Abstract
The hERG1 gene (Kv11.1) encodes a voltage-gated potassium channel. Mutations in this gene, lead to one form of the Long QT Syndrome in humans (LQTS). Promiscuous binding of drugs to hERG1 is known to alter the structure/function of the channel leading to an acquired form of the LQTS. Expectably, creation and validation of reliable 3D model of the channel has been a key target in molecular cardiology and pharmacology for the last decade. While many models were built, they all were limited to pore domain. In this work, a full model of the hERG1 channel is developed which includes all trans-membrane segments. We tested a template-driven de-novo design with ROSETTA-membrane modeling using side-chain placements optimized by subsequent molecular dynamics (MD) simulations. While backbone templates for the homology modeled parts of the pore and voltage sensors were based on the available structures of KvAP, Kv1.2 and Kv1.2–Kv2.1 chimera channels, the missing parts are modeled de-novo. The impact of several alignments on the structure of the S4 helix in the voltage-sensing domain was also tested. Herein, final models are evaluated for consistency to the reported structural elements discovered mainly on the basis of mutagenesis and electrophysiology. These structural elements include: salt bridges and close contacts in the voltage-sensor domain; and the topology of the extracellular S5-pore linker compared to that established by toxin foot-printing and NMR studies. Implications of the refined hERG1 model to binding of blockers and channels activators (potent new ligands for channel activations) are discussed.
Keywords: hERG1 channel, de-novo design, protein-drug interactions, mechanism
INTRODUCTION
hERG1 is a member of the ether a-go-go (EAG) family of genes that encode voltage-gated potassium channels1–3. More broadly, hERG1 is a member of the cyclic nucleotide binding domain (CNBD) family of channels, which includes hyperpolarization-activated channels, cyclic nucleotide gated channels, as well as numerous plant and bacterial channels. Mutations in hERG channels lead to the Long QT2 Syndrome (LQTS), associated with risk for sudden cardiac death3–5. The majority of hERG mutations causing LQTS are located in the trans-membrane (TM) domains. Most missense mutations produce a “loss-of-function” phenotype because of trafficking defects6,7. Some mutations are thought to affect physiological functions of the voltage-sensing domain (VSD) (S1 to S4), while other mutations located in the pore-domain (PD) (S5-pore-S6) can alter inactivation or permeation properties4,5,8–11.
Another dimension of hERG channels structure-function relates to drug interactions that either block or activate the channel12–14. Promiscuous binding of drugs to a molecular pocket in the distal S6 blocks conductance of the channel and leads to an acquired form of the LQTS14,15. Promiscuous binding represents a substantial challenge for rational drug design. The hERG potassium channels have been coined the “drug-killer” protein in pharmacological research.
Expectably, creation and validation of reliable 3D model of the channel has been a key target in molecular cardiology for the last decade16. While many homology models of hERG channels have been reported 16–21, most of them were designed to study drug binding to the central cavity. It was therefore adequate to build the models on templates from bacterial 2 TM channels such as KcsA or MthK, where the voltage-sensor is absent and the S5-pore helix linker is short compared to hERG1. A partial exception to this is the recent study by Tseng et al. which included the hERG1 S5-pore helix linker but not the S1–S4 TM helices of the VSD 21. Their study provided initial glimpses on how the S5-pore helix linker modifies hERG1 pore domain structure.
Further reasons to immediately consider refinements of the hERG1 S1–S6 TM domains and the extracellular components are of a great pragmatic nature. Recently a new anti-arrhythmic strategy is being developed; the creation of pharmacologic channel openers or activators that modify hERG1 function by targeting extra-cellular domains14. The targets for these drugs are thought to be in the VSD, but ambiguity in the location of these sites is yet to be resolved.
We have previously developed an integrated approach for modeling of the hERG1 PD structure that combines de novo partial folding with ROSETTA-Membrane and MD simulations17. In silica modeling of the hERG1 PD identified previously unknown structural features, including an extensive network of interactions between the S5 and the pore helix17. This prediction was confirmed experimentally, providing a basis to establish validity of the combined ROSETTA/MD approach to the refinement of close contacts in the membrane domain of hERG1.
A full model of the hERG1 channel, including both the VSD and the PD offer an indispensable template for rational drug design for openers that presumably target the extra-cellular surface of the channel and for identification of inadvertent blockers that bind to the inner cavity, as the cavity structure is dependent on the orientation of the S4–S5 linker. Our goal with present studies is an extension of the integrated approach to refinement of an S1–S6 model of hERG1. A major challenge in structural modeling of hERG1 trans-membrane (TM) segments is a proper alignment of the S4 TM helix, which contains positively charged amino-acid residues critical for voltage gating. The periodicity of charged and non-polar residues in the sequence makes it difficult to distinguish the relative quality of one alignment from the next. In order to circumvent this problem we have created several alignments for the S4 helix (Fig S1) and built a plethora of models to test them with ROSETTA modeling and molecular dynamics (MD) refinement. The resulting model of hERG1 appears to be consistent with the published literature and also some unpublished data discussed below. We have also extended our studies with the refined model to mapping of binding sites for blockers and activator of the hERG1 channel.
METHODS
Model Building
A critical step in any homology modeling is careful sequence alignment. We have used an initial alignment produced by ClustalW algorithm22. The alignment of the S5 was then adjusted manually according to our previous publication17 and the S4 alignment was varied as described below. Examples of different alignments for S4 can be found in the supplementary material section (Fig. S1). The pore domain and S6 helixes are relatively straightforward to model as the sequence similarity is reasonably high (36–45 %). One challenge in the modeling of the pore domain of the hERG1 channel is to obtain a 3D structure of the elongated flexible S5-pore linker or turret. Different approaches to model flexible elements of membrane proteins based on low-resolution data have been proposed in the past23. We used the ROSETTA-Membrane program to construct a secondary structure model of the S5P linker that preserves the global folding of the PD.24
ROSETTA-Membrane De Novo Modeling
The ROSETTA de novo prediction method has been adapted recently to study membrane protein structures24,25 and is commonly known now as ROSETTA-Membrane. The method is based on the assumption that the native state of a protein is at its global free energy minimum. A global search of the conformational space for a protein tertiary structures is carried out to select conformations that are especially low in free energy for a given amino acid sequence. Two key components of the method include: a procedure to efficiently carry out conformational searches; and a free energy function to evaluate the various conformations under differing environmental constraints (membrane associated vs. extracellular).
Conserved elements such as TM helices, backbone/side-chain fragments homologous to the template (Kv1.2 and Kv1.2-Kv2.1 Chimera26–28) were subjected to the refinement with MD simulations in presence of harmonic constraints acting on the backbone heavy atoms. Less conserved elements such as the elongated and flexible turret (S5-Pore linker), and other extra-cellular and intra-cellular loops were modeled de-novo with Rosetta-Membrane. For TM or membrane associated protein elements, a membrane-specific energy function was applied, which favors burial of small hydrophilic residues and exposure of large hydrophobic residues within the membrane hydrophobic core. In contrast, for the extracellular linker elements exposure of the hydrophobic residues to the polar environment is minimized by burial. Approximately 15000 low-resolution models were generated and clustered as described previously24,29,30. The models representing the central structure of the largest clusters were chosen as the best initial guess for the structure containing VSD, S5-Pore linker and PD. This resulted in six different cluster representatives. Different alignments for S4-helix were considered separately using a similar protocol. These protein structures were then subjected to refinement with MD simulations for optimization of side-chain packing.
Refinement of side-chain packing for ROSETTA-membrane models with MD simulations
Molecular simulation enables partial refinement of the low-resolution models created by ROSETTA. The key difference between representatives of six clusters is in the packing of the elongated S5-Pore linker (data not shown). For example, three cluster representatives show no well-defined secondary structure elements in the turret in apparent disagreement with biochemical and structural studies. The membrane-inserted elements of the structure were conserved amongst all clusters.
