Abstract
Poly(A) polymerase I (PAP I), encoded by the pcnB gene, is a major enzyme responsible for RNA polyadenylation in Escherichia coli, a process involved in the global control of gene expression in this bacterium through influencing the rate of transcript degradation. Recent studies have suggested a complicated regulation of pcnB expression, including a complex promoter region, a control at the level of translation initiation and dependence on bacterial growth rate. In this report, studies on transcription regulation of the pcnB gene are described. Results of in vivo and in vitro experiments indicated that (a) there are three σ70-dependent (p1, pB, and p2) and two σS-dependent (pS1 and pS2) promoters of the pcnB gene, (b) guanosine tetraphosphate (ppGpp) and DksA directly inhibit transcription from pB, pS1 and pS2, and (c) pB activity is drastically impaired at the stationary phase of growth. These results indicate that regulation of the pcnB gene transcription is a complex process, which involves several factors acting to ensure precise control of PAP I production. Moreover, inhibition of activities of pS1 and pS2 by ppGpp and DksA suggests that regulation of transcription from promoters requiring alternative σ factors by these effectors of the stringent response might occur according to both passive and active models.
Keywords: pcnB gene, RNA polyadenylation, Transcription initiation, Sigma factors, ppGpp, DksA
Introduction
Activity of bacterial poly(A) polymerase (PAP) was discovered almost 50 years ago (August et al. 1962). This discovery apparently has been overlooked for many years, most probably due to technical problems in demonstrating the presence of poly(A) tails at the ends of short-lived bacterial transcripts. In 1986, Lopilato et al. (1986) reported the presence of the pcnB gene in Escherichia coli. Mutants in this gene significantly influenced copy number of a ColE1-like plasmid pBR322 and its derivatives; thus, the name pcnB (for plasmid copy number) was proposed. Soderbom and Wagner (1998) found effects of the pcnB gene product on degradation of CopA, an antisense RNA involved in the regulation of R1 plasmid replication. In the meantime, Cao and Sarkar (1992) discovered that the main E. coli PAP I, an enzyme catalyzing RNA polyadenylation at the 3′ end, is encoded by pcnB.
In contrast to eukaryotic cells, RNA polyadenylation in bacteria usually leads to its faster degradation (Regnier and Arraiano 2000). This was demonstrated for various specific transcripts, whose half-lives increased significantly in pcnB mutants (O’Hara et al. 1995; Xu and Cohen 1995; Szalewska-Palasz et al. 1998; Blum et al. 1999). Since Mohanty and Kushner (2006) demonstrated that transcripts derived from over 90% of open reading frames are polyadenylated in exponentially growing cultures of E. coli, it could be suggested that expression of most of bacterial genes may be regulated by polyadenylation-dependent mechanisms. One of the first insights suggesting that this may be the case came from studies on a short transcript of bacteriophage λ, called oop RNA, which demonstrated that oop RNA is polyadenylated by PAP I (Wrobel et al. 1998) and that this modification results in its decreased stability (Szalewska-Palasz et al. 1998). In fact, oop RNA transcripts were shown to be polyadenylated more efficiently in slowly growing cells than in rapidly growing bacteria (Jasiecki and Wegrzyn 2003). Recently, Joanny et al. (2007) demonstrated the poly(A)-dependent regulation of expression of the glmS gene in E. coli. Although this regulation appears to be indirect and involves the control of stability of a small regulatory RNA encoded by glmY (Reichenbach et al. 2008; Urban and Vogel 2008), these results suggest that efficiency of polyadenylation of certain RNAs may be different under various environmental conditions, indicating an important regulatory role for this reaction (Szalewska-Palasz et al. 2007b). Since some other small regulatory RNAs were shown to be regulated by polyadenylation (Soderbom and Wagner 1998; Szalewska-Palasz et al. 1998; Wrobel et al. 1998; Viegas et al. 2007), one might speculate that this is a major mechanism for controlling gene expression by this process. Furthermore, PAP I is phosphorylated in E. coli, which significantly influences the activity of this enzyme (Jasiecki and Wegrzyn 2006a). PAP I is located in both the cytoplasm and cytoplasmic membrane (Jasiecki and Wegrzyn 2005), thus any factors affecting specific localization of molecules of the pcnB gene product may indirectly cause alterations in expression of other genes.
Since RNA polyadenylation appears to be a global regulatory process in the physiological control of gene expression in bacteria and of replication of extrachromosomal genetic elements (plasmids and phages) (for a review, see Szalewska-Palasz et al. 2007b), discovery of mechanisms controlling production of PAP I is necessary for both understanding these crucial cellular processes and possibly employing them in genetic engineering and biotechnology.
In this light, perhaps surprisingly, regulation of expression of the pcnB gene in E. coli is relatively poorly understood. Liu and Parkinson (1989), on the basis of nucleotide sequence analysis, proposed a location of the putative pcnB gene promoter (named pS1 in this report) (Fig. 1). Subsequent studies indicated, however, that this region is located downstream of the actual translation start codon (a non-canonical initiation codon AUU), and another transcription start site (called pB) was discovered by Binns and Masters (2002) (Fig. 1). It appeared that regulation of the pcnB gene expression occurs at the stage of translation initiation (Binns and Masters 2002); however, subsequent studies strongly suggested that transcription control may contribute significantly to the modulation of PAP I production (Jasiecki and Wegrzyn 2003). In fact, it was demonstrated that apart from the pB promoter (Binns and Masters 2002), there are two other promoters, designated p1 and p2 (Fig. 1), that might potentially influence pcnB expression (Jasiecki and Wegrzyn 2006b).
Fig. 1.
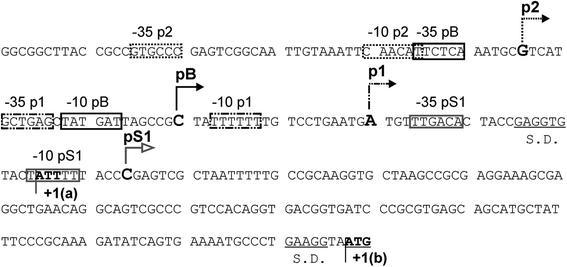
Nucleotide sequence of the pcnB promoter region. The previously reported (or proposed) transcription start sites from particular promoters (pS1, p1, pB and p2) are marked, with first transcribed nucleotides indicated by large bold letters and arrows showing the direction of transcription, and −35 and −10 boxes are shown in frames. The Shine–Dalgarno (S.D.) sequences are underlined, and translation initiation codons (ATT and ATG, called +1(a) and +1(b), respectively) are indicated by bold letters
The discovery of the dependence of pcnB expression on bacterial growth rate (Jasiecki and Wegrzyn 2003) might suggest that factors responsible for modulation of transcription in response to various nutritional conditions could be involved in the control of PAP I production. In fact, the stringent control, a bacterial response to amino acid starvation and many other nutritional and environmental stresses, is a global regulatory system that ensures optimal energy usage under unfavorable growth conditions (for recent reviews, see Potrykus and Cashel 2008; Wu and Xie 2009). Molecules recognized as alarmons of this control system have been considered to belong to the most far-reaching effectors, whose major role is an immediate response to rapidly changing environmental conditions (Szalewska-Palasz et al. 2007b).
There are two major effectors of the stringent response. The first one is a couple of two specific nucleotides, guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp), collectively called (p)ppGpp and rapidly produced in response to variety of physico-chemical and nutritional stresses (for reviews, see Magnusson et al. 2005; Potrykus and Cashel 2008). Since in E. coli ppGpp appears to be more abundant and of higher physiological importance than pppGpp, it is often mentioned as the main stringent response factor. In fact, ppGpp can directly and indirectly regulate expression of variety of genes, important for survival of bacterial cells. It interacts with RNA polymerase (Artsimovitch et al. 2004); however, its exact binding site is still controversial (Vrentas et al. 2008), and it does not introduce any long-lasting conformational alterations to this enzyme (Szalewska-Palasz 2008). The second effector of the stringent response is the DksA protein, which was found as an indispensable factor for ppGpp-mediated effects on ribosomal promoters in vivo (Paul et al. 2004). This protein interacts with RNA polymerase (Perederina et al. 2004), and it was suggested that DksA is a co-factor of the stringent response, enhancing both negative and positive effects of ppGpp on activities of promoters (Paul et al. 2005). Nevertheless, recent reports indicated that roles of ppGpp and DksA in transcription regulation can be independent and even opposing (Magnusson et al. 2007; Lyzen et al. 2009; Merrikh et al. 2009).
The major mechanism of direct ppGpp-mediated transcription regulation was proposed to be a decrease of stability of the promoter-RNA polymerase open complexes (Barker et al. 2001a, b). According to this “active model”, promoters that form unstable open complexes are inhibited by ppGpp, while those forming relatively stable complexes might be either stimulated by this nucleotide or insensitive to its presence. Although it was demonstrated that a promoter forming an extremely stable open complex with RNA polymerase can also be negatively regulated by ppGpp both in vivo and in vitro (Potrykus et al. 2002), the model assuming that ppGpp negatively regulates only promoters which form unstable open complexes is generally accepted as the best explanation of impairment of transcription from promoters dependent on the major σ factor in E. coli, σ70 (Srivatsan and Wang 2008).
