile acid binding protein (Fabp6 gene); CRBP I and II, cellular retinol binding proteins I and II; and CRABP I and II, cellular retinoic acid binding proteins I and II.
The current review focuses on L-FABP contributions to LCFA uptake, metabolism, and obesity. An integrated model suggesting the physiological roles of L-FABP is presented in Fig. 2, which incorporates functions first suggested by in vitro studies of the pure protein, supported by findings in living cultured cells overexpressing L-FABP, and finally established in vivo through the use of L-FABP gene-ablated mice. In multiple steps, L-FABP: (i) enhances cellular LCFA uptake; (ii) binds LCFAs and LCFA-CoAs to minimize toxic effects (detergent properties, inhibition of enzymes) of these poorly soluble lipids; (iii) enhances intracellular transport/diffusion through the cytoplasm; (iv) targets LCFA to peroxisomes for β-oxidation (straight-chain LCFAs) and α-oxidation (branched-chain LCFAs); (v) delivers LCFAs and LCFA-CoAs to mitochondria for oxidation; (vi) targets LCFA and LCFA-CoA to endoplasmic reticulum (ER) for transacylation to complex lipids for membrane synthesis (phosphatidic acid, phospholipids), and storage (triacylglycerides, cholesteryl esters) in multiple tissues wherein L-FABP is expressed as well as for hepatic secretion in VLDL (triacylglycerides, cholesteryl esters); (vii) transports LCFAs and LCFA-CoAs to the nucleus for regulation of nuclear receptors important in transcription of genes encoding proteins involved in LCFA and glucose metabolism [PPAR-α, HNF-4α, liver X receptor (LXR), thyroid hormone receptor (THR)]. Consistent with this model (Fig. 2), ablation of L-FABP inhibits LCFA uptake, reduces LCFA intracellular transport/diffusion, inhibits LCFA esterification, inhibits LCFA oxidation, and inhibits LCFA targeting to the nucleus to thereby redirect dietary LCFA for storage in adipose phenotypically evident as sex-, age-, and high-fat diet dependent obesity. Similarities and differences in phenotype of an independently generated L-FABP gene-ablated mouse underscore the importance of understanding the impact of GFP knock-in strategy, construct design, backcrossing, age, sex, appropriate control diets, and composition of high-fat diets.
Figure 2. Model of L-FABP functions in living cells.
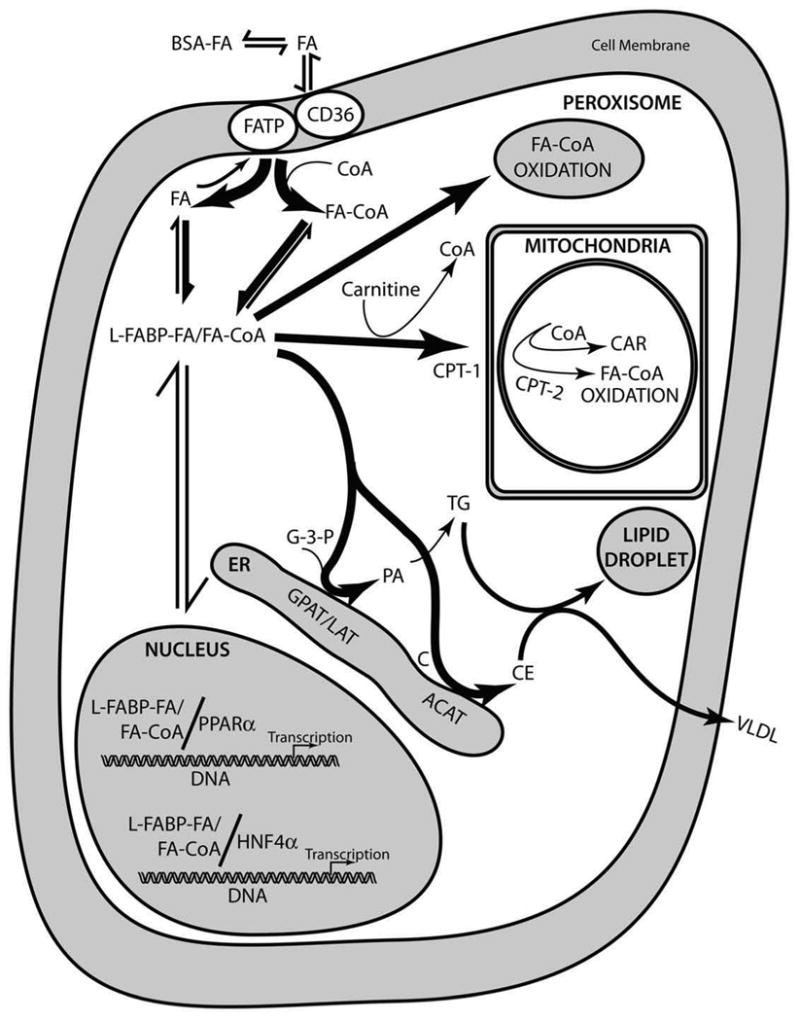
Bold arrows refer to reactions most greatly enhanced by L-FABP. Abbreviations are as follows: BSA, serum albumin; FA, long chain fatty acid; FATP, plasma membrane fatty acid transport protein; CD36, plasma membrane fatty acid translocase protein; CoA, coenzyme A; L-FABP, liver fatty acid binding protein; CPT-1, carnitine palmitoyl transferase I (outer mitochondrial membrane); CPT-2, carnitine palmitoyl transferase II (inner mitochondrial membrante); CAR, carnitine; G-3-P, glycerol-3-phosphate; GPAT, glycerol-3-phosphate acyltransferase; LAT, lysophosphatidic acid acyltransferase; PA, phosphatidic acid; TG, triacylglyceride; C, cholesterol; ACAT, acyl CoA cholesterol acyl transferase; CE, cholesteryl ester; VLDL very low density lipoprotein (applies only to the liver); ER, endoplasmic reticulum; PPARα, peroxisome proliferator activated receptor α; HNF4α, hepatocyte nuclear factor 4α.
2. L-FABP gene
2.1. Gene structure
By chromosomal mapping, the human L-FABP gene was identified and localized to the centromeric p12-q11 region of chromosome 2. The mouse and rat L-FABP genes were localized to chromosome 6 and 4, respectively [22]. Overall, the L-FABP gene structure (four exons and three introns) is identical to other members in the FABP family. For L-FABP, exon 1 encodes amino acids (aa) 1-22, exon 2 (aa 23-79), exon 3 (aa 80-112), and exon 4 (aa 113-126)] [23]. The primary structure of L-FABP in several mammalian species (rat, mouse, human, bovine) displays 79-90% amino acid identity (Fig. 3A), indicating a highly conserved function across several species.
Figure 3. Sequence homology in the liver fatty acid binding proteins (L-FABPs) from different species.
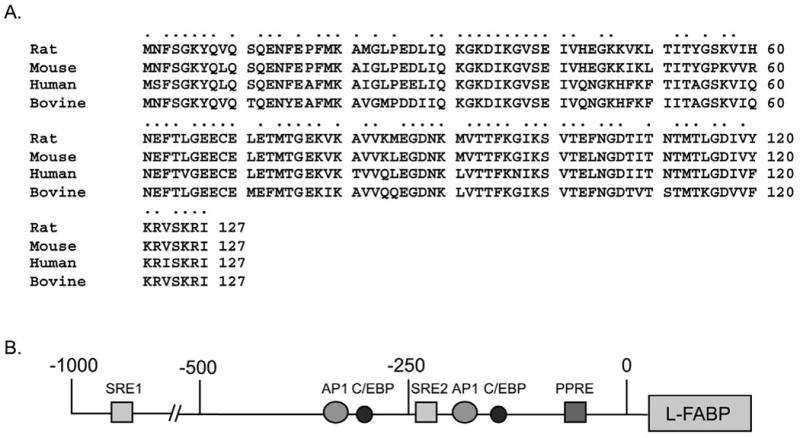
(A) Amino acid sequence alignment for L-FABP derived from rat, mouse, human, and bovine. Identical amino acids are indicated by dots. (B) Promoter region of the human L-FABP gene. Response elements within the promoter for two putative sterol response elements (SRE 1 and 2), activator protein (AP1), CCAAT/enhancer binding protein (C/EBP), and the peroxisomal proliferator response element (PPRE) are indicated.
2.2. Transcriptional regulation: peroxisome proliferator activated response element (PPRE)
The promoter region of the L-FABP gene contains several response elements involved in fat metabolism (Fig. 3B). The most well understood of these is the peroxisome proliferator activated response element (PPRE) located between nucleotides -68 and -56. This PPRE contains an imperfect direct repeat sequence that binds and is activated by peroxisome proliferator-activated receptor-α (PPAR-α) (rev. in [24, 25]. Consistent with this finding, L-FABP is upregulated by peroxisome proliferator-activated receptor-α ligands (rev. in [17]. Expression of L-FABP is regulated by dietary LCFA as well as by its intracellular activated form—i.e. LCFA-CoA. By transporting LCFAs/LCFA-CoAs to the nucleus for interaction with PPARα, L-FABP regulates its own transcription (Fig. 3B) (rev. in [15, 17, 18, 26]. Dietary LCFAs induce the expression of L-FABP in liver, but their potency is highly dependent on structure, chain-length, and unsaturation—the branched-chain LCFAs being the most potent as shown by transactivation in cultured cells and dietary studies in animals (rev. in [15, 24, 26-35]. Peroxisome proliferator drugs also bind to L-FABP and induce expression of L-FABP (rev. in [26, 34, 36-39].
While these data suggest free LCFAs (and peroxisome proliferator drugs) could regulate expression of L-FABP and LCFA oxidative enzymes through PPARα, earlier radioligand binding studies requiring separation of bound from free LCFAs yielded only very weak binding affinities of PPARα for LCFAs (Kds in the μM range)—much higher than nucleoplasmic levels of LCFAs which are in the nM range [17, 30, 31]. Thus induction of PPARα by dietary LCFAs was attributed to LCFA metabolite(s) such as LCFA-CoAs, leukotrienes, prostaglandins, and others. However, it is now known that radioligand binding assays requiring separation of bound from free fatty acids seriously underestimate affinity of binding proteins for LCFA and LCFA-CoA by 2-3 orders of magnitude (rev. in [6, 40]. To resolve this issue, more recent studies took advantage of the L-FABP intrinsic aromatic amino acid properties (fluorescence) and peptide interaction with circularly polarized light (circular dichroism, CD), fluorescent ligands (LCFAs, LCFA-CoAs), and fluorescence resonance energy transfer (FRET) (rev. in [17, 18, 30, 31, 41]. These assays, not requiring separation of bound from free LCFAs or LCFA-CoAs, established that PPARα exhibits high affinity (low nM Kds) for free LCFAs and LCFA-CoAs in a ligand-dependent manner: (i) branched-chain LCFAs are high affinity (nM Kds) PPARα ligands; (ii) unsaturated LCFAs are also high affinity (nM Kds) PPARα ligands; (iii) both saturated and unsaturated LCFA-CoAs are high affinity (nM Kds) PPARα ligands; and (iv) saturated, straight-chain LCFAs are poor PPARα ligands [17, 30, 31, 41, 42]. LCFA and LCFA-CoAs that bind with high affinity alter PPARα structure, DNA binding, coactivator recruitment, and coactivation—with the branched-chain LCFA being the most potent [17, 30, 31, 42]. In addition, nucleoplasmic levels of unesterified LCFAs and LCFA-CoAs are in the same range as PPARα levels in the nucleus of living cells (rev. in [17, 43-46]. Thus, many free LCFAs, as well as their CoA thioesters, are physiologically important endogenous ligands of PPARα which in turn induces formation of L-FABP itself in a positive feed-back loop (rev. in [17, 47-50]. The potential roles of LCFAs and LCFA-CoAs as functional ligands of other PPAR subtypes (e.g. PPARγ and PPARδ) are further addressed elsewhere [15, 51].
Taken together with the fact that L-FABP transfers LCFAs into the nucleus [43-45, 52], these findings suggest a model wherein L-FABP functions in nuclear receptor regulation by binding LCFAs to alter L-FABP conformation, trafficks into the nucleus to bind with PPARα, and transfers bound LCFA ligands to PPARα (Fig. 3) (rev. in [17]. Similar models have been proposed for other FABPs (A-FABP, E-FABP, and others) interacting with other PPAR subtypes (PPARγ, PPARδ) [15] and for other members of the fatty acid binding protein family (cellular retinoic acid binding protein-2) and retinoic acid-mediated regulation of the retinoid X receptor (RXR) in the nucleus [53, 54].
2.3. Transcriptional regulation via other response elements involved in fat metabolism
Although the L-FABP gene promoter region also contains two putative sterol response elements SRE1 and SRE2 at -801 to -790 and -245 to -233 respectively [55], L-FABP and PPAR-α expression are only slightly affected by cholesterol-rich diet [56]. In addition, the L-FABP promoter has sites for the following transactivators known to control a number of cellular processes involved in differentiation, proliferation, and apotosis: CCAAT/enhancer binding protein (C/EBP) at -318 to -305 and -177 to -163 and activator protein-1 (AP-1) at -338 to -330 and -236 to -226 (Fig. 3B). The fact that hepatic levels of L-FABP are sex-specific (higher in male than female mice; higher in female than male rats) (rev. in [24, 37, 57-59], increase during pregnancy and lactation [60], are regulated by growth hormone [61, 62], and decrease with age [62] suggests the existence of additional response elements that influence the sex, age, and obesity phenotype of mice—especially in response to potent PPARα agonists such as phytanic acid and clofibrate (rev. in [24, 37, 57-59].
2.4. Isoforms and posttranslational regulation
An Asn105/Asp105 substitution in bovine liver L-FABP (rev. in [63, 64] implies the existence of different isoforms of L-FABP, but the possibility of post-translational modification (such as deamidation) must also be considered. More recent reports have not found such isoforms in rat liver L-FABP. For example, isoelectric focusing of native L-FABP isolated from rat liver resolves several different bands that did not differ in amino acid composition but differed in the amount of bound LCFAs (rev. in [63]. Biochemical fractionation and mass spectrometry of two native rat liver L-FABP subfractions also shows differences in bound LCFA content, but not amino acid structure [65]. Interestingly, the delipidated forms of these subfractions differed markedly in structure/folding (fluorescence, CD), ligand specificity, and ability to stimulate LCFA-CoA transacylation by microsomal glycerol-3-phosphate acyltransferase [63-67]. Since recombinant L-FABP is not resolvable into two fractions differing in structure and function, these data suggest the existence of an as yet unresolved mechanism that may regulate the proportion of conforms (differ in structure/folding) for the native L-FABP in liver in vivo.
In contrast to rat liver native L-FABP, post-translational modification (S-thiolation, N-acetylation) of L-FABP has been reported for several other species—suggesting the formation of L-FABP conformers differing in structure/folding may be species dependent (rev. in [63, 65]. The functional significance of L-FABP putative isoforms or post-translational modifications is not well understood. S-thioloation of rat liver L-FABP selectively decreases affinity for unsaturated, but not saturated, LCFAs [68].
3. L-FABP protein
3.1. Surface domains
While L-FABP is largely regarded as a soluble protein that binds LCFAs within its capacious binding cavity for intracellular transfer through the cytoplasm, growing evidence indicates that L-FABP directly binds to membranes and to proteins that utilize L-FABP bound ligands. For example, L-FABP has an α–helical surface domain that interacts with anionic-phospholipid rich membranes to facilitate ligand transfer in vitro [69]. L-FABP also directly binds to the surface-exposed ligand binding domain of carnitine palmitoyl transferase 1—the key rate limiting enzyme in mitochondrial β-oxidation of LCFAs [70]. In addition, L-FABP physically interacts with PPARα [71, 72] and functionally interacts with both PPARα and PPARγ--key nuclear receptors involved in LCFA metabolism [26, 71].
3.2. Interior core ligand binding domain
The amino acid sequence of L-FABP, established over 20 yrs ago, predicted a protein with high content of β–sheet (rev. in [10, 13, 73], a prediction that was subsequently confirmed by CD and fourier transform infrared spectroscopy of native rat liver L-FABP conformers [65, 66] and recombinant L-FABPs [66, 71, 74, 75]. Low resolution tertiary structure by differential polarized phase fluorometry and time-resolved fluorescence showed that recombinant L-FABP was ellipsoidal in shape [66, 75-77]. While detailed high resolution crystal (x-ray) and solution (NMR) structures of native L-FABP have not yet been reported, recombinant L-FABP reveals a barrel-like ligand binding domain comprised of β-sheets with a cavity sufficiently large to accommodate at least two LCFAs—one oriented with a carboxyl interacting with arg and two ser residues in the interior of the binding pocket, and the other oppositely oriented with the carboxyl at the opening of the binding pocket and exposed to the aqueous environment [78, 79]. Second-derivative absorption spectra, tyrosine emission spectra, acrylamide quenching, CD, and differential polarized phase fluorometry all indicated that LCFA binding elicits subtle alterations in conformation and/or tertiary structure of L-FABP in solution [66, 75, 77]. Although the x-ray crystal structure of recombinant holo-L-FABP is known, difficulty in crystallizing the apo-L-FABP has precluded direct comparison to determine if LCFA binding alters conformation/structure by x-ray crystallography [78]. However, this issue was recently addressed by the NMR solution structures of apo-L-FABP and holo-L-FABP (containing bound LCFA) [79]. While LCFA binding did not alter the overall types and locations of secondary structural elements in L-FABP as determined from chemical shift indices, the LCFA entry portal region of L-FABP exhibits considerable conformational variability and an unusual “open cap” orientation with respect to the β-barrel [79]. The conformational flexibility of this region suggests that LCFA binding may occur through adjustments in the helix-turn-helix motif to open or close the top of the β-barrel [79].
Finally, it is important to note that the volume of the L-FABP ligand binding pocket is the largest of any of the intracellular fatty acid binding protein family, 440 Å3 [78]. Consequently, L-FABP is rather promiscuous in not only being able to accommodate LCFAs and LCFA-CoAs, but also other lipids that also impact development of obesity.
3.3. L-FABP binds up to two LCFAs or LCFA-CoAs—key lipidic ligands in fatty acid metabolism and obesity
L-FABP is unique among the FABP family in its ability to bind more than one LCFA/LCFA-CoA and in the size of ligand that can accommodate. L-FABP contains a high affinity site (Kds 4-60 nM) and a lower affinity binding site (Kds 0.3-12 μM) [34, 66, 80-82]. In the higher affinity site, the LCFA is oriented with carboxyl buried deep in the interior, interacting with Arg122 [78, 79]. In the weaker affinity site, an LCFA is oriented with carboxyl facing the surface of the protein binding pocket opening [78, 79]. Occupancy of the lower affinity LCFA binding site may depend upon prior binding of an LCFA to the higher affinity site. L-FABP exhibits modestly higher affinity (2-4 fold) for unsaturated (kinked-chain) LCFAs as compared to their saturated (straight-chain) counterparts [34, 66, 80, 81, 83].