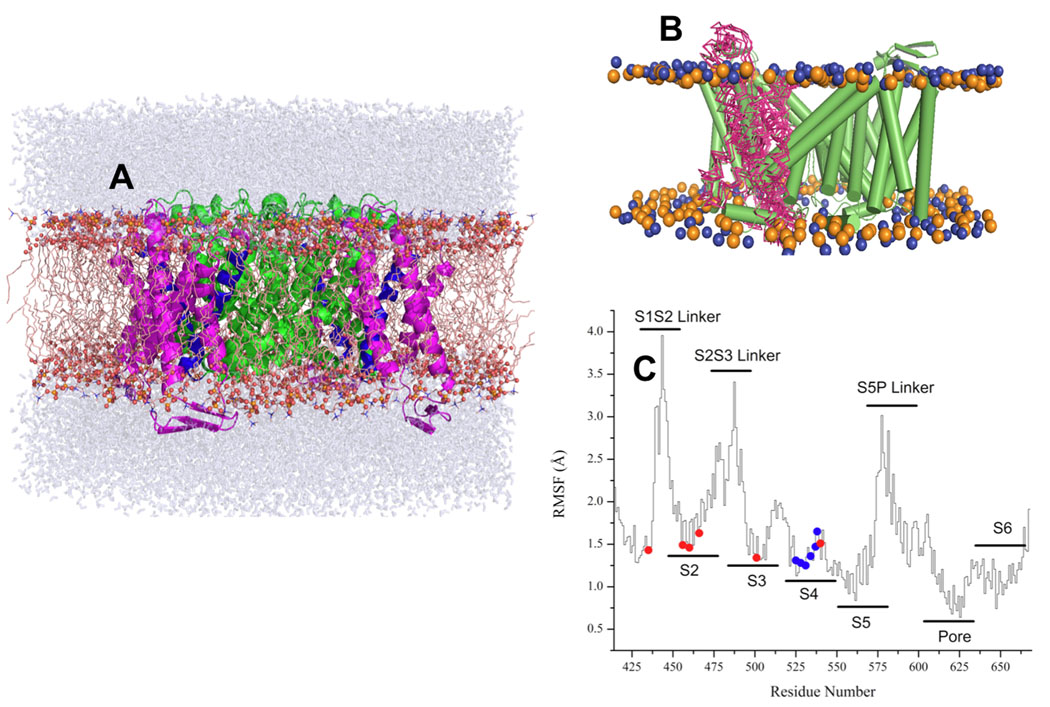
The most stable orientation and packing of the turret with two short ampipathic helices have been determined in our previous work on pore-domain.17 In good agreement with simulations done on the PD alone, only one packing of the turret produces stable MD simulations. An all-atom MD simulation was carried out on the six models representing the different clusters found by ROSETTA modeling with each of the different S4 alignments. S0:S2:S4 ion occupancy of the selectivity filter was used. This occupancy of the selectivity filter is known to be stable in numerous MD simulations performed on other potassium channels with known crystal structure31. The simulation box contained 1 hERG1 channel, 3 bound potassium, 28–30 water molecules in the intra-cellular cavity, and a 232 palmitoyl-oleoyl phosphatidyl choline (POPC) molecules, solvated by a neutralizing 100 mM KCl aqueous salt solution. The simulation system comprising a total of about ~100000 atoms is shown in Fig 1A. The simulation box containing protein, lipids and explicit water was constructed for each model with different S4 alignment (referred in the text as herg+0, herg+3, herg+6 and herg+9). All structures were relaxed and equilibrated with gradually decreasing harmonic constraints for over 2 ns and then were subjected to an 8 ns production run. All computations were carried out using the CHARMM program version c35b1. The simulation methodology is similar to that used previously to study ion selectivity in the KcsA K+ channel 31. Briefly, all simulations were carried at constant pressure (1 atm) and constant temperature (315 K) using the periodic boundary conditions of NPT ensemble. Electrostatic interactions were treated using a particle mesh Ewald (PME) algorithm similar to that of previous MD simulations of K-channels.
Figure 1. MD simulation setup and RMSD Analysis.
(A) The structure of ROSETTA-TM herg+3 model after 10 ns equilibration. S5–S6 domains are shown in a green color, S4 is blue and S1–S3 is in magenta (B) the lateral fluctuation of positions for S1–S2 segment along the MD trajectory for ROSETTA-Membrane herg+3 model. (C) RMSD scans along MD trajectory for equilibrated ROSETTA-Membrane herg+3 model. The RMS fluctuations (RMSF) for the pore domain and helical elements are around 2 Å, whereas most flexible elements (EC and IC linkers) display larger fluctuations of up to 4 Å from average structure.
Molecular Docking Studies
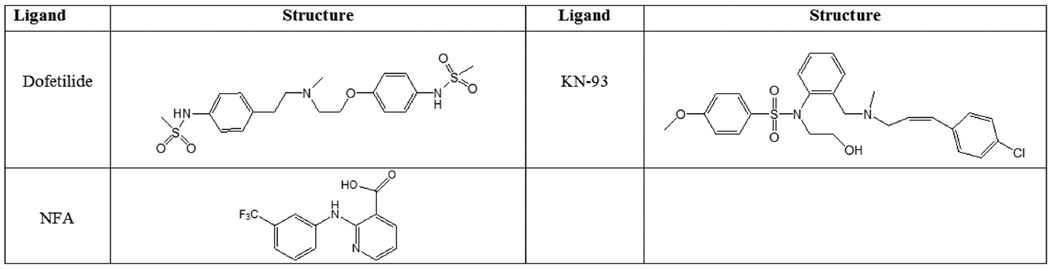
The structures of known hERG blockers and activators (dofetilide, KN-93 and NFA) were initially optimized using Gaussian-0332 with density functional theory (DFT) and the following basis set: B3LYP/6–31G* (Fig. 2). This approach is known to reproduce geometry efficiently. The derived coordinates of drugs were docked to the channel using the AUTODOCK (v. 4.0)33 and GOLD (v. 4.1.1) docking softwares34. The GOLD docking program allows partial side chain flexibility of receptor residues and may help to improve the predictive power of the docking procedures. In this algorithm, each flexible side chain is allowed to undergo torsional rotation around one or more of its acyclic bonds during docking. The rotamers flexed by 10° increments to cover a full 360° rotation. In order to avoid the docking poses at the receptor-membrane interface this region was filled with dummy atoms aimed to represent lipid-occupied volume. Initially the ligands were docked blindly to the entire protein. Cluster analysis of docking poses at a 2.0 Å tolerance failed to achieve distinct clustering for both docking programs. However, at an 8–10 Å tolerance, the clusters spatially overlapped particularly within the confined space of the cavity. In order to focus the binding poses, the channel was mapped using a cross-section of one of four VSDs together with the PD region (Table S1 and Figure S2). This approach achieved distinct clustering of ligand binding. Parameters used for docking are collected in Table S1 of Supplementary Materials.
Figure 2. Chemical Structures of the ligands used for molecular docking studies.
Evaluation of constructed hERG1 model and elucidation of possible binding sites have been performed by molecular docking simulations. Dofetilide and KN-93 were used as representatives of hERG1 blockers and NFA was used as an activator.
RESULTS AND DISCUSSION
We used MD simulations to ensure proper packing of side-chains and to eliminate unstable ROSETTA models. The algorithm of ROSETTA modeling is aimed at the identification of local or global minima for the entire structure. The MD simulations however led to discrimination of several models based on the broken helicality in voltage-sensor bundle. This is a reasonable assumption. While it was reported that S4-helix of VSD is very flexible and can undergo conformational transitions between alpha-helical to 310 forms when opened by strong depolarizing potentials35–38, helical elements in all of the known VSD are essentially super-imposable. The average root mean square fluctuations (RMSF) of the channel backbone heavy atoms relative to the initial structure were used to judge the stability of the models (Figures 1B and 1C). The RMS fluctuations per-residue are shown for the most stable model (herg+3) in Figure 1 (Figures 1B and 1C). The RMSF in positions of the Cα-atoms forming PD are less than 2.3 Å and the selectivity filter is very stable throughout the simulations, with RMSF of ~1.5–2.0 Å. The magnitude of the fluctuations of the heavy backbone atoms in the S5P linker is relatively large, on the order of 3–3.5 Å. The most flexible elements (EC and IC linkers) display larger fluctuations of up to 4 Å from average structure. The residues forming voltage sensor display RMSF at the order of 2.8–3.2 Å. This range of fluctuations for the mobile element of the structure is not surprising. Jogini and Roux39 reported similar numbers for the dynamics of the flexible parts of voltage-sensor domain in the Kv1.2 channel with known crystal structure, emphasizing importance of lateral movements in voltage sensor relative to the membrane plane (shown in Figure 1B for hERG1). These results on RMSF are also in accord with previous simulations done on Kv1.2 and KvAP channels39–41. The initial ROSETTA modeling for the missing elements of structure and subsequent evaluation of protein dynamics was done in the absence of experimentally derived constraints and thus may be considered a blind experiment. Information about the structure-function of hERG1 channels is abundant in the literature and herein we compare the main features of this simulated model to data available from existent experimental studies.