Another mechanism is proposed to explain effects of ppGpp and DksA on transcription from promoters dependent on alternative σ factors. The stringent response effectors have been reported as positive regulators of various promoters recognized by RNA polymerase holoenzymes containing σ subunits known to operate under specific environmental or physiological conditions, e.g. σ54 (σN), σ38 (σS) and σ24 (σE) (for a review, see Srivatsan and Wang 2008). Thus, the “passive model” of ppGpp- and DksA-mediated transcription regulation was proposed (summarized by Srivatsan and Wang 2008). According to this model, stimulation of transcription by ppGpp and DksA under stress conditions is a global consequence of strong negative effects of these effectors on functions of stringently regulated powerful σ70-dependent promoters. Namely, dramatic down-regulation of transcription from rRNA promoters leads to a significant increase in cellular availability of the core of RNA polymerase for association with alternative σ factors. Such a type of regulation has been experimentally supported mainly for σN-dependent promoters (Bernardo et al. 2006, 2009; Szalewska-Palasz et al. 2007a); nevertheless, it is generally accepted for promoters recognized by other alternative σ factors, especially in the light of several examples of ppGpp- and DksA-mediated stimulation of transcription from σS-dependent promoters (reviewed by Srivatsan and Wang 2008). On the other hand, σ28-dependent promoters can be inhibited rather than stimulated by ppGpp and DksA (Lemke et al. 2009), indicating that the above-presented mechanism must be more complicated.
In this report, we present results of our studies on the control of pcnB gene transcription. Our experiments indicated that this control is complex, and involves activities of several factors, including σ70 and σS subunits of RNA polymerase, ppGpp and DksA.
Materials and methods
Bacterial strains, plasmids and growth conditions
E. coli K-12 strain MG1655 (Xiao et al. 1991) and its rpoD800 (Grossman et al. 1983), relA spoT (ppGpp0) (Szalewska-Palasz et al. 2007a), dksA::kan (DksA0) (Kang and Craig 1990) and ΔpcnB::kan (pcnB) (Wrobel et al. 1998) derivatives were used. The relA spoT dksA (ppGpp0 DksA0) mutant was constructed by P1vir transduction of the ΔdksA::tet allele from TE8114 (Brown et al. 2002) into the above-described relA spoT strain. Due to frequent accumulation of suppressor mutations by ppGpp0 and DksA0 strains, bacteria used for experiments were checked for specific phenotypes, like amino acid auxotrophy. The rpoS mutant was constructed by P1vir transduction of the rpoS359::Tn10 from RH90 (Lange and Hengge-Aronis 1991) into MG1655. The rpoS relA spoT strain was constructed by P1vir transduction of rpoS359::Tn10 into ppGpp0.
Plasmid pTE103 (Elliott and Geiduschek 1984) was employed. For construction of the plasmid pTE103-F4-fanti, a PCR fragment, flanked by primers Fuzja 4 (5′AGA ATT CTC ATT CAT CGC CGT GAT G) and fanti (5′-AGG ATC CAA ATT AGC GAC TCG GGT) (Jasiecki and Wegrzyn 2006b) was cloned into BamHI and EcoRI sites of pTE103. This construct was verified by DNA sequencing.
Bacteria were cultured either in LB medium (Sambrook et al. 1989) or in a minimal medium MM (Jasiecki and Wegrzyn 2003) supplemented with various carbon sources: 0.2% glucose (MMGlu), 0.2% glycerol (MMGly), 0.6% sodium succinate (MMSuc) or 0.8% sodium acetate (MMAce).
Proteins and ppGpp
DksA protein, and RNA polymerase core and various σ factors were purified according to previously reported procedures (Gamer et al. 1992; Janaszak et al. 2007; Szalewska-Palasz et al. 2007a). The ΔHTH-PspF protein was purified as already described (Jovanovic et al. 1999). His-tagged proteins were purified by Ni-affinity chromatography using the BioLogic LP chromatography system (Bio-Rad). ppGpp was purified as described previously (Lyzen et al. 2009). The σS and σ54 factors were provided by Dr. Anna Janaszak (Medical University of Gdańsk, Poland). The σE factor was provided by Ewa Stec (University of Gdańsk, Poland). The E. coli RNA polymerase core, the Eσ70 holoenzyme, and the DksA protein were purified as described previously by Lyzen et al. (2009).
RNA isolation
Total RNA was isolated from either exponentially growing bacterial cultures (samples were withdrawn at A 575 of 0.4) or cultures being at the stationary phase (at the beginning of the plateau value of A 575 in the bacterial growth curve). 10 ml of the culture was centrifuged (3,000g, 10 min) and RNA was isolated from bacterial cells using the Total RNA kit (A&A Biotechnology, Gdynia, Poland), according to manufacturer’s instruction. Quality of RNA samples was verified electrophoretically and spectrophotometrically.
Primer extension analysis
The primer extension reactions were performed essentially as described by Janaszak et al. (2007). Briefly, either 50 μg of total RNA or 1.5 μg of specific pcnB mRNA (generated by in vitro transcription), was mixed with 0.5 pmol of 32P-labeled primer. Different primers were used to detect various transcripts: Pr11 (5′-CAG CCT CGC TTT CCT CGC GGC TTA GC) (Jasiecki and Wegrzyn 2006b), pcnTR2 (5′-ACT GTC TCA GTG AAC TCA TCC AT), PS1.rev (5′-TAC ATT ACC TTC AGG GCA TTT TCA), PS2.rev (5′-TTG CTC GAC GCT GAA ATC CTG CCA), OmpA (5′-GCA CCA GTG TAC CAG GTG TTA TC), ompArev (5′-CGG TGA AGG ATT TAA CCG TG), Rsd.1 (5′-ATA TCG TGA CGC CGC TGC TGT TGT), Rsd.rev (5′-TGA CGC GCT CCG TCA GGT TAT CGA G), Fim.B1 (5′-ACT GCG CTC CAT GAA ATA GCC AT), FimB.rev (5′ACG TTG CCA TAC AAA CGC CAT GCT). The mixture was incubated at 85°C for 20 min. Following addition of the M-MulLV buffer (Fermentas, Vilnius, Lithuania), a primer was hybridized to RNA at optimal temperature for 60 min. Then, samples were immediately put into ice-bath, and 1 μl of 25 mM solution of four dNTPs, 1 μl of RevertAid M-MuLV Reverse Transcriptase (Fermentas) and RNase inhibitor (1 unit per μl) were added. The reaction was conducted for 60 min at 42°C, and stopped by addition of 5 μl of the denaturation solution and incubation at 95°C for 5 min. The reaction products were separated during electrophoresis in 7% polyacrylamide gel with 8 M urea; products of the sequencing reaction, with the use of the same primer, were separated in the same gel. The bands were visualized by autoradiography and analyzed using the Quantity One software (Bio-Rad).
Each primer extension experiment was repeated at least three times to assess reproducibility.
In vitro transcription
The in vitro transcription reactions in the presence of ppGpp and/or DksA were performed as described previously (Lyzen et al. 2009), with minor modifications. The following templates were used: for pcnB promoters, plasmid pTE103-F4-fanti; for measurement of p R activity, pTE103-derived plasmid described by Lyzen et al. (2009); and for studies on p L activity, plasmid pVI901 (Szalewska-Palasz et al. 2007a). Plasmid DNA was purified by CsCl density gradient ultracentrifugation and chromatography, using P-30 columns (Bio-Rad). Reactions were conducted in the final volume of 20 μl, in the transcription buffer (50 mM Tris–HCl pH 8, 10 mM MgCl2, 10 mM β-mercaptoethanol, 150 mM KCl, 10 μg/ml BSA). Following preliminary experiments, in which optimal concentrations of the template and RNA polymerase were estimated, further experiments were performed with either 4 or 8 nM template and either 30 or 60 nM RNA polymerase holoenzyme. DNA template, RNA polymerase holoenzyme and indicated amounts of ppGpp and/or DksA were incubated for 10 min at 37°C. Following addition of a nucleotide mix (to final concentrations: 1 mM ATP, 150 μM GTP, 150 μM CTP, 15 μM UTP, and 1 μCi [α-32P]UTP, 3,000 Ci/mmol) the mixture was incubated for 12 min at 37°C. Then, heparin was added (to 0.1 mg/ml) and the incubation was continued for 5 min. The reaction was stopped by addition of 5 μl of the denaturation solution (150 mM EDTA, 1.05 M NaCl, 7 M urea, 10% glycerol, 0.0375% xylene cyanol, 0.0375% bromophenol blue) and incubation for 5 min at 95°C. The reaction products were separated by electrophoresis in a 4% polyacrylamide gel containing 8 M urea in the TBE buffer (Sambrook et al. 1989), and visualized by autoradiography using PhosphorImager (Bio-Rad). The results were quantified densitometrically, employing Quantity One software (Bio-Rad).
In vitro transcription with RNA polymerase holoenzymes containing different σ factors: σ70, σ54, σ32, σE or σS (Eσ70, Eσ54, Eσ32, EσE or EσS, respectively) was performed in either the TN buffer (50 mM Tris–HCl pH 7.5, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.1 mM EDTA), for Eσ70, Eσ32, EσE and EσS, or the STA buffer (25 mM Tris–acetate pH 8.0, 8 mM magnesium acetate, 10 mM KCl, 1 mM DTT, 3.5% PEG 6000), for Eσ54, in the presence of BSA (100 μg/ml) at 37°C. Before the transcription reactions, σ factors were preincubated with the RNA polymerase core enzyme (at molar ratio 1:4) for 10 min at 37°C. The final volume of the reaction samples was 20 μl, and contained 8 nM template and 30 nM appropriate RNA polymerase holoenzyme. When Eσ54 was used, an unspecific activator protein ΔHTH-PspF, which can stimulate transcription from σ54-dependent promoters independently of the DNA sequence (Jovanovic et al. 1999; Janaszak et al. 2007; Wigneshweraraj et al. 2003), was added to 1 μM together with 2.5 mM ATP, and incubation was continued for 10 min (in experiments with other holoenzymes, this stage was omitted). Following addition of nucleotide mixture (as described above), the reaction was conducted for 12 min, and after addition of heparin, for another 5 min. The reaction was stopped by addition of the denaturation solution and separated electrophoretically (as described above).
