Abstract
The process of crop domestication has long been a topic of active research for biologists, anthropologists and others. Genetic data have proved a powerful resource for drawing inferences on questions regarding the geographical origins of crops, the numbers of independent domestication events for a given crop species, the specific molecular changes underlying domestication traits, and the nature of artificial selection during domestication and subsequent crop improvement. We would argue that these genetic inferences are fundamentally compatible with recent archaeological data that support a view of domestication as a geographically diffuse, gradual process. In this review, we summarize methodologies ranging from QTL mapping to resequencing used in genetic analyses of crop evolution. We also highlight recent major insights regarding the timing and spatial patterning of crop domestication and the distinct genetic underpinnings of domestication, diversification, and improvement traits.
Captivating crops
Domesticated plants have provided excellent study systems for many fields of plant biology, from molecular biology to physiology to population genetics, as well as other disciplines such as archaeology and ethnobotany. There is also a strong human interest in plant domestication because of the important role crops have played in shaping current societies, and, indeed, allowing many more humans to exist on the planet than would otherwise have been possible. Although this broad interest in plant domestication is an enviable position compared with what is faced by those studying under-appreciated systems, it can sometimes mean that there are lags in communication between the different disciplines studying this process, so that important findings are not always fully recognized.
In this review, we explore some of the recent findings in the area of plant domestication genetics, a field in which new advances are being made at a rapid pace. We describe how these genetic findings can be interpreted in light of apparently conflicting inferences from other scientific disciplines, particularly archaeology. We explore what can be synthesized from the expanding pool of studies about the nature of genetic changes that occur under artificial selection. Finally, we discuss the future of plant domestication genetics in an era of readily accessible genomic data for non-model plant species.
Plant domestication: timing and number of origins
There are two main questions about the origin of domesticated species that can be addressed using both archaeological and evolutionary genetic techniques: the origin of a crop (including whether there is evidence for multiple, geographically distinct domestication events); and the tempo of the domestication event(s). Genetic studies that examine the geographical origins of a crop typically rely on genome-wide neutral markers, which are used to assess allele frequencies in a crop compared with populations of its wild relatives. Due to the strong genetic bottlenecks that occur during the domestication process, the allelic diversity in the crop is expected to be a subset of that found in the wild population(s) from which it was derived. Thus, if populations of the wild progenitor are extant in the geographical location(s) where domestication occurred, the geographical origin(s) of domestication can potentially be pinpointed to a particular population or region. Genetic data have indicated single domestication events for many, but not all, crops [1] (discussed below).
Similarly, genetic data can be used to address the tempo of plant domestication. The duration of the domestication event can potentially be inferred from the severity of the genome-wide bottleneck, and by whether a selectively favored ‘domestication allele’ (see Box 1 and Table 1) spread to fixation quickly or slowly, as measured by the size of the selective sweep (region of reduced variation) in the surrounding genomic region [2–5]. Supporting information for the potential speed of a domestication event can also be gained from the genetic architecture of domestication traits, as inferred from quantitative trait locus (QTL) maps; in this case, it is assumed that domestication phenotypes under simple genetic control can be achieved more quickly than traits with a complex genetic basis [6–8].
Box 1. Domestication: definitions.
The process of plant domestication has been aptly described as a continuum of increasing codependence between plants and people [109]. At the starting point, plants are free-living, and humans invest little in plant survival and production. At the other extreme, plants cannot reproduce or survive without the investment of a large amount of human labor, and humans have shifted to an agricultural economy, giving up other modes of subsistence. This continuum can make it difficult to know when some plants are ‘domesticated’, and therefore difficult to categorically classify traits and underlying genes as being directly related to domestication. For simplicity, domesticated plants are generally classified as such because they possess at least a subset of a suite of traits constituting the ‘domestication syndrome’ [110]. This includes an increase in fruit or grain size compared with the wild progenitor, more determinate growth and/or apical dominance, robust stature, and particularly a loss of dispersal mechanisms; these traits should be found in nearly every variety of a domesticated species. Following domestication, there is also a process of crop improvement and diversification, such as selection for grain quality (changes in starch and other compounds), fruit or grain color, fruit or grain shape, flowering time (synchronization or loss of photoperiod sensitivity), and plant height [111]. These changes can be dramatic, but they are of a nature that the plant would still be considered domesticated in the absence of such traits, and the traits are often found in only a subset of domesticates.
In Table 1, we have listed domestication genes from multiple crops. However, we have only listed improvement and diversification genes from rice due to the large number of plant improvement and diversification genes that have now been identified.
Table 1.
Crop domestication, diversification and improvement genes
| Gene | Crop | Trait | Causative change | Classification | Sel’na | Prevalence | Refs |
|---|---|---|---|---|---|---|---|
| Domestication genes | |||||||
| Vrs1 (six-rowed spike 1) | Barley | Inflorescence structure | Premature stop (insertion, deletion, or AA change) | Domestication | N.T. | Subset of domesticates | [92] |
| tb1 (teosinte branched1) | Maize | Plant and inflorescence structure | Regulatory change | Domestication | Yes | All domesticates | [16,93] |
| tga1 (teosinte glume architecture 1) | Maize | Seed casing | AA change | Domestication | Yes | All domesticates | [94] |
| sh4 (QTL 4 responsible for the reduction of grain shattering) | Rice | Shattering | Regulatory and AA change | Domestication | Yes | All domesticates | [69,71] |
| PROG1 (PROSTRATE GROWTH 1) | Rice | Plant structure | AA change | Domestication | Yesb | All domesticates | [18,19] |
| qSH1 (QTL for seed shattering on chromosome 1) | Rice | Shattering | Regulatory change | Domestication and improvement | No | Subset of domesticates | [70,71] |
| Rc (red pericarp) | Rice | Grain color | Premature stop (deletion or AA change) | Domestication and improvement | Yes | Subset of domesticates (most modern) | [30,64] |
| Sdr4 (Seed dormancy 4) | Rice | Seed dormancy | Regulatory change | Domestication | N.T. | Subset of domesticates | [95] |