Critical Analysis of Different S4 alignments
The sequence alignment for the S4 of hERG1 to crystallized potassium channels is ambiguous. The S4 domain of hERG1, similar to other voltage-gated K-channels, contains a periodicity of positively charged residues separated by 2 hydrophobic residues starting with K525. Based on the gating charge of hERG1 side-chains 42, we estimate that the membrane-inserted portion of the S4 helix contains around 4–5 charged side-chains. There is no consensus on the exact number of amino acids comprising its S4. Therefore, we produced and evaluated 4 models for the S4 helix based on 3 residue shifts of the alignment of the periodic charged group elements relative to similar sequences in potassium channels that have been crystallized (i.e. in Fig. S1 these alignments have been represented for herg+0 and herg+3 alignments). There is no significant sequence similarity in the S3–S4 and S4–S5 linkers. Therefore these structures were modeled de novo first and then subjected to short MD simulations up to 2 ns in the explicit lipid bilayers. Subsequent MD simulations for 5–10 ns resulted in discarding of the alignments based on the +6 and +9 sequence displacements. Their S4 helices were severely distorted leading to destabilization of the overall voltage-sensor. Only two models, with shifts of 0 and +3 residues, resulted in stable structures with preserved helicity of the S4 domain. The close contacts and interactions for these models are summarized in Table 1 as averaged from last 5 ns of MD simulations.
Table 1.
Intra and Inter Subunits Salt-Bridge and Hydrogen-Bonding Statistics* as per residue for S4 voltage sensor.
| N1res | N1res | Overall Lifetime (ps) | Inter vs. Intra** |
|---|---|---|---|
| K525 | E435 | 130.4 | intra |
| I521 | 70.8 | intra | |
| R528 | 5.4 | intra | |
| L529 | 29.2 | intra | |
| E575 | 10.0 | inter | |
| Q576 | 57.5 | inter | |
| R528 | D456 | 95.6 | intra |
| D460 | 200.3 | intra | |
| L524 | 82.2 | intra | |
| K525 | 5.4 | intra | |
| R531 | 34.7 | intra | |
| R531 | S428 | 328.3 | intra |
| D460 | 121.8 | intra | |
| L529 | 5.6 | intra | |
| R534 | 19.4 | intra | |
| V535 | 5.4 | intra | |
| R534 | D466 | 178.1 | intra |
| W497 | 5.0 | intra | |
| D501 | 706.4 | intra | |
| A504 | 10.0 | intra | |
| R531 | 19.4 | intra | |
| R537 | 6.0 | intra | |
| R538 | 9.4 | intra | |
| R537 | D466 | 456.7 | intra |
| N470 | 21.3 | intra | |
| K495 | 5.0 | intra | |
| D501 | 114.5 | intra | |
| V533 | 20.7 | intra | |
| R534 | 6.9 | intra | |
| V535 | 5.0 | intra | |
| L539 | 6.7 | intra | |
| D540 | 12.8 | intra | |
| R538 | V535 | 9.4 | intra |
| L539 | 11.8 | intra | |
| D540 | 5.4 | intra |
The production run of 3000 ps have been used to average interactions between amino-acid residues every 5 ps.
Interaction within same chain is marked as “intra” and between two different chains “inter”
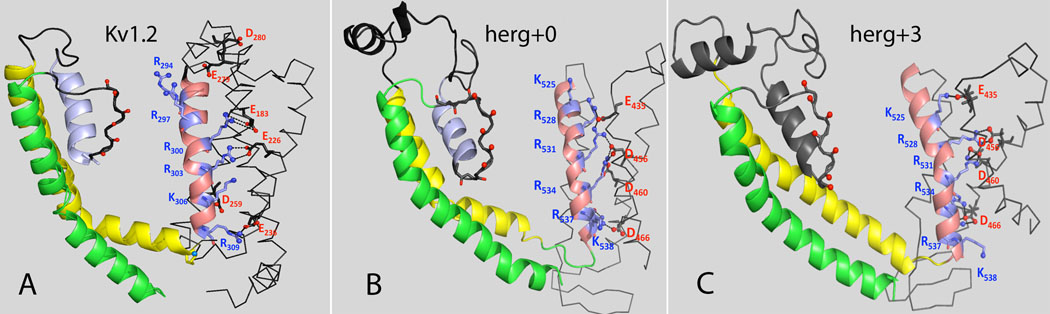
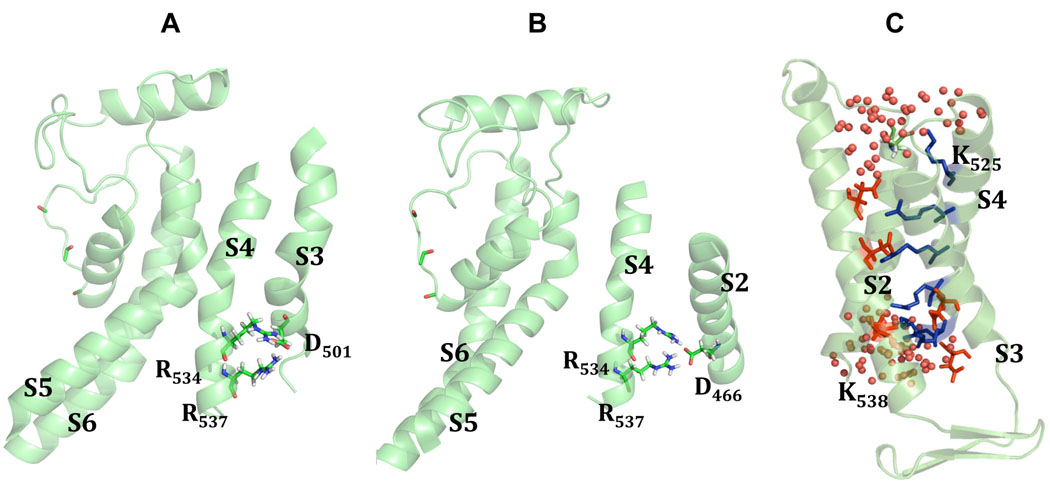
The impact of the 0 versus +3 alignments (Fig. S1) on the orientation of positively charged residues in the S4 domain is notable (Figs. 3 and 4). In simulations, a +3 shift in the alignment led to a tighter packing of the hydrophobic residues at the tips of the S4 and S5 helices improving stability of the whole VSD. The initial ROSETTA-built herg+0 and herg+3 models had a inter-subunit salt bridge forming between the charged side-chains of K525 on the top portion of the S4 helix and E575 near the tip of S5 helix of the other hERG1 subunit. Experimentally, after K525 is mutated to C, the channel can be further modified by MTSET leading to slowing of channel deactivation42,43. This finding suggests that K525 is located close to the water/membrane interface in agreement with its initial location in both herg+0 and herg+3 models.
Figure 3. Close contacts in the VSDs for different models of hERG1 and Kv1.2.
The orientations and packing of positively charged residues in S4 domain for (A) Open State of the Kv1.2 channel26,30, (B) MD-refined herg+0 model and (C) MD refined herg+3 model. The following color-code for TM-hellices was used: S4 – salmon-red, S5 – yellow, S6 – green, P-helix – steel blue, S5-Pore linker – dark grey. Interacting residues in the VSD are shown are balls and sticks. Salt-bridges between S4 and S1/S2 are show as dotted lines.
Figure 4. Tentative Cd2+ binding site in hERG1 models.
(A) herg+0 and (B) herg+3
In order to further discriminate between the herg+0 and the herg+3 models, we compared predicted charged interactions to the published literature. Divalent cations such as cadmium are known to modulate hERG gating by targeting sites in the S2 and S3 helices45. The binding site for Cd2+ is thought to be formed by D509 in S3, D456 and D460 in S245. Fernandez et al. 45 showed that single mutations of residues D456, D460 or D509 reduced the Cd2+-induced change in Vact from Δ+36 mV to +2.5 mV, but did not eliminated Cd2+ induced changes on inactivation. At the same time combined mutations of two or three of these key aspartic acid residues nearly eliminated the Cd2+-induced shifts in activation.
The proposed location of the binding site is in line with orientation of these side-chains from ROSETTA-Membrane modeling. In the herg+3 model, the carboxylate groups of D460 and D509 nicely frame tentative Cd2+ site. In the herg+0 model, D456 is facing in the same direction as D460 (Figs. 4A and Fig. 4B) and their side-chains are salt-bridged to the positively charged R534 (Figs. 4A and Fig. 4B) and thus would not be oriented correctly to bind with Cd2+. Furthermore, interactions presented in model built on herg+0 alignment are not supported by the study of Zhang et al.43. Contrary to herg+0 alignment, in the herg+3 model D509 turns away from the center of the voltage sensor domain and both D460 and D456 side-chains form stable interactions with R528 and R531 (Fig. 4B). The same experimental study 43 confirms direct interaction between D456 and R528 found only in herg+3 alignment (Fig. 4B).