L-FABP is also unique among this protein family in that it can bind two LCFA-CoAs instead of two LCFAs [66]. While the two LCFA-CoA binding sites differ about 12-18 fold in Kds, L-FABP bound both with relatively high affinity, as indicated by Kds of 8-10 nM and 97-180 nM for the high and low affinity sites, respectively [66]. Since the CoA moiety of LCFA-CoAs is too large and polar to be accommodated within the L-FABP binding cavity volume, modeling studies indicate that most likely both acyl chains are buried and both CoA moieties are oriented at the surface opening of the binding pocket (rev. in [84]. This orientation is ideal for presenting the thioester linkage of the L-FABP bound LCFA-CoA to enzymes that transacylate LCFA-CoA to phospholipids and cholesteryl esters.
3.4. L-FABP binds intermediates in LCFA anabolism and catabolism
L-FABP binds a variety of intermediates in LCFA oxidation (e.g. fatty acyl-carnitines) [64, 85-87] and glyceride synthesis (e.g. 1-oleoylglycerol) or up to 2 lysophospholipids (e.g. lysophosphatidylcholine), with Kds in of 0.08-1 μM, depending upon the specific lysophospholipid species and assay [85, 86, 88].
3.5. L-FABP binds a variety of steroid-like ligands in fatty acid metabolism and storage
Surprisingly, L-FABP binds several types of ligands with a steroid-like nucleus (cholesterol, bile acids)—apparently at a single binding site. Because of the very poor solubility of cholesterol in water (critical micelle concentration near 30 nM), initial studies using high concentrations of cholesterol in displacement assays or in the Lipidex 1000 assay (requires separation of bound from free) did not detect cholesterol binding to L-FABP [80, 89]. In contrast, many laboratories using more sensitive fluorescence and photo-crosslinking techniques have now demonstrated that L-FABP clearly binds cholesterol and fluorescent cholesterol analogues (NBD-cholesterol, DHE, photoactivatable FCBP) [90-98]. As is the case of LCFA/LCFA-CoA, cholesterol binding also alters L-FABP conformation [98]. Due the higher solubility of bile acids (oxidation products of cholesterol), a variety of methods could readily demonstrate that bile acids, important in both LCFA and cholesterol metabolism, are bound by L-FABP at a single site [88, 89, 99]. However, bile acids were more weakly bound by L-FABP (Kds 4-50 μM) [88].
3.6. L-FABP binds ligands involved in lipid signaling, growth regulation, and fat storage
The L-FABP binding site (440 Å3) is sufficiently large to accommodate a variety of lipidic molecules with roles in signaling [78]. L-FABP has high affinity for LCFA-CoAs [66]—ligands that regulate vesicular budding from the Golgi [100, 101]. The physiological significance of L-FABP in vesicle budding is demonstrated by studies with L-FABP gene-ablated mice in which the lack of L-FABP reduced budding of pre-chylomicron transport vesicles from intestinal ER [102]. L-FABP exhibits high affinity for products of phospholipid hydrolysis (1-oleoylglycerol, lysophospholipids) released by lipases during signaling (Kds ∼0.08-1 μM) [85, 86, 88], and it also binds a variety of other fatty acid metabolites (prostaglandins, lipoxygenase products, retinoids) that are involved in lipid signaling [64, 87, 103]. L-FABP reduces 15-lipoxygenase-induced oxygenation of linoleic acid and arachidonic acid [104]. L-FABP also binds glycerides such as 1-oleoylglycerol and up to 2 lysophospholipids (e.g. lysophosphatidylcholine) with Kds ∼0.08-1 μM, depending upon the specific lysophospholipid species and assay used [85, 86, 88].
L-FABP also binds a variety of other unrelated ligands including heme and its degradation product bilirubin [4, 105, 106], as well as warfarin [107], carcinogens [108-110], and the dietary trace metal selenium [111]. While the physiological significance of these findings is not completely clear, studies in transfected cells in culture indicate that L-FABP binding of carcinogens and certain LCFAs can regulate mitogenesis and carcinogenesis [112-116].
In summary, the finding that L-FABP binds a variety of lipidic molecules involved in signaling suggests additional routes whereby L-FABP may impact lipid metabolism, cell grown, and obesity.
4. L-FABP directly and indirectly regulates enzymes involved in LCFA metabolism
4.1. L-FABPdirectly facilitates LCFA desorption from membranes in vitro
Although cellular membranes exhibit high affinity for LCFAs and LCFA-CoAs, high levels of these ligands inhibit the activities of many membrane bound enzymes/signaling proteins involved in lipid metabolism (rev. in [4]. It has been proposed that, by removing these ligands from membranes and solubilizing them in the cytosol, L-FABP can prevent the potent inhibitory/detergent effects of LCFA/LCFA-CoA and facilitate their utilization inside the cell. Consistent with this possibility, L-FABP markedly enhanced desorption of LCFAs and LCFA-CoAs from membranes (rev. in [6, 63, 67, 117, 118]. Based on the high affinity of L-FABP for these ligands and the known concentration of L-FABP in hepatic cytosol, the vast majority of unesterified LCFAs and LCFA-CoAs are likely to be bound by cytosolic proteins such as L-FABP.
4.2. Role of L-FABP in activation of LCFAs to LCFA-CoAs
Once taken up into the cell or membrane, LCFAs are rapidly (<1 min) converted to LCFA-CoAs by ubiquitous fatty acyl CoA synthases, membrane-associated enzymes present in plasma membrane, ER, mitochondria, peroxisomes, and nuclear membranes [119-121]. Earlier in vitro studies with L-FABP isolated from liver suggest that L-FABP directly interacts with fatty acyl CoA synthases to donate bound LCFAs for conversion to LCFA-CoAs (rev. in [6, 14]. However, later studies with recombinant L-FABP showed that acyl CoA binding protein (ACBP), a likely contaminant of early native L-FABP preparations, stimulated long chain fatty acyl CoA synthases while highly purified L-FABP does not (rev. in [6]. Nevertheless, L-FABP may still play a role in this stimulation via increasing LCFA-CoAs by inhibiting LCFA-CoA hydrolysis. The thioester linkage in unbound LCFA-CoAs is subject to auto-hydrolysis in aqueous environments as well as to degradation by cellular hydrolases [118, 122, 123]. L-FABP-bound LCFA-CoAs are protected from microsomal hydrolysis (rev. in [6, 63, 67, 117, 118].
4.3. L-FABP facilitates intermembrane transfer of LCFAs/LCFA-CoAs and cholesterol
Consistent with its ability to interact with membranes (see 3.1) L-FABP stimulates the transfer of bound LCFAs and LCFA-CoAs from model membranes and from the plasma membrane to other membranes (ER) in vitro (rev. in [4, 6, 124]. L-FABP also stimulates transfer of bound cholesterol from the plasma membrane to ER in vitro (rev. in [125-127].
4.4. L-FABP facilitates mcirosomal LCFA esterification
L-FABP stimulates two key LCFA-CoA transacylation reactions: 1) by increasing activity of several LCFA anabolic enzymes involved in the synthesis of triglycerides and cholesteryl esters—lipids that accumulate to pathological levels in obesity; and ii) enhancing the rate limiting step in phosphatidic acid synthesis mediated by glycerol-3-phosphate-acyltransferase (Fig. 2, GPAT) (rev. in [63, 67, 117, 118, 128]. Phosphatidic acid is the precursor of a variety of membrane phospholipids as well as triacyglycerols either stored in lipid droplets in multiple cells expressing L-FABP or secreted by the liver in very low density lipoproteins (Fig. 2). L-FABP also increases the synthesis of cholesteryl esters mediated by cholesterol acyl CoA acyltransferase (ACAT) and targeted for storage in lipid droplets in multiple cells expressing L-FABP or secreted by the liver in very low density lipoproteins (Fig. 2) [129].
4.5. L-FABP stimulates mitochondrial and peroxisomal oxidation of LCFAs
Purified L-FABP removes substrate inhibition of carnitine palmitoyltransferase-1 (CPT-1, the rate limiting step in mitochondrial LCFA oxidation) by palmitoyl CoA [130, 131]. L-FABP functions as a LCFA donor protein for mitochondrial LCFA oxidation [132]. To determine if L-FABP directly binds CPT-1 or simply shuttles bound LCFA-CoAs to CPT-1 located in the outer mitochondrial membrane, a fluorescence resonance energy transfer (FRET) study was performed. FRET is a sensitive technique effective in measuring intermolecular distances between proteins 1-100 Å apart [71, 94]. L-FABP directly interacted with CPT-1, exhibited saturable binding for CPT-1 with 1:1 stoichiometry, and bound CPT-1 with high affinity (2.1 ± 0.3nM Kd) [70]. The intermolecular distance between L-FABP and CPT-1 was calculated as 53.1 ± 0.9Å [70]. Thus, data were consistent with a close molecular interaction. To determine if L-FABP binding to CPT-1 altered protein conformation, the two proteins were examined separately (Fig. 4A) and together (Fig. 4B) by CD. The proportion of different secondary structures measured experimentally in the complex differed significantly from the theoretical prediction if the proteins did not interact (Fig. 4C)—indicating that L-FABP directly interacted with CPT-1 These studies thus suggest a model wherein L-FABP binds with CPT-1 to undergo a conformational change that transfers bound LCFA-CoA to CPT-1. Consistent with a potential role for L-FABP in LCFA transfer to mitochondria, both subcellular fractionation and immunogold electron microscopy (EM) detect small amounts of L-FABP associated with mitochondria [128]. L-FABP also serves as a LCFA donor protein for peroxisomal LCFA oxidation [133]. Consistent with a functional role for L-FABP in LCFA transfer to peroxisomes, small amounts of L-FABP have been detected associated with purified peroxisomes and by immunogold EM [134].
Figure 4. Interaction of L-FABP with carnitine palmitoyl transferase I (CPT1) determined by circular dichroism.
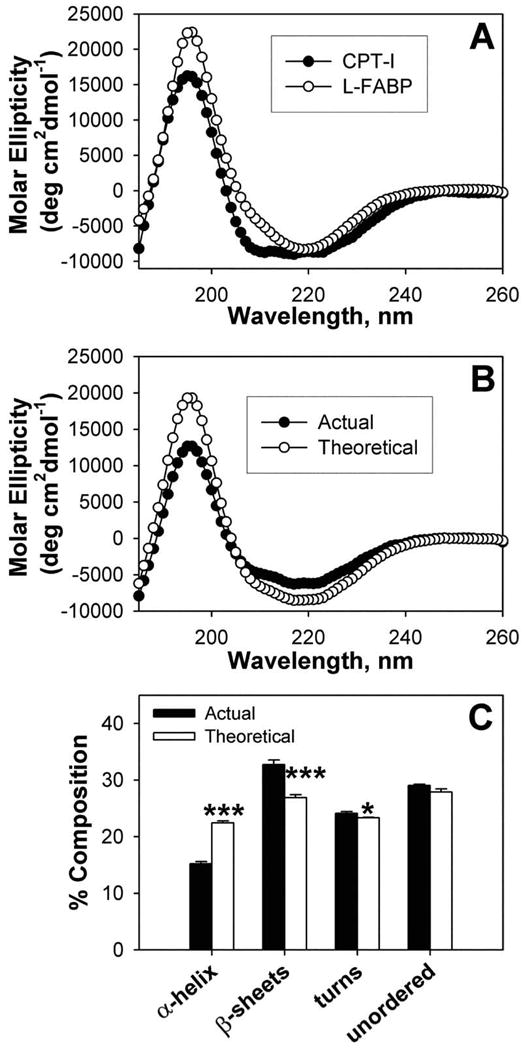
(A) Individual far-UV circular dichroic (CD) spectra of WT CPTI C-terminal 89-residue peptide (filled circles) and an equal amino acid molarity of L-FABP (open circles). (B) Comparison of the far-UV CD spectra of an equal amino acid molarity mixture of WT CPT peptide and L-FABP obtained experimentally (actual, filled circles) and the theoretically expected spectrum (theoretical, open circles) if no conformational change occurred (i.e. the average of the two proteins). (C) Proportion of secondary structures (e.g. α–helix, β-sheet, turn, unordered) in equal molarity mixtures of CPT peptide and L-FABP obtained experimentally (actual, filled bars) and the theoretically expected (theoretical, open bars). Asterisks represent significant differences between the actual and theoretical for each compositional component; * P < 0.05; *** P < 0.001.
4.6. L-FABP stimulates LCFA transfer into purified nuclei
LCFAs alone poorly enter purified nuclei, while L-FABP facilitates the entry of LCFAs by binding and cotransporting the bound LCFA into the nucleus [52]. Several immunofluorescence and immunogold EM studies detected L-FABP in the nucleoplasm of hepatocytes and in cultured cells overexpressing L-FABP [43, 44, 72, 128].
5. Effect of L-FABP overexpression and ablation on LCFA uptake and transport in cultured cells
5.1. L-FABP overexpression enhances while L-FABP ablation inhibits LCFA and cholesterol uptake in living cells
L-FABP enhances uptake of LCFAs and cholesterol from the medium of transfected L-cells overexpressing L-FABP, hepatoma cells expressing increasing amounts of L-FABP, and primary cultured hepatocytes [84, 135-141]. Conversely, L-FABP gene ablation decreases uptake of LCFAs in cultured primary hepatocytes and in freshly isolated hepatocytes [142, 143]. Whether L-FABP facilitates LCFA uptake by enhancing desorption of LCFAs from the plasma membrane, from plasma membrane LCFA translocases, or simply acts as an acceptor/cytoplasmic sink for these LCFAs is not clear.
5.2. L-FABP overexpression enhances while L-FABP ablation inhibits intracellular LCFA transport/diffusion in cell culture
L-FABP exhibits high affinity for a fluorescent LCFA analogue, NBD-stearic acid—a poorly metabolizable LCFA [43, 44, 144, 145]. Advantage was taken of this property to determine if L-FABP enhances LCFA cytoplasmic transport/diffusion measured in real-time in living cells by fluorescence recovery after photobleaching (FRAP) imaging. In hepatocytes differing in L-FABP expression (male versus female) and in cultured hepatoma cells, L-FABP significantly enhanced LCFA cytoplasmic diffusion [146, 147]. Similarly, in transfected L-cell fibroblasts overexpressing L-FABP, L-FABP increased the cytoplasmic diffusion of this fluorescent LCFA [6, 148]. Cultured primary hepatocytes from L-FABP gene-ablated mice had a significantly decreased rate of diffusion of NBD-stearic acid through the cytoplasm [142]. Since L-FABP (mw 14,286 Da) is much larger than a typical LCFA (300 Da), binding of the LCFA to the L-FABP should slow down, rather than speed up the diffusion of the LCFA. While not intuitively obvious, one explanation for this conundrum can be based on the fact that: (i) the cytoplasmic concentration of unesterified LCFA is very low, (ii) the viscosity of cytoplasm is at least an order of magnitude higher than that of aqueous buffers, and (iii) the path length of LCFA through the cytoplasm is much more ‘tortuous’ due to the presence not only of many other proteins but even more so of a high concentration of membranous organelles and microfilaments that restrict motion [149-151]. The finding that cytosol from hepatocytes of L-FABP null mice has markedly diminished capacity for binding LCFA and LCFA-CoA suggests that L-FABP may enhance cytoplasmic diffusion of LCFA at least in part by facilitating desorption of membrane bound LCFA to increase cytoplasmic concentration of unesterified LCFA [152, 153].
6. Effect of L-FABP overexpression and ablation on LCFA metabolic targeting and nuclear regulatory targeting in cultured cells
6.1. Targeting LCFA to ER for esterification
Since intracellular accumulation of unesterified LCFAs and LCFA-CoAs (both potent detergents) and cholesterol (crystallizes even at low concentration) is deleterious, a major function of FABPs is thought to be the removal of these ligands by enhancing their anabolic and catabolic metabolism. A role for L-FABP in anabolic removal of LCFAs is supported by studies with transfected cells overexpressing L-FABP which show that L-FABP facilitates transport of LCFA and cholesterol from the plasma membrane to ER for esterification by ACAT, increasing cellular cholesteryl ester mass [135, 154-156]. L-FABP overexpression markedly increased incorporation of LCFA into triglycerides nearly 3-fold, albeit without increasing triglyceride mass—suggesting other mechanisms (e.g. lipase/hydrolase activity) contribute to maintain mass constant [137, 155, 157]. In transfected L-cells overexpressing L-FABP, incorporation of LCFA into phospholipids increased, but in addition phospholipid mass increased 1.7-fold [155, 157, 158]. Cultured primary hepatocytes and freshly isolated hepatocytes from L-FABP null mice had reduced incorporation of LCFAs (palmitic acid, oleic acid, phytanic acid) into total lipids—especially the neutral lipids (triglycerides, diglycerides, and/orcholesteryl esters) [142, 143]. Total mass of triglycerides was decreased 40-60% while phospholipid and cholesteryl esters were increased or unaltered/spared in these cells [142, 143]. Thus, L-FABP plays an important role in LCFA anabolic metabolism to facilitate incorporation into triacylglycerols important for LCFA storage or secretion (VLDL in liver, chylomicrons in intestine). In triglyceride secretion, increased expression of L-FABP in transfected hepatoma cells increased secretion of apoB-100 and decreased cellular biosynthesis and secretion, and increased PPARα mRNA levels—indicating that L-FABP may interact with PPARα to amplify the effects of endogenous PPARα agonists on the assembly of VLDL [159]. Taken together, these data suggest a potential role for L-FABP in LCFA esterification at the ER
6.2. Targeting LCFA for oxidation (mitochondria, peroxisomes) in cultured cells
L-FABP facilitates removal of LCFAs by catabolic metabolism, stimulating oxidation of straight-chain LCFAs such as palmitic acid (primarily mitochondrial β-oxidation) in cultured fibroblasts overexpressing L-FABP [139]. Likewise, L-FABP overexpression enhanced oxidation of branched-chain LCFAs such as phytanic acid (primarily peroxisomal α- and β-oxidation) in these fibroblasts [139]. Conversely, L-FABP gene ablation inhibits oxidation of both palmitic acid (mitochondrial) and even more so phytanic acid (peroxisomal) in cultured primary hepatocytes and freshly isolated hepatocytes from L-FABP null mice [139, 160]. Thus, L-FABP plays an important role in directing LCFAs toward oxidation pathways in cell culture.