| Style2.1 (QTL for style length on chromosome 2) | Tomato | Autogamy | Regulatory change | Domestication | N.T. | All domesticates | [96] |
| fw2.2 (QTL for fruit weight on chromosome 2) | Tomato | Fruit weight (fruit size) | Regulatory change | Domestication and improvement | N.T. | Subset of domesticates (most modern) | [66] |
| fas (fasciated) | Tomato | Locule number (fruit size) | Regulatory change | Domestication and improvement | N.T. | Subset of domesticates (most modern) | [97] |
| Q | Wheat | Shattering and free-threshing | Regulatory and AA change | Domestication | N.T. | All domesticates | [17] |
| Improvement and diversification genes in rice | |||||||
| GIF1 (GRAIN INCOMPLETE FILLING 1) | Rice | Grain filling | Regulatory change | Improvement | Yes | All domesticates (survey not complete) | [98] |
| GS3 (QTL for grain size and length on chromosome 3) | Rice | Grain size and length | Premature stop (deletion) | Improvement | Yes | Subset of domesticates | [99] |
| qSW5 (QTL for seed width on chromosome 5) | Rice | Grain width | Deletion | Improvement | Yesb | Subset of domesticates | [100] |
| GW2 (QTL for grain weight on chromosome 2) | Rice | Grain width and weight | Premature stop (deletion) | Improvement | N.T. | Subset of domesticates (survey incomplete) | [101] |
| BADH2 (BETAINE ALDEHYDE DEHYDROGENASE 2) | Rice | Fragrance | Premature stop (deletion or AA change) | Diversification | Yes | Subset of domesticates | [57,58] |
| Ghd7 (QTL for grain number, plant height, and heading date) | Rice | Grain number, plant structure, and flowering date | Several unique alleles with different effects; some premature stop and deletion alleles | Improvement | N.T. | Subset of domesticates | [102] |
| Phr1 (Phenol reaction 1) | Rice | Grain discoloration (oxidation) | Premature stop (insertion or deletion) | Diversification | Yes | Subset of domesticates | [103,104] |
| Waxy | Rice | Grain quality (starch) | Intron splicing defect (non-functional) | Diversification | Yes | Subset of domesticates | [29,53] |
| Gn1a (QTL for grain number on chromosome 1, a) | Rice | Grain number | Premature stop (deletion) | Improvement | N.T. | Subset of domesticates | [68] |
| sd1 (semidwarf1) | Rice | Plant structure | Premature stop (deletion) or AA change | Improvement | Yes | Subset of domesticates | [105–108] |
Evidence of positive selection based on patterns of genetic variation and/or population genetic tests; N.T. indicates that selection was not tested.
Inferred based on prevalence of an allele, not based on patterns of genetic variation or population genetic tests.
Since the 1990s, the application of QTL mapping, followed by the characterization of functional mutations at some domestication genes, has revealed that the suite of changes associated with domestication (Box 1 and Table 1) can, in many plants, have a relatively simple genetic basis [8–14] (but see Refs [7,15]), and that some major morphological transitions can be achieved via changes at a single locus [16–19]. These genetic approaches complement a limited number of domestication experiments where artificial selection has been shown to rapidly change the phenotypes of wild or crop–wild hybrids, and to achieve domesticate-like forms in less than 20 generations [20–23]. Together, these lines of evidence have suggested that domestication need not be a slow or gradual process, and that it could potentially be fairly rapid, given strong selective pressures and an appropriate genetic architecture. Similarly, evidence of severe genome-wide reductions in variation (due to population bottlenecks in combination with selection) [24–28] and large selective sweeps [29–32] have suggested rapid domestication and/or spread of domestication traits in some crop species.
Recently, archaeological evidence has surfaced to suggest that the process of plant domestication might have been a more gradual process, at least for cereal crops in the Fertile Crescent. Supporting evidence has included widespread indications of pre-domestication cultivation of wild plants, archaeological evidence of multiple domestication ‘trials’ in geographically distinct locations, and the gradual appearance in the archaeobotanical record of some classical domestication traits, such as the non-shattering phenotype (Box 2) [33,34]. It has been argued that these findings contradict or should substantially alter our understanding of the process of plant domestication [35,36]. We would instead argue that findings from genetic and archaeological studies represent complementary perspectives on domestication, each highlighting a different facet of this complex process.
Box 2. Why were we so slow in selecting for non-shattering grains?
Although limited in number, artificial domestication experiments have played an important role in our understanding of crop evolution. These studies (all, to our knowledge, conducted in cereals) have shown that classical domestication traits, such as the loss of shattering and loss of seed dormancy, can arise and increase in frequency over a short time period when subjected to strong selection [20–23]. In contrast, Old World archaeological data indicate that the appearance of non-shattering grains was probably gradual, at least in barley, wheat, and rice [33,34,112]. In these crops, the non-shattering phenotype appears only after an increase in grain size, a trait that itself reflects selection for germination under active cultivation conditions (e.g., tillage and greater sowing depth). Thus, although the loss of shattering would be expected to greatly facilitate the harvesting of grains in planted fields, the phenotype does not actually appear in the initial stages of active cultivation and selection [33].
One explanation proposed for this pattern in rice is that strong selection did not occur until after an optimal, reduced-shattering phenotype had arisen [71]. Mutations conferring a complete loss of shattering might facilitate harvesting, but they would also make the subsequent threshing process more laborious. By contrast, a reduction in shattering, as occurs with the rice sh4 domestication allele [69], would provide a better balance between harvesting and threshing. Therefore, the increase in non-shattering phenotypes would be slow until the appearance of this optimal phenotype (potentially not discernable in the archeological record), at which time it would be under strong selection. Alternatively, conscious selection might simply have been ineffective in the face of gene flow from wild or weedy relatives (demonstrated by [113]).
Quantification of the strength of selection on diverse domestication genes might also provide insights into this question. The selection coefficient (s) for a domestication allele can potentially be inferred from the size of the selective sweep in the surrounding genomic region [29]. If s values for non-shattering alleles were found to be lower overall than those of other domestication-related alleles, this would support the hypothesis that the strength of selection for this phenotype has been weaker than for other domestication-related traits. To our knowledge, there is currently only one non-shattering allele for which a selective coefficient has been quantified (rice sh4 [71]), and there are only a few genes to which the value can be compared. The value for s for sh4 is between 0.187 and 0.235; this value is lower than the s for the waxy allele (4.59-4.24), but higher than what is seen for the tb1 allele (s = 0.05) [29]. Additional measures of s and comparative analyses across diverse domestication genes (see Box 1 and Table 1) are needed to fully test this hypothesis.