The herg+3 model predicts a salt bridge between D501 and R534. We have found that mutation of D501 to K eliminated hERG1 expression (n=13) (unpublished). However, double mutation D501K/R534E did express hERG1 activity albeit with altered voltage-dependence of activation (n=14). The single mutation R534E expressed hERG1 activity with near normal characteristics (unpublished). The ability to partially rescue function is not a clear indication of direct interactions, but these data suggest that the salt-bridge found between D501 and R534 in the herg+3 model might be correct.
Close Contacts in S1–S4 Helix Bundle
To further clarify the bonding network for charged residues within the S4 helix, the interaction energy analysis was performed for the herg+3 model. The results in Table 1 summarize statistics for the most stable salt bridges and hydrogen bonds through the trajectory. The results of this monitoring for the interactions energies of K525, R528, R531, R534 and R537 along the trajectory are collected in Table 2. The initially built ROSETTA-Membrane model for herg+3 predicts salt bridging of K525 in the S4 with E575 near the top of the S5 helix. Remaining positively charged R528, R531, R534 and R537 residues are oriented toward negatively charged residues in the S2 and S3 domains forming the network of interactions that stabilizes the voltage-sensor.
Table 2.
Interaction energies between positively charged residues in the voltage-sensing domain and remaining residues in the monomer of hERG1 channel.
| N1res | N2res | Eint (kcal/mol) PEPA |
Eint (kcal/mol) PEPB |
Eint (kcal/mol) PEPC |
Eint (kcal/mol) PEPD |
|---|---|---|---|---|---|
| K525 | E435 | −67.5 | −20.6 | −22.35 | −63.01 |
| I521 | −6.28 | −6.06 | −5.9 | −6.08 | |
| R528 | D456 | −62 | −44 | −51.3 | −57.5 |
| D460 | −61 | −63 | −56.5 | −64.5 | |
| L529 | −6.9 | −7.1 | −7.1 | −7.5 | |
| R531 | 19.8 | 11.8 | 24.5 | 19.8 | |
| R531 | D456 | −18.3 | −14.6 | −59.1 | −45.1 |
| D460 | −55.9 | −61 | −61.3 | −69.3 | |
| A504 | −13.6 | −4.9 | 0 | 0 | |
| A505 | −11.2 | −5.1 | 0 | 0 | |
| R528 | 19.8 | 11.8 | 24.5 | 19.8 | |
| R534 | D466 | −46.2 | −23.9 | −48.5 | −56.5 |
| D501 | −51.1 | −44.9 | −46.7 | −47.3 | |
| R537 | 11.2 | 19.79 | 22.8 | 10.4 | |
| R537 | D466 | −13.3 | −11.2 | −15.9 | −45.6 |
| D501 | −52.8 | −39.8 | −64.5 | −62.3 | |
| R534 | 11.2 | 19.7 | 22.8 | 10.1 | |
| D540 | −11 | 0 | −9.8 | −7.9 | |
| K538 | D466 | −5.9 | |||
| V535 | −5.5 | −6.7 | |||
| L539 | −9.1 | −7.59 | −9.2 | −11.3 |
The energy cutoff was set to be 5 kcal/mol. The production run of 10 ns have been used to average interactions between amino-acid residues every 5 ps.
Most salt-bridges presented in the starting structures are still preserved after MD simulations, with one notable exception, the inter-subunit K525 - E575 interaction. The data on salt-bridges and close contacts collected in the Figure 3, Tables 1 and 2 indicate that K525 from the S4 helix is capable of forming an intra-subunit salt bridge with E435 in the S1–S2 linker, breaking away from bridging to E575. To confirm/disapprove these salt bridges we have made K525D and K525E mutants and characterized their basic electrophysiological properties (unpublished). K525D and K525E mutants were constitutively open at a holding potential of −80 mV. However E575K, E575R, E435K and E435R mutants are nearly indistinguishable from the normal channel and none of the 8 double mutant combinations (K525 with E575 or E435) rescued the constitutively open phenotype of the K525 mutants, indicating minimal salt-bridging of K525 with either E575 or E435 (unpublished). These seemingly contradictory results can be better understood following results from MD simulations. These simulations predict an occurrence rate of 0.3 % and 30 % for salt bridges between K525 and E575 or E435, respectively (Table 1). Gea-Ny Tseng in a personal communication has revealed that K525 may interact with D456, indicating an orientation of K525 towards the S2 helix. Further refinements of our model will help to characterize this dynamic interface.
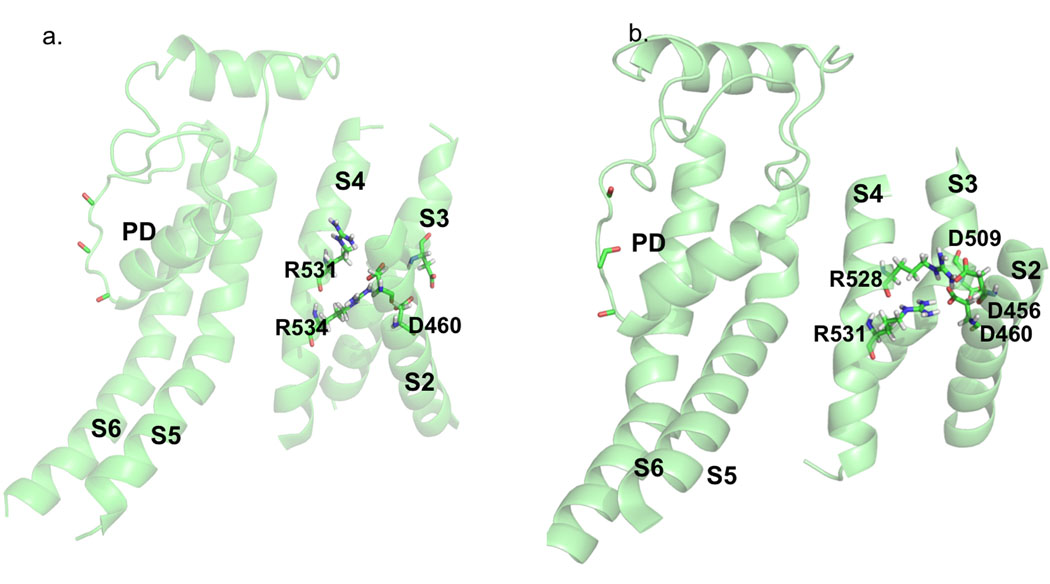
Our model (herg+3) indicates that the positively charged side chains of R528 and R531 interact with the negatively charged side chains of D460 and D456 in the S2 helix (Fig. 4B). The monitoring of time-series for interaction energies suggests a bifurcating character of salt-bridges, which are present for about 50 % of the time (Table 1). The analysis for R534/R537 indicates that these two residues may form bifurcating salt-bridges with D501 and D466 (Fig. 5A and 5B). The trajectory analysis for close-contact patterns of residues located at the intra-cellular tips of S4 (K537 and K538) suggest that they are very mobile (especially K538) and accessible to water penetration into the voltage sensor.
Figure 5. Critical Interactions in VS domain in herg+3 model.
The analysis of interaction energies for R534/R537 suggests that these two residues may form bifurcating salt-bridges with D501 (A) and D466 (B). Panel C shows water accessibility of the S4 helix averaged from MD simulations of the herg+3 model. Two lysines (K525 and K538) were found to be exposed to the lipid-water interface, while other four arginines (R528, R531, R534, R537) are buried inside of voltage-sensor bundle forming stable contacts to negatively charged residues from S1 and S2 TM helices.
The orientation of charged residues in relation to hydrophobic interior of the lipid bilayer has been hotly debated27,36,37. Tryptophan and glutamine scanning mutagenesis of hERG1 reported by Subbiah et al.46 led to the conclusion that that some of positively charged residues in voltage-sensor of hERGs might be exposed to the hydrophobic region in the lipid bilayer. In our model only two lysines K525 and K538 are exposed to the lipid-water interface (see Fig. 5C), while other four arginines are buried inside of voltage-sensor bundle (R528, R531, R534, R537) forming stable contacts to negatively charged side-chains from S1 and S2 TM helices with no direct exposure to the hydrophobic core.
To characterize the environment of S4 gating residues, the association of water molecules with nearby lipid head-groups as well as with these residues was monitored over the last 5 ns of MD simulations. Notably, the side chains of the two outermost residues, K525 and K538 become hydrated rapidly (see Fig. 5C). K525, R537 and K538 adopt a well-defined interfacial orientation. None of these charged residues penetrate into the core of lipid bilayer, rather they salt-bridge to negatively charged residues from S1 to S3. Water molecules rapidly (in 1–3 ns) diffuse from intracellular and extracellular milieu to occupy any cavities inside the S1–S4 helical bundle.