6.3. Targeting LCFA to nuclei for interaction with PPARα
By real-time confocal and multiphoton imaging of several non- or poorly-metabolizable fluorescent LCFAs, overexpression of L-FABP increased the distribution of synthetic (NBD-stearic acid, BODIPY-C12, BODIPY-C16) and naturally-occurring (parinaric acid) fluorescent LCFAs to the nucleoplasm in cultured L-cells overexpressing L-FABP [43, 44]. Conversely, lack of L-FABP decreased the distribution of LCFAs into the nucleoplasm of cultured primary hepatocytes from L-FABP null mice [45]. As shown by immunocoprecipitation, CD, and FRET in vitro, L-FABP binds PPARα with high affinity [26, 71]. L-FABP also directly interacts with PPARα in fixed cultured cells and cultured primary hepatocytes as shown by immunofluroescence and immunogold EM [44, 71]. Additionally, transactivation and gene ablation studies show L-FABP overexpression enhances while L-FABP gene ablation inhibits PPARα transcription of genes coding for LCFA oxidative enzymes–thereby showing the functional and physiological significance of L-FABP/PPARα interaction [26, 71].
7. In vivo function of L-FABP in long chain fatty acid uptake, oxidation, and esterification
7.1. Generation of L-FABP null mice
The first reported L-FABP null (-/-) C57Bl/6 mice mouse line was generated using a neo construct, ablating most of the 5′ non-coding (promoter) region and all 4 exons of the small L-FABP gene, as well as part of the 3′ noncoding region [152]. This design precluded the possibility of expressing L-FABP fragments (small peptides, truncated protein) [152]. The gene deletion was verified by long PCR and confirmed by nested PCR. On the protein level, Western blotting confirmed there were no detectable L-FABP protein or L-FABP-derived peptides in these L-FABP null mice [152]. The N2 generation L-FABP null (-/-) C57Bl/6 mice were further backcrossed to C57BL/6N mice from Charles River Laboratories (Wilmington, MA) obtained through the National Cancer Institute (Frederick Cancer Research and Development Center, Frederick, MD) in order to obtain at least 99.9% homogeneity (N10) backcross generation. Herein, these mice are referred to as the “L-FABP null” mouse line.
A subsequently reported different L-FABP null mouse line was generated by use of a neo and GFP knock-in that left intact the 5′ noncoding (promoter) region, all of exons 3 and 4, and the 3′ noncoding region [143]. Thus, while Northern blotting did not reveal the presence of the L-FABP transcript, based on the construct design and the results presented, the existence of smaller peptides(s) encoded by exons 3-4 (aa80-126—part of a ligand binding domain) could not be ruled out, especially since the full Northern blot was not shown [124, 143, 161]. While western blotting revealed the absence of the 14 kDa L-FABP, the blot represented only the 14 kDa band and did not indicate whether or not any smaller fragments were present. The N2 generation of these L-FABP null (-/-) C57Bl/6 mice were further backcrossed to C57BL/6J mice from Jackson Laboratories (Bar Harbor, ME)—a substrain much more susceptible high fat diet induced obesity than the C57BL/6N substrain to which the original “L-FABP null” mouse line (see above) was backcrossed as shown in JAX NOTES, Issue 511, Fall 2008 (http://jaxmice.jax.org/jaxnotes/511/511n.html) from the Jackson Laboratory (Bar Harbor, ME). GFP was prominently overexpressed in these mice are herein referred to as the “L-FABP null/GFP overexpressor” mouse line.
7.2. L-FABP gene ablation does not affect food consumption or intestinal lipid absorption
As compared to wild-type controls, neither L-FABP null mouse line exhibited any significant alterations in consumption of commercial or defined control diets or in energy expenditure [56, 59, 97, 98, 143, 152, 162, 163]. While the finding that intestinal enterocytes from the L-FABP null/GFP overexpressor line exhibit reduced budding of prechylomicron transport vesicles from the ER—the rate limiting step in transit of absorbed dietary fat across the enterocyte [21, 102], studies with L-FABP null/GFP overexpressor mice fed control chow, high fat diet, or cholestatic diet did not detect any effect on intestinal lipid absorption [163-165].
7.3. L-FABP enhances hepatic uptake and oxidation of long chain fatty acids
Based on the above cultured transfected cells, and cultured primary hepatocytes studies performed in vitro, it was expected that L-FABP should enhance LCFA uptake and oxidation in liver. Indeed, hepatic uptake of 14C-oleic acid from serum was reduced not only in fed, but even more so fasted L-FABP null mice [152]. A role for L-FABP in hepatic LCFA oxidation was initially based direct correlation of rat liver L-FABP content with LCFA oxidative capacity [5, 166] in that incubation of 14C-palmitic acid with liver homogenates from L-FABP null mice decreased total oxidation (measured a 14C in CO2 + acid soluble products) and β-hydroxybutyrate production by 34% and 36%, respectively [160]. L-FABP gene ablation also inhibits hepatic oxidation of LCFAs, but not medium chain fatty acids such as octanoic acid (not bound by L-FABP, freely permeable into cells and mitochondria) in vivo as shown by direct correlation of L-FABP expression (Fig. 5B) with serum β-hydroxybutyrate levels (Fig. 5A) in a variety of genetically engineered fasted mice [143, 160, 167]. In contrast, serum β-hydroxybutyrate levels (Fig. 5A) were inversely correlated with SCP-2 expression (Fig. 5C) and did not correlate with expession of SCP-x (Fig. 5D)—proteins more involved in peroxisomal than mitochondrial LCFA oxidation. Serum β-hydroxybutyrate levels in L-FABP null mice were decreased even further after prolonged fasting (48h), feeding a ketogenic diet, or fasting after a ketogenic diet [160].
Figure 5. Correlation of LCFA oxidation with hepatic L-FABP levels in vivo.
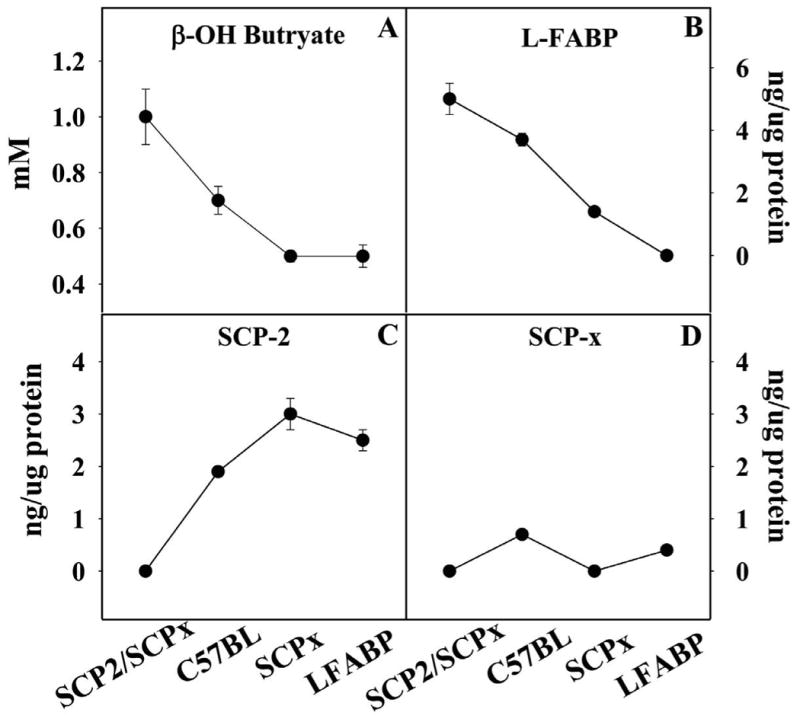
Levels of serum β-hydroxybutyrate (A) as well as L-FABP (B), SCP-2 (C), and SCP-x (D) in liver homogenates were determined in male SCP2/SCP-x null, C57BL6N wild-type, SCP-x null, and L-FABP null mice fed control standard rodent chow diet fed ad libitum.
7.4. L-FABP enhances hepatic esterification of long chain fatty acids
L-FABP gene ablation in the “L-FABP null” mouse line (see 7.1) decreased hepatic triglyceride accumulation in fasted male, but not female, mice fed standard commercial or defined control diets [56, 59, 97, 143, 152, 162], while increasing hepatic cholesteryl ester accumulation (and in most cases unesterified cholesterol) in fasted female, but not male mice fed standard commercial or defined control diet at all ages examined [59, 97, 152, 152]. While serum triglyceride levels were unaltered in aged L-FABP null mice, serum triglyceride and cholesteryl ester were decreased in younger males fed a defined control chow, while those in females were increased [142].
Taken together, these studies indicate that L-FABP functions in hepatic LCFA uptake, oxidation, esterification, and secretion.
8. Age- and sex- dependent weight gain and obesity in L-FABP null mice
8.1. Potential confounding effects of phytol and phytoestrogen in standard, low fat commercial rodent chow on weight gain and obesity
Based on the decreased LCFA uptake, intrahepatocyte transport, oxidation, and esterification reported in L-FABP null mice fed normal commercial rodent chow, it was predicted that loss of L-FABP would redirect LCFAs toward utilization by tissues other than liver (muscle, adipose). Whether the modestly decreased intestinal lipid absorption in L-FABP null/GFP overexpressor mice (observed only with control chow, but not high fat chow fed null mice) counteracts this expectation is unclear, because a confounding factor is the normal variation in amount of dietary phytol [58, 59, 168-170] and phytoestrogen [171, 172] in standard commercial rodent chows and, even more so in commercial high fat diets (i.e., breeder chow), which also vary in the source of dietary fat (e.g., butter and lard are much higher in phytol content as compared to vegetable oils) [173]. Metabolites of phytol (e.g. phytanic acid, pristanic acid) are potent activators of PPARα (and other PPARs) which in turn regulate transcription of L-FABP and LCFA oxidative enzymes (rev. in [15, 17]. Phytol metabolism is sex-dependent, and accumulation of high levels of phytol metabolites is toxic in animals [58, 59, 139, 142, 168-170], including humans [173-175]. Phytoestrogens exert estrogenic effects in mice [171, 172]. To understand these confounding issues, weight gain and whole body phenotype (fat tissue mass, lean tissue mass) were determined in L-FABP null mice fed two types of control fat (5%) rodent chows—standard commercial versus defined (phytol-free, phytoestrogen-free, low-fat) rodent chow.
8.2. Weight gain and obesity in L-FABP gene-ablated mice fed phytol-free, phytoestrogen-free, low fat rodent chow
Early back-cross generation L-FABP null fed different commercial control rodent chows ad libitum did not exhibit any significant difference in weight gain or final body weight as compared to age- and sex- matched wild-type counterparts [143, 152]. However, after 6 back-cross generations (N6), important age- and sex-dependent differences emerged (Table 1). After 6 mo of age, female and male L-FABP null mice exhibited increased body weight and weight gain as compared to age- and sex- matched wild-type counterparts [176, 177]. This phenotype emerged at even earlier age when the L-FABP null mice were pair-fed a defined (phytol-free, phytoestrogen-free, low fat) control chow diet (Table 1). When 2 mo-old male and female L-FABP null of this null line (≥N6 back-cross generation) were fed for a short time (18 days) on a defined (phytol-free, phytoestrogen-free, low-fat) control chow diet, weight gain and final body weight did not differ from their age- and sex-matched wild-type counterparts [59, 167]. However, after ≥42 days of pair-feeding this defined diet, females (but not males) exhibited increased weight gain and increased final body weight as compared to age- and sex-matched littermates [56, 162, 167].
Table 1. Effect of L-FABP gene ablation on weight gain in mice fed control chow ad libitum or pair-fed defined (phytol-free, phytoestrogen-free) control chow diets.a.
| Type of control chow diet | Feeding | Age (mo) | Sex | Body Weight (g) | |
|---|---|---|---|---|---|
| Rate of gain | Final Weight | ||||
| Standard rodent chow | ad libitum | < 6 mo | M, F | No change | No change |
| Standard rodent chow | ad libitum | > 6 mo | M, F | Increase | Increase |
| Defined rodent chow | pair-fed | 2.5 mo | M, F | No change | No change |
| Defined rodent chow | pair-fed | 3.5 mo | F | Increase | Increase |
| Defined rodent chow | pair-fed | 3.5 mo | M | No change | No change |
L-FABP null mice, age- and sex-matched (up to N10) to wild-type littermates by heterozygote/heterozygote breeding, were fed continuously on control (Rodent Diet 8604, standard low fat, 5% of energy from fat) chow from Harlan Teklad, Madison, WI. Alternately, mice were fed on this control chow for 2 mo and then pair-fed for 18 or ≥42 days on defined control (AIN-76A phytol-free, phytoestrogen-free, 5% calories from fat) chow from Research Diets, New Brunswick, NJ. Food consumption and mouse weight were determined every other day. There were no differences in food consumption in response to L-FABP gene ablation.
To determine if the age- and sex-dependent increase of weight gain in standard commercial or defined diets was associated with obesity, whole body mass as fat tissue (FTM) and lean tissue (LTM) as well as their relative distributions (i.e. %FTM, %LTM) in L-FABP null mice (≥N6 back-cross generation) were determined by dual emission x-ray absorptiometry (DEXA). L-FABP null mice exhibited significant age- and sex-dependent changes in whole body phenotype FTM and lean tissue mass LTM (Table 2). After 6 mo of age, females fed a standard commercial rodent chow ad libitum exhibited increases in FTM (but not LTM), while the LTM was unchanged and the % LTM decreased as compared to age- and sex- matched counterparts [176, 177]. A basically similar, but milder phenotype was observed in the males (Table 2) [176, 177]. The obese phenotype emerged at even earlier age when females and males were pair-fed a defined control phytol-free, phytoestrogen-free, low-fat chow diet (Table 2). When 2 mo old wild-type and L-FABP null females (≥N6 back-cross generation) were fed for a short time (18 days) on a this defined chow, neither the mass nor % of FTM and LTM were altered as compared to age- and sex-matched littermates [59, 167]. However, after ≥42 days of pair-feeding of this same diet, the null females had increased their FTM and % FTM (less so in LTM and %LTM) as compared to their age- and sex-matched counterparts [56, 162, 167]. Under the same conditions, males also had significant but lower increases in FTM and % FTM, while LTM and % LTM were unchanged or decreased as compared to age- and sex-matched counterparts [56, 162, 167]. Whole body FTM and LTM in L-FABP null/GFP overexpressor mice are not yet available for any type of feeding study.
Table 2. Effect of L-FABP gene ablation on whole body fat tissue mass (FTM) and lean tissue mass (LTM) in mice fed control chow ad libitum or pair-fed defined (phytol-free, phytoestrogen-free, 5% fat) control chow diets.a.
| Type of control rodent chow diet | Feeding | Age (mo) | Sex | Change in Tissue Mass | |||
|---|---|---|---|---|---|---|---|
| FTM (g) | LTM (g) | FTM (%) | LTM (%) | ||||
| Standard | ad libitum | < 6 | M, F | 0 | 0 | 0 | 0 |
| Standard | ad libitum | ≥ 6 | F | +++ | 0 | +++ | - - |
| Standard | ad libitum | ≥ 6 | M | ++ | + | ++ | - |
| Defined | pair-fed | 2.5 | M, F | 0 | 0 | 0 | 0 |
| Defined | pair-fed | 3.5 | F | +++ | ++ | +++ | + |
| Defined | pair-fed | 3.5 | M | +/0 | 0/- | +/0 | 0/- |
L-FABP null mice, age- and sex-matched (up to N10) to wild-type littermates by heterozygote/heterozygote breeding, were fed continuously on control (Rodent Diet 8604, standard low fat, 5% of energy from fat) chow from Harlan Teklad, Madison, WI. Alternately, mice were fed on this control chow for 2 mo and then pair-fed for 18 or ≥42 days on defined control (AIN-76A phytol-free, phytoestrogen-free, 5% calories from fat) chow from Research Diets, New Brunswick, NJ. Whole body fat tissue mass (FTM) and lean tissue mass (LTM) were determined by dual emission x-ray absorptiometry. 0, +, and − refer to no difference, increase, or decrease versus wild-type mice.
Thus L-FABP null mice exhibit an age- and sex-dependent weight gain and obesity when fed either of two lower fat (5%) diets—commercial standard chow ad libitum or a pair-fed, phytol-free, phytoestrogen-free, low fat defined diet. Null females appeared obese, significantly gaining the most weight as compared to wild-type females, primarily as increased mass and proportion of fat.
9. Whole body phenotype of L-FABP gene-ablated mice: high-fat diet induced weight gain and obesity
9.1. Response of L-FABP null mice to a high-fat diet
Thus L-FABP enhances hepatic uptake of LCFAs, intrahepatic LCFA transport, intrahepatic LCFA targeting for oxidation (peroxisomes, mitochondria) and esterification (ER), and LCFA targeting to the nucleus for regulation of nuclear receptors important in transcription of genes encoding proteins involved in LCFA and glucose metabolism (PPAR-α, HNF-4α, LXR, THR). Since ablation of L-FABP inhibits LCFA uptake, reduces LCFA intracellular transport/diffusion, inhibits LCFA esterification, inhibits LCFA oxidation, and inhibits LCFA targeting to the nucleus, L-FABP null mice fed a high fat diet should redirect LCFAs for utilization in other tissues. However, none of the studies reported to date indicate any increased activity/LCFA use by muscle. Consequently, is would be expected that the excess LCFAs are redirected to adipose tissue for storage and result in an obese phenotype—especially in response to high-fat diet.
To prevent any complication due to mouse palatability preference for fat, L-FABP null mice and age and sex-matched wild-type counterparts were fed a defined control low fat diet or a high fat (24% fat) diet based upon the consumption of control mice the previous day for 12 wks (i.e., pair-feeding) [167]. Body weight as well as food consumption were measured every 2 days for the entire study [167]. The amount of high fat diet consumed (whether expressed as total g, total kcal, g/day, or kcal/day) over the entire study did not differ between L-FABP null and wild-type mice because of this pair-feeding method [167]. As expected, high-fat fed L-FABP null males and females both had reduced serum β-hydroxybutyrate levels as compared to high fat fed controls—consistent with reduced ability of L-FABP null hepatocytes to take up and/or oxidize LCFA under a high LCFA load [167]. L-FABP gene ablation did not protect either males or females from high-fat induced weight gain and obesity [167]. On the contrary, female (and less so male) L-FABP null mice increased in body weight as compared to controls fed the same high fat diet (Table 3). Dual x-ray emission absorptiometry showed that the modestly higher weight in L-FABP null males was associated with increases in both fat tissue mass (FTM) and lean tissue mass (LTM), but when expressed on a % basis, the % FTM increased more than the % LTM (Table 3) [167]. In contrast, the much greater increase in weight of L-FABP null females was also associated with increased FTM as well as LTM, but when expressed on a % basis, the % FTM increased more than % LTM (Table 3) [167]. Thus in pair-fed mice, the ablation of the L-FABP gene did not protect the mice from high-fat diet induced weight gain and relative obesity as compared to wild-type controls.