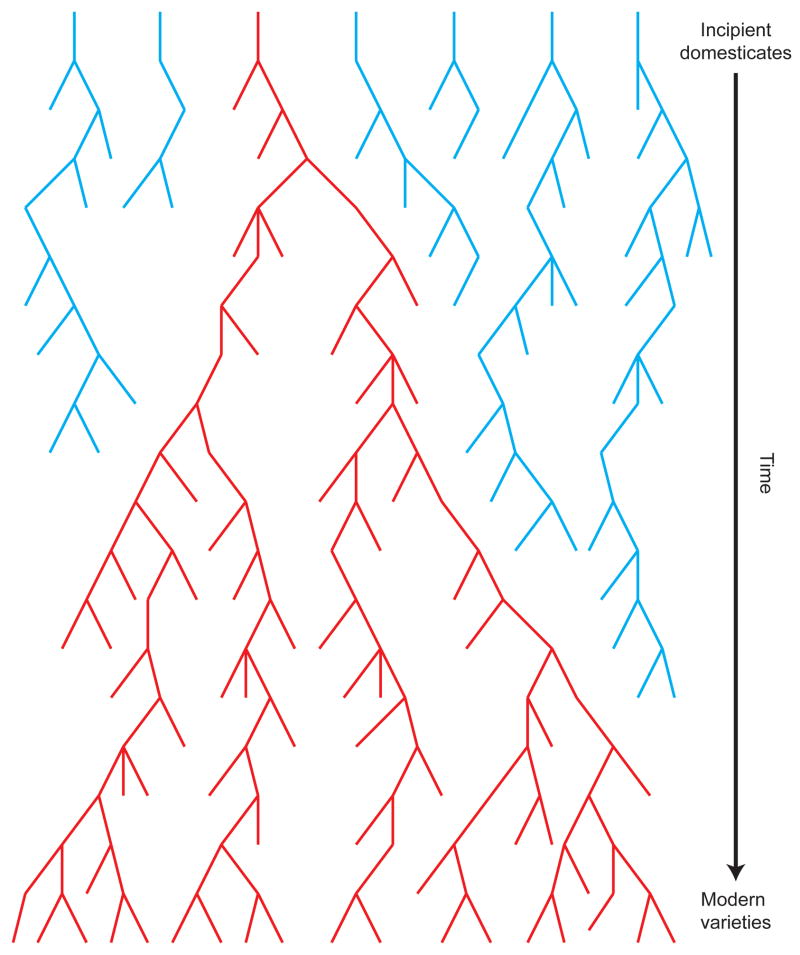
In this context, it is important to draw a distinction between the domestication processes inferred by sampling the present-day representatives of a crop species, and those inferred by considering the archaeological record. The genetic evidence that can be extracted from living crops cannot provide information about any lineage that did not contribute to the currently extant domesticate (Figure 1). Even studies that use ancient DNA (e.g., Ref. [37]) capture only a small subset of the historical genetic diversity of a crop. Thus, it is possible that there could be numerous early ‘experiments’ in the domestication of many crop species, but that few of these lineages have persisted to contribute to the genetic diversity of contemporary germplasm [33,38]. Indeed, genetic studies of species in the incipient stages of domestication have revealed multiple domestication origins or high ongoing gene flow between wild and cultivated varieties [39,40], which is consistent with this scenario.
Figure 1.
Why archaeology and genetics do not match. This figure shows the genealogy of seven lineages over time. Genetic approaches applied to the modern crop can only detect the lineage shown in red. By contrast, all lineages can potentially be detected using archaeological techniques.
Even with the necessarily limited historical picture obtained by genetic sampling of present-day germplasm, it is interesting to note the number of crop species for which genetic data have revealed multiple origins. For example, genetic analysis has confirmed the multiple origins of barley (Hordeum vulgare) [41], Phaseolus beans [42,43], and Asian rice (Oryza sativa) [44]; notably, these are species for which morphological or archaeological evidence alone was insufficient to confirm multiple origins. Evolutionary genetics have also been used to explore details of the multiple origins of peppers (Capsicum spp.) [45], squash (Cucurbita spp.) [46], and African rice (Oryza glaberrima) [47]; these are all species for which obvious geographical or morphological differences already strongly suggested multiple origins. These findings would appear to stand in contrast to assertions that molecular evolutionary studies “…have invariably assumed rapid, single origins for domestic species” [2].
Domesticated plants: models for convergent and parallel evolution
Convergent phenotypic evolution (the appearance of the same trait in independent evolutionary lineages) and parallel phenotypic evolution (the appearance of the same trait in closely related or potentially interbreeding lineages) can both occur in crop domestication. Convergent evolution is manifest in the repeated evolution of classical domestication traits, such as the loss of seed shattering and seed dormancy seen in most cereal crops. Parallel evolution occurs as the appearance of the same domestication trait in multiple, independent origins of a single domesticated crop (although this pattern can also reflect selective introgression of a single favored allele across crop varieties; Box 3). Both types of evolution pose the same intriguing question: what is the genetic basis of phenotypic changes that have evolved repeatedly in response to human selective pressures?
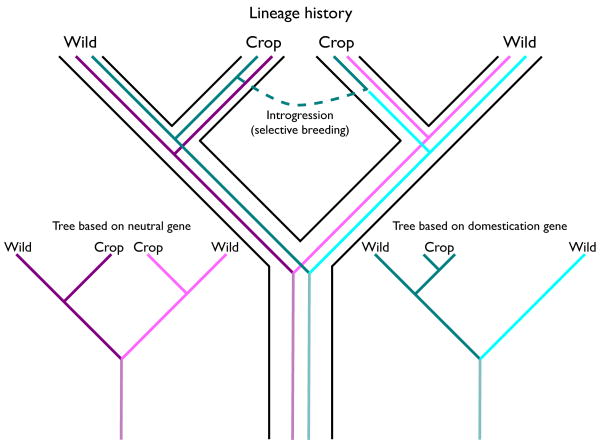
Box 3. Multiple origins and mixed signals.
Multiple origins of a single species (domesticated or wild) can be discerned from the relationship of the alleles in the derived lineage relative to the progenitor. A lineage with multiple origins will show a polyphyletic pattern, whereas a lineage with a single origin will appear monophyletic. However, not all genes will reflect the history of a crop accurately. In particular, domestication alleles that result in desirable traits can easily be moved from one domesticate to another through hybridization and selective breeding (see Figure I, shown in the large tree). Thus, although the majority of the genes in the genome will represent the true history of a domesticated lineage (Figure I, the tree based on neutral genes), domestication genes might falsely indicate that the crop has a single origin (Figure I, the tree based on domestication genes). The difference between gene trees and lineage histories (usually called species trees) is reviewed in [114]. Overall, even though two unique domestication alleles for the same trait is strongly supportive of multiple origins (as is the case for the brittle rachis loci in barley, where the two domestication alleles have unique origins [52]), a single origin for a domestication allele does not necessarily indicate a single origin for a crop (as is the case for shattering in rice, where the domestication allele originated in the japonica group and spread to the indica group through selective breeding [69,71]).