Implications of the current model to gating mechanism in hERGs
The voltage-sensor is a flexible element of the structure particularly during state transitions from closed (energetically favorable) to the open state (energetically unfavorable) on a microsecond time interval. The absence of applied voltage and the nano-second time scale of simulations in this work limit discussions on plausible gating mechanisms of hERG1. However, several important observations can be made based on the literature. Transformations between open and closed states in hERG channels are thought to be associated with the dynamics of the salt-bridges involving interactions of D456 sequentially with R528 and R53143. For herg+3 model the salt-bridge between R531 and D456 (Fig. 3C and Fig. 4B) are in a good accord with studies by Subbiah et al.46–48 They showed that mutation at the position 531 stabilizes a closed state of the channel. It was shown that R531Q mutation shifts voltage dependence of activation substantially to depolarized potentials. Experimentally it was shown that R531 becomes uncoupled from D411 and D466 or D509 upon inactivation gating49. It may be concluded that contacts between R531 and D456/D460 are functionally important and current model provides structural evidence for it. The fact that they are bifurcating bridges may explain their relative instability leading to transition between open and closed states. According to our interaction energy analysis (Table 2), R531 strongly interacts with D456 and D460 in the course of MD simulations and there is no notable interaction or contacts made to either D411 or D466 or D509, suggesting that the model may represent an inactivated state of the channel.
Structure of the S5-pore-Linker (Turret)
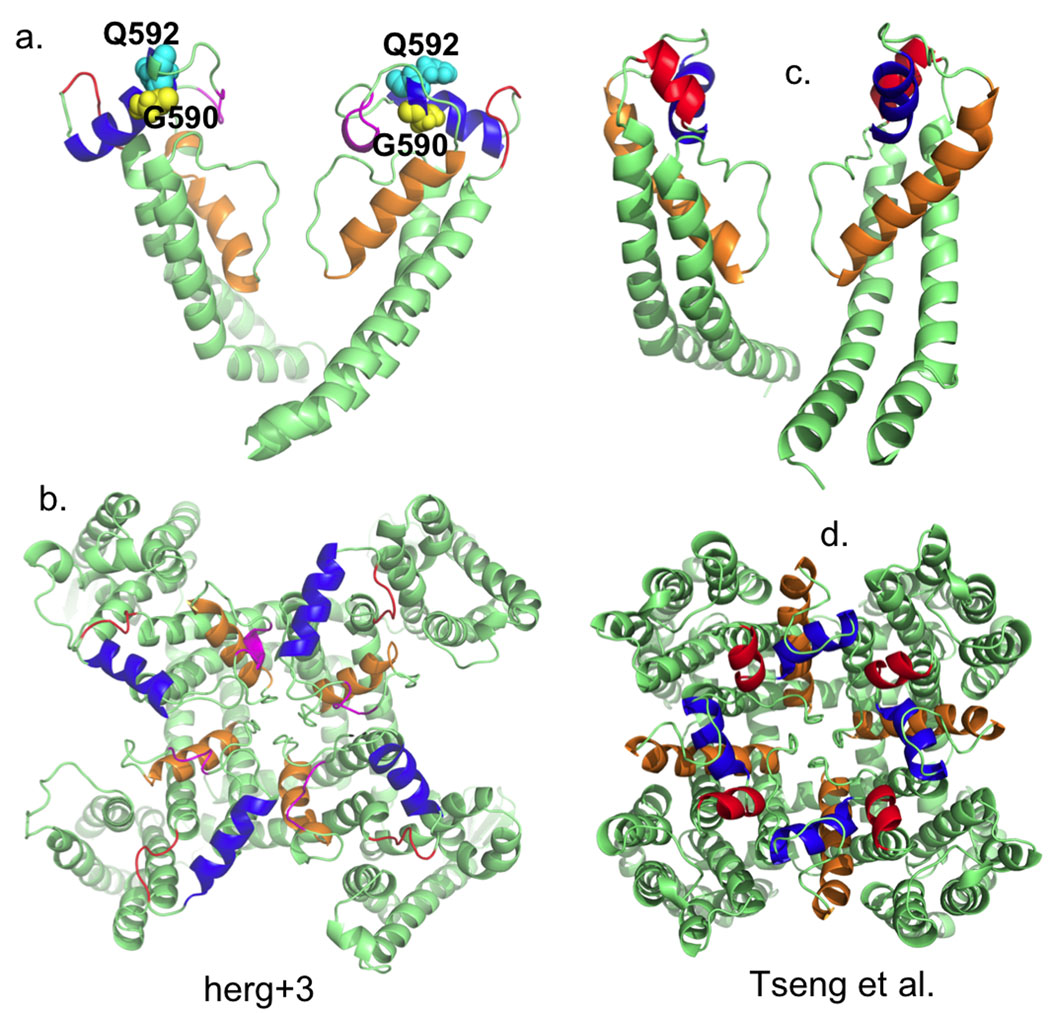
The vast majority of models for the pore domain of hERG1 exclude the S5-P loop fragment16. It is important to take into account this region of hERG1 due to its involvement in inactivation. Accordingly, we have modeled the extracellular turret. Our Rosetta-membrane model shares common features with that reported recently by Tseng at al. derived from experimental constraints21. The de-novo model of the turret has a helical segment, residues 582–593 (Fig. 6a) in agreement with NMR data50, showing two helical regions at W585-I593 and G604-Y61120. Another NMR study suggests presence of two helices at I583-G590 and D591-I593 region51. Our initial MD production model had a short helix (I571 - E575) just outside of S5. After further MD simulations some helical elements were unstable and two regions (in red color at Fig. 6a,b) with some helical curvature were located at S599-G603 and E575-M579. This is different from Tseng et al. model (also in red color Fig. 6c,d), although, their model also has a short helix as well (S5-P helix). However, the first helix in the model by Tseng et al. appears in the region P577-I583 and it is seven residues long. The second amphipathic α-helix (shown in blue in Fig. 6) is conserved for both our model and that of Tseng et al.21. The helix is formed by the residues W585-G594/R582-I593 in both models. The remaining structure of the pore domain is very similar for the ROSETTA-Membrane/MD modeling and experimentally driven model of Tseng et al. It displays all the key features of the hERG1 pore domain for example a 17 residues long pore helix and the SVGFG sequence constitutes the selectivity filter.
Figure 6. Outer mouth pore arrangement for two hERG models: Rosetta herg+3 model and by Tseng et al.
MD-refined model display a short helix (I571 - E575) just outside of S5.
One particularly interesting feature of all hERG1 models produced in this work is positioning of the G590 residue within a turret helix. This amino-acid residue is on the inner side of the helix in direct proximity to the selectivity filter (Fig. 6). The replacement of glycine in this position by cysteine leads to abolishment of the C-type inactivation in hERG1 and remove almost completely selectivity for K+ ions21. A similar mutation in a position 592 (Q/C) does not produce non-selective channel and retain C-type inactivation21. In our model, Q592 is pointing towards the outside water vestibule and in good agreement with experimental observations that it is unlikely to affect dynamics of the selectivity filter, but may be involved in inhibition of toxin binding. That is Q592C has been shown experimentally to be alter high-affinity toxin binding to hERG21.
The model of the pore domain for the hERG1 channel refined with similar approach was already published by our group17. The characteristic feature of the model is the presence of the hydrogen-bonding network between S5 helix and pore domain of hERG1. Modeling combined with electrophysiological recordings was used to show that selectivity and likely permeation will be altered severely by mutations at the position of H562 (S5) or matching hydrophilic side-chains from the pore helix. Independent experimental study reported in 2009 from the group of J. Vandenberg20 has reached similar conclusion about existence of functionally important network of interactions between the S5 and pore helices. Therefore, this leads to a conclusion that extended hydrogen-bonding between H residue in S5 and polar residues in the apex of pore helix is functionally important and compliments already existing network of interactions between S4 and S5.
In review, the herg+3 model is most consistent with the experimental data. Some of the salt-bridges (most notably K525 to E575) predicted by ROSETTA-membrane modeling were proven wrong by subsequent MD simulations, which agrees with experimental data. The shortcomings of ROSETTA-membrane modeling are understandable in this case. The K525 residue is located at the lipid-solvent interface with complex dynamics. An explicit treatment of its hydration may be necessary to evaluate the most stable arrangement for this residue.