When the in vitro transcription reaction products were used subsequently for primer extension experiments, unlabeled UTP (150 μM) was added instead of [α-32P]UTP, and the reaction volume was 100 μl. Furthermore, the RNA products were purified using the phenol–chloroform extraction and ethanol precipitation (Sambrook et al. 1989).
DNase I footprinting
Primer Pr10 (5′-AGC ACC TTG CGG CAA AAA TTA GCG AC) (Jasiecki and Wegrzyn 2006b) was end-labeled with [γ-32P]ATP and T4 polynucleotide kinase (Fermentas). DNA fragments were PCR amplified using 32P end-labeled Pr10 primer and unlabeled Przedpcn primer (5′-AGA ATC ATG CGC CTG CGT TGC). DNase I footprinting reactions were performed as described previously (Janaszak et al. 2007), in a total volume of 10 μl of the TN buffer (50 mM Tris–HCl pH 7.5, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.1 mM EDTA). The labeled DNA fragments (at final concentration of 10 nM) were incubated for 15 min at 30°C with Eσ70. DNase I (5 × 10−4 units) was added, and after incubation for 1 min at 37°C the reaction was stopped by the addition of EDTA to the final concentration of 50 nM, followed by heating for 5 min at 95°C in the urea loading dye. The reaction products were separated by electrophoresis in a 7% polyacrylamide gel containing 8 M urea, together with the products of DNA sequencing reactions performed using the Pr10 primer and the fmol DNA Cycle Sequencing System (Promega). Each footprinting experiment was repeated at least three times to assess reproducibility.
Results
pB is the major promoter of the pcnB gene
Previous studies indicated that there are at least three promoters located upstream of the pcnB coding sequence in E. coli MG1655 genome, called p1, pB and p2 (see Fig. 1; Jasiecki and Wegrzyn 2006b). These promoters were found by primer extension experiments, in which relatively gentle reaction conditions were used (Jasiecki and Wegrzyn 2006b). Such conditions were allowed to obtain comparable strengths of signals from all these promoters. To distinguish actual activities of p1, pB and p2, we have performed primer extension experiments using significantly more stringent reaction conditions that should cause impairment of signals caused by annealing of the primer to mRNA molecules which occur in low abundance in cells. Relative to the previously reported conditions (Jasiecki and Wegrzyn 2006b), the following changes were made in the protocol used in this work: (a) higher amount of total RNA (50 μg vs. 10–500 ng in the previously reported study) and lower amount of the primer (0.5 vs. 15–20 pm) were used, (b) the denaturation step was longer (20 vs. 5 min) and performed at higher temperature (80 vs. 65°C), and (c) the annealing step was added [incubation for 1 h at the temperature equal to T m of the primer was introduced rather than starting the primer extension reaction immediately after cooling the mixture (composed of RNA and primer molecules) to 42°C, and addition of other components of the reaction].
Under these experimental conditions (i.e. under the “stringent” conditions), a highly predominant signal corresponded to the product of the pB promoter activity (Fig. 2a). The p1- and p2-derived transcripts could also be detected, but only after significantly longer exposure of the electrophoretic gels with labeled reaction products (Fig. 2b). Such a picture of pB was observed when RNA was isolated from bacteria growing in a rich medium (LB), as well as in a minimal medium (MMGlu), but the weaker promoter signals were detected only in samples from the minimal medium (Fig. 2). This is important as previously described experiments were performed either only in LB (when pB was discovered; Binns and Masters 2002) or only in MMGlu (when p1 and p2 were discovered; Jasiecki and Wegrzyn 2006b), thus, suggesting the source of ostensible discrepancies between the results. Nevertheless, it appears that pB is the major promoter for pcnB expression under various growth conditions of E. coli cultures.
Fig. 2.
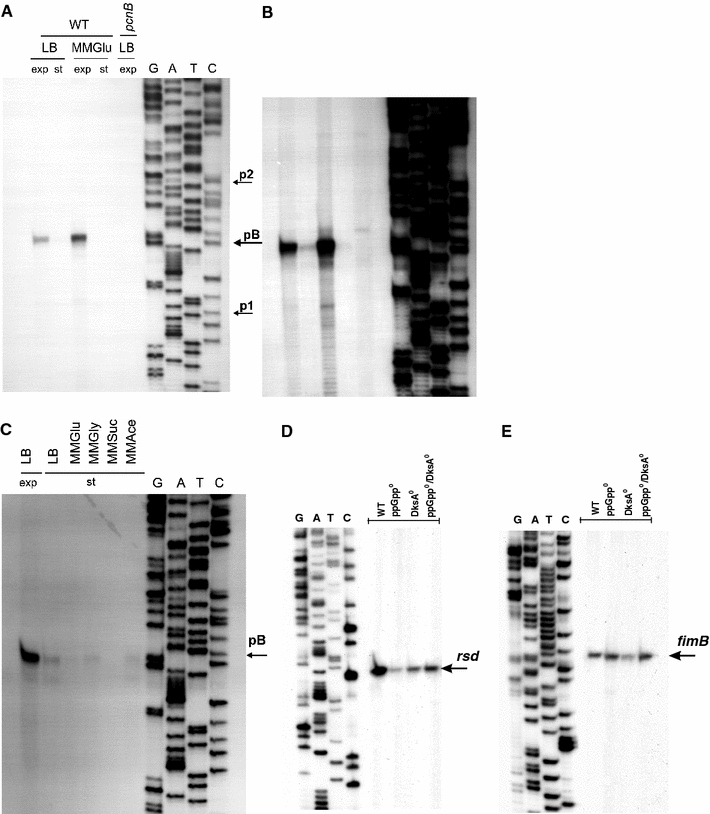
Transcription from the pcnB (a–c), rsd (d) and fimB (e) promoter regions under various growth conditions of bacterial cultures. In experiments shown in a and b, wild-type (WT) E. coli or the pcnB (ΔpcnB::kan) mutant was grown in LB or MMGlu medium, to exponential (exp) or stationary (st) phase of growth at 37°C. Primer extension experiments were performed with primer Pr11 as described in “Materials and methods”, and the products of the reactions were separated during polyacrylamide gel electrophoresis, with the products of the sequencing reaction (performed using the same primer) run at the same gel (lanes G, A, T and C). The position of the products of primer extension reactions, corresponding to transcripts originating from the pB promoter, is shown in a. Positions previously reported as p1 and p2 transcription start sites are also marked by arrows. Apart from results of the experiment performed using the standard procedure (shown in a), results of a significantly longer exposition of the same gel during autoradiography are demonstrated (b) to show products corresponding to activities of other promoters. Analogous experiments, but with E. coli cells cultured at the exponential phase in LB medium (LB exp) and at the stationary phase (st) in various media (LB, MMGlu, MMGly, MMSuc, MMAce) are shown in c. In control experiments, levels of the rsd (d) and fimB (e) transcripts in E. coli wt cells (WT), and ppGpp0 (relA spoT double mutant), DksA0 (dksA::kan mutant) and ppGpp0 DksA0 (relA spoT dksA triple mutant) derivatives cultured at the stationary phase (d) or the exponential phase (e) in LB medium. Primer extension experiments were performed with Rsd.rev and FimB.rev primers, and the products of the reactions were separated during polyacrylamide gel electrophoresis, with the products of the sequencing reaction (performed using the same primer) run at the same gel (lanes G, A, T and C)
Activity of the pB promoter in the stationary phase of growth
Although the pB promoter was active in bacteria cultured in both rich (LB) and minimal (MMGlu) media during the exponential growth (Fig. 2a), we found that its activity is drastically reduced at the stationary phase of growth (Fig. 2c). This impairment of the pB activity was independent of the growth medium, as similar results were observed in LB and MMGlu, as well as in other minimal media, in which various carbon sources were employed (Fig. 2c).
Since we observed a lack or drastically decreased level of particular signals on gels in samples derived from stationary phase bacterial cultures (Fig. 2), one might argue that this effect could result from a putative general process occurring under specific experimental conditions and causing either impaired transcription or enhanced RNA degradation. However, in control experiments, we were able to detect other transcripts, specific for rsd (Fig. 2d) or fimB (Fig. 2e) genes which were demonstrated previously to occur in starved E. coli cells (Jishage and Ishihama 1999) or to be differentially regulated by ppGpp and DksA (Aberg et al. 2008), respectively, in the same samples which were employed for analyses of pcnB transcription. These experiments served also as internal controls for RNA preparation quality.
ppGpp and DksA inhibit transcription from pB
Under conditions of nutritional starvation, which occur also in bacteria being in the stationary phase of growth, the level of ppGpp, a mediator of the stringent response of bacterial metabolism to starvation conditions, increases significantly (for a recent review, see Potrykus and Cashel 2008). It was demonstrated that ppGpp interacts directly with RNA polymerase. Recent studies indicated that the DksA protein plays an important role in modulation of the ppGpp action on transcription efficiency from various promoters (for reviews, see Szalewska-Palasz et al. 2007b; Potrykus and Cashel 2008). Therefore, we have estimated levels of pB-derived transcripts in wild-type cells as well as in the relA spoT mutant (unable to produce ppGpp, called ppGpp0), dksA mutant (called DksA0), and the relA spoT dksA strain (called ppGpp0 DksA0).