Figure I.
Lineage history and gene trees for a crop with multiple origins. The lineage history is the large tree, shown in black, and reflects the fact that there are two domestication events. Neutral and domestication gene trees are shown in color; the same trees are shown both inside the lineage tree and independently in the lower left and right hand sides.
For the case of convergent evolution, it is interesting to note that domestication QTL that map to syntenic regions across multiple crop species do not necessarily involve the same underlying loci. For example, although broad similarities in the genetic architecture of domestication traits of cereal crops initially suggested that the same loci might be responsible across the grass family [11], fine mapping and cloning of the underlying genes has not borne this out. Instead, more precise QTL maps have revealed that the same traits in different cereals are controlled by distinct constellations of loci [48,49]. In the Solanaceae, some fruit weight QTL in tomato (Solanum lycopersicum) and pepper (Capsicum spp.) do map to the same locations [50], suggesting that this prediction might hold true for a subset of traits in this family. However, overall, domestication genes that have been cloned and characterized to date have not been of major importance in multiple domesticates. This is illustrated by the well-studied domestication gene tb1, which is responsible for major plant architectural changes in maize (Zea mays) [16], but which has only a slight effect on the branching architecture of domesticated foxtail millet (Setaria italica) [51]. More telling, perhaps, is the finding that even multiple domestication events within the same species can result from changes at different loci, as is the case for shattering in barley, which is controlled by unique loci in the two domesticated lineages [52]. These findings all indicate that there are many ways to ‘make’ domesticated plants. It is also notable that most of the domestication alleles characterized to date are not loss-of-function mutations (Box 1 and Table 1); thus, unique domestication genes are not simply the product of ‘breaking’ a given pathway at different points in each species.
In contrast to domestication genes per se, recent studies of genes selected upon after the initial domestication process (Box 1) are revealing more instances of repeated evolution affecting the same genes both within and among species. ‘Diversification’ genes are the target of selection for phenotypic variation among varieties of a crop, such as different types of starch or flavor. One classic example of a trait under diversifying selection is the glutinous or sticky phenotype of cooked cereal grains, reflecting the absence (or near absence) of the starch amylose in the endosperm. The glutinous phenotype is favored in select varieties of rice (primarily a subset of japonica varieties of O. sativa) [29,53], maize [54], and foxtail millet [55,56], and is controlled by unique mutations at the Waxy gene in all these crops. This pattern has also been shown within a single species, specifically in the context of diversifying selection for aroma qualities in cultivated rice. The rice BADH2 gene underlies variation in the production of 2-acetyl-1-pyrroline, a primary determinant of aromatic qualities in rice [57]. A survey of the BADH2 gene in aromatic rice accessions from around the world has shown that although one aromatic allele is by far the most common, the aromatic phenotype has also been generated via a variety of mutations at the same gene [58].
Some traits that are widespread in domesticated crops are important because they change the quality of the harvested crop, although they do not strongly affect cultivation in terms of ease of harvest or sowing. These traits represent the effect of selection on ‘improvement’ genes following the initial stages of domestication. Grain or kernel color has been a target of selection for improved quality in wheat (Triticum aestivum), maize, and rice [59–63]. In rice, the genetic basis for the widespread white (nonpigmented) grains in domesticated varieties (compared with the pigmented grains of wild rice) has been revealed to be loss-of-function mutations at the Rc gene, which encodes a regulatory protein in the proanthocyanidin synthesis pathway [64]. Rc is often considered as both a domestication gene and an improvement gene (see Box 1 and Table 1) because it is closely linked to QTL that underlie variation in seed dormancy [65]. The majority of nonpigmented rice cultivars (>97%) carry a single loss-of-function allele as a result of selective introgression of this allele across variety groups (Box 3); however, a few landraces (<3%) carry an independently evolved loss-of-function allele instead [30]. Unlike genes selected upon during the initial domestication process, ‘diversification’ and ‘improvement’ phenotypes in rice appear more likely to result from a variety of mutations at the same gene (5 out of 11, if Rc is counted as an improvement gene; Box 1 and Table 1). In addition, these are generally loss-of-function mutations, making the pattern somewhat counter-intuitive, because it would seem that there might be many ways to ‘break’ a particular pathway other than repeated mutations in the same gene. It is unclear whether this early trend will hold as more genes are cloned and characterized in a variety of cultivars within and among species.
Finding domestication, diversification, and improvement genes
Quantitative trait locus (QTL) mapping conducted in crop–wild crosses was one of the first techniques applied to understand the genetic basis of domestication traits [66,67]. As described above, the limited number of QTL related to domestication traits suggested that many plants have been domesticated via changes at relatively few loci [1]. QTL mapping is necessarily limited in resolution, and each QTL peak can potentially span more than 100 genes; nevertheless, the apparent simplicity of genetic transitions required to produce a domestication phenotype is still impressive. In addition, the same crop–wild crosses used for QTL mapping have often provided the recombinant mapping populations that led the way to map-based (or positional) cloning of domestication genes. Map-based cloning has provided the majority of cloned and characterized domestication genes to date (Box 1 and Table 1), and have confirmed the predictions that individual domestication genes themselves (and not just QTL) could have major phenotypic effects.
QTL mapping and map-based cloning have also been conducted using crosses other than crop–wild combinations. These alternatives include wild–weedy crosses and crosses between different varieties within a domesticate [65,68]. All these crosses can provide valuable information, but it is important to recognize that the QTL (and eventually the genes) identified in a particular cross might not be important in other crosses or environments. One interesting demonstration of this phenomenon is the two shattering genes that have so far been cloned in rice, sh4 [69] and qSH1 [70]. The sh4 gene was cloned from a crop–wild mapping population, whereas qSH1 was cloned from a cross between the two independently domesticated subspecies of Asian rice, indica and japonica. Surveys of the occurrence of these two genes in a world-wide sample of domesticated and wild rice showed that the sh4 domestication allele has been fixed in all cultivated rice, whereas the qSH1 domestication allele is found in only a subset of temperate japonica varieties [71,72]. This distribution of domestication alleles at the two different shattering loci is entirely congruent with the mapping populations used to the discover them; sh4 is a locus that differentiates wild from domesticated rice, whereas qSH1 is probably more important for improvement or modification within the temperate japonica variety group. Researchers should carefully consider the nature of the traits and genes they are interested in characterizing when they choose the basis of their mapping populations.