Table 3. Effect of L-FABP gene ablation and high fat diet on mouse whole body phenotype.a.
| Sex | Change in Body Weight (g) | Change in Tissue Mass | |
|---|---|---|---|
| FTM (%) | LTM (%) | ||
| M | 0/+ | + | 0 |
| F | + | + + | + |
L-FABP null mice, age- and sex-matched to wild-type littermates by heterozygote/heterozygote breeding, (N6 back-cross generation) were fed for 7 wks standard (4.5 gm% fat) standard Rodent Diet 8604 chow from Harlan Teklad, Madison, WI. Thereafter, the mice were individually housed and switched to a defined low fat (4.3% fat) control diet (Diet D12450, phytol-free, phytoestrogen-free) from Research Diets, New Brunswick, NJ. After 1 week on this diet, half of the mice in each group continued on the defined low fat control diet while the other half were pair-fed a high fat (24% fat) isocaloric diet based on the same defined control diet. Food consumption and mouse weight were measured every 2 days for 12 weeks. Food consumption (g or kcal) did not differ in response to L-FABP gene ablation. Whole body fat tissue mass (FTM) and lean tissue mass (LTM) were determined by dual emission x-ray absorptiometry at the beginning and end of the dietary study. 0, +, and − refer to no difference, increase, or decrease vs age-and sex-matched littermate wild-type L-FABP +/+ mice.
9.2. Response of L-FABP null mice to a lithogenic diet
By mapping susceptibility loci in response to diet-induced gallstone formation, genetic studies have mapped a locus with L-FABP as a positional candidate from a quantitative trait locus near D6Mit123 on chromosome 6 [165, 178, 179]. Consistent with this possibility, significant upregulation of L-FABP (5-6 fold) in SCP-x/SCP-2 gene-ablated mice was associated with biliary hypersecretion of cholesterol [180, 181]. These findings suggested that L-FABP may play a role in lipid absorption and whole body phenotype in response to a lithogenic diet. Therefore, the effects of a 4-week defined, lithogenic diet (Research Diet D0208081, containing 4.9% fat, 1.25% cholesterol, and 0.5% cholic acid) on 8 week-old male and female L-FABP null mice was examined. Despite similar food consumption (Fig. 6A), and much like results from the cholesterol diet [56, 162], lithogenic diet fed wild-type females but not males exhibited increased weight gain (Fig. 6B), primarily as fat tissue mass (Fig. 6C), as compared to control fed wild type females. Control diet fed L-FABP null female mice, but not males, exhibited increased weight gain (Fig. 6B) as compared to their control fed wild-type counterparts. In contrast, lithogenic diet fed L-FABP null females as well as males exhibited decreased weight gain (Fig. 6B), primarily as decreased fat tissue mass in females (Fig. 6C). There was little to no effect observed with the lean tissue mass when comparing each feeding group (Fig. 6D). It is important to note, however, that while less weight gainoccurred in the L-FABP null than wild-type females on the lithogenic diet, there was still a 6% weight gain in the L-FABP null females. Thus, L-FABP gene ablation reduced, but did not protect, the female mice from lithogenic diet induced weight gain.
Figure 6. Effect of lithogenic diet on body weight, fat tissue mass, and lean tissue mass.
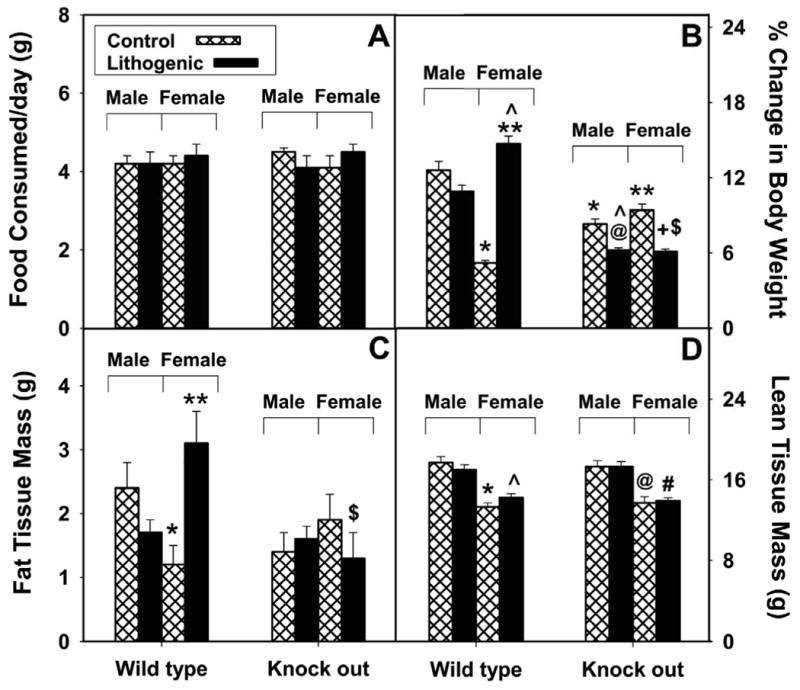
Average daily food consumption (A), percent change in body weight (B), fat tissue mass (C), and lean tissue mass (D) was determined in male and female mice fed a control (hatched bar) and lithogenic (solid bar) diet for 4 weeks. Values represent the mean ± SEM, n=5-7. Statistical analysis was as follows: * p≤ 0.04 vs male L-FABP +/+ on control-diet; * * p≤ 0.03 vs female L-FABP +/+ on control-diet; @ p≤ 0.004 vs male L-FABP -/- on control-diet; + p≤ 0.001 vs female L-FABP -/- on control-diet; ˆ p≤ 0.005 vs male L-FABP +/+ on lithogenic diet; # p≤ 0.004 vs male L-FABP -/- on lithogenic diet; $ p≤ 0.02 vs female L-FABP +/+ on lithogenic diet.
9.3. Response of “L-FABP null/GFP overexpressor” mice to high fat and lithogenic diets
The “L-FABP null/GFP overexpressor” mouse line shares in common with the L-FABP null mouse line reduced hepatic uptake of fatty acids, reduced hepatic fatty acid oxidation, and showed no difference in weight gain or final body weight in response to feeding different commercial control rodent chows ad libitum [143, 152]. However, the phenotype of “L-FABP null/GFP overexpressor” mice differed significantly from that of L-FABP null mouse strain in response to several different types of high fat diet.
“L-FABP null/GFP overexpressor” mice were fed ad libitum several high fat diets from different vendors, body weight was measured weekly, food consumption was measured only for a 3 day period toward the end of the study, fat absorption was measured for a short time toward the end of the study, and whole body fat tissue mass and lean tissue mass were not determined. In an earlier study, L-FABP null/GFP overexpressor mice (N5 backcross generation) were fed ad libitum a high-saturated fat, high cholesterol Western diet (21% milkfat, 0.15% cholesterol) from Harlan Teklad (Madison, WI) for 10-12 wk [163]. Food consumption and fat absorption did not differ between L-FABP null/GFP overexpressor females and C57BL/6J controls (Jackson Labs, Bar Harbor, ME). Despite the fact that fat oxidation (measured as serum β-hydroxybutyrate) was reduced nearly 5-fold in L-FABP null/GFP overexpressor females fed this high fat diet, L-FABP null/GFP overexpressor null females and males exhibited reduced weight gain as compared to their C57BL/6J controls also fed the same high fat diet ad libitum (i.e., not pair-fed) [163, 182]. In a subsequent study, L-FABP null/GFP overexpressor females (apparently N5 backcross generation) control C57BL/6 females (Jackson Labs, Bar Harbor, ME) were fed ad libitum either a high saturated fat diet (20% coconut oil) or a high polyunsaturated fat diet (20% safflower oil) from MP Biomedicals (Solon, OH) for 10-12 wks [182]. While L-FABP gene ablation did not alter food consumption or fat absorption with either diet, L-FABP null/GFP overexpressor mice fed the high saturated fat diet (but not high polyunsaturated fat diet) ad libitum exhibited a reduced weight gain as compared to C57BL/6 controls [182]. In another study, L-FABP null/GFP overexpressor females (N-10 backcross generation) were again fed ad libitum either a high saturated fat diet (20% coconut oil) or a high polyunsaturated fat diet (20% safflower oil) from MP Biomedicals (Solon, OH) for 8-24 wks (not pair fed) [164]. While fat absorption was unaltered with either high fat diet, there was a trend towards decreased food consumption in L-FABP null/GFP overexpressor females fed the high saturated fat diet, but not the high polyunsatured fat diet [164]. The L-FABP null/GFP overexpressor females had a reduced weight gain in response to high saturated fat, but not to high polyunsaturated fat [164]. Thus, when fed high fat diets ad libitum, L-FABP null/GFP overexpressor females (males were not reported) exhibited a reduced weight gain or relative weight loss rather than an increased weight gain. This reduced weight gain was somewhat surprising in the face of reduced serum β-hydroxybutyrate (indicative of reduced hepatic uptake/oxidation of LCFAs) and no upregulation of LCFA oxidation in skeletal muscle, which together with the unaltered food consumption and unaltered fat absorption, would have suggested redirection of LCFAs to adipose for storage/obesity.
While the mechanistic basis for the variation in high-dietary fat responses exhibited by “L-FABP null/GFP overexpressor” as compared to L-FABP null mouse lines is not completely clear, detailed comparisions reveal multiple potential factors that may contribute (Table 4). Of these, major differences in construct design, overexpression of GFP, backcrossing to a C57BL6J substrain that is more susceptible to high-fat diet induced obesity, and feeding high fat diets ad libitum are very likely contributors. The “L-FABP null/GFP overexpressor” mouse studies used the C57BL/6J (more susceptible to high fat diet induced obesity) mouse strain for backcrossing and as controls and fed high fat diets ad libitum. In contrast, L-FABP null mice were backcrossed to C57BL/6N (less susceptible to high fat diet induced obesity) mouse strain, performed with age- and sex-matched littermate controls, and fed high fat diets in a pair feeding regimen. Ad libitum feeding high fat diet resulted in 20-37% higher (p<0.01) higher total food consumption [163, 164, 182] as compared to pair-feeding a high fat diet [167]. Finally, it is important to note that dietary LCFAs induce transcriptional activity of PPARα, which in turn increases L-FABP transcription and even more LCFA uptake (rev. in [17, 18, 47, 48, 51, 183]. Ad libitum consumption of diets high in fat increases hepatic L-FABP levels (rev. in [4, 15, 24, 26-35]. Thus, the higher genetic susceptibility of the background C57BL/6J mouse strain, ad libitum feeding of high fat diet, and induction of L-FABP in the wild-type L-FABP +/+ control (but not the L-FABP null) mice together likely resulted in dramatically higher total fat consumption and absorption by the control wild-type mice (express L-FABP but not GFP) than the “L-FABP null/GFP overexpressor” mice—thereby perhaps accounting for the observed ‘protection’ of “L-FABP null/GFP overexpressor” mice from high fat diet induced obesity.
Table 4. Potential factors contributing to observation of somewhat different obese phenotype noted with “L-FABP null/GFP overexpressor” mice.
Differences were derived by comparisons of methodology reported for “L-FABP null/GFP overexpressor mice” [143, 163-165, 182, 197] with those for the original “L-FABP null mice” mice [45, 56, 59, 71, 98, 142, 152, 153, 162, 167, 176, 177]. Additional references in the table refer to other publications demonstrating such differences in factors/parameters can affect mouse phenotype/obesity/lipid metabolism.
| Factor | Parameter | Affects Mouse Phenotype/Lipid Metabolism/Obesity | Additional References |
|---|---|---|---|
| Construct | Lipid phenotype | Yes | [198-200] |
| Peptide fragments? | Yes | [201-208, 208, 209, 209-215] | |
| GFP overexpression | Yes | [216-221] | |
| Backcrossing | C57BL/J substrain more obesity susceptible | Yes | [222], http://jaxmice.jax.org/jaxnotes/511/511n.html |
| Backcross generation number of L-FABP null | No | [98] | |
| Control mice | Vendor supplied vs wild-type littermates | Yes | |
| Age | Yes | [223-225] | |
| Sex | Yes | [58, 169] | |
| Diets | Vendor source, non-defined, not phytol free, not phytoestrogen free | Yes | [58, 168, 171, 172] |
| Ad libitum feeding, length of time on high fat diet | Yes | ||
| Intestinal microflora | Obesity | Yes | [226-231] |
9.2. Response of “L-FABP null/GFP overexpressor” mice to a lithogenic diet
In similar fashion to the L-FABP null mice, 8-10 wk old male L-FABP null/GFP overexpressor mice were fed ad libitum for 2-8 wks on a high fat lithogenic diet (Research Diet 960393 containing 18.8% fat, 1.25% cholesterol, and 0.5% cholic acid). In contrast to results with male wild-type mice fed lithogenic diet in the preceding study, wild-type mice fed high-fat lithogenic diet exhibited weight loss [165]. Similarly, “L-FABP null/GFP overexpressor” males fed high-fat lithogenic diet also exhibited weight loss, but with the final body weight lower and weight loss greater than wild-type [165]. Male “L-FABP null/GFP overexpressor” mice exhibited increased susceptibility to high-fat lithogenic diet induced gallstone formation [165]. There were no significant differences in fat absorption, cholesterol absorption, and food intake between male wild type and L-FABP null/GFP overexpressor mice [165]. However, the effect of high-fat lithogenic diet on female “L-FABP null/GFP overexpressor” mice was not reported. While it was concluded from these data that L-FABP may partially protect male mice from a high-fat lithogenic diet-induced weight loss, this conclusion was complicated by the fact that the wild type control mice (express L-FABP but not GFP) fed high-fat lithogenic diet developed 3-fold hepatic upregulation of L-FABP concomitant with a 3-fold decreased intestinal L-FABP expression [165].
Taken together, these findings underscore the fact that even in wild-type mice, key differences in the fat content of lithogenic diet elicit markedly different effects on weight gain. While this complicates direct comparisons between the 2 different L-FABP null mouse lines fed different types of lithogenic diets, in both cases L-FABP gene ablation exacerbated weight loss or reduced weight gain induced by lithogenic diets. These findings suggest agreement in at least this aspect of diet-induced phenotype between the 2 different L-FABP null lines.
10. Physiological significance of genetic modifications in L-FABP in humans: the Thr94Ala mutation
Since to date there have been no reports of complete loss of the L-FABP protein in humans, it is not yet possible to directly correlate the studies of obesity in L-FABP null mice with complete loss of L-FABP in humans and associated pathology. However, some insights may be gained from single nucleotide polymorphisms (SNPs) of L-FABP. While SNPs of L-FABP are fairly common in humans, they have not resulted in loss of the L-FABP but rather a mutant L-FABP protein is expressed. Unfortunately, it is not yet known if the mutant L-FABP differs in structure/function from the native L-FABP or if the expression of the L-FABP protein is altered by the SNP. While either possibility might yield an altered phenotype, neither equates to complete loss of L-FABP as in the L-FABP null mouse model. Furthermore, the genotype of humans is much more heterogeneous than that of inbred mice such that SNPs resulting on a specific phenotype in one population may not necessarily yield the same phenotype in another population. These possibilities are described by examples from SNPs in the human L-FABP gene as well as in SNPs in the related intestinal fatty acid binding protein (I-FABP).
For example, a single nucleotide missense mutation resulting in substitution of threonine 94 (Thr94) with alanine (Ala94) in L-FABP has been reported in two groups of Caucasians: French Canadians [184, 185] and Germans [186, 187]. This point mutation is apparently common in these Caucasians, wherein 64% are homozygous for Thr94, 23% are Thr94/Ala heterozygotes, and 13% are homozygous for Ala94/Ala [184-187]. The fact that 36% of the human population carries the Ala94 substitution in L-FABP suggests that there is not a very strong selection for one or the other allele and that, at least by about 50 yrs of age (the age of the individuals in these 2 studies), this mutation alone is not sufficient to be pathologic. Correlative examination, however, showed that the body mass index (BMI) of both male and female humans with the Ala94/Ala polymorphism was 4-6% lower than that of the wild-type Thr94/Thr counterparts [186, 187]. There was also a significant association of increased serum LDL-cholesterol and triglycerides in females (but not males) with the Ala94/Ala polymorphism [186]. Covariance analysis suggested that the wild type Thr94/Thr has higher serum apoB levels while the Ala94 carriers seem to be protected against high apoB levels when consuming a high fat and saturated fat diet [184]. Other covariance analyses indicate that the Thr94/Ala substitution may influence obesity indices as well as the risk to exhibit residual hypertriglyceridemia following lipid lowering therapy with fenofibrate [185]. In addition, a study of a limited number of subjects indicates that the Thr94/Ala substitution appears to reduce hepatic glycogenolysis and blunted elevation of plasma glucose levels in lipid-exposed subjects [187]. While the above studies show modest effects of the Thr94/Ala substitution in L-FABP of humans, it should be noted that none of the above studies reported any western blotting or rtPCR data showing if the Thr94/Ala substitution altered the expression of L-FABP in human liver or other L-FABP expressing tissues (e.g. intestine, kidney, etc.). Further, it is not known whether the Thr94/Ala substitution alters the structure and thereby function of L-FABP. In the T94A mutation of L-FABP (FABP1), at codon 94 there is a replacement substitution of an alanine for the threonine [184-187]. Examination of the crystal structure of L-FABP shows that there is a pair of threonines (T93 and T94) which are located near the high affinity ligand binding pocket [78]. While the hydroxyl group in the T93 side chain is oriented into the binding cavity, its nearest neighbor T94 has its hydroxyl group oriented outward into the solvated environment [184]. Although binding affinities could be affected as in the case of the intestinal fatty acid binding protein I-FABP (also called FABP2) [188], it would be more likely that effects on L-FABP interactions with other proteins (e.g. CPT1, PPARα) and thus mitochondrial oxidative activity (e.g. via L-FABP/CPT1 interaction) or expression regulation (e.g. via L-FABP/PPARα interaction) would be more pronounced.
Lessons learned from an analogous I-FABP SNP mutation in humans are instructive. In I-FABP (FABP2), an A54T mutation first reported in Pima Indians and subsequently shown to occurr in most populations with an allelic frequency near 30% [189]. The A54T mutation enhances ligand binding affinity approximately two-fold, increases intestinal fatty acid absorption, and is associated with increased fatty acid oxidation and insulin resistance [188, 190, 191]. It is important to note that there are only two promoter alleles associated with the 54 codon whether coded for the alanine or the variant threonine [192]. In vitro studies using luciferase activity showed that the promoter carried on the Thr54 allele resulted in a threefold decrease in expression as compared to the promoter carried on the Ala54 allele [192]. Taken together, the studies with the I-FABP Ala54/Thr mutation indicate that further work needs to be done to clarify the functional consequence of the L-FABP Thr94/Ala mutation, especially the effect on expression of this protein in the liver. Furthermore, while analyses of L-FABP structural models suggest that the Thr94/Ala substitution should influence, but not likely abolish, function of L-FABP, there are as yet no reported data directly examining the effect of the Thr94/Ala substitution on L-FABP structure, ligand specificity, ligand affinity, interaction with other proteins (e.g. PPARα, CPT1, etc.), function in vitro or function in cultured cells.