Since expression of the ompA gene is maintained at constant levels in relA strains (Durfee et al. 2008), we have estimated abundance of ompA transcripts (Fig. 3a) in the tested samples to normalize results according to this internal control. Increased levels of transcripts derived from pB were observed in exponentially growing bacteria devoid of ppGpp, DksA or both (Fig. 3b). However, even more dramatic changes in abundance of these transcripts could be observed when RNA samples from stationary phase cells were analyzed. The signal in the sample from the dksA mutant was significantly higher than that from the wild-type strain, and levels of the studied transcript derived from both ppGpp0 and ppGpp0 DksA0 strains were comparable to those estimated for exponentially growing wild-type cells (Fig. 3c). Therefore, we suggest that a combined action of ppGpp and DksA is responsible for dramatically decreased transcription from the pB promoter during stationary phase of bacterial culture growth.
Fig. 3.
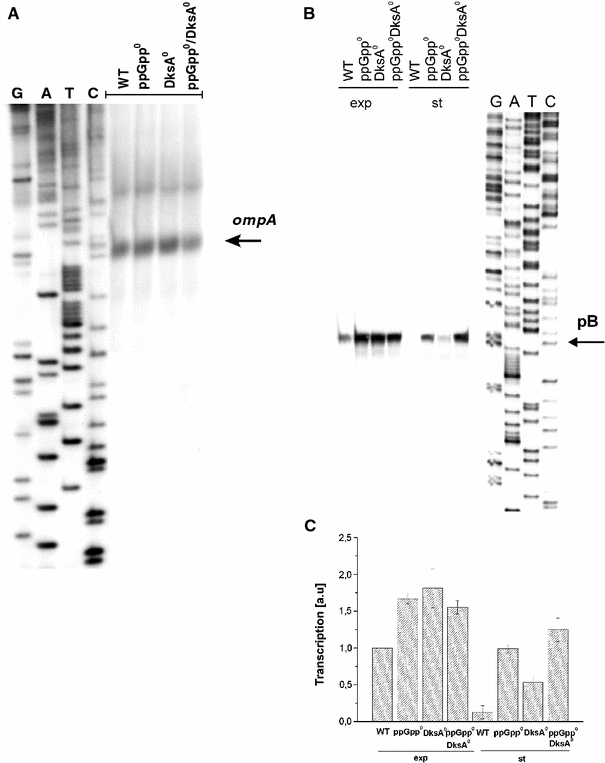
Effects of ppGpp and DksA on the ompA gene expression (a) and the pB promoter activity (b, c). Wild-type (WT) E. coli or ppGpp0 (relA spoT double mutant), DksA0 (dksA::kan mutant) and ppGpp0 DksA0 (relA spoT dksA triple mutant) strains were cultured in the LB medium at 37°C to exponential (a and exp in b) or stationary (st in b) phase of growth. Primer extension experiments were performed as described in “Materials and methods”, and the products of the reactions were separated during polyacrylamide gel electrophoresis, with the products of the sequencing reaction (performed using the same primer) run at the same gel (lanes G, A, T and C). a and b show representative autoradiograms (positions of the products of primer extension reactions, corresponding to transcripts originating from ompA and pB promoters, are shown). c demonstrates quantification (by densitometric analysis) of the results exemplified in b (mean values from three independent experiments are shown in c with error bars indicating SD)
Results of the in vivo experiments, suggesting inhibition of pB activity by ppGpp and DksA, were confirmed in the in vitro transcription reactions, in which production of pB-initiated transcripts in the presence or absence of ppGpp, DksA or both, was tested. DksA weakly inhibited this transcription, while significantly more pronounced impairment of pB activity was caused by ppGpp (Fig. 4). Importantly, a dramatic decrease in the level of pB-derived transcripts was observed when both DksA and ppGpp were present in the reaction mixture (Fig. 4). In control experiments, we have demonstrated that effects of ppGpp and DksA on the activity of the λp R promoter were very different from those found for pB (Fig. 4), indicating specificity of the effects on pB and expected activity of purified reaction compounds.
Fig. 4.
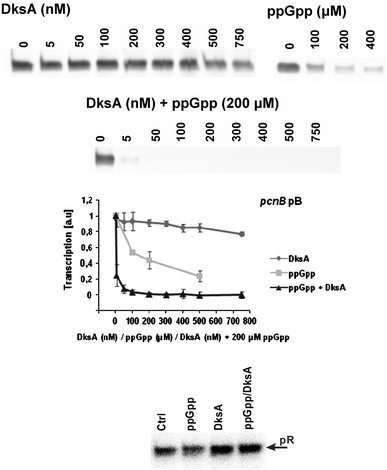
Effects of DksA and ppGpp on in vitro transcription from the pB promoter. Representative results are shown on autoradiograms, and the summary of the results (mean values from three experiments with error bars indicating SD) is shown at the diagram (the presence of following factors in the reaction mixture is shown: diamonds DksA, squares ppGpp, triangles DksA and ppGpp). In control experiments, analogous reactions were performed with the λp R promoter (the bottom panel), with either no additional factors (Ctrl), ppGpp (200 μM), DksA (400 nM) or ppGpp and DksA (200 μM and 400 nM, respectively)
σ70- and σS-dependent transcription of pcnB
The pB promoter sequence, proposed by Binns and Masters (2002), may correspond to the sequence recognized by RNA polymerase holoenzyme containing the σ70 subunit (Eσ70) (Fig. 1). However, since some E. coli σ factors may be responsible for recognizing similar promoter sequences, we tested whether pB is a σ70-dependent promoter indeed.
The rpoD800 mutation results in production of the σ70 subunit whose activity is impaired at elevated temperatures (Liebke et al. 1980; Lowe et al. 1981). We found that abundance of the pB-derived transcripts increased in wild-type bacteria shortly (15 min) after the transfer of cultures from 37 to 45°C, while the signals were comparable in the rpoD800 mutant before and after the temperature shift (Fig. 5).
Fig. 5.
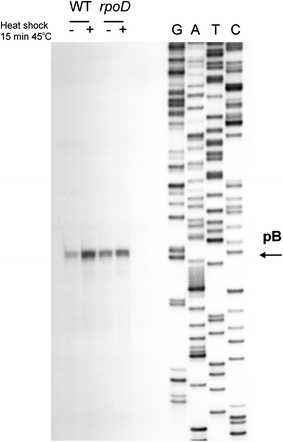
Effects of heat shock on levels of pB-derived transcripts in E. coli wild-type (WT) bacteria and the rpoD800 (rpoD) mutant. Bacteria were cultured to A 575 of 0.4 (exponential phase) and then one half of the culture was transferred to 45°C (+) while the second half remained at 37°C (−). Following further cultivation for 15 min, RNA was isolated and primer extension experiments were performed as described in “Materials and methods”. The position of the products of primer extension reactions, corresponding to transcripts originating from the pB promoter, is shown
DNA footprinting experiments, performed with Eσ70 and the template encompassing the pB promoter region, indicated that the RNA polymerase holoenzyme binds to this DNA fragment, though apparently the promoter was not fully occupied with the protein, which may support previous suggestions (Binns and Masters 2002; Jasiecki and Wegrzyn 2006b) that pB is not a strong promoter (Fig. 6).
Fig. 6.
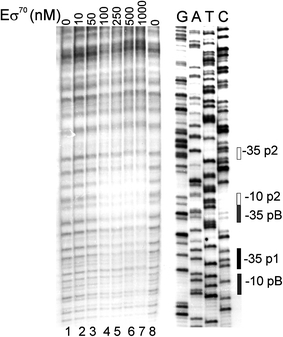
Interaction of Eσ70 with the promotor region of the pcnB gene as assessed by DNase I footprinting. A 32P-labeled 0.3 kb DNA fragment was incubated with 0 nM (negative control, lanes 1 and 8), 10 nM (lane 2), 50 nM (lane 3), 100 nM (lane 4), 250 nM (lane 5), 500 nM (lane 6) or 1,000 nM (lane 7) Eσ70. The footprinting experiments were performed as described in “Materials and methods”. The −10 and −35 sequences of p1, pB and p2 are indicated
The results described above strongly suggested that pB is a Eσ70-dependent promoter. However, to verify this hypothesis, we have performed in vitro transcription reactions with the DNA template containing the pB promoter sequence and RNA polymerase holoenzymes containing different σ subunits, namely: σ70, σ54, σ32, σS, and σE. To identify precisely products of these reactions, the obtained transcripts were subjected to primer extension reactions. As demonstrated in Fig. 7, specific products were obtained when Eσ70 or EσS was used in the in vitro transcription reaction. No products could be detected in experiments with Eσ54, Eσ32, and EσE (data not shown). Interestingly, it appears that apart from pB, which is a σ70-dependent promoter but can also be weakly recognized by EσS (Fig. 7), there is another promoter, whose activity is weak in the presence of Eσ70, but which is active in the presence of EσS (Fig. 7). The transcription start site of this promoter, which we named pS1, corresponds exactly to the localization of the putative promoter of the pcnB gene, proposed previously (Liu and Parkinson 1989) (compare Figs. 1 and 7). That proposal was subsequently suggested to be erroneous (Jasiecki and Wegrzyn 2006b), in the light of the work by Binns and Masters (2002). Nevertheless, our results (Fig. 7) indicated that pS1 may be a functional promoter, but recognized considerably more effectively by EσS than by Eσ70.
Fig. 7.