Despite the large amount of information gained from QTL mapping and map-based cloning to date, this approach suffers from the problem of bias in human perception of important traits; that is, we tend to investigate what we already think is important. Genomic scans (also referred to as hitchhiking mapping), where diversity at molecular markers in wild and domesticated populations is compared to identify reductions in variation consistent with selection [5,73,74], are unbiased about the type of locus that might be identified as being important in domestication. Genome scans are becoming easier to implement because of the relative accessibility of genomic data from which markers can be developed, although large sample sizes and appropriate data analysis are still challenges. These scans have mainly been implemented to identify the proportion of genes under selection during domestication, and have provided lists of candidate domestication genes for further consideration [3,75–77]. As with QTL mapping, the choice of sampling is key to the conclusions that can be drawn from a study–a comparison of wild plants and domesticated landraces can be used to identify domestication genes, whereas a comparison of landraces to elite cultivars can provide a list of candidate genes important in crop improvement and diversification [78–80]. Interestingly, genomic scans in sunflower (Helianthus annuus) and maize have revealed the signature of selection at genes involved in amino acid synthesis in the domesticates, suggesting similar patterns of change at the molecular level that might not have detected based on investigations starting at the phenotypic level [3,78].
Plant biologists are also beginning to pursue association mapping, an approach closely related to QTL mapping. In both methods, the goal is to statistically associate segregating allelic variation with a phenotype of interest; however, with association mapping the associations are inferred using population samples of potentially unrelated individuals rather than the progeny of an artificial cross [81]. Human geneticists pioneered this technique, in part because of the obvious constraints of genetic mapping in humans [82,83]. Association mapping offers an advantage compared with any other technique in potentially identifying a restricted genomic region or even the causal single-nucleotide polymorphism (SNP) controlling the trait of interest owing to the long history of recombination in natural populations, compared with the few generations usually represented in mapping populations [84–86]. However, association mapping in selfing species (which are common among domesticated plants) does have a drawback because the build-up of linkage disequilibrium (LD) in these lineages due to population structure can lead to spurious associations [87]. Interestingly, crop–wild hybrid zones, which are fairly common for some domesticated plants, might offer a source of material for association mapping approaches, provided the hybrid zones are persistent rather than ephemeral [88,89].
Finally, high-throughput resequencing techniques now offer a complement to (or simply a faster implementation of) the approaches outlined above. One obvious application is the resequencing of a candidate domestication gene or of a diversification or improvement gene in population-level samples of the wild or domesticated species. This approach can be used to test for the presence of a selective sweep [6], and, if combined with a phenotypic assay, can potentially enable a functional polymorphism to be pinpointed through association mapping [90]. In addition, easily obtained sequence data can provide a wealth of new markers for fine-mapping in a non-model species. Alternatively, for domesticates that have experienced a severe genetic bottleneck (which makes the detection of selective sweeps difficult [91]), these resequencing surveys can identify regions of high divergence between the crop and the wild species, which are also candidate regions for changes associated with domestication. In the future, it should be possible to combine genome scans and association mapping in many species, along with QTL mapping in species that are amenable to crosses.
Future research
Our understanding of plant domestication is constantly being reshaped by new discoveries in a variety of disciplines, and many of the patterns that have emerged for the tempo of domestication, the origins of cultivated plants, and the nature of genes underlying domestication traits are still subject to change. Indeed, because cereal crops have so dominated research into plant domestication (owing to their economic importance and genetic tractability), additional information from other crops will be particularly important for confirming or rethinking the current paradigms. Over the next ten years, the application of genome resequencing, genome scans, and selective sweep mapping should greatly expand the pool of available evidence for non-model domesticates. It will be interesting to see whether this new information will bolster or reshape our current understanding of the pattern and process of plant domestication.
Acknowledgments
Special thanks to Genevieve K. Croft, Jared L. Strasburg, and Katherine E. Waselkov and for their advice and critical review of an earlier version of this manuscript. B.L.G. was supported by a National Institutes of Health Ruth L. Kirschstein Postdoctoral Fellowship (1F32GM082165). K.M.O. was supported by a National Science Foundation PGRP award (DBI-0638820).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burger JC, et al. Molecular insights into the evolution of crop plants. Am J Bot. 2008;95:113–122. doi: 10.3732/ajb.95.2.113. [DOI] [PubMed] [Google Scholar]
- 2.Purugganan MD, Fuller DQ. The nature of selection during plant domestication. Nature. 2009;457:843–848. doi: 10.1038/nature07895. [DOI] [PubMed] [Google Scholar]
- 3.Wright SI, et al. The effects of artificial selection on the maize genome. Science. 2005;308:1310–1314. doi: 10.1126/science.1107891. [DOI] [PubMed] [Google Scholar]
- 4.Tenaillon MI, et al. Selection versus demography: A multilocus investigation of the domestication process in maize. Mol Biol Evol. 2004;21:1214–1225. doi: 10.1093/molbev/msh102. [DOI] [PubMed] [Google Scholar]
- 5.Maynard Smith J, Haigh J. Hitch-hiking effect of a favorable gene. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- 6.Tian F, et al. Tracking footprints of maize domestication and evidence for a massive selective sweep on chromosome 10. Proceedings of the National Academy of Sciences. 2009;106:9979–9986. doi: 10.1073/pnas.0901122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burke JM, et al. Genetic analysis of sunflower domestication. Genetics. 2002;161:1257–1267. doi: 10.1093/genetics/161.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doebley J, Stec A. Inheritance of the morphological differences between maize and teosinte: Comparison of results for two F2 populations. Genetics. 1993;134:559–570. doi: 10.1093/genetics/134.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doebley J, Stec A. Genetic analysis of the morphological differences between maize and teosinte. Genetics. 1991;129:285–295. doi: 10.1093/genetics/129.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai HW, Morishima H. QTL clusters reflect character associations in wild and cultivated rice. Theor Appl Genet. 2002;104:1217–1228. doi: 10.1007/s00122-001-0819-7. [DOI] [PubMed] [Google Scholar]