It is important to note that the alterations in L-FABP phenotype due to Thr94/Ala substitution in Caucasians may not necessarily extend to other human populations, as has been the case for the Ala54/Thr mutation in I-FABP [188, 190, 191, 193]. The Ala54/Thr substitution in I-FABP is associated with increased BMI, body fat, and fasting plasma triglycerides in aboriginal Canadians and with higher fasting lipid oxidation rate, higher fasting plasma HDL and LDL triglycerols, and increased postprandial lipemic response in Finns (rev. in [193]. In contrast, in healthy young Europeans, the Ala54/Thr substitution is not associated with postprandial responses to fat and glucose tolerance tests-suggesting that in addition to the differences in promoter makeup there may be environmental factors, other genetic factors, and selection of study population that may also explain the difference between this and earlier studies of the I-FABP Ala54/Thr polymorphism [193].
In summary, while the studies with SNPs resulting on single amino acid substitutions in L-FABP (or I-FABP) in some human populations are informative, they may not necessarily extend to other human populations, and may be difficult to directly correlate at this point with the effects of total ablation of L-FABP in mice—a much more homogenous inbred population than humans.
11. Conclusions regarding the role of L-FABP in obesity
While some discrepancies exist, the overall experimental data obtained in vitro, with cultured cells, and L-FABP gene-ablated mice strongly support the hypothesis that L-FABP may play an important role in LCFA utilization by liver versus adipose tissue. L-FABP enhances LCFA uptake, intracellular LCFA transport, and targets bound LCFA and/or LCFA-CoA to intracellular organelles esterification (endoplasmic reticulum), storage (lipid droplets), secretion (VLDL), or most importantly, oxidation (mitochondria, peroxisomes) (rev. in [6, 139, 142, 146, 148, 151, 152]. Thus loss of L-FABP was predicted to redirect LCFAs towards storage in adipose tissue and induce weight gain/obesity—especially in response to high fat diet. This possibility was supported by studies with L-FABP null mice generated by complete deletion of all 4 exons of the L-FABP gene, backcrossing to the C57BL/6N substrain (less susceptible to diet induced obesity). Increased weight gain and obesity was observed in L-FABP null mice fed control low fat (4.3 %) chow for >6 mo, pair-fed defined control (phytol-free and phytoestrogen-free) chow for shorter time, or pair-fed defined isocaloric high fat diet. Under all of these conditions, L-FABP gene ablation inhibited LCFA oxidation and promoted weight gain and/or obesity, especially in L-FABP null females and less so males fed high dietary fat. Thus, loss of L-FABP did not protect mice from the deleterious effects of fat in the diet. A similar phenotypic pattern was also exhibited by adipocyte fatty acid binding protein (A-FABP) null mice, which showed increased weight gain as compared with their wild-type counterparts when fed a high-fat diet [194]. I-FABP null mice also exhibit a higher weight gain than their wild-type counterparts when fed either a low-fat or a high-fat diet [195].
The physiological significance of studies with L-FABP null mice compared to humans is underscored by the fact that L-FABP accounts for as much as 7-11% of cytosolic protein in normal human liver [196], nearly 2-fold more than in normal mouse liver wherein L-FABP comprises 3-5% of cytosolic protein (rev. in [4, 6]. L-FABP levels are very responsive to high fat diet (rev. in [15, 24, 26-35] and to peroxisome proliferators and lipid lowering drugs such as fibrates (rev. in [26, 34, 36-39].
The pathological significance of studies with L-FABP null mice in humans is indicated by the finding that human genetic variations in the L-FABP gene impact blood lipoprotein/lipid levels, response to lipid-lowering therapy with fenofibrate (a cholesterol synthesis inhibitor) and glycogenolysis [216-219]. Decreased expression of L-FABP (as well as I-FABP) occurs in the fat-laden enterocytes of the proximal intestine of humans with genetic lipid malabsorption syndromes such as abetalipoproteinemia and Anderson's disease [228]. Conversely, hepatic L-FABP levels are increased nearly 2-fold to 20% of cytosolic protein in patients with Reyes syndrome—a condition characterized in part by visceral fatty degeneration, block of fatty acid oxidation, and peroxisomal proliferation [227]. Thus, while direct comparisons between the human mutations and L-FABP null mouse cannot yet be accomplished, the impact of further research to understand the role of L-FABP in obesity is becoming increasingly evident.
Acknowledgments
This work was supported in part by the United States Public Health Service National Institutes of Health grants DK41402 (FS and ABK), GM31651 (FS and ABK), and DK70965 (BPA).
References
- 1.Ockner RK, Manning JA, Poppenhausen RB, Ho WK. A binding protein for fatty acids in cytosol of intestinal mucosa, liver, myocardium, and other tissues. Science. 1972;177:56–58. doi: 10.1126/science.177.4043.56. [DOI] [PubMed] [Google Scholar]
- 2.Glatz JF, Veerkamp JH. Intracellular fatty acid-binding proteins. [Review] Int J Biochem. 1985;17:13–22. doi: 10.1016/0020-711x(85)90080-1. [DOI] [PubMed] [Google Scholar]
- 3.Glatz JFC, Van der Vusse GJ, Veerkamp JH. Fatty acid-binding proteins and their physiological significance. New Physiol Sci. 1988;3:41–43. [Google Scholar]
- 4.Paulussen RJA, Veerkamp JH. Intracellular fatty acid-binding proteins characteristics and function. In: Hilderson HJ, editor. Subcellular Biochemistry. New York: Plenum Press; 1990. pp. 175–226. [DOI] [PubMed] [Google Scholar]
- 5.Veerkamp JH, van Moerkerk HT. Fatty acid-binding protein and its relation to fatty acid oxidation. Mol Cell Biochem. 1993;123:101–106. doi: 10.1007/BF01076480. [DOI] [PubMed] [Google Scholar]
- 6.McArthur MJ, Atshaves BP, Frolov A, Foxworth WD, Kier AB, Schroeder F. Cellular uptake and intracellular trafficking of long chain fatty acids. J Lipid Res. 1999;40:1371–1383. [PubMed] [Google Scholar]
- 7.Kaikaus RM, Bass NM, Ockner RK. Functions of fatty acid binding proteins. Experientia. 1990;46:617–630. doi: 10.1007/BF01939701. [DOI] [PubMed] [Google Scholar]
- 8.Ockner RK, Manning JA, Kane JP. Fatty acid binding protein. Isolation from rat liver, characterization, and immunochemical quantification. J Biol Chem. 1982;257:7872–7878. [PubMed] [Google Scholar]
- 9.Ockner RK, Kaikaus RM, Bass NM. Fatty acid binding proteins: recent concepts of regulation and function. [Review] Prog Clin Biol Res. 1992;375:189–204. [PubMed] [Google Scholar]
- 10.Banaszak L, Winter N, Xu Z, Bernlohr DA, Cowan S, Jones TA. Lipid-binding proteins: A family of fatty acid and retinoid transport proteins. Adv Protein Chem. 1994;45:89–151. doi: 10.1016/s0065-3233(08)60639-7. [DOI] [PubMed] [Google Scholar]
- 11.Hanhoff T, Lucke C, Spener F. Insights into binding of fatty acids by fatty acid binding proteins. Mol Cell Biochem. 2002;239:45–54. [PubMed] [Google Scholar]
- 12.Veerkamp JH, Maatman RG. Cytoplasmic fatty acid-binding proteins: their structure and genes. [Review] Prog Lipid Res. 1995;34:17–52. doi: 10.1016/0163-7827(94)00005-7. [DOI] [PubMed] [Google Scholar]
- 13.Bernlohr DA, Simpson MA, Hertzel AV, Banaszak L. Intracellular lipid binding proteins and their genes. Annu Rev Nutr. 1997;17:277–303. doi: 10.1146/annurev.nutr.17.1.277. [DOI] [PubMed] [Google Scholar]
- 14.Storch J, Thumser AE. The fatty acid transport functions of fatty acid binding proteins. Biochim Biophys Acta. 2000;1486:28–44. doi: 10.1016/s1388-1981(00)00046-9. [DOI] [PubMed] [Google Scholar]
- 15.Adida A, Spener F. Intracellular lipid binding proteins and nuclear receptors inolved in branched-chain fatty acid signaling. Prost Leukot Essen Fatty Acids. 2002;67:91–98. doi: 10.1054/plef.2002.0404. [DOI] [PubMed] [Google Scholar]
- 16.Makowski L, Hotamisligil GS. Fatty acid binding proteins--the evolutionary crossroads of inflammatory and metabolic responses. J Nutr. 2004;134:2464S–2468S. doi: 10.1093/jn/134.9.2464S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schroeder F, Petrescu AD, Huang H, Atshaves BP, McIntosh AL, Martin GG, Hostetler HA, Vespa A, Landrock K, Landrock D, Payne HR, Kier AB. Role of fatty acid binding proteins and long chain fatty acids in modulating nuclear receptors and gene transcription. Lipids. 2008;43:1–17. doi: 10.1007/s11745-007-3111-z. [DOI] [PubMed] [Google Scholar]
- 18.Hostetler HA, McIntosh AL, Petrescu AD, Huang H, Atshaves BP, Murphy EJ, Kier AB, Schroeder F. Fluorescence methods to assess the impact of lipid binding proteins on ligand mediated activation of gene expression. In: Murphy EJ, Rosenberger TA, editors. Lipid-mediated signaling. Boca Raton, FL: CRC Press-Taylor and Francis Group, LLC; 2009. pp. 299–348. [Google Scholar]
- 19.Haunerland NH, Spener F. Fatty acid binding proteins--insights from genetic manipulations. Prog Lipid Res. 2004;43:328–349. doi: 10.1016/j.plipres.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Binas B, Erol E. FABPs as determinants of myocellular and hepatic fuel metabolism. Mol Cell Biochem. 2007;299:75–84. doi: 10.1007/s11010-005-9043-0. [DOI] [PubMed] [Google Scholar]
- 21.Storch J, Corsico B. The emerging functions and mechanisms of mammalian fatty acid binding proteins. Annu Rev Nutr. 2008;28:18.1–18.23. doi: 10.1146/annurev.nutr.27.061406.093710. [DOI] [PubMed] [Google Scholar]
- 22.Zimmerman AW, Veerkamp JH. New insights into the structure and function of fatty acid binding proteins. Cell Mol Life Sci. 2002;59:1096–1116. doi: 10.1007/s00018-002-8490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matarese V, Stone RL, Waggoner DW, Bernlohr DA. Intracellular fatty acid trafficking and the role of cytosolic lipid binding proteins. Prog Lipid Res. 1989;28:245–272. doi: 10.1016/0163-7827(89)90001-5. [DOI] [PubMed] [Google Scholar]
- 24.Borchers T, Spener F. Fatty acid binding proteins. In: Hoekstra D, editor. Current Topics in Membranes. Orlando, FL: Academic Press Inc.; 1994. pp. 261–294. [Google Scholar]
- 25.Schachtrup C, Emmler T, Bleck B, Sandqvist A, Spener F. Functional analysis of peroxisome proliferator response element motifs in genes of fatty acid binding proteins. Biochem J. 2004;382:239–245. doi: 10.1042/BJ20031340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolfrum C, Borrmann CM, Borchers T, Spener F. Fatty acids and hypolipidemic drugs regulate PPARalpha and PPARgamma gene expresion via L-FABP: a signaling path to the nucleus. Proc Natl Acad Sci. 2001;98:2323–2328. doi: 10.1073/pnas.051619898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bass NM. The cellular fatty acid binding proteins: aspects of structure, regulation, and function. Int Rev Cytol. 1988;111:143–184. doi: 10.1016/s0074-7696(08)61733-7. [DOI] [PubMed] [Google Scholar]
- 28.Kaikaus RM, Chan WK, Ortiz de Montellano PR, Bass NM. Mechanisms of regulation of liver fatty acid-binding protein. [Review] Mol Cell Biochem. 1993;123:93–100. doi: 10.1007/BF01076479. [DOI] [PubMed] [Google Scholar]
- 29.Ellinghaus P, Wolfrum C, Assmann G, Spener F, Seedorf U. Phytanic acid activates the peroxisome proliferator-activated receptor alpha (PPARalpha) in sterol carrier protein-2-/sterol carrier protein x-deficient mice. J Biol Chem. 1999;274:2766–2772. doi: 10.1074/jbc.274.5.2766. [DOI] [PubMed] [Google Scholar]
- 30.Hostetler HA, Petrescu AD, Kier AB, Schroeder F. Peroxisome proliferator activated receptor alpha (PPARalpha) interacts with high affinity and is conformationally responsive to endogenous ligands. J Biol Chem. 2005;280:18667–18682. doi: 10.1074/jbc.M412062200. [DOI] [PubMed] [Google Scholar]
- 31.Hostetler HA, Kier AB, Schroeder F. Very-long-chain and branched-chain fatty acyl CoAs are high affinity ligands for the peroxisome proliferator-activated receptor alpha (PPARalpha) Biochemistry. 2006;45:7669–7681. doi: 10.1021/bi060198l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanhoff T, Wolfrum C, Ellinghaus P, Seedorf U, Spener F. Pristanic acid is activator of PPARalpha. Eur J Lipid Sci. 2001;103:75–80. [Google Scholar]
- 33.Wolfrum C, Ellinghaus P, Fobker M, Seedorf U, Assmann G, Borchers T, Spener F. Phytanic acid is ligand and transcriptional activator of murine liver fatty acid binding protein. J Lipid Res. 1999;40:708–714. [PubMed] [Google Scholar]
- 34.Wolfrum C, Borchers T, Sacchettini JC, Spener F. Binding of fatty acids and perooxisome proliferators to orthologous fatty acid binding proteins from human, murine, and bovine liver. Biochemistry. 2000;39:1469–1474. doi: 10.1021/bi991638u. [DOI] [PubMed] [Google Scholar]
- 35.Wolfrum C, Spener F. Fatty acids as regulators in lipid metabolism. Eur J Lip Sci Technol. 2000;102:746–762. [Google Scholar]
- 36.Karam WG, Ghanayem BI. Induction of replicative DNA synthesis and PPARα-dependent gene transcription by Wy-14 643 in primary rat hepatocyte and non-parenchymal cell co-cultures. Carcinogenesis. 1997;18:2077–2083. doi: 10.1093/carcin/18.11.2077. [DOI] [PubMed] [Google Scholar]
- 37.Bass NM, Manning JA, Ockner RK, Gordon JI, Seetharam S, Alpers DH. Regulation of the biosynthesis of two distinct fatty acid binding proteins in rat liver and intestine. Influence of sex difference and of clofibrate. J Biol Chem. 1985;260:1432–1436. [PubMed] [Google Scholar]
- 38.Brandes R, Kaikaus RM, Lysenko N, Ockner RK, Bass NM. Induction of fatty acid binding protein by peroxisome proliferators in primary hepatocyte cultures and its relationship to the induction of peroxisomal beta-oxidation. Biochim Biophys Acta. 1990;1034:53–61. doi: 10.1016/0304-4165(90)90152-m. [DOI] [PubMed] [Google Scholar]
- 39.Kaikaus RM, Chan WK, Lysenko N, Ray R, Ortiz de Montellano PR, Bass NM. Induction of peroxisomal fatty acid β oxidation and liver fatty acid binding protein by peroxisome proliferators. J Biol Chem. 1993;268:9593–9603. [PubMed] [Google Scholar]
- 40.Gossett RE, Frolov AA, Roths JB, Behnke WD, Kier AB, Schroeder F. Acyl Co A binding proteins: multiplicity and function. Lipids. 1996;31:895–918. doi: 10.1007/BF02522684. [DOI] [PubMed] [Google Scholar]
- 41.Lin Q, Ruuska SE, Shaw NS, Dong D, Noy N. Ligand selectivity of the peroxisome proliferator-activated receptor α. Biochem. 1999;38:185–190. doi: 10.1021/bi9816094. [DOI] [PubMed] [Google Scholar]
- 42.Hostetler HA, Syler LR, Hall LN, Zhu G, Schroeder F, Kier AB. A novel high-throughput screening assay for putative anti-diabetic agents through PPARa interactions. J Biomol Screening. 2008;13:855–861. doi: 10.1177/1087057108323127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang H, Starodub O, McIntosh A, Kier AB, Schroeder F. Liver fatty acid binding protein targets fatty acids to the nucleus: real-time confocal and multiphoton fluorescence imaging in living cells. J Biol Chem. 2002;277:29139–29151. doi: 10.1074/jbc.M202923200. [DOI] [PubMed] [Google Scholar]
- 44.Huang H, Starodub O, McIntosh A, Atshaves BP, Woldegiorgis G, Kier AB, Schroeder F. Liver fatty acid binding protein colocalizes with peroxisome proliferator receptor alpha and enhances ligand distribution to nuclei of living cells. Biochemistry. 2004;43:2484–2500. doi: 10.1021/bi0352318. [DOI] [PubMed] [Google Scholar]
- 45.McIntosh AL, Atshaves BP, Hostetler HA, Huang H, Davis J, Lyuksyutova OI, Landrock D, Kier AB, Schroeder F. Liver type fatty acid binding protein (L-FABP) gene ablation reduces nuclear ligand distribution and peroxisome proliferator activated receptor-alpha activity in cultured primary hepatocytes. Arch Biochem Biophys. 2009;485:160–173. doi: 10.1016/j.abb.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Faergeman NJ, Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem J. 1997;323:1–12. doi: 10.1042/bj3230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Desvergne B, Wahli W. Peroxisome proliferator activated receptors: nuclear control of metabolism. Endocrine Rev. 1999;20:649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 48.Desvergne B, Michalik L, Wahli W. Be fit or be sick: peroxisome proliferator-activated receptors are down the road. Mol Endocrinology. 2004;18:1321–1332. doi: 10.1210/me.2004-0088. [DOI] [PubMed] [Google Scholar]