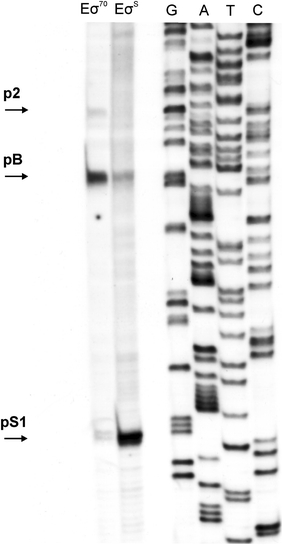
Effects of the presence of different (σ70 or σS) sigma factors on in vitro transcription from pcnB promoters. Primer extension experiments were performed with primer pcnTR2 (as described in Fig. 2) using templates obtained in the in vitro transcription reactions, and either Eσ70 or EσS. Positions corresponding to transcripts derived from pS1, pB and p2 promoters are marked by arrows
Two σS-dependent promoters are located in the region upstream of the pcnB gene
By using the in vitro transcription system allowing to obtain longer transcripts, we found that in the region of the pcnB gene there are two promoters (rather than only one) whose activities are evident in the presence of EσS (Fig. 8a). Apart from the σ70-dependent transcription signal from pB, and the two σS-dependent signals (pS1 and pS2), no other transcription initiation signals could be detected in this region with the use of all other tested σ factors (Fig. 8a).
Fig. 8.
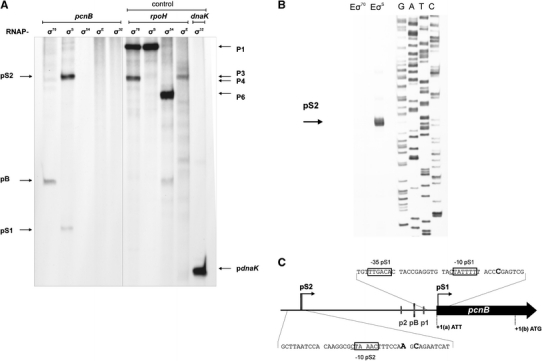
In vitro transcription from the region of the pcnB gene (a) and mapping of the pS2 transcription start sites (b). In experiments shown in a, E. coli RNA polymerase holoenzymes bearing different σ factors (marked above particular lanes) were used in the reactions performed as described in “Materials and methods”. Positions of transcripts derived from pB, pS1 and pS2 promoters are indicated. In the control experiment, activities of all holoenzymes were demonstrated by employing DNA templates containing promoters specific for various σ factors (templates with the rpoH gene promoter region and the dnaK gene promoter region, described by Janaszak et al. 2007, were used); positions of bands corresponding for transcripts originating from particular promoters are indicated. In experiments shown in b, primer extension experiments with primer PS2.rev were performed using the products of in vitro transcription reactions as templates. In c, proposed localization of pS1 and pS2 promoters in the region upstream of the coding sequence of the pcnB gene is shown
The region of transcription initiation from the second σS-dependent promoter for pcnB, called pS2, was determined in primer extension experiments, in which RNAs obtained in in vitro transcription experiments were employed as templates (Fig. 8b). Note that there are perhaps at least two alternative start sites from pS2, located in the region between positions −263 and −265 relative to the translation start codon ATT.
The regions of the pS1 and pS2 promoters are shown in Fig. 8c. While putative −10 and −35 boxes can be easily predicted for pS1, in the case of pS2, it is possible to predict only −10 element (TAAACT), whereas there is no obvious −35 box. On the other hand, both pS1 and pS2 sequences bear the C residue in the neighborhood of the −10 element. Such a residue is known to be crucial for selective recognition of promoters dependent on EσS (Lacour et al. 2003).
Activity of the σS-dependent promoters of the pcnB gene in vivo
We asked whether the σS-dependent promoters detected in in vitro experiments are active in vivo. To address this question, primer extension experiments were performed. Wild-type bacteria as well rpoS and ppGpp0 mutants were cultured in LB broth, and samples were withdrawn at the exponential and stationary phases of growth. Following RNA isolation, the primer extension experiments revealed no detectable signal from the pS1 promoter in exponentially growing cells, as expected. Surprisingly, also no such a signal could be observed in samples from stationary phase wild-type bacteria (Fig. 9). However, a signal corresponding to the pS1-initiated transcription was evident in the ppGpp0 mutant. This transcription was σS-dependent as it was totally impaired in the rpoS mutant, irrespective of the presence or absence of ppGpp (Fig. 9).
Fig. 9.
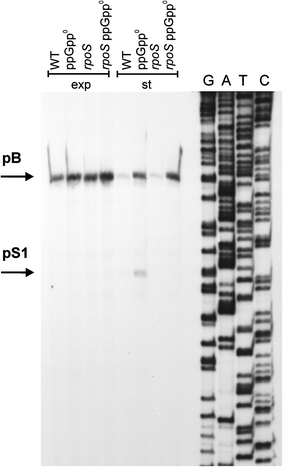
Activity of the pS1 promoter in vivo and its dependence on the rpoS gene function. Wild-type (WT) E. coli or ppGpp0 (relA spoT double mutant), rpoS and ppGpp0 rpoS (relA spoT rpoS triple mutant) strains were cultured in the LB medium at 37°C to exponential (exp) or stationary (st) phase of growth. Primer extension experiments were performed with primer PS1.rev as described in “Materials and methods”, and the products of the reactions were separated during polyacrylamide gel electrophoresis, with the products of the sequencing reaction (performed using the same primer) run at the same gel (lanes G, A, T and C)
Results analogous to those described above were obtained when the pS2 promoter was tested. Its activity could be detected only in the stationary phase of bacterial growth in the absence of ppGpp and in the presence of σS activity (Fig. 10).
Fig. 10.
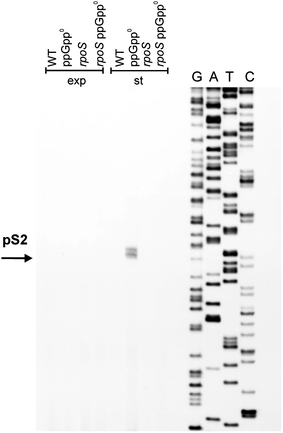
Activity of the pS2 promoter in vivo and its dependence on the rpoS gene function. Wild-type (WT) E. coli or ppGpp0 (relA spoT double mutant), rpoS and ppGpp0 rpoS (relA spoT rpoS triple mutant) strains were cultured in the LB medium at 37°C to exponential (exp) or stationary (st) phase of growth. Primer extension experiments were performed with primer PS2.rev as described in “Materials and methods”, and the products of the reactions were separated during polyacrylamide gel electrophoresis, with the products of the sequencing reaction (performed using the same primer) run at the same gel (lanes G, A, T and C)
The pS1 and pS2 promoters are negatively regulated by ppGpp in vivo
Since pS1 and pS2 activities appeared to be negatively regulated by ppGpp, we have tested their responses to lack of either one or two major effectors of the stringent response, ppGpp and DksA, in bacteria from stationary phase of growth. Both promoters were inactive in the presence of ppGpp, irrespective of the activity of DksA (Fig. 11). These results suggest that ppGpp is the main negative regulator of pS1 and pS2 in vivo.
Fig. 11.
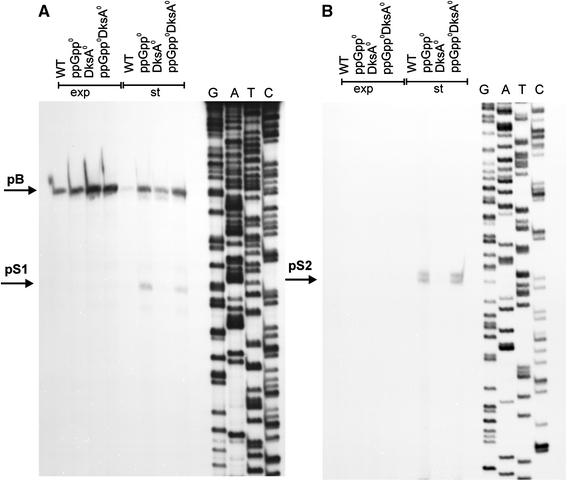
Effects of ppGpp and DksA on the pS1 (a) and pS2 (b) promoter activity. Wild-type (WT) E. coli or ppGpp0 (relA spoT double mutant), DksA0 (dksA::kan mutant) and ppGpp0 DksA0 (relA spoT dksA triple mutant) strains were cultured in the LB medium at 37°C to exponential (exp) or stationary (st) phase of growth. Primer extension experiments were performed as described in “Materials and methods”, with the use of PS1.rev primer (left panel) and PS2.rev primer (right panel), and the products of the reactions were separated during polyacrylamide gel electrophoresis, with the products of the sequencing reaction (performed using the same primers) run at the same gel (lanes G, A, T and C). Positions corresponding to pB, pS1 and pS2 transcription start sites are shown
The pS1 and pS2 promoters are negatively regulated by ppGpp and DksA in vitro
To test whether the ppGpp-mediated negative regulation of activities of pS1 and pS2 in vivo is direct or indirect, we have performed in vitro transcription experiments. Again, both promoters responded in the same way to the presence of ppGpp and/or DksA in the reaction mixture. The DksA protein alone had little, if any, effect on transcription from both tested promoters (Fig. 12a–d). A marked and concentration-dependent impairment of pS1 and pS2 activities could be detected in the presence of ppGpp. These results indicate that the effects of this nucleotide on transcription from these two promoters are direct. Although DksA alone was not able to inhibit activities of pS1 and pS2 significantly, this protein enhanced the inhibitory effects of ppGpp, when present together with this nucleotide in the reaction mixture (Fig. 12a–d).
Fig. 12.