- 11.Paterson AH, et al. Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science. 1995;269:1714–1718. doi: 10.1126/science.269.5231.1714. [DOI] [PubMed] [Google Scholar]
- 12.Koinange EMK, et al. Genetic control of the domestication syndrome in common bean. Crop Sci. 1996;36:1037–1045. [Google Scholar]
- 13.Keim P, et al. RFLP mapping in soybean: Association between marker loci and variation in quantitative traits. Genetics. 1990;126:735–742. doi: 10.1093/genetics/126.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doganlar S, et al. Conservation of gene function in the Solanaceae as revealed by comparative mapping of domestication traits in eggplant. Genetics. 2002;161:1713–1726. doi: 10.1093/genetics/161.4.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wills DM, Burke JM. Quantitative trait locus analysis of the early domestication of sunflower. Genetics. 2007;176:2589–2599. doi: 10.1534/genetics.107.075333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doebley J, et al. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- 17.Simons KJ, et al. Molecular characterization of the major wheat domestication gene Q. Genetics. 2006;172:547–555. doi: 10.1534/genetics.105.044727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin J, et al. Genetic control of rice plant architecture under domestication. Nat Genet. 2008;40:1365–1369. doi: 10.1038/ng.247. [DOI] [PubMed] [Google Scholar]
- 19.Tan L, et al. Control of a key transition from prostrate to erect growth in rice domestication. Nat Genet. 2008;40:1360–1364. doi: 10.1038/ng.197. [DOI] [PubMed] [Google Scholar]
- 20.Hillman GC, Davies MS. Domestication rates in wild-type wheats and barley under primitive cultivation. Biol J Linn Soc. 1990;39:39–78. [Google Scholar]
- 21.Hillman GC, Davies MS. Measured domestication rates in wild wheats and barley under primitive cultivation, and their archaeological implications. Journal of World Prehistory. 1990;4:157–222. [Google Scholar]
- 22.Hilu KW, Wet JMJD. Effect of artificial selection on grain dormancy in Eleusine (Gramineae) Syst Bot. 1980;5:54–60. [Google Scholar]
- 23.Oka HI, Morishima H. The dynamics of plant domestication: Cultivation experiments with Oryza perennis and its hybrid with O. sativa. Evolution. 1971;25:356–364. doi: 10.1111/j.1558-5646.1971.tb01889.x. [DOI] [PubMed] [Google Scholar]
- 24.Caicedo A, et al. Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 2007;3:e163. doi: 10.1371/journal.pgen.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haudry A, et al. Grinding up wheat: A massive loss of nucleotide diversity since domestication. Mol Biol Evol. 2007;24:1506–1517. doi: 10.1093/molbev/msm077. [DOI] [PubMed] [Google Scholar]
- 26.Hyten DL, et al. Impacts of genetic bottlenecks on soybean genome diversity. Proceedings of the National Academy of Sciences. 2006;103:16666–16671. doi: 10.1073/pnas.0604379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu A, Burke JM. Patterns of nucleotide diversity in wild and cultivated sunflower. Genetics. 2006;173:321–330. doi: 10.1534/genetics.105.051110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller JC, Tanksley SD. RFLP analysis of phylogenetic relationships and genetic variation in the genus Lycopersicon. Theor Appl Genet. 1990;80:437–448. doi: 10.1007/BF00226743. [DOI] [PubMed] [Google Scholar]
- 29.Olsen KM, et al. Selection under domestication: Evidence for a sweep in the rice Waxy genomic region. Genetics. 2006;173:975–983. doi: 10.1534/genetics.106.056473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sweeney MT, et al. Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genet. 2007;3:e133. doi: 10.1371/journal.pgen.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark RM, et al. Pattern of diversity in the genomic region near the maize domestication gene tb1. Proceedings of the National Academy of Sciences. 2004;101:700–707. doi: 10.1073/pnas.2237049100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palaisa K, et al. Long-range patterns of diversity and linkage disequilibrium surrounding the maize Y1 gene are indicative of an asymmetric selective sweep. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9885–9890. doi: 10.1073/pnas.0307839101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuller DQ. Contrasting patterns in crop domestication and domestication rates: Recent archaeobotanical insights from the Old World. Ann Bot. 2007;100:903–924. doi: 10.1093/aob/mcm048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuller DQ, et al. The domestication process and domestication rate in rice: Spikelet bases from the lower Yangtze. Science. 2009;323:1607–1610. doi: 10.1126/science.1166605. [DOI] [PubMed] [Google Scholar]
- 35.Allaby RG, et al. The genetic expectations of a protracted model for the origins of domesticated crops. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13982–13986. doi: 10.1073/pnas.0803780105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown TA, et al. The complex origins of domesticated crops in the Fertile Crescent. Trends Ecol Evol. 2009;24:103–109. doi: 10.1016/j.tree.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 37.Jaenicke-Despres V, et al. Early allelic selection in Maize as revealed by ancient DNA. Science. 2003;302:1206–1208. doi: 10.1126/science.1089056. [DOI] [PubMed] [Google Scholar]
- 38.Smith BD. Origins of agriculture in Eastern North America. Science. 1989;246:1566–1571. doi: 10.1126/science.246.4937.1566. [DOI] [PubMed] [Google Scholar]
- 39.Otero-Arnaiz A, et al. Genetic variation and evolution of Polaskia chichipe (Cactaceae) under domestication in the Tehuacàn Valley, central Mexico. Mol Ecol. 2005;14:1603–1611. doi: 10.1111/j.1365-294X.2005.02494.x. [DOI] [PubMed] [Google Scholar]
- 40.Miller A, Schaal B. Domestication of a Mesoamerican cultivated fruit tree, Spondias purpurea. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12801–12806. doi: 10.1073/pnas.0505447102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morrell PL, Clegg MT. Genetic evidence for a second domestication of barley (Hordeum vulgare) east of the Fertile Crescent. Proceedings of the National Academy of Sciences. 2007;104:3289–3294. doi: 10.1073/pnas.0611377104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gepts P, et al. Phaseolin-protein variability in wild forms and landraces of the common bean (Phaseolus vulgaris): Evidence for multiple centers of domestication. Econ Bot. 1986;40:451–468. [Google Scholar]