Implications of Current Model to Rational Drug Design
A goal for our structural modeling is an attempt to identify plausible drug binding pockets for drugs known to interact with hERG. Results of docking studies done on the full hERG1 model are summarized below for hERG1 blockers and activators. We have focused our assessment of drug binding only to the herg+3 model. Rational drug design aims at avoiding the creation of new drugs that inadvertent bind to the intra-cavitary molecular pocket of hERG114. Although drug binding to hERG1 has been extensively studied for homology models of the pore domain11,13,52, no theoretical studies of drug targeting full hERG1 channel is reported to date.
In silico Docking Studies of hERG1 Blockers
In order to evaluate hERG1 model and elucidate possible binding sites, we have performed extensive docking of the dofetilide and KN-93 blockers (Fig. 2). The docking was done in the two steps. This step might be expected to help identify a large number of potential binding poses and preferable locations. This method was unsuccessful in detecting distinct clustering, likely related to the coarse grid spacing, which missed crucial interactions. Accordingly we focused attention on smaller regions with small grid spacing. The cross-sections of only one of four voltage sensor domains that included the selectivity filter and central cavity fields was mapped first (Figure S2).
The binding pockets for the highest-ranked clusters identified in docking were formed by L622, S649, T623, S624, Y652, A653, G626, F656, M645, G657 and V625 amino-acid residues for dofetilide and KN93 ligands (Supporting Material, Tables S3 and S4 and Figure S2). Analysis of the clustering for dofetilide and KN-93 indicated that these two hERG blockers would target primarily the central cavity (93% and 58%, respectively by AUTODOCK; and 94% and 86%, respectively by GOLD), (Supporting Material, Tables S3 and S4). It must be noted that comparison of docking pose populations for blockers at the outer mouth of the selectivity filter has shown that KN-93 has a higher number of docking poses compared to dofetilide (38%, and 5% respectively by AUTODOCK; and 14% and 6%, respectively by GOLD), (Supporting Material, Tables S3 and S4). Our theoretical analysis of drug-protein interaction energies suggests that the S/T rich region of the pore helix (T618 to S624) might be important for maintaining of the stable hydrogen bonding with the blockers. Aromatic residues from the S6 helix enable proper alignment of the ligands within the cavity and fast dehydration within the inner cavity region. Acting together polar residues of the pore helix and aromatic residues of the S6 align blockers within the binding pocket. This result is in good agreement with previous docking and free energy simulations studies done on Kv1.5 voltage-gated potassium channel53 and various hERG models13 as well with experimental studies13,53–57.
GOLD fitness scores for dofetilide and KN93 are found to be very similar to that of AutoDock. Both programs predict that KN-93 displays a slightly higher affinity to hERG1 than dofetilide (Supporting Material, Tables S3 and S4). This result is inconsistent with experimental binding affinity values of dofetilide (IC50=7–10 nM) and KN-93 (IC50=110 nM). More sophisticated and computationally extensive approaches to computations of substrate binding affinity, such as free energy simulations may help to overcome this problem18,53,58.
Docking Studies of hERG1 Activators
A new class of compounds with potential to eliminate hERG-related arrhythmias has been introduced recently. These compounds are classified as hERG1 channel agonist, sometimes being coined as hERG1 openers. One of the earliest identified hERG1 agonist is niflumic acid (NFA) (Fig. 2). Its potential for the drug development is circumvented by the promiscuous binding to a broad range of channels from Shaker family, structural principles underlying mechanism of action are of a great interest to future drug development. A turning point in the identification of potential targets for openers binding was a recent string of publications combining electrophysiology, binding assays and mutagenesis59–61.
The experimental studies suggest that many of hERG1 openers tend to bind to the EC S3–S4 or S5-pore linkers with high specificity and affinity impacting gating/inactivation properties of the channel59. It is yet unclear if any of these compounds exhibit other modes of binding. In this study we chose to assess docking of niflumic acid to both the pore and voltage sensor domains. In our simulations, the two different protonation states of niflumic acid (charged and neutral carboxylate group (COOH and COO−) were docked. Both neutral and anionic forms showed similar results. All of the top five low energy clusters for neutral and anionic forms mapped to the extracellular surface of the hERG1 channel. (Supporting Material, Tables S3 and S4 and Figure S3). Although AUTODOCK docking results for the neutral form of niflumic acid have shown slightly better binding score than the anionic form (by ~1 kcal/mol); crucial amino acids for binding are similar for these two forms (binding scores by GOLD for neutral and anionic forms are very close to each other (−6.41 kcal/mol and −6.37 kcal/mol, respectively), (Supporting Material, Tables S3 and S4). Several residues were found to be very important for niflumic acid binding to hERG1, most notably K525 and T436 (Fig. S3). The location of the binding site for NFA around EC tip of the S4 is in accord with experimental data from Sanguinetti lab59 placing this binding pocket at or near S3–S4 linker. Almost all of these residues are involved in the gating process of hERG1 channel and plays an important role in the packing of S1:S4 helix bundle. The theoretical results support the proposition that NFA targets extracellular surface or in the proximity of VS domain rather than targeting a common intra-cellular binding site found in all K-channels.
A cautious note on limitations of docking studies
Previous theoretical work53,62–63 on the membrane protein-drug interaction studies with free energy and MD simulations emphasized importance of the correct accounting for the conformational dynamics of the substrate in the bulk phase and the receptor site. The lack in conformational sampling for degrees of freedom in solute and receptor, oversimplified treatment of environmental effects and lack of the robust strategy for computations of the entropic components in affinities compromise accuracy of predicted binding53,62. There are also ambiguities related to the standard states of the drug62, usually solid phase – powder in experimental measurements for IC50 and gas-phase in many theoretical studies. Therefore exact binding affinities estimated from docking studies are hard to parallel with the one measured in experiment. Despite all of the limitations in molecular docking, the binding scores, while they are not exact representative of experimental measurements, may still help to establish an indispensable insight into potential organization of the binding pocket. Here, two independent docking algorithms were able to distinguish the drugs as hERG1 blockers and activators, identify the potential location of binding sites for blockers and activator distinguished successfully and these findings are in good agreement with experimental data.
CONCLUSIONS
Our goal was to develop a robust step-by-step algorithm for de novo modeling of a potassium channel with great medical importance. We were able to test several models based on different alignments and to refine the side-chain packing with short MD simulations. These were not refined on the basis of experimental data and thus this work should be considered a blinded experiment. The template-driven de novo Rosetta modeling combined with atomistic MD simulations led to a model of the full hERG1 structure capable of accommodating variety of sometime conflicting experimental information. The modeling has provided validation or structural explanation to a broad experimental data accumulated to date and particularly on the interaction within the VSD and the pore. Docking simulations were able to distinguish the potential binding pockets for blockers and activator. The location of the binding pocket for NFA (external sites predicted by experimental studies) at the tip of S4 helix provides insights into plausible mechanism of hERG1 activation.
Supplementary Material
ACKNOWLEDGEMENTS
We are gratefully acknowledged contribution of Dr. Gea-Ny Tseng (VCU) for helpful comments and discussions of her data on toxin-foot printing, refinement of the S5-pore linker and 3-D model of the pore domain. This work was supported by the NIMH Career Development Research Grant K01 MH67625 (to V.Y-Y.), Canadian Institutes of Health Research (MOP-186232) to SN and HJD, Heart and Stroke Foundation of Alberta (HJD). SN is CIHR New Investigator and an Alberta Heritage Foundation for Medical Research Scholar and HJD is an Alberta Heritage Foundation for Medical Research Medical Scientist. The computational support for this work was provided by the West-Grid Canada through resource allocation award to SN.