- 49.Hihi AK, Michalik L, Wahli W. PPARs: transcriptional effectors of fatty acids and their derivatives. Cell Mol Life Sci. 2002;59:645–650. doi: 10.1007/s00018-002-8467-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patsouris D, Mandard S, Voshol PJ, Escher P, Tan NS, Havekes LM, Koenig W, Maerz W, Tafuri S, Wahli W, Mueller M, Kersten S. PPARalpha governs glycerol metabolism. J Clin Invest. 2004;114:94–103. doi: 10.1172/JCI20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan NS, Shaw NS, Vinckenbosch N, Liu P, Yasmin R, Desvergne B, Wahli W, Noy N. Selective cooperation between fatty acid binding proteins and peroxisome proliferator activated receptors in regulating transcription. Mol Cell Biol. 2002;22:5114–5127. doi: 10.1128/MCB.22.14.5114-5127.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lawrence JW, Kroll DJ, Eacho PI. Ligand dependent interaction of hepatic fatty acid binding protein with the nucleus. J Lipid Res. 2000;41:1390–1401. [PubMed] [Google Scholar]
- 53.Budhu AS, Noy N. Direct channeling of retinoic acid between cellular retinoic acid binding protein II and retinoic acid receptor sensitizes mammary carcinoma cells to retinoic acid induced growth arrest. Mol Cell Biol. 2002;22:2632–2641. doi: 10.1128/MCB.22.8.2632-2641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dong D, Ruuska SE, Levinthal DJ, Noy N. Distinct roles for cellular retinoic acid binding proteins I and II in regulating signaling by retinoic acid. J Biol Chem. 1999;274:23695–23698. doi: 10.1074/jbc.274.34.23695. [DOI] [PubMed] [Google Scholar]
- 55.Landrier JF, Thomas C, Grober J, Duez H, Percevault F, Souidi M, Linard C, Staels B, Besnard P. Statin induction of liver fatty acid binding protein (L-FABP) gene expression is peroxisome proliferator activated receptor dependent. J Biol Chem. 2004;279:45512–45518. doi: 10.1074/jbc.M407461200. [DOI] [PubMed] [Google Scholar]
- 56.Martin GG, Atshaves BP, McIntosh AL, Mackie JT, Kier AB, Schroeder F. Liver fatty acid binding protein (L-FABP) gene ablation potentiates hepatic cholesterol accumulation in cholesterol-fed female mice. Am J Physiol. 2006;290:G36–G48. doi: 10.1152/ajpgi.00510.2004. [DOI] [PubMed] [Google Scholar]
- 57.Pignon JP, Bailey NC, Baraona E, Lieber CS. Fatty acid-binding protein: a major contributor to the ethanol-induced increase in liver cytosolic proteins in the rat. Hepatology. 1987;7:865–871. doi: 10.1002/hep.1840070512. [DOI] [PubMed] [Google Scholar]
- 58.Atshaves BP, Payne HR, McIntosh AL, Tichy SE, Russell D, Kier AB, Schroeder F. Sexually dimorphic metabolism of branched chain lipids in C57BL/6J mice. J Lipid Res. 2004;45:812–830. doi: 10.1194/jlr.M300408-JLR200. [DOI] [PubMed] [Google Scholar]
- 59.Atshaves BP, McIntosh AL, Payne HR, Mackie J, Kier AB, Schroeder F. Effect of branched-chain fatty acid on lipid dynamics in mice lacking liver fatty acid binding protein gene. Am J Physiol. 2005;288:C543–C558. doi: 10.1152/ajpcell.00359.2004. [DOI] [PubMed] [Google Scholar]
- 60.Besnard P, Foucaud L, Mallordy A, Berges C, Kaikaus RM, Bernard A, Bass NM, Carlier H. Expression of fatty acid binding protein in the liver during pregnancy and lactation in the rat. Biochim Biophys Acta. 1995;1258:153–158. doi: 10.1016/0005-2760(95)00114-r. [DOI] [PubMed] [Google Scholar]
- 61.Berry SA, Yoon JB, List J, Seelig S. Hepatic fatty acid-binding protein mRNA is regulated by growth hormone. J Am Coll Nutr. 1993;12:638–642. doi: 10.1080/07315724.1993.10718354. [DOI] [PubMed] [Google Scholar]
- 62.Singer SS, Henkels K, Deucher A, Barker M, Singer J, Trulzsch DV. Growth hormone and aging change rat liver fatty acid binding protein levels. J Am Coll Nutr. 1996;15:169–174. doi: 10.1080/07315724.1996.10718584. [DOI] [PubMed] [Google Scholar]
- 63.Schroeder F, Jolly CA, Cho TH, Frolov AA. Fatty acid binding protein isoforms: structure and function. Chem Phys Lipids. 1998;92:1–25. doi: 10.1016/s0009-3084(98)00003-6. [DOI] [PubMed] [Google Scholar]
- 64.Santiago M, Pietro D, Esteban C, Dell A, Veerkamp JH, Sterin-Speziale N, Santome JA. Amino acid sequence, binding properties and evolutionary relationships of the basic liver fatty-acid-binding protein from the catfish Rhamdia sapo. Eur J Biochem. 1997;249:510–517. doi: 10.1111/j.1432-1033.1997.00510.x. [DOI] [PubMed] [Google Scholar]
- 65.Murphy EJ, Edmondson RD, Russell DH, Schroeder F. Isolation and characterization of two distinct forms of liver fatty acid binding protein from the rat. Biochim Biophys Acta. 1999;1436:413–425. doi: 10.1016/s0005-2760(98)00150-7. [DOI] [PubMed] [Google Scholar]
- 66.Frolov A, Cho TH, Murphy EJ, Schroeder F. Isoforms of rat liver fatty acid binding protein differ in structure and affinity for fatty acids and fatty acyl CoAs. Biochemistry. 1997;36:6545–6555. doi: 10.1021/bi970205t. [DOI] [PubMed] [Google Scholar]
- 67.Jolly CA, Murphy EJ, Schroeder F. Differential influence of rat liver fatty acid binding protein isoforms on phospholipid fatty acid composition: phosphatidic acid biosynthesis and phospholipid fatty acid remodeling. Biochim Biophys Acta. 1998;1390:258–268. doi: 10.1016/s0005-2760(97)00186-0. [DOI] [PubMed] [Google Scholar]
- 68.Hitomi M, Odani S, Ono T. Glutathione-protein mixed disulfide decreases the affinity of rat liver fatty acid-binding protein for unsaturated fatty acid. Eur J Biochem. 1990;187:713–719. doi: 10.1111/j.1432-1033.1990.tb15358.x. [DOI] [PubMed] [Google Scholar]
- 69.Corsico B, Cistola DP, Frieden C, Storch J. The helical domain of intestinal fatty acid binding protein is critical for collisional transfer of fatty acids to phospholipid membranes. Proc Natl Acad Sci. 1998;95:12174–12178. doi: 10.1073/pnas.95.21.12174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hostetler HA, Lupas D, Dai J, Woldegiorgis G, Kier AB, Schroeder F. Characterization of a C-terminal 89 residue peptide in liver carnitine palmitoyltransferase I (L-CPTI) important for ligand and ligand-transfer protein binding. Arch Biochem Biophys. 2010 submitted. [Google Scholar]
- 71.Hostetler HA, McIntosh AL, Atshaves BP, Storey SM, Payne HR, Kier AB, Schroeder F. Liver type Fatty Acid Binding Protein (L-FABP) interacts with peroxisome proliferator activated receptor-α in cultured primary hepatocytes. J Lipid Res. 2009;50:1663–1675. doi: 10.1194/jlr.M900058-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schroeder F, Atshaves BP, Starodub O, Boedeker AL, Smith R, Roths JB, Foxworth WB, Kier AB. Expression of liver fatty acid binding protein alters growth and differentiation of embryonic stem cells. Mol Cell Biochem. 2001;219:127–138. doi: 10.1023/a:1010851130136. [DOI] [PubMed] [Google Scholar]
- 73.Lowe JB, Sacchettini JC, Laposta M, McQuillan JJ, Gordon JI. Expression of rat intestinal fatty acid binding protein in E. coli. Purificaiton and comparison of ligand binding characteristics with that of E. coli derived rat liver fatty acid binding protein. J Biol Chem. 1987;262:5931–5937. [PubMed] [Google Scholar]
- 74.Muga A, Cistola DP, Mantsch HH. A comparative study of the conformational properties of Escherichia coli-derived rat intestinal and liver fatty acid binding proteins. Biochim Biophys Acta. 1993;1162:291–296. doi: 10.1016/0167-4838(93)90293-z. [DOI] [PubMed] [Google Scholar]
- 75.Frolov AA, Schroeder F. Time-resolved fluorescence of intestinal and liver fatty acid binding proteins: Role of fatty acyl CoA and fatty acid. Biochem. 1997;36:505–517. doi: 10.1021/bi961392i. [DOI] [PubMed] [Google Scholar]
- 76.Nemecz G, Hubbell T, Jefferson JR, Lowe JB, Schroeder F. Interaction of fatty acids with recombinant rat intestinal and liver fatty acid-binding proteins. Arch Biochem Biophys. 1991;286:300–309. doi: 10.1016/0003-9861(91)90044-j. [DOI] [PubMed] [Google Scholar]
- 77.Nemecz G, Jefferson JR, Schroeder F. Polyene fatty acid interactions with recombinant intestinal and liver fatty acid binding proteins. J Biol Chem. 1991;266:17112–17123. [PubMed] [Google Scholar]
- 78.Thompson J, Winter N, Terwey D, Bratt J, Banaszak L. The crystal structure of the liver fatty acid-binding protein. J Biol Chem. 1997;272:7140–7150. doi: 10.1074/jbc.272.11.7140. [DOI] [PubMed] [Google Scholar]
- 79.He Y, Yang X, Wang H, Estephan R, Francis F, Kodukula S, Storch J, Stark RE. Solution-state molecular structure of apo and oleate-liganded liver fatty acid binding protein. Biochemistry. 2007;46:12543–12556. doi: 10.1021/bi701092r. [DOI] [PubMed] [Google Scholar]
- 80.Rolf B, Oudenampsen-Kruger E, Borchers T, Faergeman NJ, Knudsen J, Lezius A, Spener F. Analysis of the ligand binding properties of recombinant bovine liver-type fatty acid binding protein. Biochim Biophys Acta. 1995;1259:245–253. doi: 10.1016/0005-2760(95)00170-0. [DOI] [PubMed] [Google Scholar]
- 81.Richieri GV, Ogata RT, Kleinfeld AM. Equilibrium constants for the binding of fatty acids with fatty acid binding proteins from adipocyte, intestine, heart, and liver measured with the flourscent probe ADIFAB. J Biol Chem. 1994;269:23918–23930. [PubMed] [Google Scholar]
- 82.Norris AW, Spector AA. Very long chain n-3 and n-6 polyunsaturated fatty acids bind strongly to liver fatty acid binding protein. J Lipid Res. 2002;43:646–653. [PubMed] [Google Scholar]
- 83.Richieri GV, Ogata RT, Zimmerman AW, Veerkamp JH, Kleinfeld AM. Fatty acid binding proteins from different tissues show distinct patterns of fatty acid interaction. Biochemistry. 2000;39:7197–7204. doi: 10.1021/bi000314z. [DOI] [PubMed] [Google Scholar]
- 84.Schroeder F, Jefferson JR, Powell D, Incerpi S, Woodford JK, Colles SM, Myers-Payne S, Emge T, Hubbell T, Moncecchi D, Prows DR, Heyliger CE. Expression of rat L-FABP in mouse fibroblasts: role in fat absorption. Mol Cell Biochem. 1993;123:73–83. doi: 10.1007/BF01076477. [DOI] [PubMed] [Google Scholar]
- 85.Thumser AE, Wilton DC. The binding of natural and fluorescent lysophospholipids to wild- type and mutant rat liver fatty acid-binding protein and albumin. Biochem J. 1995;307:305–311. doi: 10.1042/bj3070305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thumser AE, Voysey JE, Wilton DC. The binding of lysophospholipids to rat liver fatty acid-binding protein and albumin. Biochem J. 1994;301:801–806. doi: 10.1042/bj3010801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Raza H, Pongubala JR, Sorof S. Specific high affinity binding of lipoxygenase metabolites of arachidonic acid by liver fatty acid binding protein. Biochem Biophys Res Commun. 1989;161:448–455. doi: 10.1016/0006-291x(89)92619-3. [DOI] [PubMed] [Google Scholar]
- 88.Richieri GV, Ogata RT, Kleinfeld AM. Thermodynamic and kinetic properties of fatty acid interactions with rat liver fatty acid-binding protein. J Biol Chem. 1996;271:31068–31074. doi: 10.1074/jbc.271.49.31068. [DOI] [PubMed] [Google Scholar]
- 89.Thumser AE, Wilton DC. The binding of cholesterol and bile salts to recombinant rat liver fatty acid-binding protein. Biochem J. 1996;320:729–733. doi: 10.1042/bj3200729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fischer RT, Cowlen MS, Dempsey ME, Schroeder F. Fluorescence of delta 5,7,9(11),22-ergostatetraen-3 beta-ol in micelles, sterol carrier protein complexes, and plasma membranes. Biochemistry. 1985;24:3322–3331. doi: 10.1021/bi00334a037. [DOI] [PubMed] [Google Scholar]
- 91.Sams GH, Hargis BM, Hargis PS. Identification of two lipid binding proteins from liver of Gallus domesticus. Comp Biochem Physiol. 1991;99B:213–219. doi: 10.1016/0305-0491(91)90032-9. [DOI] [PubMed] [Google Scholar]
- 92.Schroeder F, Frolov A, Starodub O, Russell W, Atshaves BP, Petrescu AD, Huang H, Gallegos A, McIntosh A, Tahotna D, Russell D, Billheimer JT, Baum CL, Kier AB. Pro-sterol carrier protein-2: role of the N-terminal presequence in structure, function, and peroxisomal targeting. J Biol Chem. 2000;275:25547–25555. doi: 10.1074/jbc.M000431200. [DOI] [PubMed] [Google Scholar]
- 93.Stolowich NJ, Frolov A, Petrescu AD, Scott AI, Billheimer JT, Schroeder F. Holo-sterol carrier protein-2: 13C-NMR investigation of cholesterol and fatty acid binding sites. J Biol Chem. 1999;274:35425–35433. doi: 10.1074/jbc.274.50.35425. [DOI] [PubMed] [Google Scholar]
- 94.Martin GG, Hostetler HA, McIntosh AL, Tichy SE, Williams BJ, Russell DH, Berg JM, Spencer TA, Ball JA, Kier AB, Schroeder F. Structure and function of the sterol carrier protein-2 (SCP-2) N-terminal pre-sequence. Biochem. 2008;47:5915–5934. doi: 10.1021/bi800251e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Avdulov NA, Chochina SV, Igbavboa U, Warden CH, Schroeder F, Wood WG. Lipid binding to sterol carrier protein-2 is inhibited by ethanol. Biochim Biophys Acta. 1999;1437:37–45. doi: 10.1016/s0005-2760(98)00178-7. [DOI] [PubMed] [Google Scholar]
- 96.Schroeder F, Frolov A, Schoer J, Gallegos A, Atshaves BP, Stolowich NJ, Scott AI, Kier AB. Intracellular sterol binding proteins, cholesterol transport and membrane domains. In: Chang TY, Freeman DA, editors. Intracellular Cholesterol Trafficking. Boston: Kluwer Academic Publishers; 1998. pp. 213–234. [Google Scholar]
- 97.Martin GG, Atshaves BP, Huang H, McIntosh AL, Williams BW, Russell DH, Kier AB, Schroeder F. Hepatic phenotype of liver latty acid binding protein (L-FABP) gene ablated mice. Am J Physiol. 2009 doi: 10.1152/ajpgi.00116.2009. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Martin GG, Atshaves BP, Huang H, McIntosh AL, Williams BW, Pai PJ, Russell DH, Kier AB, Schroeder F. Hepatic phenotype of liver fatty acid binding protein gene ablated mice. Am J Physiol. 2009;297:G1053–G1065. doi: 10.1152/ajpgi.00116.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Miller KR, Cistola DP. Titration calorimetry as a binding assay for lipid-binding proteins. Mol Cell Biochem. 1993;123:29–37. doi: 10.1007/BF01076472. [DOI] [PubMed] [Google Scholar]
- 100.Pfanner N, Glick BS, Arden SR, Rothman JE. Fatty acylation promotes fusion of transport vesicles with Golgi cisternae. J Cell Biol. 1990;110:955–961. doi: 10.1083/jcb.110.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pfanner N, Orci L, Glick BS, Amherdt M, Arden SR, Malhotra V, Rothman JE. Fatty acyl CoA is required for budding of transport vesicles from Golgi cisternae. Cell. 1989;59:95–102. doi: 10.1016/0092-8674(89)90872-6. [DOI] [PubMed] [Google Scholar]
- 102.Neeli I, Siddiqi SA, Siddiqi S, Mahan J, Lagakos WS, Binas B, Gheyi T, Storch J, Mansbach CM. Liver fatty acid binding protein initiates budding of pre-chylomicron transport vesicles from intestinal endoplasmic reticulum. J Biol Chem. 2007;282:17974–17984. doi: 10.1074/jbc.M610765200. [DOI] [PubMed] [Google Scholar]
- 103.Khan SH, Sorof S. Preferential binding of growth inhibitory prostaglandins by the target protein of a carcinogen. Proc Natl Acad Sci. 1990;87:9401–9405. doi: 10.1073/pnas.87.23.9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ek BA, Cistola DP, Hamilton JA, Kaduce TL, Spector AA. Fatty acid binding proteins reduced 15-lipoxygenase-induced oxygenation of linoleic acid and arachidonic acid. Biochim Biophys Acta. 1997;1346:75–85. doi: 10.1016/s0005-2760(97)00021-0. [DOI] [PubMed] [Google Scholar]
- 105.Wilton DC. Studies on fatty-acid-binding proteins. The purification of rat liver fatty-acid-binding protein and the role of cysteine-69 in fatty acid binding. Biochem J. 1989;261:273–276. doi: 10.1042/bj2610273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Thumser AE, Voysey J, Wilton DC. Mutations of recombinant rat liver fatty acid-binding protein at residues 102 and 122 alter its structural integrity and affinity for physiological ligands. Biochem J. 1996;314:943–949. doi: 10.1042/bj3140943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Myszka DG, Swenson RP. Identification by photoaffinity labeling of fatty acid-binding protein as a potential warfarin receptor in rat liver. J Biol Chem. 1991;266:20725–20731. [PubMed] [Google Scholar]