- 43.Sonnante G, et al. Evolution of genetic diversity during the domestication of common-bean (Phaseolus vulgaris L.) Theor Appl Genet. 1994;89:629–635. doi: 10.1007/BF00222458. [DOI] [PubMed] [Google Scholar]
- 44.Londo JP, et al. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestication of cultivated rice Oryza sativa. Proc Natl Acad Sci USA. 2006;103:9578–9583. doi: 10.1073/pnas.0603152103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pickersgill B. Genetic resources and breeding of Capsicum spp. Euphytica. 1997;96:129–133. [Google Scholar]
- 46.Sanjur OI, et al. Phylogenetic relationships among domesticated and wild species of Cucurbita (Cucurbitaceae) inferred from a mitochondrial gene: Implications for crop plant evolution and areas of origin. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:535–540. doi: 10.1073/pnas.012577299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Semon M, et al. The population structure of African cultivated rice Oryza glaberrima (Stued): Evidence for elevated levels of linkage disequilibrium caused by admixture with O sativa and ecological adaptation. Genetics. 2005;169:1639–1647. doi: 10.1534/genetics.104.033175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W, Gill B. Multiple genetic pathways for seed shattering in the grasses. Functional & Integrative Genomics. 2006;6:300–309. doi: 10.1007/s10142-005-0015-y. [DOI] [PubMed] [Google Scholar]
- 49.Sood S, et al. The major threshability genes soft glume (sog) and tenacious glume (Tg), of diploid and polyploid wheat, trace their origin to independent mutations at non-orthologous loci. Theor Appl Genet. 2009;119:341–351. doi: 10.1007/s00122-009-1043-0. [DOI] [PubMed] [Google Scholar]
- 50.Paran I, van der Knaap E. Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper. J Exp Bot. 2007;58:3841–3852. doi: 10.1093/jxb/erm257. [DOI] [PubMed] [Google Scholar]
- 51.Doust AN, et al. Genetic control of branching in foxtail millet. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9045–9050. doi: 10.1073/pnas.0402892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Azhaguvel P, Komatsuda T. A phylogenetic analysis based on nucleotide sequence of a marker linked to the brittle rachis locus indicates a diphyletic origin of barley. Ann Bot. 2007;100:1009–1015. doi: 10.1093/aob/mcm129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olsen KM, Purugganan MD. Molecular evidence on the origin and evolution of glutinous rice. Genetics. 2002;162:941–950. doi: 10.1093/genetics/162.2.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fan L, et al. Molecular evidence for post-domestication selection in the Waxy gene of Chinese waxy maize. Mol Breed. 2008;22:329–338. [Google Scholar]
- 55.Fukunaga, et al. Structural variation in the Waxy gene and differentiation in foxtail millet [Setaria italica (L.) P. Beauv.]: implications for multiple origins of the waxy phenotype. Mol Genet Genomics. 2002;268:214–222. doi: 10.1007/s00438-002-0728-8. [DOI] [PubMed] [Google Scholar]
- 56.Kawase M, et al. Diverse origins of waxy foxtail millet crops in East and Southeast Asia mediated by multiple transposable element insertions. Mol Genet Genomics. 2005;274:131–140. doi: 10.1007/s00438-005-0013-8. [DOI] [PubMed] [Google Scholar]
- 57.Bradbury LMT, et al. The gene for fragrance in rice. Plant Biotechnol J. 2005;3:363–370. doi: 10.1111/j.1467-7652.2005.00131.x. [DOI] [PubMed] [Google Scholar]
- 58.Kovach MJ, et al. The origin and evolution of fragrance in rice (Oryza sativa L.) Proceedings of the National Academy of Sciences. 2009;106:14444–14449. doi: 10.1073/pnas.0904077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hanson MA, et al. Evolution of anthocyanin biosynthesis in maize kernels: The role of regulatory and enzymatic loci. Genetics. 1996;143:1395–1407. doi: 10.1093/genetics/143.3.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Himi E, Noda K. Red grain colour gene (R) of wheat is a Myb-type transcription factor. Euphytica. 2005;143:239–242. [Google Scholar]
- 61.Metzger R, Silbaugh B. Location of genes for seed coat color in hexaploid wheat, Triticum aestivum L. Crop Sci. 1970;10:495–496. [Google Scholar]
- 62.Robbins ML, et al. A Mutator transposon insertion is associated with ectopic expression of a tandemly repeated multicopy Myb gene pericarp color1 of maize. Genetics. 2008;178:1859–1874. doi: 10.1534/genetics.107.082503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Furukawa T, et al. The Rc and Rd genes are involved in proanthocyanidin synthesis in rice pericarp. The Plant Journal. 2007;49:91–102. doi: 10.1111/j.1365-313X.2006.02958.x. [DOI] [PubMed] [Google Scholar]
- 64.Sweeney MT, et al. Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell. 2006;18:283–294. doi: 10.1105/tpc.105.038430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gu XY, et al. Multiple loci and epistases control genetic variation for seed dormancy in weedy rice (Oryza sativa) Genetics. 2004;166:1503–1516. doi: 10.1534/genetics.166.3.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frary A, et al. fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- 67.Doebley J, et al. Genetic and morphological analysis of a maize-teosinte F2 population: Implications for the origin of maize. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:9888–9892. doi: 10.1073/pnas.87.24.9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ashikari M, et al. Cytokinin oxidase regulates rice grain production. Science. 2005;309:741–745. doi: 10.1126/science.1113373. [DOI] [PubMed] [Google Scholar]
- 69.Li C, et al. Rice domestication by reducing shattering. Science. 2006;311:1936–1939. doi: 10.1126/science.1123604. [DOI] [PubMed] [Google Scholar]
- 70.Konishi S, et al. An SNP caused loss of seed shattering during rice domestication. Science. 2006;312:1392–1396. doi: 10.1126/science.1126410. [DOI] [PubMed] [Google Scholar]
- 71.Zhang LB, et al. Selection on grain shattering genes and rates of rice domestication. New Phytol. 2009;184:708–720. doi: 10.1111/j.1469-8137.2009.02984.x. [DOI] [PubMed] [Google Scholar]
- 72.Lin Z, et al. Origin of seed shattering in rice (Oryza sativa L.) Planta. 2007;226:11–20. doi: 10.1007/s00425-006-0460-4. [DOI] [PubMed] [Google Scholar]
- 73.Harr B, et al. Hitchhiking mapping: a population-based fine-mapping strategy for adaptive mutations in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:12949–12954. doi: 10.1073/pnas.202336899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Storz JF. Using genome scans of DNA polymorphism to infer adaptive population divergence. Mol Ecol. 2005;14:671–688. doi: 10.1111/j.1365-294X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- 75.Vigouroux Y, et al. Identifying genes of agronomic importance in maize by screening microsatellites for evidence of selection during domestication. Proceedings of the National Academy of Science. 2002;99:9650–9655. doi: 10.1073/pnas.112324299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Whitt SR, et al. Genetic diversity and selection in the maize starch pathway. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:12959–12962. doi: 10.1073/pnas.202476999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Casa A, et al. Diversity and selection in sorghum: simultaneous analyses using simple sequence repeats. Theor Appl Genet. 2005;111:23–30. doi: 10.1007/s00122-005-1952-5. [DOI] [PubMed] [Google Scholar]