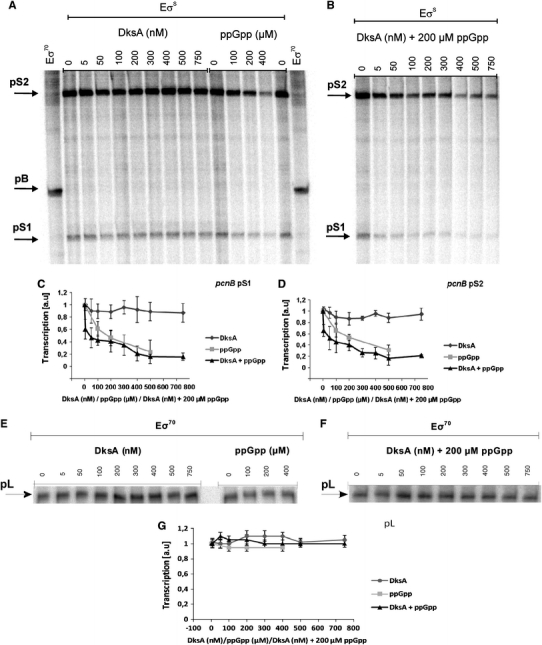
Effects of DksA and ppGpp on in vitro transcription from the pS1, pS2 (a–d) and λp L (e–g) promoters. Representative results are shown on autoradiograms (a, b, e and f). The presence and concentrations of particular factors in the reaction mixtures are indicated and positions of bands corresponding to certain transcripts are shown. Summaries of the results (mean values from three experiments with error bars indicating SD) are shown at the diagrams (c, d and g). The presence of following factors in the reaction mixtures is shown: DksA (diamonds), ppGpp (squares), DksA and ppGpp (triangles)
To test whether the effects of ppGpp and DksA are specific to pS1 and pS2, we have performed control experiments with a template containing a promoter demonstrated previously to be unaffected by these factors. The λp L promoter appeared to be a good candidate for such a control. Indeed, we found no significant effects of ppGpp and/or DksA on pL-initiated transcription (Fig. 12e–g). Therefore, we conclude that there is a specific negative regulation of pS1 and pS2 by ppGpp and DksA.
Discussion
Since polyadenylation of bacterial RNA plays a role in the regulation of expression of as many as 90% of genes (for a review, see Regnier and Hajnsdorf 2009), it is clear that determination of mechanisms leading to different efficiency of adding poly(A) tails at the 3′ ends of transcripts is required to understand this specific control process. On the other hand, regulation of RNA polyadenylation efficiency in E. coli is relatively poorly understood. It was demonstrated that both production of PAP I and polyadenylation reaction are inversely correlated to bacterial growth rate (Jasiecki and Wegrzyn 2003). However, only basic information on expression of the pcnB gene, coding for PAP I, was available to date. It appeared that there are at least three promoters located upstream of the pcnB coding sequence (Jasiecki and Wegrzyn 2006b) and that synthesis of PAP I may also be regulated at the level of translation initiation (Binns and Masters 2002). Moreover, PAP I was found to be a substrate for phosphorylation, and this modification may influence the enzyme activity (Jasiecki and Wegrzyn 2006a).
Results demonstrated in this report indicated that among all pcnB promoters, pB is the major one in bacteria growing in both rich and minimal media. We also conclude that p1 and p2 may have only minor roles in the control of pcnB expression, though they are still active promoters (compare Figs. 2a and 7). The differences in relative activities of pB, p1 and p2 reported in three papers, this one and published previously (Binns and Masters 2002; Jasiecki and Wegrzyn 2006b), may be due to various growth conditions used in different studies (rich vs. minimal media) and various conditions (“stringent” vs. “relaxed”) of primer extension reactions. One should also note that activities of the fusions of pB, p1 and p2 with lacZ, reported previously (Jasiecki and Wegrzyn 2006b), reflected collective activities of different promoters, rather than those of individual promoters, due to close proximity of their sequences. Moreover, these fusions contained only short fragments of sequences located beyond −10 and −35 regions of tested promoters; thus, they might lack important regulatory sequences. This may explain why only weak activity of pB could be detected when a fusion containing only −10 and −35 boxes of this promoter was tested in bacteria grown in minimal media (Jasiecki and Wegrzyn 2006b).
Interestingly, transcription of the pcnB gene was found to be drastically inhibited at the stationary phase of growth. The stringent control alarmone, ppGpp, and the DksA protein are responsible for this inhibition as mutations in genes that lead to the absence of ppGpp and DksA caused restoration of transcription from pB in cells from the stationary phase to the level estimated in exponentially growing wild-type bacteria. Moreover, pB activity was also impaired by ppGpp and DksA in a purified in vitro transcription system. This is interesting in the light of previous findings that both pcnB transcription and PAP I levels are inversely proportional to bacterial growth rate (Jasiecki and Wegrzyn 2003). Since ppGpp levels increase moderately in slowly growing E. coli cells (see Potrykus and Cashel 2008, for a review), one might speculate that effects opposite to those described in this report could occur at the stationary phase of growth. However, it appears that physiological processes may be controlled differentially in slowly growing and non-growing bacteria. We suppose that physiological significance of such a regulation may reflect the control of energy resources in bacterial cells. In slowly growing cells, enhanced expression of certain genes might be required, and under poor nutritional conditions, nucleotides necessary for transcription of these genes could be obtained from rapidly degraded transcripts. Thus, an increased level of pcnB transcription (and expression), and resultant more efficient RNA polyadenylation and its more rapid degradation, might provide sufficient pool of nucleotides. However, when cell growth is totally inhibited, like at the stationary phase, production of an excess of PAP I, and subsequent intensive RNA polyadenylation, would be a waste of energy. Moreover, PAP I was proposed to be toxic for cells when occurring at significantly elevated levels (Binns and Masters 2002); thus, an impaired transcription of pcnB might be a mechanism, which together with a specific regulation of translation from a non-canonical start codon (Binns and Masters 2002) could protect the cell from PAP I toxicity.
The localization of the functional pcnB gene promoter was controversial for several years. Early description of such a promoter, on the basis of nucleotide sequence analysis (Liu and Parkinson 1989), was subsequently criticized as a erroneous, due to demonstration that pcnB translation starts from the AUU codon located upstream of the promoter mentioned above, and identification of another functional promoter (Binns and Masters 2002). Perhaps surprisingly, we have found that the promoter described by Liu and Parkinson (1989), although located downstream of the translation start codon AUU, is also active in vitro, but requires EσS for maximal activity (contrary to pB, which is Eσ70-dependent). This promoter functions also in vivo, but only under specific conditions (inhibition of bacterial culture growth in the absence of ppGpp). As suggested by Liu and Parkinson (1989), there are putative Shine-Dalgarno and AUG sequences that could potentially promote translation of the shorter mRNA produced on the pcnB gene template. It is intriguing that there is a common picture described in the literature, showing that after electrophoretic separation of E. coli proteins and Western blotting, two protein bands react strongly with anti-PAP I antibodies (Jasiecki and Wegrzyn 2003; Mohanty et al. 2004; Mohanty and Kushner 2006). Comparison of migrations of these two proteins during SDS-PAGE indicates that it is possible that there are two variants of PAP I, one whose translation starts from the AUU codon and the second, initiated at the AUG codon proposed by Liu and Parkinson (1989) (compare Fig. 1). The question whether such a putative N-terminally truncated PAP I is really produced in cells and might function in RNA polyadenylation remains to be answered, especially because one might argue that this is unlikely as pS1 requires specific conditions for its activity.
Although it is reasonable to assume that expression of the pcnB gene must be precisely regulated to avoid PAP I overproduction, one should ask about a physiological role for σS-dependent expression of pcnB. We speculate that there might be specific environmental conditions, under which the cells would benefit from enhanced synthesis of PAP I, resulting in liberation of nucleotides from rapidly degraded transcripts, even when cell’s growth is halted. This might facilitate expression of specific genes required for bacterial survival, but if so, transcription of the pcnB gene from σS-dependent promoter(s) would be required. However, as demonstrated in this report, pS1 and pS2 promoters are active only in the absence of ppGpp. In fact, conditions that inhibit cell growth but do not provoke production of ppGpp can occur, indeed, and may be exemplified by sub-lethal concentrations of toxins or antibiotics. In this light, it is worth reminding that treatment of wild-type E. coli cells with chloramphenicol induces the relaxed response (Baracchini and Bremer 1988), which is defined as a lack of ppGpp production under conditions of protein synthesis inhibition.
Results presented in this report may also shed some light on the mechanism(s) of the transcription regulation under conditions of the stringent control. In the generally accepted models of ppGpp/DksA-mediated regulation of transcription, σ70-dependent promoters are either inhibited (if they form unstable open complexes) or stimulated (if they form stable open complexes), and transcription from promoters requiring one of alternative σ factors can be enhanced according to the passive model, due to releasing RNA polymerase core from σ70-dependent promoters and facilitating the use of other σ factors (for a review, see Srivatsan and Wang 2008). In fact, there are examples of σN- and σE-dependent promoters stimulated under condition of the stringent response in vivo (see, e.g., Bernardo et al. 2006; Costanzo and Ades 2006; Szalewska-Palasz et al. 2007a; Costanzo et al. 2008). Stimulation of σN-dependent promoters by ppGpp/DksA observed in vivo could not be detected in in vitro transcription systems with only one type of the RNA polymerase holoenzyme, EσN; however, the in vitro competition experiments could support the passive model for transcription regulation by ppGpp and DksA (Laurie et al. 2003; Szalewska-Palasz et al. 2007a).