- 108.Bassuk JA, Tsichlis PN, Sorof S. Liver fatty acid binding protein is the mitosis-associated polypeptide target of a carcinogen in rat hepatocytes. Proc Natl Acad Sci. 1987;84:7547–7551. doi: 10.1073/pnas.84.21.7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Khan SH, Sorof S. Liver fatty acid-binding protein: Specific mediator of the mitogenesis induced by two classes of carcinogenic peroxisome proliferators. Proc Natl Acad Sci. 1994;91:848–851. doi: 10.1073/pnas.91.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Munir KM, Blackburn GR, Sorof S. Late target protein of the carcinogen N-2-fluorenylacetamide in rat liver. Cancer Res. 1988;48:6745–6752. [PubMed] [Google Scholar]
- 111.Bansal MP, Cook RG, Danielson KG, Medina D. A 14-kilodalton selenium-binding protein in mouse liver is fatty acid-binding protein. J Biol Chem. 1989;264:13780–13784. [PubMed] [Google Scholar]
- 112.Custer RP, Sorof S. Target polypeptide of a carcinogen is associated with normal mitosis and carcinogen-induced hyperplasias in adult hepatocytes. Proc Natl Acad Sci. 1984;81:6738–6742. doi: 10.1073/pnas.81.21.6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Keler T, Barker CS, Sorof S. Specific growth stimulation by linoleic acid in hepatoma cell lines transfected with the target protein of a liver carcinogen. Proc Natl Acad Sci. 1992;89:4830–4834. doi: 10.1073/pnas.89.11.4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Keler T, Sorof S. Growth promotion of transfected hepatoma cells by liver fatty acid binding protein. J Cell Physiol. 1993;157:33–40. doi: 10.1002/jcp.1041570105. [DOI] [PubMed] [Google Scholar]
- 115.Keler T, Khan SH, Sorof S. Liver fatty acid binding protein and mitogenesis in transfected hepatoma cells. Adv Exper Med Biol. 1997;400A:517–524. doi: 10.1007/978-1-4615-5325-0_70. [DOI] [PubMed] [Google Scholar]
- 116.Sorof S. Modulation of mitogenesis by liver fatty acid binding protein. Cancer Metastasis Rev. 1994;13:317–336. doi: 10.1007/BF00666102. [DOI] [PubMed] [Google Scholar]
- 117.Jolly CA, Hubbell T, Behnke WD, Schroeder F. Fatty acid binding protein: Stimulation of microsomal phosphatidic acid formation. Arch Biochem Biophys. 1997;341:112–121. doi: 10.1006/abbi.1997.9957. [DOI] [PubMed] [Google Scholar]
- 118.Jolly CA, Wilton DA, Schroeder F. Microsomal fatty acyl CoA transacylation and hydrolysis: fatty acyl CoA species dependent modulation by liver fatty acyl CoA binding proteins. Biochim Biophys Acta. 2000;1483:185–197. doi: 10.1016/s1388-1981(99)00170-5. [DOI] [PubMed] [Google Scholar]
- 119.Coleman RA, Lewin TM, Van Horn CG, Gonzalez-Baro MR. Do long chain acyl CoA synthetases regulate fatty acid entry into synthetic vs degradative pathways? J Nutr. 2002;132:2123–2126. doi: 10.1093/jn/132.8.2123. [DOI] [PubMed] [Google Scholar]
- 120.Lewin TM, Kim JH, Granger DA, Vance JE, Coleman RA. Acyl CoA synthetase isoforms 1, 4, and 5 are present in different subcellular membranes in rat liver and can be inhibited independently. J Biol Chem. 2001;276:24674–24679. doi: 10.1074/jbc.M102036200. [DOI] [PubMed] [Google Scholar]
- 121.Mashek DG, Coleman RA. Cellular fatty acid uptake: the contribution of metabolism. Curr Opin Lipidol. 2006;17:274–278. doi: 10.1097/01.mol.0000226119.20307.2b. [DOI] [PubMed] [Google Scholar]
- 122.Schroeder F, Huang H, Hostetler HA, Petrescu AD, Hertz R, Bar-Tana J, Kier AB. Stability of fatty acyl CoA thioester ligands of hepatocyte nuclear factor -4alpha and peroxisome proliferator-activated receptor alpha. Lipids. 2005;40:559–568. doi: 10.1007/s11745-005-1416-y. [DOI] [PubMed] [Google Scholar]
- 123.Jolly CA, Chao H, Kier AB, Billheimer JT, Schroeder F. Sterol carrier protein-2 suppresses microsomal acyl CoA hydrolysis. Mol Cell Biochem. 2000;205:83–90. doi: 10.1023/a:1007001614939. [DOI] [PubMed] [Google Scholar]
- 124.Davies JK, Thumser Alfred EA, Wilton DA. Binding of recombinant rat liver fatty acid binding protein to small anionic phospholipid vesicles results in ligand release: a model for interfacial binding and fatty acid targeting. Biochemistry. 1999;38:16932–16940. doi: 10.1021/bi991926q. [DOI] [PubMed] [Google Scholar]
- 125.Frolov A, Woodford JK, Murphy EJ, Billheimer JT, Schroeder F. Spontaneous and protein-mediated sterol transfer between intracellular membranes. J Biol Chem. 1996;271:16075–16083. doi: 10.1074/jbc.271.27.16075. [DOI] [PubMed] [Google Scholar]
- 126.Frolov AA, Woodford JK, Murphy EJ, Billheimer JT, Schroeder F. Fibroblast membrane sterol kinetic domains: modulation by sterol carrier protein 2 and liver fatty acid binding protein. J Lipid Res. 1996;37:1862–1874. [PubMed] [Google Scholar]
- 127.Woodford JK, Behnke WD, Schroeder F. Liver fatty acid binding protein enhances sterol transfer by membrane interaction. Mol Cell Biochem. 1995;152:51–62. doi: 10.1007/BF01076463. [DOI] [PubMed] [Google Scholar]
- 128.Bordewick U, Heese M, Borchers T, Robenek H, Spener F. Compartmentation of hepatic fatty-acid-binding protein in liver cells and its effect on microsomal phosphatidic acid biosynthesis. Biol Chem Hoppe-Seyler. 1989;370:229–238. doi: 10.1515/bchm3.1989.370.1.229. [DOI] [PubMed] [Google Scholar]
- 129.Nemecz G, Schroeder F. Selective binding of cholesterol by recombinant fatty acid-binding proteins. J Biol Chem. 1991;266:17180–17186. [PubMed] [Google Scholar]
- 130.Woldegiorgis G, Bremer J, Shrago E. Substrate inhibition of carnitine palmitoyltransferase by palmitoyl-CoA and activation by phospholipids and proteins. Biochim Biophys Acta. 1985;837:135–140. doi: 10.1016/0005-2760(85)90236-x. [DOI] [PubMed] [Google Scholar]
- 131.Bhuiyan AKMJ, Pande SV. Carnitine palmitoyltransferase activities: effects of serum albumin, acyl-CoA binding protein and fatty acid binding protein. Mol Cell Biochem. 1994;139:109–116. doi: 10.1007/BF01081733. [DOI] [PubMed] [Google Scholar]
- 132.Rasmussen JT, Faergeman NJ, Kristiansen K, Knudsen J. Acyl-CoA-binding protein (ACBP) can mediate intermembrane acyl- CoA transport and donate acyl-CoA for beta-oxidation and glycerolipid synthesis. Biochem J. 1994;299:165–170. doi: 10.1042/bj2990165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Reubsaet FA, Veerkamp JH, Bruckwilder ML, Trijbels JM, Monnens LA. The involvement of fatty acid-binding protein in peroxisomal fatty acid oxidation. FEBS Lett. 1990;267:229–230. doi: 10.1016/0014-5793(90)80931-8. [DOI] [PubMed] [Google Scholar]
- 134.Antonenkov VD, Sormunen RT, Ohlmeier S, Amery L, Fransen M. Localization of a portion of the liver isoform of fatty acid binding protein (L-FABP) to peroxisomes. Biochem J. 2006;394:475–484. doi: 10.1042/BJ20051058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Murphy EJ, Prows DR, Jefferson JR, Schroeder F. Liver fatty acid binding protein expression in transfected fibroblasts stimulates fatty acid uptake and metabolism. Biochim Biophys Acta. 1996;1301:191–198. doi: 10.1016/0005-2760(96)00024-0. [DOI] [PubMed] [Google Scholar]
- 136.Murphy EJ, Prows DR, Jefferson JR, Incerpi S, Hertelendy ZI, Heiliger CE, Schroeder F. Cis-parinaric acid uptake in L-cells. Arch Biochem Biophys. 1996;335:267–272. doi: 10.1006/abbi.1996.0507. [DOI] [PubMed] [Google Scholar]
- 137.Prows DR, Murphy EJ, Schroeder F. Intestinal and liver fatty acid binding proteins differentially affect fatty acid uptake and esterification in L-Cells. Lipids. 1995;30:907–910. doi: 10.1007/BF02537481. [DOI] [PubMed] [Google Scholar]
- 138.Wolfrum C, Buhlman C, Rolf B, Borchers T, Spener F. Variation of liver fatty acid binding protein content in the human hepatoma cell line HepG2 by peroxisome proliferators and antisense RNA affects the rate of fatty acid uptake. Biochim Biophys Acta. 1999;1437:194–201. doi: 10.1016/s1388-1981(99)00008-6. [DOI] [PubMed] [Google Scholar]
- 139.Atshaves BP, Storey S, Huang H, Schroeder F. Liver fatty acid binding protein expression enhances branched-chain fatty acid metabolism. Mol Cell Biochem. 2004;259:115–129. doi: 10.1023/b:mcbi.0000021357.97765.f2. [DOI] [PubMed] [Google Scholar]
- 140.Moncecchi DM, Murphy EJ, Prows DR, Schroeder F. Sterol carrier protein-2 expression in mouse L-cell fibroblasts alters cholesterol uptake. Biochim Biophys Acta. 1996;1302:110–116. doi: 10.1016/0005-2760(96)00044-6. [DOI] [PubMed] [Google Scholar]
- 141.Burczynski FJ, Zhang MN, Pavletic P, Wang GQ. Role of fatty acid binding protein on hepatic palmitate uptake. Can J Physiol Pharmacol. 1997;75:1350–1355. [PubMed] [Google Scholar]
- 142.Atshaves BP, McIntosh AL, Lyuksyutova OI, Zipfel WR, Webb WW, Schroeder F. Liver fatty acid binding protein gene ablation inhibits branched-chain fatty acid metabolism in cultured primary hepatocytes. J Biol Chem. 2004;279:30954–30965. doi: 10.1074/jbc.M313571200. [DOI] [PubMed] [Google Scholar]
- 143.Newberry EP, Xie Y, Kennedy S, Buhman KK, Luo J, Gross RW, Davidson NO. Decreased hepatic triglyceride accumulation and altered fatty acid uptake in mice with deletion of the liver fatty acid binding protein gene. J Biol Chem. 2003;278:51664–51672. doi: 10.1074/jbc.M309377200. [DOI] [PubMed] [Google Scholar]
- 144.Avdulov NA, Chochina SV, Myers-Payne S, Hubbell T, Igbavboa U, Schroeder F, Wood WG. Expression and lipid binding of sterol carrier protein-2 and liver fatty acid binding proteins: differential effects of ethanol in vivo and in vitro. In: Riemersma RA, A R, K RW, W R, editors. Essential Fatty Acids and Eicosanoids: Invited Papers from the Fourth International Congress. Champaign, IL: American Oil Chemists Society Press; 1998. pp. 324–327. [Google Scholar]
- 145.Schroeder F, Myers-Payne SC, Billheimer JT, Wood WG. Probing the ligand binding sites of fatty acid and sterol carrier proteins: effects of ethanol. Biochemistry. 1995;34:11919–11927. doi: 10.1021/bi00037a033. [DOI] [PubMed] [Google Scholar]
- 146.Luxon BA, Weisiger RA. Sex differences in intracellular fatty acid transport: role of cytoplasmic binding proteins. Am J Physiol. 1993;265:G831–41. doi: 10.1152/ajpgi.1993.265.5.G831. [DOI] [PubMed] [Google Scholar]
- 147.Luxon BA, Milliano MT. Cytoplasmic codiffusion of fatty acids is not specific for fatty acid binding protein. Am J Physiol. 1997;273:C859–C867. doi: 10.1152/ajpcell.1997.273.3.C859. [DOI] [PubMed] [Google Scholar]
- 148.Murphy EJ. L-FABP and I-FABP expression increase NBD-stearate uptake and cytoplasmic diffusion in L-cells. Am J Physiol. 1998;275:G244–G249. doi: 10.1152/ajpgi.1998.275.2.G244. [DOI] [PubMed] [Google Scholar]
- 149.Phelps-Luby K, Weisiger RA. Role of Cytoarchitecture in Cytoplasmic Transport. Comp Biochem Physiol. 1996;115B:295–306. [Google Scholar]
- 150.Weisiger RA. Cytoplasmic transport of lipids: Role of binding proteins. Comp Biochem Physiol. 1996;115B:319–331. [Google Scholar]
- 151.Weisiger RA. Cytosolic fatty acid binding proteins catalyze two distinct steps in intracellular transport of their ligands. Mol Cell Biochem. 2005;239:35–42. [PubMed] [Google Scholar]
- 152.Martin GG, Danneberg H, Kumar LS, Atshaves BP, Erol E, Bader M, Schroeder F, Binas B. Decreased liver fatty acid binding capacity and altered liver lipid distribution in mice lacking the liver fatty acid binding protein (L-FABP) gene. J Biol Chem. 2003;278:21429–21438. doi: 10.1074/jbc.M300287200. [DOI] [PubMed] [Google Scholar]
- 153.Martin GG, Huang H, Atshaves BP, Binas B, Schroeder F. Ablation of the liver fatty acid binding protein gene decreases fatty acyl CoA binding capacity and alters fatty acyl CoA pool distribution in mouse liver. Biochem. 2003;42:11520–11532. doi: 10.1021/bi0346749. [DOI] [PubMed] [Google Scholar]
- 154.Jefferson JR, Slotte JP, Nemecz G, Pastuszyn A, Scallen TJ, Schroeder F. Intracellular sterol distribution in transfected mouse L-cell fibroblasts expressing rat liver fatty acid binding protein. J Biol Chem. 1991;266:5486–5496. [PubMed] [Google Scholar]
- 155.Jefferson JR, Powell DM, Rymaszewski Z, Kukowska-Latallo J, Schroeder F. Altered membrane structure in transfected mouse L-Cell fibroblasts expressing rat liver fatty acid-binding protein. J Biol Chem. 1990;265:11062–11068. [PubMed] [Google Scholar]
- 156.Murphy EJ, Schroeder F. Sterol carrier protein-2 mediated cholesterol esterification in transfected L-cell fibroblasts. Biochim Biophys Acta. 1997;1345:283–292. doi: 10.1016/s0005-2760(97)00003-9. [DOI] [PubMed] [Google Scholar]
- 157.Prows DR, Murphy EJ, Moncecchi D, Schroeder F. Intestinal fatty acid-binding protein expression stimulates fibroblast fatty acid esterification. Chem Phys Lipids. 1996;84:47–56. doi: 10.1016/s0009-3084(96)02619-9. [DOI] [PubMed] [Google Scholar]
- 158.Murhpy EJ, Prows DR, Stiles T, Schroeder F. Liver and intestinal fatty acid binding protein expression increases phospholipid content and alters phospholipid fatty acid composition in L-cell fibroblasts. Lipids. 2000;35:729–738. doi: 10.1007/s11745-000-0579-x. [DOI] [PubMed] [Google Scholar]
- 159.Linden D, Lindberg K, Oscarsson J, Claesson C, Asp L, Li L, Gustafsson M, Boren J, Oloffson SO. Influence of peroxisome proliferator-activated receptor alpha agonists on the intracellular turnover and secretion of apolipoprotein (apo) B-100 and apoB-48. J Biol Chem. 2002;277:23044–23053. doi: 10.1074/jbc.M110416200. [DOI] [PubMed] [Google Scholar]
- 160.Erol E, Kumar LS, Cline GW, Shulman GI, Kelly DP, Binas B. Liver fatty acid-binding protein is required for high rates of hepatic fatty acid oxidation but not for the action of PPAR-a in fasting mice. FASEB J. 2004;18:347–349. doi: 10.1096/fj.03-0330fje. [DOI] [PubMed] [Google Scholar]
- 161.Thompson J, Ory J, Reese-Wagoner A, Banaszak L. The liver fatty acid binding protein-comparison of cavity properties of intracellular lipid binding proteins. Mol Cell Biochem. 1999;192:9–16. [PubMed] [Google Scholar]
- 162.Martin GG, Atshaves BP, McIntosh AL, Mackie JT, Kier AB, Schroeder F. Liver fatty acid binding protein (L-FABP) gene ablation alters liver bile acid metabolism in male mice. Biochem J. 2005;391:549–560. doi: 10.1042/BJ20050296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Newberry EP, Xie Y, Kennedy SM, Luo J, Davidson NO. Protection against western diet-induced obesity and hepatic steatosis in liver fatty acid binding protein knockout mice. Hepatology. 2006;44:1191–1205. doi: 10.1002/hep.21369. [DOI] [PubMed] [Google Scholar]
- 164.Newberry EP, Kennedy SM, Xie Y, Sternard BT, Luo J, Davidson NO. Diet-induced obesity and hepatic steatosis in L-FABP-/- mice is abrogated with SF, but not PUFA, feeding and attenuated after cholesterol supplementation. Am J Physiol Gastrointest and Liver Phys. 2008;294:G307–G314. doi: 10.1152/ajpgi.00377.2007. [DOI] [PubMed] [Google Scholar]
- 165.Xie Y, Newberry EP, Kennedy SM, Luo J, Davidson NO. Increased susceptibility to diet-induced gallstones in liver fatty acid binding protein knockout mice. J Lipid Res. 2009;50:977–987. doi: 10.1194/jlr.M800645-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Veerkamp JH, van Moerkerk HT. The fatty acid-binding protein content and fatty acid oxidation capacity of rat tissues. Prog Clin Biol Res. 1992;375:205–210. [PubMed] [Google Scholar]
- 167.Atshaves BP, McIntosh AL, Storey SM, Landrock KK, Kier AB, Schroeder F. High dietary fat exacerbates weight gain and obesity in female liver fatty acid binding protein gene ablated mice. Lipids. 2010;45:97–1. doi: 10.1007/s11745-009-3379-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Seedorf U, Raabe M, Ellinghaus P, Kannenberg F, Fobker M, Engel T, Denis S, Wouters F, Wirtz KWA, Wanders RJA, Maeda N, Assmann G. Defective peroxisomal catabolism of branched fatty acyl coenzyme A in mice lacking the sterol carrier protein-2/sterol carrier protein-x gene function. Genes and Development. 1998;12:1189–1201. doi: 10.1101/gad.12.8.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mackie JT, Atshaves BP, Payne HR, McIntosh AL, Schroeder F, Kier AB. Phytol-induced hepatotoxicity in mice. Toxicol Pathol. 2009;37:201–208. doi: 10.1177/0192623308330789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Atshaves BP, McIntosh AL, Landrock D, Payne HR, Mackie J, Maeda N, Ball JM, Schroeder F, Kier AB. Effect of SCP-x gene ablation on branched-chain fatty acid metabolism. Am J Physiol. 2007;292:939–951. doi: 10.1152/ajpgi.00308.2006. [DOI] [PubMed] [Google Scholar]
- 171.Thigpen JE, Setchell KD, Ahlmark KB, Kocklear J, Spahr T, Caviness GF, Goelz MF, Haseman JK, Newbold RR, Forsythe DB. Phytoestrogen content of purified, open- and closed-formula laboratory animal diets. Lab An Science. 1999;49:530–536. [PubMed] [Google Scholar]