- 78.Chapman MA, et al. A genomic scan for selection reveals candidates for genes involved in the evolution of cultivated sunflower (Helianthus annuus) Plant Cell. 2008;20:2931–2945. doi: 10.1105/tpc.108.059808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yamasaki M, et al. A large-scale screen for artificial selection in maize identifies candidate agronomic loci for domestication and crop improvement. The Plant Cell. 2005;17:2859–2872. doi: 10.1105/tpc.105.037242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hufford KM, et al. Patterns of selection and tissue-specific expression among maize domestication and crop improvement loci. Plant Physiol. 2007;144:1642–1653. doi: 10.1104/pp.107.098988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Myles S, et al. Association mapping: Critical considerations shift from genotyping to experimental design. Plant Cell. 2009;21:2194–2202. doi: 10.1105/tpc.109.068437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 83.Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–731. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- 84.Thornsberry JM, et al. Dwarf8 polymorphisms associate with variation in flowering time. Nature. 2001;28:286–289. doi: 10.1038/90135. [DOI] [PubMed] [Google Scholar]
- 85.Harjes CE, et al. Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science. 2008;319:330–333. doi: 10.1126/science.1150255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aranzana MaJ, et al. Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet. 2005;1:e60. doi: 10.1371/journal.pgen.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhao K, et al. An Arabidopsis example of association mapping in structured samples. PLoS Genet. 2007;3:e4. doi: 10.1371/journal.pgen.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rieseberg LH, Buerkle CA. Genetic mapping in hybrid zones. Am Nat. 2002;159:S36–S50. doi: 10.1086/338371. [DOI] [PubMed] [Google Scholar]
- 89.Buerkle CA, Lexer C. Admixture as the basis for genetic mapping. Trends Ecol Evol. 2008;23:686–694. doi: 10.1016/j.tree.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 90.Jones H, et al. Population-based resequencing reveals that the flowering time adaptation of cultivated barley originated east of the Fertile Crescent. Mol Biol Evol. 2008;25:2211–2219. doi: 10.1093/molbev/msn167. [DOI] [PubMed] [Google Scholar]
- 91.Hamblin MT, et al. Challenges of detecting directional selection after a bottleneck: Lessons from Sorghum bicolor. Genetics. 2006;173:953–964. doi: 10.1534/genetics.105.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Komatsuda T, et al. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proceedings of the National Academy of Sciences. 2007;104:1424–1429. doi: 10.1073/pnas.0608580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang RL, et al. The limits of selection during maize domestication. Nature. 1999;398:236–239. doi: 10.1038/18435. [DOI] [PubMed] [Google Scholar]
- 94.Wang H, et al. The origin of the naked grains of maize. Nature. 2005;436:714–719. doi: 10.1038/nature03863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sugimoto K, et al. Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proceedings of the National Academy of Sciences. 2010 doi: 10.1073/pnas.0911965107. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen KY, et al. Changes in regulation of a transcription factor lead to autogamy in cultivated tomatoes. Science. 2007;318:643–645. doi: 10.1126/science.1148428. [DOI] [PubMed] [Google Scholar]
- 97.Cong B, et al. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat Genet. 2008;40:800–804. doi: 10.1038/ng.144. [DOI] [PubMed] [Google Scholar]
- 98.Wang E, et al. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat Genet. 2008;40:1370–1374. doi: 10.1038/ng.220. [DOI] [PubMed] [Google Scholar]
- 99.Takano-Kai N, et al. Evolutionary history of GS3, a gene conferring grain length in rice. Genetics. 2009;182:1323–1334. doi: 10.1534/genetics.109.103002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shomura A, et al. Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet. 2008;40:1023–1028. doi: 10.1038/ng.169. [DOI] [PubMed] [Google Scholar]
- 101.Song XJ, et al. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet. 2007;39:623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- 102.Xue W, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- 103.Yu Y, et al. Independent losses of function in a polyphenol oxidase in rice: Differentiation in grain discoloration between subspecies and the role of positive selection under domestication. Plant Cell. 2008;20:2946–2959. doi: 10.1105/tpc.108.060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gross BL, et al. Novel Phr1 mutations and the evolution of phenol reaction variation in US weedy rice (Oryza sativa) New Phytol. 2009;184:842–850. doi: 10.1111/j.1469-8137.2009.02957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sasaki A, et al. Green revolution: A mutant gibberellin-synthesis gene in rice. Nature. 2002;416:701–702. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- 106.Monna L, et al. Positional cloning of rice semidwarfing gene, sd-1: Rice “Green Revolution Gene” encodes a mutant enzyme involved in gibberellin synthesis. DNA Res. 2002;9:11–17. doi: 10.1093/dnares/9.1.11. [DOI] [PubMed] [Google Scholar]
- 107.Spielmeyer W, et al. Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proceedings of the National Academy of Sciences. 2002;99:9043–9048. doi: 10.1073/pnas.132266399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nagano H, et al. Genealogy of the “Green Revolution” gene in rice. Genes and Genetic Systems. 2005;80:351–356. doi: 10.1266/ggs.80.351. [DOI] [PubMed] [Google Scholar]
- 109.Zeder MA. Central questions in the domestication of plants and animals. Evolutionary Anthropology: Issues, News, and Reviews. 2006;15:105–117. [Google Scholar]
- 110.Hammer K. Das Domestikationssyndrom. Kulturpflanze. 1984;32:11–34. [Google Scholar]
- 111.Doebley JF, et al. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 112.Tanno K-i, Willcox G. How fast was wild wheat domesticated? Science. 2006;311:1886. doi: 10.1126/science.1124635. [DOI] [PubMed] [Google Scholar]
- 113.Barnaud A, et al. A weed-crop complex in sorghum: The dynamics of genetic diversity in a traditional farming system. Am J Bot. 2009;96:1869–1879. doi: 10.3732/ajb.0800284. [DOI] [PubMed] [Google Scholar]
- 114.Nichols R. Gene trees and species trees are not the same. Trends in Ecology and Evolution. 2001;16:358–364. doi: 10.1016/s0169-5347(01)02203-0. [DOI] [PubMed] [Google Scholar]