There are also many examples of positive regulation of transcription from σS-dependent promoters under conditions of the stringent response and/or stationary phase of growth. Efficient expression of genes coding for integration host factor (Aviv et al. 1994), transketolase A (Harinarayanan et al. 2008), OpgG and OpgH (Costa et al. 2009), cyclopropane fatty acid synthase (Eichel et al. 1999), and glutaredoxin 2 (Potamitou et al. 2002) requires both ppGpp and EσS in vivo. In fact, expression of the rpoS gene (which is under control of a σ70-dependent promoter), coding for σS, is stimulated by ppGpp and DksA (Gentry et al. 1993; Lange et al. 1995; Brown et al. 2002; Bougdour and Gottesman 2007). Thus, one might assume that most of the above-mentioned effects are indirect, and result from an increase in the level of this alternative σ factor. However, among the promoters requiring ppGpp and EσS, there are some which are apparently regulated by ppGpp independently from EσS. Moreover, it was suggested that σS-dependent promoters may require ppGpp for induction even in the presence of high levels of this σ factor (Kvint et al. 2000). Therefore, one might propose that apart from the passive model of the transcription stimulation by ppGpp/DksA under stress conditions, the active model may also operate through direct activation of promoters dependent on alternative σ factors (or, more precisely, at least on σS) by these effectors of the stringent response. The weak point of such a hypothesis would be a fact that only activation of transcription of σS-dependent promoters by ppGpp/DksA was reported to date. Clearly, a lack of the negative regulation would not be expected in the active model.
To our knowledge, the results presented in this report demonstrate for the first time an experimental evidence for direct inhibition of transcription from σS-dependent promoters by ppGpp/DksA. Therefore, they support the proposal that apart from the passive model of ppGpp/DksA-mediated regulation of transcription initiated by RNA polymerase bearing alternative σ factors, the active model may also be operating, at least in the case of EσS.
In summary, results reported in this article indicate that regulation of transcription of the pcnB gene in E. coli is a complex process, controlled by various factors, including different σ subunits of RNA polymerase (σ70 and σS) and the stringent response factors (ppGpp and DksA). It appears that this complex regulation of transcription, together with the control of the pcnB expression at the translation initiation stage, described previously (Binns and Masters 2002), ensures production of PAP I in precisely desired amounts, depending on bacterial growth conditions. Moreover, we demonstrated for the first time that activities of σS-dependent promoters may be inhibited by ppGpp and DksA. This discovery supports the hypothesis that regulation of transcription from promoters requiring alternative σ factors by these effectors of the stringent response might occur according to both passive and active models. Such a regulation may have a physiological importance for survival of bacteria under specific environmental conditions.
Acknowledgments
We thank Katarzyna Brzozowska for her assistance at the early stages of this project. The authors are grateful to Dr. A. Janaszak for providing the σS and σ54 proteins, and to E. Stec for providing the σE protein. We thank Dr. Bernd Bukau for providing the E. coli strain BB2060 (pDMI,1, pUHE211-1) for σ32 overproduction. This work was supported by the Ministry of Science and Higher Education (Poland) (project grants no. NN301 192439 and NN301 161635, to AW and AS-P, respectively).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Aberg A, Shingler V, Balsalobre C. Regulation of the fimB promoter: a case of differential regulation by ppGpp and DksA in vivo. Mol Microbiol. 2008;67:1223–1241. doi: 10.1111/j.1365-2958.2008.06115.x. [DOI] [PubMed] [Google Scholar]
- Artsimovitch I, et al. Structural basis for transcription regulation by alarmone ppGpp. Cell. 2004;117:299–310. doi: 10.1016/S0092-8674(04)00401-5. [DOI] [PubMed] [Google Scholar]
- August JT, Ortiz PJ, Hurwitz J. Ribonucleic acid-dependent ribonucleotide incorporation. I. Purification and properties of the enzyme. J Biol Chem. 1962;237:3786–3793. [PubMed] [Google Scholar]
- Aviv M, Giladi H, Schreiber G, Oppenheim AB, Glaser G. Expression of the genes coding for the Escherichia coli integration host factor are controlled by growth phase, rpoS, ppGpp and by autoregulation. Mol Microbiol. 1994;14:1021–1031. doi: 10.1111/j.1365-2958.1994.tb01336.x. [DOI] [PubMed] [Google Scholar]
- Baracchini E, Bremer H. Stringent and growth control of rRNA synthesis in Escherichia coli are both mediated by ppGpp. J Biol Chem. 1988;263:2597–2602. [PubMed] [Google Scholar]
- Barker MM, Gaal T, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J Mol Biol. 2001;305:689–702. doi: 10.1006/jmbi.2000.4328. [DOI] [PubMed] [Google Scholar]
- Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- Bernardo LM, Johansson LU, Solera D, Skarfstad E, Shingler V. The guanosine tetraphosphate (ppGpp) alarmone, DksA and promoter affinity for RNA polymerase in regulation of sigma-dependent transcription. Mol Microbiol. 2006;60:749–764. doi: 10.1111/j.1365-2958.2006.05129.x. [DOI] [PubMed] [Google Scholar]
- Bernardo LM, Johansson LU, Skarfstad E, Shingler V. Sigma 54-promoter discrimination and regulation by ppGpp and DksA. J Biol Chem. 2009;284:828–838. doi: 10.1074/jbc.M807707200. [DOI] [PubMed] [Google Scholar]
- Binns N, Masters M. Expression of the Escherichia coli pcnB gene is translationally limited using an inefficient start codon: a second chromosomal example of translation initiated at AUU. Mol Microbiol. 2002;44:1287–1298. doi: 10.1046/j.1365-2958.2002.02945.x. [DOI] [PubMed] [Google Scholar]
- Blum E, Carpousis AJ, Higgins CF. Polyadenylation promotes degradation of 3′-structured RNA by the Escherichia coli mRNA degradosome in vitro. J Biol Chem. 1999;274:4009–4016. doi: 10.1074/jbc.274.7.4009. [DOI] [PubMed] [Google Scholar]
- Bougdour A, Gottesman S. ppGpp regulation of RpoS degradation via anti-adaptor protein IraP. Proc Natl Acad Sci USA. 2007;104:12896–12901. doi: 10.1073/pnas.0705561104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L, Gentry D, Elliott T, Cashel M. DksA affects ppGpp induction of RpoS at a translational level. J Bacteriol. 2002;184:4455–4465. doi: 10.1128/JB.184.16.4455-4465.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao GJ, Sarkar N. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc Natl Acad Sci USA. 1992;89:10380–10384. doi: 10.1073/pnas.89.21.10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa CS, Pizarro RA, Anton DN. Influence of RpoS, cAMP-receptor protein, and ppGpp on expression of the opgGH operon and osmoregulated periplasmic glucan content of Salmonella enterica serovar Typhimurium. Can J Microbiol. 2009;55:1284–1293. doi: 10.1139/W09-086. [DOI] [PubMed] [Google Scholar]
- Costanzo A, Ades SE. Growth phase-dependent regulation of the extracytoplasmic stress factor, sigma E, by guanosine 3′,5′-bispyrophosphate (ppGpp) J Bacteriol. 2006;188:4627–4634. doi: 10.1128/JB.01981-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo A, Nicoloff H, Barchinger SE, Banta AB, Gourse RL, Ades SE. ppGpp and DksA likely regulate the activity of the extracytoplasmic stress factor sigma E in Escherichia coli by both direct and indirect mechanisms. Mol Microbiol. 2008;67:619–632. doi: 10.1111/j.1365-2958.2007.06072.x. [DOI] [PubMed] [Google Scholar]
- Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichel J, Chang YY, Riesenberg D, Cronan JE., Jr Effect of ppGpp on Escherichia coli cyclopropane fatty acid synthesis is mediated through the RpoS sigma factor (sigma S) J Bacteriol. 1999;181:572–576. doi: 10.1128/jb.181.2.572-576.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott T, Geiduschek EP. Defining a bacteriophage T4 late promoter: absence of a “−35” region. Cell. 1984;36:211–219. doi: 10.1016/0092-8674(84)90091-6. [DOI] [PubMed] [Google Scholar]
- Gamer J, Bujard H, Bukau B. Physical interaction between heat shock proteins DnaK, DnaJ, and GrpE and the bacterial heat shock transcription factor sigma 32. Cell. 1992;69:833–842. doi: 10.1016/0092-8674(92)90294-M. [DOI] [PubMed] [Google Scholar]
- Gentry DR, Hernandez VJ, Nguyen LH, Jensen DB, Cashel M. Synthesis of the stationary-phase sigma factor sigma S is positively regulated by ppGpp. J Bacteriol. 1993;175:7982–7989. doi: 10.1128/jb.175.24.7982-7989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman AD, Burgess RR, Walter W, Gross CA. Mutations in the Ion gene of E. coli K12 phenotypically suppress a mutation in the sigma subunit of RNA polymerase. Cell. 1983;32:151–159. doi: 10.1016/0092-8674(83)90505-6. [DOI] [PubMed] [Google Scholar]
- Harinarayanan R, Murphy H, Cashel M. Synthetic growth phenotypes of Escherichia coli lacking ppGpp and transketolase A (tktA) are due to ppGpp-mediated transcriptional regulation of tktB. Mol Microbiol. 2008;69:882–894. doi: 10.1111/j.1365-2958.2008.06317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janaszak A, Majczak W, Nadratowska B, Szalewska-Palasz A, Konopa G, Taylor A. A sigma54-dependent promoter in the regulatory region of the Escherichia coli rpoH gene. Microbiology. 2007;153:111–123. doi: 10.1099/mic.0.2006/000463-0. [DOI] [PubMed] [Google Scholar]