REFERENCES
- 1.Curran ME, Splawski I, Timothy KW, Vincent GM, Green ED, Keating MT. A Molecular-Basis for Cardiac-Arrhythmia - Herg Mutations Cause Long Qt Syndrome. Cell. 1995;80(5):795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 2.Tseng GN. I(Kr): the hERG channel. J Mol Cell Cardiol. 2001;33(5):835–849. doi: 10.1006/jmcc.2000.1317. [DOI] [PubMed] [Google Scholar]
- 3.Vandenberg JI, Walker BD, Campbell TJ. HERG K+ channels: friend and foe. TrendsPharmSci. 2001;22(5):240–246. doi: 10.1016/s0165-6147(00)01662-x. [DOI] [PubMed] [Google Scholar]
- 4.Splawski I, Shen JX, Timothy KW, Lehmann MH, Priori S, Robinson JL, Moss AJ, Schwartz PJ, Towbin JA, Vincent GM, Keating MT. Spectrum of mutations in long-QT syndrome genes KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation. 2000;102(10):1178–1185. doi: 10.1161/01.cir.102.10.1178. [DOI] [PubMed] [Google Scholar]
- 5.Witchel HJ, Hancox JC. Familial and acquired long QT syndrome and the cardiac rapid delayed rectifier potassium current. Clin Exp Pharm Physiol. 2000;27(10):753–766. doi: 10.1046/j.1440-1681.2000.03337.x. [DOI] [PubMed] [Google Scholar]
- 6.Saenen JB, Paulussen ADC, Jongbloed RJ, Marcelis CL, Gilissen RAHJ, Aerssens J, Snyders DJ, Raes AL. A single hERG mutation underlying a spectrum of acquired and congenital long QT syndrome phenotypes. J Mol Cell Cardiol. 2007;43(1):63–72. doi: 10.1016/j.yjmcc.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 7.Seebohm G, Strutz-Seebohm N, Ureche ON, Henrion U, Baltaev R, Mack AF, Korniychuk G, Steinke K, Tapken D, Pfeufer A, Kaab S, Bucci C, Attali B, Merot J, Tavare JM, Hoppe UC, Sanguinetti MC, Lang F. Long QT Syndrome-Associated Mutations in KCNQ1 and KCNE1 Subunits Disrupt Normal Endosomal Recycling of I-Ks Channels. Circ Res. 2008;103(12) doi: 10.1161/CIRCRESAHA.108.177360. 1451-U1217. [DOI] [PubMed] [Google Scholar]
- 8.Huang FD, Chen J, Lin M, Keating MT, Sanguinetti MC. Long-QT syndrome-associated missense mutations in the pore helix of the HERG potassium channel. Circulation. 2001;104(9):1071–1075. doi: 10.1161/hc3501.093815. [DOI] [PubMed] [Google Scholar]
- 9.Lees-Miller JP, Duan YJ, Teng GQ, Thorstad K, Duff HJ. Novel gain-of-function mechanism in K+ channel-related Long-QT syndrome: Altered gating and selectivity in the HERG1 N629D mutant. Circ Res. 2000;86(5):507–513. doi: 10.1161/01.res.86.5.507. [DOI] [PubMed] [Google Scholar]
- 10.Teng GQ, Lees-Miller JP, Duan YJ, Li BT, Li P, Duff HJ. [K+](o)-dependent change in conformation of the HERG1 long QT mutation N629D channel results in partial reversal of the in vitro disease phenotype. Cardiovasc Res. 2003;57(3):642–650. doi: 10.1016/s0008-6363(02)00778-2. [DOI] [PubMed] [Google Scholar]
- 11.Sanguinetti MC, Mitcheson JS. Predicting drug-hERG channel interactions that cause acquired long QT syndrome. Trens Pharm Sci. 2005;26(3):119–124. doi: 10.1016/j.tips.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Perry M, de GM, Helliwell R, Leishman D, Tristani-Firouzi M, Sanguinetti MC, Mitcheson J. Structural determinants of HERG channel block by clofilium and ibutilide. Mol Pharmacol. 2004;66:240–249. doi: 10.1124/mol.104.000117. [DOI] [PubMed] [Google Scholar]
- 13.Stansfeld PJ, Gedeck P, Gosling M, Cox B, Mitcheson JS, Sutcliffe MJ. Drug block of the hERG potassium channel: Insight from modeling. Proteins. 2007;68(2):568–580. doi: 10.1002/prot.21400. [DOI] [PubMed] [Google Scholar]
- 14.Nattel S, Carlsson L. Innovative approaches to anti-arrhythmic drug therapy. Nature Rev Drug Disc. 2006;5(12):1034–1049. doi: 10.1038/nrd2112. [DOI] [PubMed] [Google Scholar]
- 15.Sanguinetti MC, Tristani-Firouzi M. hERG potassium channels and cardiac arrhythmia. Nature. 2006;440(7083):463–469. doi: 10.1038/nature04710. [DOI] [PubMed] [Google Scholar]
- 16.Stary A, Wacker SJ, Boukharta L, Zachariae U, Karimi-Nejad Y, Aqvist J, Vriend G, de Groot BL. Toward a Consensus Model of the hERG Potassium Channel. ChemMedChem. 2010;5(3):455–467. doi: 10.1002/cmdc.200900461. [DOI] [PubMed] [Google Scholar]
- 17.Lees-Miller JP, Subbotina JO, Guo J, Yarov-Yarovoy V, Noskov SY, Duff HJ. Interactions of H562 in the S5 helix with T618 and S621 in the pore helix are important determinants of hERG1 potassium channel structure and function. Biophys J. 2009;96(9):3600–3610. doi: 10.1016/j.bpj.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osterberg F, Aqvist J. Exploring blocker binding to a homology model of the open hERG K+ channel using docking and molecular dynamics methods. FEBS Letters. 2005;579(13):2939–2944. doi: 10.1016/j.febslet.2005.04.039. [DOI] [PubMed] [Google Scholar]
- 19.Kutteh R, Vandenberg JI, Kuyucak S. Molecular dynamics and continuum electrostatics studies of inactivation in the HERG potassium channel. J Phys Chem B. 2007;111(5):1090–1098. doi: 10.1021/jp066294d. [DOI] [PubMed] [Google Scholar]
- 20.Ju P, Pages G, Riek RP, Chen PC, Torres AM, Bansal PS, Kuyucak S, Kuchel PW, Vandenberg JI. The pore domain outer helix contributes to both activation and inactivation of the HERG K+ channel. J Biol Chem. 2009;284(2):1000–1008. doi: 10.1074/jbc.M806400200. [DOI] [PubMed] [Google Scholar]
- 21.Tseng GN, Sonawane KD, Korolkova YV, Zhang M, Liu J, Grishin EV, Guy HR. Probing the outer mouth structure of the HERG channel with peptide toxin footprinting and molecular modeling. Biophys J. 2007;92(10):3524–3540. doi: 10.1529/biophysj.106.097360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 23.Elofsson A, von Heijne G. Membrane protein structure: Prediction versus reality. Annual Rev Biochem. 2007;76:125–140. doi: 10.1146/annurev.biochem.76.052705.163539. [DOI] [PubMed] [Google Scholar]
- 24.Yarov-Yarovoy V, Schonbrun J, Baker D. Multipass membrane protein structure prediction using Rosetta. Proteins. 2006;62(4):1010–1025. doi: 10.1002/prot.20817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barth P, Wallner B, Baker D. Prediction of membrane protein structures with complex topologies using limited constraints. Proc Natl Acad Sci USA. 2009;106(5):1409–1414. doi: 10.1073/pnas.0808323106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long SB, Campbell EB, MacKinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309(5736):897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 27.Long SB, Campbell EB, MacKinnon R. Voltage sensor of Kv1.2: Structural basis of electromechanical coupling. Science. 2005;309(5736):903–908. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- 28.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450(7168) doi: 10.1038/nature06265. 376-U373. [DOI] [PubMed] [Google Scholar]
- 29.Yarov-Yarovoy V, Baker D, Catterall WA. Voltage sensor conformations in the open and closed states in ROSETTA structural models of K+ channels. Proc Natl Acad Sci USA. 2006;103(19):7292–7297. doi: 10.1073/pnas.0602350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pathak MM, Yarov-Yarovoy V, Agarwal G, Roux B, Barth P, Kohout S, Tombola F, Isacoff EY. Closing in on the resting state of the shaker K+ channel. Neuron. 2007;56(1):124–140. doi: 10.1016/j.neuron.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 31.Noskov SY, Berneche S, Roux B. Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands. Nature. 2004;431(7010):830–834. doi: 10.1038/nature02943. [DOI] [PubMed] [Google Scholar]
- 32.Gaussian 03 RCMJ, Frisch GW, Trucks HB, Schlegel GE, Scuseria MA, Robb JR, Cheeseman JA, Montgomery T, Jr, Vreven KN, Kudin JC, Burant JM, Millam SS, Iyengar J, Tomasi V, Barone B, Mennucci M, Cossi G, Scalmani N, Rega GA, Petersson H, Nakatsuji M, Hada M, Ehara K, Toyota R, Fukuda J, Hasegawa M, Ishida T, Nakajima Y, Honda O, Kitao H, Nakai M, Klene X, Li JE, Knox HP, Hratchian JB, Cross V, Bakken C, Adamo J, Jaramillo R, Gomperts RE, Stratmann O, Yazyev AJ, Austin R, Cammi C, Pomelli J, WOchterski PY, Ayala K, Morokuma GA, Voth P, Salvador JJ, Dannenberg VG, Zakrzewski S, Dapprich AD, Daniels MC, Strain O, Farkas DK, Malick AD, Rabuck K, Raghavachari JB, Foresman JV, Ortiz Q, Cui AG, Baboul S, Clifford J, Cioslowski BB, Stefanov G, Liu A, Liashenko P, Piskorz I, Komaromi RL, Martin DJ, Fox T, Keith MA, Al-Laham CY, Peng A, Nanayakkara M, Challacombe PMW, Gill B, Johnson W, Chen MW, Wong C, Gonzalez JA. Wallingford CT: Pople, Gaussian, Inc; 2004. [Google Scholar]