- 172.Thigpen JE, Setchell KD, Goelz MF, Forsythe DB. The phytoestrogen content of rodent diets. Envron Health Persp. 1999;107:A182–A183. doi: 10.1289/ehp.107-1566530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Steinberg D. Refsums disease. In: Scriver CR, Beaudet AL, Sly WS, Vallee D, editors. The Metabolic and Molecular Basis of Inherited Disease. New York, NY: McGraw-Hill; 1995. pp. 2351–2370. [Google Scholar]
- 174.Ferdinandusse S, Denis S, van Berkel E, Dacremont G, Wanders RJ. Peroxisomal fatty acid oxidation disorders and 58 kDa sterol carrier protein-x (SCP=x): activity measurements in liver and fibroblasts using a newly developed method. J Lipid Res. 2000;41:336–342. [PubMed] [Google Scholar]
- 175.Wanders RJA, Vreken P, Ferdinandusse S, Jansen GA, Waterham HR, van Roermund CW, van Grunsven EG. Peroxisomal fatty acid alpha and beta oxidation in humans: enzymology, peroxisomal metabolite transporters and peroxisomal diseases. Biochem Soc Trans. 2001;29:250–267. doi: 10.1042/0300-5127:0290250. [DOI] [PubMed] [Google Scholar]
- 176.Martin GG, Atshaves BP, McIntosh AL, Mackie JT, Kier AB, Schroeder F. Liver Fatty Acid Binding Protein GeneAblated Female Mice Exhibit Increased AgeDependent Obesity. J Nutr. 2008;138:1859–1865. doi: 10.1093/jn/138.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Martin GG, Atshaves BP, McIntosh AL, Mackie JT, Kier AB, Schroeder F. Liver fatty acid binding protein gene ablation enhances age-dependent weight gain in male mice. Mol Cell Biochem. 2009;324:101–115. doi: 10.1007/s11010-008-9989-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lyons MA, Wittenburg H, Li R, Walsh KA, Leonard MR, Korstanje R, Churchill GA, Carey MC, Paigen B. Lith6:a new QTL for cholesterol gallstones from an intercross of CAST/Ei and DBA/2J mouse strains. J Lipid Res. 2003;44:1763–1771. doi: 10.1194/jlr.M300149-JLR200. [DOI] [PubMed] [Google Scholar]
- 179.Lyons MA, Wittenburg H. Cholesterol gallstone susceptibility loci: a mouse map, candidate gene evaluation, and guide to human LITH genes. Gastroenterology. 2006;131:1943–1970. doi: 10.1053/j.gastro.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 180.Fuchs M, Lammert F, Wang DQH, Paigen B, Carey MC, Cohen DE. Sterol carrier protein-2 participates in hypersecretion of biliary cholesterol during cholesterol gallstone formation in genetically gallstone susceptible mice. Biochem J. 1998;336:33–37. doi: 10.1042/bj3360033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Fuchs M, Hafer A, Muench C, Kannenberg F, Teichmann S, Scheibner J, Stange EF, Seedorf U. Disruption of the sterol carrier protein 2 gene in mice impairs biliary lipid and hepatic cholesterol metabolism. J Biol Chem. 2001;276:48058–48065. doi: 10.1074/jbc.M106732200. [DOI] [PubMed] [Google Scholar]
- 182.Newberry EP, Kennedy SM, Xie Y, Luo J, Davidson NO. Diet-induced alterations in intestinal and extrahepatic lipid metabolism in liver fatty acid binding protein knockout mice. Mol Cell Biochem. 2009;326:79–86. doi: 10.1007/s11010-008-0002-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Feige JN, Gelman L, Michalik L, Desvergne B, Wahli W. From molecular action to physiological outputs: PPARs are nuclear receptors at the crossroads of key cellular functions. Prog Lipid Res. 2006;45:120–159. doi: 10.1016/j.plipres.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 184.Robitaille J, Brouillette C, Lemieux S, Perusse L, Gaudet D, Vohl MC. Plasma concentrations of apolipoprotein B are modulated by a gene-diet interaction effect between the L-FABP T94A polymorphism and dietary fat intake in French-Canadian men. Mol Gen and Metab. 2004;82:296–303. doi: 10.1016/j.ymgme.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 185.Brouillette C, Bose Y, Perusse L, Gaudet D, Vohl MC. Effect of liver fatty acid binding protein (FABP) T94A missense mutation on plasma lipoprotein responsiveness to treatment with fenofibrate. J Hum Gen. 2004;49:424–432. doi: 10.1007/s10038-004-0171-2. [DOI] [PubMed] [Google Scholar]
- 186.Fisher E, Weikert C, Klapper M, Lindner I, Mohlig M, Spranger J, Boeing H, Schrezenmeir J, Doring F. L-FABP T94A is associated with fasting triglycerides and LDL-cholesterol in women. Mol Gen and Metab. 2007;91:278–284. doi: 10.1016/j.ymgme.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 187.Weikert MO, Loeffelholz Cv, Roden M, Chandramouli V, Brehm A, Nowotny P, Osterhoff MA, Isken F, Spranger J, Landau BR, Pfeiffer A, Mohlig M. A Thr94Ala mutation in human liver fatty acid binding protein contributes to reduced hepatic glycogenolysis and blunted elevation of plasma glucsoe levels in lipid-exposed subjects. Am J Physiol Endocrinol Metab. 2007;293:E1078–E1084. doi: 10.1152/ajpendo.00337.2007. [DOI] [PubMed] [Google Scholar]
- 188.Baier LJ, Sacchettini JC, Knowler WC, Eads J, Paolisso G, Tataranni PA, Mochizuki H, Bennett PH, Bogardus C, Prochazka M. An amino acid substitution in the human intestinal fatty acid binding protein is associated with increased fatty acid binding, increased fat oxidation, and insulin resistance. J Clin Inv. 1995;95:1281–1287. doi: 10.1172/JCI117778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.de Luis DA, Sagrado MG, Aller R, Izaola O, Conde R. Influence of ALA54THR polymorphism of fatty acid binding protein 2, role on insulin resistance and cardiovascular risk facotrs in presurgical morbid obesity patients. Horm Metab Res. 2007;11:10892–10896. [Google Scholar]
- 190.Baier LJ, Bogardus C, Sacchettini JC. A polymorphism in the human intestinal fatty acid binding protein alters fatty acid transport across Caco-2 cells. J Biol Chem. 1996;271:10892–10896. doi: 10.1074/jbc.271.18.10892. [DOI] [PubMed] [Google Scholar]
- 191.Tataranni PA, Baier LJ, Paolisso G, Howard BV, Ravussin E. Role of lipids in development of noninsulin-dependent diabetes mellitus: Lessons learned from Pima Indians. Lipids. 1996;31:S267–S270. doi: 10.1007/BF02637088. [DOI] [PubMed] [Google Scholar]
- 192.Formanack ML, Baier LJ. Variation in the FABP2 promoter affects gene expression: implications for prior association studies. Diabetologia. 2004;47:349–351. doi: 10.1007/s00125-003-1289-z. [DOI] [PubMed] [Google Scholar]
- 193.Tahvanainen E, Molin M, Vainio S, Tiret L, Nicaud V, Farinaro E, Masana L, Ehnholm C. Intestinal fatty acid binding protein polymorphism at codon 54 is not associated with postprandial responses to fat and glucose tolerance tests in healthy young Europeans. Results from EARS II participants. Atherosclerosis. 2000;152:317–325. doi: 10.1016/s0021-9150(99)00488-8. [DOI] [PubMed] [Google Scholar]
- 194.Hotamisligl GS, Johnson RS, Distel RJ, Ellis RF, Papaioannou VE, Spiegelman BM. Uncoupling of obesity from insulin resistance through a targeted mutation in aP2, the adipocyte fatty acid binding protein. Science. 1996;274:1377–1379. doi: 10.1126/science.274.5291.1377. [DOI] [PubMed] [Google Scholar]
- 195.Vassileva G, Huwyler L, Poirer K, Agellon LB, Toth MJ. The intestinal fatty acid binding protein is not essential for dietary fat absorption in mice. FASEB J. 2000;14:2040–2046. doi: 10.1096/fj.99-0959com. [DOI] [PubMed] [Google Scholar]
- 196.Vergani L, Fanin M, Martinuzzi A, Galassi A, Appi A, Carrozzo R, Rosa M, Angelini C. Liver fatty acid-binding protein in two cases of human lipid storage. Mol Cell Biochem. 1990;98:225–230. doi: 10.1007/BF00231388. [DOI] [PubMed] [Google Scholar]
- 197.Newberry EP, Kennedy SM, Xie Y, Luo J, Stanley SE, Semenkovich CF, Crooke RM, Graham MJ, Davidson NO. Altered hepatic triglyceride content after partial hepatectomy without impaired liver regeneration in multiple muring genetic models. Hepatology. 2008;48:1097–1105. doi: 10.1002/hep.22473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Fex M, Lucas S, Winzell MS, Ahren B, Holm C, Mulder H. β-Cell lipases and insulin secretion. Diabetes. 2006;55:S24–S31. [Google Scholar]
- 199.Tansey JT, Sztalryd C, Gruia-Gray J, Roush DL, Zee JV, Gavrilova O, Reitman ML, Deng CX, Li C, Kimmel AR, Londos C. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc Natl Acad Sci USA. 2001;98:6494–6499. doi: 10.1073/pnas.101042998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Martinez-Botas J, Anderson JB, Tessier D, Lapillonne A, Chang BHJ, Quast MJ, Gorenstein D, Chen KH, Chan L. Absence of perilipin results in leanness and reverses obesity in Leprdb/db mice. Nature Genetics. 2000;26:474–479. doi: 10.1038/82630. [DOI] [PubMed] [Google Scholar]
- 201.Huang H, Ball JA, Billheimer JT, Schroeder F. The sterol carrier protein-2 amino terminus: a membrane interaction domain. Biochemistry. 1999;38:13231–13243. doi: 10.1021/bi990870x. [DOI] [PubMed] [Google Scholar]
- 202.Huang H, Ball JA, Billheimer JT, Schroeder F. Interaction of the N-terminus of sterol carrier protein-2 with membranes: role of membrane curvature. Biochem J. 1999;344:593–603. [PMC free article] [PubMed] [Google Scholar]
- 203.Huang H, Gallegos A, Zhou M, Ball JM, Schroeder F. Role of sterol carrier protein-2 N-terminal membrane binding domain in sterol transfer. Biochemistry. 2002;41:12149–12162. doi: 10.1021/bi0260536. [DOI] [PubMed] [Google Scholar]
- 204.Gallegos AM, Atshaves BP, Storey SM, Starodub O, Petrescu AD, Huang H, McIntosh A, Martin G, Chao H, Kier AB, Schroeder F. Gene structure, intracellular localization, and functional roles of sterol carrier protein-2. Prog Lipid Res. 2001;40:498–563. doi: 10.1016/s0163-7827(01)00015-7. [DOI] [PubMed] [Google Scholar]
- 205.Stolowich NJ, Petrescu AD, Huang H, Martin G, Scott AI, Schroeder F. Sterol carrier protein-2: structure reveals function. Cell Mol Life Sci. 2002;59:193–212. doi: 10.1007/s00018-002-8416-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Seedorf U, Ellinghaus P, Nofer JR. Sterol carrier protein-2. Biochim Biophys Acta. 2000;1486:45–54. doi: 10.1016/s1388-1981(00)00047-0. [DOI] [PubMed] [Google Scholar]
- 207.Knudsen J. Acyl-CoA-binding and transport, an alternative function for diazepam binding inhibitor (DBI), which is identical with acyl- CoA-binding protein. Neuropharmacology. 1991;30:1405–1410. doi: 10.1016/s0028-3908(11)80009-2. [DOI] [PubMed] [Google Scholar]
- 208.Rouet-Smih F, Tonon MC, Pelletier G, Vaudry H. Characterization of endozepine-related peptides in the central nervous system and in peripheral tissues of the rat. Peptides. 1992;13:1219–1225. doi: 10.1016/0196-9781(92)90032-x. [DOI] [PubMed] [Google Scholar]
- 209.Slobodyansky E, Antkiewicz-Michaluk L, Martin B. Purification of a novel DBI processing product, DBI39-75, and characterization of its binding site in rat brain. Regul Pept. 1994;50:29–35. doi: 10.1016/0167-0115(94)90188-0. [DOI] [PubMed] [Google Scholar]
- 210.Papadopoulos V, Brown AS. Role of the peripheral-type benzodiazepine receptor and the polypeptide diazepam binding inhibitor in steroidogenesis. J Steroid Biochem Mol Biol. 1995;53:103–110. doi: 10.1016/0960-0760(95)00027-w. [DOI] [PubMed] [Google Scholar]
- 211.Papadopoulos V, Amri H, Li H, Boujrad N, Vidic B, Garnier M. Peripheral benzodiazepine receptor in cholesterol transport and steroidogenesis. J Biol Chem. 1997;272:32129–32135. doi: 10.1074/jbc.272.51.32129. [DOI] [PubMed] [Google Scholar]
- 212.Borboni P, Magnaterra R, Porzio O, Fusco A, Sesti G, Bertoli A, Lauro R, Marlier LNJL. DBI mRNA is expressed in endocrine pancreas and its post-translational product DBI33-50 inhibits insulin release. Endocrine J UK. 1995;3:267–271. doi: 10.1007/BF03021404. [DOI] [PubMed] [Google Scholar]
- 213.Sjoholm A, Funakoshi A, Efendic S, Ostenson CG, Hellerstrom C. Long term inhibitory effects of pancreastatin and diazepam binding inhibitor on pancreatic beta-cell deoxyribonucleic acid replication, polyamine content, and insulin secretion. Endocrinology. 1991;128:3277–3282. doi: 10.1210/endo-128-6-3277. [DOI] [PubMed] [Google Scholar]
- 214.De Stefanis P, Impagnatiello F, Berkovich A, Guidotti A. Inhibitory effect of ODN, a naturally occurring processing product of diazepam binding inhibitor, on secretagogues-induced insulin secretion. Regul Pept. 1995;56:153–165. doi: 10.1016/0167-0115(95)00002-s. [DOI] [PubMed] [Google Scholar]
- 215.Marquardt H, Todaro GJ, Shoyab M. Complete amino acid sequences of bovine and human endozepines. Homology with rat diazepam binding inhibitor. J Biol Chem. 1986;261:9727–9731. [PubMed] [Google Scholar]
- 216.Liu HS, Jan MS, Chou CK, Chen PH, Ke NJ. Is the green fluroescent protein toxic to the living cells? Biochem Biophy Res Comm. 1999;260:712–717. doi: 10.1006/bbrc.1999.0954. [DOI] [PubMed] [Google Scholar]
- 217.Lamhonwah AM, Tein I. GFP-human high-affinity carnitine transporter OCTN2 protein: subcellular localization and functional restoration of carnitine uptake in mutant cell lines with carnitine transporter defect. Biochem Biophy Res Comm. 1999;264:909–914. doi: 10.1006/bbrc.1999.1560. [DOI] [PubMed] [Google Scholar]
- 218.Baens M, Noels H, Broeckx V, Hagens S, Fevery S, Billiau AD, Vankelecom H, Marynen P. The dark side of EGFP: defective polyubiquitination. PLoS ONE. 2006;1:e54. doi: 10.1371/journal.pone.0000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Huang WY, Aramburu J, Douglas PS, Izumo S. Transgenic expression of green fluroescence protein can cause dilated cardiomyopathy. Nature Medicine. 2000;6:582–583. doi: 10.1038/74914. [DOI] [PubMed] [Google Scholar]
- 220.Devgan V, Rao MRS, Seshagiri PB. Impact of embryonic expression of enhanced green fluorescent protein on early mouse development. Biochem Biophy Res Comm. 2004;313:1030–1036. doi: 10.1016/j.bbrc.2003.11.184. [DOI] [PubMed] [Google Scholar]
- 221.Bogdanov AM, Mishin AS, Yampolsky IV, Belousov VV, Chudakov DM, Subach FV, Verkhusha VV, Lukyanov S, Lukyonov KA. Green fluorescent proteins are light-induced electron donors. Nature Chemical Biology. 2009 doi: 10.1038/nchembio.174. Published online April 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Silva AJ, Simpson EM, Takahashi JS, Lipp HP, Nakanishi S, Wehner JM, Giese KP, Tully T, Abel T, Chapman PF, Fox K, Grant S, Itohara S, Lathe R, Mayford M, McNamara JO, Morris RJ, Piccioto M, roder J, Shin HS, Slesinger PA, Storm DR, Stryker MP, Tonegawa S, Wang Y, Wolfer DP. Mutant mice and neuroscience: recommendations concerning genetic background. Neuraon. 1997;19:755–759. doi: 10.1016/s0896-6273(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 223.Kersten S, Seydoux J, Peters JM, Gonzalex FJ, Desvergne B, Wahli W. PPARalpha mediates the adaptive response to fasting. J Clin Inv. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Leone TC, Weinheimer CJ, Kelly DP. A critical role of for the peroxisome proliferator activated receptor alpha (PPARalpha) in the cellular fasting response: The PPARalpha-null mouse as a model for fatty acid oxidation disorders. Proc Natl Acad Sci USA. 1999;96:7473–7478. doi: 10.1073/pnas.96.13.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Hashimoto T, Cook WS, Qi C, Yeldandi AV, Reddy JK, Rao MS. Defect in peroxisome proliferator activated receptor alpha-inducible fatty acid oxidation determines the severity of hepatic steatosis in response to fasting. J Biol Chem. 2000;275:28918–28928. doi: 10.1074/jbc.M910350199. [DOI] [PubMed] [Google Scholar]
- 226.Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Tumbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host and Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Nakanishi Y, Murashima K, Ohara H, Suzuki T, Hayashi H, Sakamoto M, Fukasawa T, Kubota H, Hosono A, Kono T, Kaminogawa S, Benno Y. Increase in terminal restriction fragments of Bacteriodes-derived 16S rRNA genes after administration of short-chain fructooligosaccharides. Applied and Environmental Microbiology. 2006;72:6271–6276. doi: 10.1128/AEM.00477-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Human gut microbes associated with obesity. Nature. 2006;44:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 230.Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, Cantarel BL, Coutinho PM, Henrissat B, Crock LW, Russell A, Verberkmoes NC, Hettich RL, Gordon JI. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci USA. 2009;106:5859–5864. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Science Translational Medicine. 2009;1:10. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]