- Jasiecki J, Wegrzyn G. Growth-rate dependent RNA polyadenylation in Escherichia coli. EMBO Rep. 2003;4:172–177. doi: 10.1038/sj.embor.embor733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasiecki J, Wegrzyn G. Localization of Escherichia coli poly(A) polymerase I in cellular membrane. Biochem Biophys Res Commun. 2005;329:598–602. doi: 10.1016/j.bbrc.2005.02.025. [DOI] [PubMed] [Google Scholar]
- Jasiecki J, Wegrzyn G. Phosphorylation of Escherichia coli poly(A) polymerase I and effects of this modification on the enzyme activity. FEMS Microbiol Lett. 2006;261:118–122. doi: 10.1111/j.1574-6968.2006.00340.x. [DOI] [PubMed] [Google Scholar]
- Jasiecki J, Wegrzyn G. Transcription start sites in the promoter region of the Escherichia coli pcnB (plasmid copy number) gene coding for poly(A) polymerase I. Plasmid. 2006;55:169–172. doi: 10.1016/j.plasmid.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Jishage M, Ishihama A. Transcriptional organization and in vivo role of the Escherichia coli rsd gene, encoding the regulator of RNA polymerase sigma D. J Bacteriol. 1999;181:3768–3776. doi: 10.1128/jb.181.12.3768-3776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joanny G, et al. Polyadenylation of a functional mRNA controls gene expression in Escherichia coli. Nucleic Acids Res. 2007;35:2494–2502. doi: 10.1093/nar/gkm120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic G, Rakonjac J, Model P. In vivo and in vitro activities of the Escherichia coli σ54 transcription activator, PspF, and its DNA-binding mutant, PspFΔHTH. J Mol Biol. 1999;285:469–483. doi: 10.1006/jmbi.1998.2263. [DOI] [PubMed] [Google Scholar]
- Kang PJ, Craig EA. Identification and characterization of a new Escherichia coli gene that is a dosage-dependent suppressor of a dnaK deletion mutation. J Bacteriol. 1990;172:2055–2064. doi: 10.1128/jb.172.4.2055-2064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvint K, Farewell A, Nystrom T. RpoS-dependent promoters require guanosine tetraphosphate for induction even in the presence of high levels of sigma S. J Biol Chem. 2000;275:14795–14798. doi: 10.1074/jbc.C000128200. [DOI] [PubMed] [Google Scholar]
- Lacour S, Kolb A, Landini P. Nucleotides from −16 to −12 determine specific promoter recognition by bacterial σS-RNA polymerase. J Biol Chem. 2003;278:37160–37168. doi: 10.1074/jbc.M305281200. [DOI] [PubMed] [Google Scholar]
- Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- Lange R, Fischer D, Hengge-Aronis R. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the sigma S subunit of RNA polymerase in Escherichia coli. J Bacteriol. 1995;177:4676–4680. doi: 10.1128/jb.177.16.4676-4680.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie AD, et al. The role of the alarmone (p)ppGpp in sigma N competition for core RNA polymerase. J Biol Chem. 2003;278:1494–1503. doi: 10.1074/jbc.M209268200. [DOI] [PubMed] [Google Scholar]
- Lemke JJ, Durfee T, Gourse RL (2009) DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol Microbiol 74:1368–1379 [DOI] [PMC free article] [PubMed]
- Liebke H, Gross C, Walter W, Burgess R. A new mutation rpoD800, affecting the sigma subunit of E. coli RNA polymerase is allelic to two other sigma mutants. Mol Gen Genet. 1980;177:277–282. doi: 10.1007/BF00267439. [DOI] [PubMed] [Google Scholar]
- Liu JD, Parkinson JS. Genetics and sequence analysis of the pcnB locus, an Escherichia coli gene involved in plasmid copy number control. J Bacteriol. 1989;171:1254–1261. doi: 10.1128/jb.171.3.1254-1261.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopilato J, Bortner S, Beckwith J. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduce plasmid copy number of pBR322 and its derivatives. Mol Gen Genet. 1986;205:285–290. doi: 10.1007/BF00430440. [DOI] [PubMed] [Google Scholar]
- Lowe PA, Aebi U, Gross C, Burgess RR. In vitro thermal inactivation of a temperature-sensitive sigma subunit mutant (rpoD800) of Escherichia coli RNA polymerase proceeds by aggregation. J Biol Chem. 1981;256:2010–2015. [PubMed] [Google Scholar]
- Lyzen R, Kochanowska M, Wegrzyn G, Szalewska-Palasz A. Transcription from bacteriophage lambda pR promoter is regulated independently and antagonistically by DksA and ppGpp. Nucleic Acids Res. 2009;37:6655–6664. doi: 10.1093/nar/gkp676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson LU, Farewell A, Nystrom T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Magnusson LU, Gummesson B, Joksimovic P, Farewell A, Nystrom T. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J Bacteriol. 2007;189:5193–5202. doi: 10.1128/JB.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrikh H, Ferrazzoli AE, Lovett ST. Growth phase and (p)ppGpp control of IraD, a regulator of RpoS stability, in Escherichia coli. J Bacteriol. 2009;191:7436–7446. doi: 10.1128/JB.00412-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty BK, Kushner SR. The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 2006;34:5695–5704. doi: 10.1093/nar/gkl684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- O’Hara EB, Chekanova JA, Ingle CA, Kushner ZR, Peters E, Kushner SR. Polyadenylation helps regulate mRNA decay in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:1807–1811. doi: 10.1073/pnas.92.6.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BJ, et al. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Berkmen MB, Gourse RL. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci USA. 2005;102:7823–7828. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perederina A, et al. Regulation through the secondary channel—structural framework for ppGpp–DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Potamitou A, Neubauer P, Holmgren A, Vlamis-Gardikas A. Expression of Escherichia coli glutaredoxin 2 is mainly regulated by ppGpp and sigma S. J Biol Chem. 2002;277:17775–17780. doi: 10.1074/jbc.M201306200. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Wegrzyn G, Hernandez VJ. Multiple mechanisms of transcription inhibition by ppGpp at the lambda p(R) promoter. J Biol Chem. 2002;277:43785–43791. doi: 10.1074/jbc.M208768200. [DOI] [PubMed] [Google Scholar]
- Regnier P, Arraiano CM. Degradation of mRNA in bacteria: emergence of ubiquitous features. Bioessays. 2000;22:235–244. doi: 10.1002/(SICI)1521-1878(200003)22:3<235::AID-BIES5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Regnier P, Hajnsdorf E. Poly(A)-assisted RNA decay and modulators of RNA stability. Prog Mol Biol Transl Sci. 2009;85:137–185. doi: 10.1016/S0079-6603(08)00804-0. [DOI] [PubMed] [Google Scholar]
- Reichenbach B, Maes A, Kalamorz F, Hajnsdorf E, Görke B. The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli. Nucleic Acids Res. 2008;36:2570–2580. doi: 10.1093/nar/gkn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Soderbom F, Wagner EG. Degradation pathway of CopA, the antisense RNA that controls replication of plasmid R1. Microbiology. 1998;144(Pt 7):1907–1917. doi: 10.1099/00221287-144-7-1907. [DOI] [PubMed] [Google Scholar]
- Srivatsan A, Wang JD. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr Opin Microbiol. 2008;11:100–105. doi: 10.1016/j.mib.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Szalewska-Palasz A. Properties of Escherichia coli RNA polymerase from a strain devoid of the stringent response alarmone ppGpp. Acta Biochim Pol. 2008;55:317–323. [PubMed] [Google Scholar]
- Szalewska-Palasz A, Wrobel B, Wegrzyn G. Rapid degradation of polyadenylated oop RNA. FEBS Lett. 1998;432:70–72. doi: 10.1016/S0014-5793(98)00834-5. [DOI] [PubMed] [Google Scholar]
- Szalewska-Palasz A, et al. Properties of RNA polymerase bypass mutants: implications for the role of ppGpp and its co-factor DksA in controlling transcription dependent on sigma 54. J Biol Chem. 2007;282:18046–18056. doi: 10.1074/jbc.M610181200. [DOI] [PubMed] [Google Scholar]
- Szalewska-Palasz A, Wegrzyn G, Wegrzyn A. Mechanisms of physiological regulation of RNA synthesis in bacteria: new discoveries breaking old schemes. J Appl Genet. 2007;48:281–294. doi: 10.1007/BF03195225. [DOI] [PubMed] [Google Scholar]
- Urban JH, Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas SC, Pfeiffer V, Sittka A, Silva IJ, Vogel J, Arraiano CM. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 2007;35:7651–7664. doi: 10.1093/nar/gkm916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrentas CE, et al. Still looking for the magic spot: the crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J Mol Biol. 2008;377:551–564. doi: 10.1016/j.jmb.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigneshweraraj SR, et al. Enhancer-dependent transcription by bacterial RNA polymerase: the beta subunit downstream lobe is used by sigma 54 during open promoter complex formation. Methods Enzymol. 2003;370:646–657. doi: 10.1016/S0076-6879(03)70053-6. [DOI] [PubMed] [Google Scholar]
- Wrobel B, Herman-Antosiewicz A, Szalewska-Palasz S, Wegrzyn G. Polyadenylation of oop RNA in the regulation of bacteriophage lambda development. Gene. 1998;212:57–65. doi: 10.1016/S0378-1119(98)00127-9. [DOI] [PubMed] [Google Scholar]
- Wu J, Xie J. Magic spot: (p) ppGpp. J Cell Physiol. 2009;220:297–302. doi: 10.1002/jcp.21797. [DOI] [PubMed] [Google Scholar]
- Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- Xu F, Cohen SN. RNA degradation in Escherichia coli regulated by 3′ adenylation and 5′ phosphorylation. Nature. 1995;374:180–183. doi: 10.1038/374180a0. [DOI] [PubMed] [Google Scholar]