- 33.Morris GM, Goodsell DS, Huey R, Olson AJ. Distributed automated docking of flexible ligands to proteins: Parallel applications of AutoDock 2.4. J Comp-Aid Mol Des. 1996;10(4):293–304. doi: 10.1007/BF00124499. [DOI] [PubMed] [Google Scholar]
- 34.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267(3):727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 35.Bjelkmar P, Niemela PS, Vattulainen I, Lindahl E. Conformational Changes and Slow Dynamics through Microsecond Polarized Atomistic Molecular Simulation of an Integral Kv1.2 Ion Channel. PLOS Comp Biol. 2009;5(2):e1000289. doi: 10.1371/journal.pcbi.1000289. doi:10.1371/journal.pcbi.1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bezanilla F. Voltage Sensor: Function and Structure. J Physiol Science. 2009;59:90. [Google Scholar]
- 37.Villalba-Galea CA, Sandtner W, Starace DM, Bezanilla F. S4-based voltage sensors have three major conformations. Proc Natl Acad Sci USA. 2008;105(46):17600–17607. doi: 10.1073/pnas.0807387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chakrapani S, Sompornpisut P, Intharathep P, Roux B, Perozo E. The activated state of a sodium channel voltage sensor in a membrane environment. Proc Natl Acad Sci USA. 2010;107(12):5435–5440. doi: 10.1073/pnas.0914109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jogini V, Roux B. Dynamics of the Kv1.2 voltage-gated K+ channel in a membrane environment. Biophys J. 2007;93(9):3070–3082. doi: 10.1529/biophysj.107.112540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khalili-Araghi F, Tajkhorshid E, Schulten K. Dynamics of K+ ion conduction through Kv1.2. Biophys J. 2007:616A–619A. doi: 10.1529/biophysj.106.091926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freites JA, Tobias DJ, White SH. A voltage-sensor water pore. Biophys J. 2006;91(11):L90–L92. doi: 10.1529/biophysj.106.096065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang M, Liu J, Tseng GN. Gating charges in the activation and inactivation processes of the hERG channel. J Gen Physiol. 2004;124(6):703–718. doi: 10.1085/jgp.200409119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang M, Liu J, Jiang M, Wu DM, Sonawane K, Guy HR, Tseng GN. Interactions between charged residues in the transmembrane segments of the voltage-sensing domain in the hERG channel. J Membr Biol. 2005;207(3):169–181. doi: 10.1007/s00232-005-0812-1. [DOI] [PubMed] [Google Scholar]
- 44.Liu J, Zhang M, Jiang M, Tseng GN. Negative charges in the transmembrane domains of the HERG K channel are involved in the activation- and deactivation-gating processes. J Gen Physiol. 2003;121(6):599–614. doi: 10.1085/jgp.200308788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandez D, Ghanta A, Kinard KI, Sanguinetti MC. Molecular mapping of a site for Cd2+-induced modification of human ether-a-go-go-related gene (hERG) channel activation. J Physiol-London. 2005;567(3):737–755. doi: 10.1113/jphysiol.2005.089094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subbiah RN, Kondo M, Campbell TJ, Vandenberg JI. Tryptophan scanning mutagenesis of the HERG K+ channel: the S4 domain is loosely packed and likely to be lipid exposed. J Physiol-London. 2005;569(2):367–379. doi: 10.1113/jphysiol.2005.097386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subbiah RN, Clarke CE, Smith DJ, Zhao J, Campbell TJ, Vandenberg JI. Molecular basis of slow activation of the human ether-a-go-go related gene potassium channel. J Physiol. 2004;558:417–431. doi: 10.1113/jphysiol.2004.062588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subbiah RN, Clarke CE, Smith DJ, Zhao JT, Vandenberg JI, Campbell TJ. Alteration of voltage-dependence and activation kinetics of the HERG potassium channel by mutations in the S4 sequence. Biophys J. 2004;86(1):541a–541a. [Google Scholar]
- 49.Piper DR, Rupp J, Sachse FB, Sanguinetti MC, Tristani-Firouzi M. Cooperative interactions between R531 and acidic residues in the voltage sensing module of hERG1 channels. Cell Physiol Biochem. 2008;21(1–3):37–46. doi: 10.1159/000113745. [DOI] [PubMed] [Google Scholar]
- 50.Torres AM, Bansal PS, Sunde M, Clarke CE, Bursill JA, Smith DJ, Bauskin A, Breit SN, Campbell TJ, Alewood PF, Kuchel PW, Vandenberg JI. Structure of the HERG K+ channel S5P extracellular linker: role of an amphipathic alpha-helix in C-type inactivation. J Biol Chem. 2003;278(43):42136–42148. doi: 10.1074/jbc.M212824200. [DOI] [PubMed] [Google Scholar]
- 51.Jiang M, Zhang M, Maslennikov IV, Liu J, Wu DM, Korolkova YV, Arseniev AS, Grishin EV, Tseng GN. Dynamic conformational changes of extracellular S5-P linkers in the hERG channel. J Physiol. 2005;569(Pt 1):75–89. doi: 10.1113/jphysiol.2005.093682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stansfeld PJ, Grottesi A, Sands ZA, Sansom MS, Gedeck P, Gosling M, Cox B, Stanfield PR, Mitcheson JS, Sutcliffe MJ. Insight into the mechanism of inactivation and pH sensitivity in potassium channels from molecular dynamics simulations. Biochem. 2008;47(28):7414–7422. doi: 10.1021/bi800475j. [DOI] [PubMed] [Google Scholar]
- 53.Ander M, Luzhkov VB, Aqvist J. Ligand binding to the voltage-gated Kv1.5 potassium channel in the open state - Docking and computer simulations of a homology model. Biophys J. 2008;94(3):820–831. doi: 10.1529/biophysj.107.112045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perry M, de Groot MJ, Helliwell R, Leishman D, Tristani-Firouzi M, Sanguinetti MC, Mitcheson J. Structural determinants of HERG channel block by clofilium and ibutilide. Mol Pharmacol. 2004;66(2):240–249. doi: 10.1124/mol.104.000117. [DOI] [PubMed] [Google Scholar]
- 55.Lees-Miller JP, Duan YJ, Teng GQ, Duff HJ. Molecular determinant of high-affinity dofetilide binding to HERG1 expressed in Xenopus oocytes: Involvement of S6 sites. Mol Pharmacol. 2000;57(2):367–374. [PubMed] [Google Scholar]
- 56.Seebohm G, Chen J, Strutz N, Culberson C, Lerche C, Sanguinetti MC. Molecular determinants of KCNQ1 channel block by a benzodiazepine. Mol Pharmacol. 2003;64(1):70–77. doi: 10.1124/mol.64.1.70. [DOI] [PubMed] [Google Scholar]
- 57.Mitcheson J, Perry M, Stansfeld P, Sanguinetti MC, Witchel H, Hancox J. Structural determinants for high-affinity block of hERG potassium channels. Novartis Found Symp. 2005;266:136–150. [PubMed] [Google Scholar]
- 58.Aqvist J, Luzhkov VB, Brandsdal BO. Ligand binding affinities from MD simulations. Acc Chem Res. 2002;35(6):358–365. doi: 10.1021/ar010014p. [DOI] [PubMed] [Google Scholar]
- 59.Fernandez D, Sargent J, Sachse FB, Sanguinetti MC. Structural basis for ether-a-go-go-related gene K+ channel subtype-dependent activation by niflumic acid. Mol Pharmacol. 2008;73(4):1159–1167. doi: 10.1124/mol.107.043505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Casis O, Olesen SP, Sanguinetti MC. Mechanism of action of a novel human ether-a-go-go-related gene channel activator. Mol Pharmacol. 2006;69:658–665. doi: 10.1124/mol.105.019943. [DOI] [PubMed] [Google Scholar]
- 61.Seebohm G. Activators of cation channels: Potential in treatment of channelopathies. Mol Pharm. 2005;67(3):585–588. doi: 10.1124/mol.104.010173. [DOI] [PubMed] [Google Scholar]
- 62.Mobley DL, Dill KA. Binding of Small-Molecule Ligands to Proteins: "What You See" Is Not Always "What You Get". Structure. 2009;17(4):489–498. doi: 10.1016/j.str.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Noskov SY. Molecular mechanism of substrate specificity in the bacterial neutral amino acid transporter LeuT. Proteins. 2008;73(4):851–863. doi: 10.1002/prot.22108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.