Abstract
The availability of high-affinity agonists for peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) has led to significant advances in our understanding of the functional role of PPARβ/δ. In this study, a new PPARβ/δ antagonist, 4-chloro-N-(2-{[5-trifluoromethyl)-2-pyridyl]sulfonyl}ethyl)benzamide (GSK3787), was characterized using in vivo and in vitro models. Orally administered GSK3787 caused antagonism of 4-[2-(3-fluoro-4-trifluoromethyl-phenyl)-4-methyl-thiazol-5-ylmethylsulfanyl]-2-methyl-phenoxy}-acetic acid (GW0742)-induced up-regulation of Angptl4 and Adrp mRNA expression in wild-type mouse colon but not in Pparβ/δ-null mouse colon. Chromatin immunoprecipitation (ChIP) analysis indicates that this correlated with reduced promoter occupancy of PPARβ/δ on the Angptl4 and Adrp genes. Reporter assays demonstrated antagonism of PPARβ/δ activity and weak antagonism and agonism of PPARγ activity but no effect on PPARα activity. Time-resolved fluorescence resonance energy transfer assays confirmed the ability of GSK3787 to modulate the association of both PPARβ/δ and PPARγ coregulator peptides in response to ligand activation, consistent with reporter assays. In vivo and in vitro analysis indicates that the efficacy of GSK3787 to modulate PPARγ activity is markedly lower than the efficacy of GSK3787 to act as a PPARβ/δ antagonist. GSK3787 antagonized GW0742-induced expression of Angptl4 in mouse fibroblasts, mouse keratinocytes, and human cancer cell lines. Cell proliferation was unchanged in response to either GW0742 or GSK3787 in human cancer cell lines. Results from these studies demonstrate that GSK3787 can antagonize PPARβ/δ in vivo, thus providing a new strategy to delineate the functional role of a receptor with great potential as a therapeutic target for the treatment and prevention of disease.
Introduction
There is considerable interest in targeting nuclear receptors for the treatment and prevention of diseases because of their ability to specifically modulate the transcription of regulatory pathways that influence the cause of diseases ranging from metabolic syndrome to cancer. This is in part because of the successful development and application of nuclear receptor agonists as therapeutic drugs. For example, the fibrate class of hypolipidemic drugs activate peroxisome proliferator-activated receptor-α (PPARα), causing up-regulation of target genes that increase fatty-acid catabolism causing decreased serum lipids and increased insulin sensitivity (Staels et al., 1998). Likewise, rosiglitazone (Avandia; GlaxoSmithKline, Research Triangle Park, NC) and pioglitazone (Actos; Takeda Pharmaceuticals, Deerfield, IL) both activate PPARγ and effectively enhance insulin sensitivity and decrease serum glucose, which is the basis for their use in the treatment of type II diabetes (Gross and Staels, 2007). There is evidence supporting the development of PPARβ/δ agonists for the treatment of metabolic syndrome, diabetes, and obesity, because activating PPARβ/δ increases fatty-acid catabolism, ameliorates insulin resistance, and decreases serum glucose (Billin, 2008). However, targeting PPARβ/δ has been met with significant issues related to clinical safety because of controversial reports surrounding the role of PPARβ/δ in cancer, with some suggesting that activating PPARβ/δ potentiates tumorigenesis whereas others suggest that activating PPARβ/δ attenuates tumorigenesis or has no effect (Peters et al., 2008; Peters and Gonzalez, 2009).
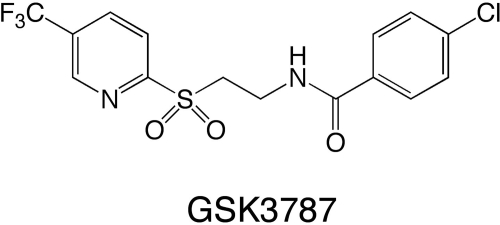
A number of tools have been developed in the last 10 years that have significantly advanced our understanding of the role of PPARβ/δ, in particular the generation of Pparβ/δ-null mouse models (Peters et al., 2000; Barak et al., 2002; Nadra et al., 2006) and high-affinity ligands that are more selective for PPARβ/δ (Shearer and Hoekstra, 2003). Coupling null-mouse models with high-affinity ligands is an excellent approach for delineating the biological function of PPARβ/δ, but there are considerable differences in responses in the different models found reported in the literature. Thus, there is a distinct need to develop alternative approaches to begin to address many of the reported disparities. Toward this goal, the recent identification of GSK0660 (Shearer et al., 2008) and SR13904 (Zaveri et al., 2009) as PPARβ/δ antagonists was a step in the right direction. Unfortunately, these antagonists have limited application because GSK0660 is not bioavailable, and the bioavailability of SR13904 has not been evaluated (Shearer et al., 2008; Zaveri et al., 2009). In contrast, the recently described PPARβ/δ antagonist 4-chloro-N-(2-{[5-trifluoromethyl)-2-pyridyl]sulfonyl}ethyl)benzamide (GSK3787; Fig. 1) exhibited more suitable pharmacokinetic properties, because a maximal concentration (Cmax) of 2.2 ± 0.4 μM with a half-life of 2.5 ± 1.1 h was attainable in mouse serum after oral administration (10 mg/kg) (Shearer et al., 2010). Moreover, GSK3787 is an irreversible antagonist of PPARβ/δ because it forms a covalent bond with a cysteine residue in the ligand binding domain of PPARβ/δ (Shearer et al., 2010). The present study provides further characterization of this new PPARβ/δ antagonist by assessing the ability of GSK3787 to antagonize PPARβ/δ function in vivo, examining the specificity of GSK3787 to antagonize PPARβ/δ using null mouse models, and by determining the effect of GSK3787 on PPARβ/δ function and cell growth in a panel of human cancer cell lines.
Fig. 1.
Chemical structure of GSK3787.
Materials and Methods
Materials.
4-[2-(3-Fluoro-4-trifluoromethyl-phenyl)-4-methyl-thiazol-5-ylmethylsulfanyl]-2-methyl-phenoxy}-acetic acid (GW0742) (Sznaidman et al., 2003), GSK0660 (Shearer et al., 2008), and GSK3787 (Shearer et al., 2010) were synthesized by GlaxoSmithKline. Acetic acid, (2-methyl-4-(((4-methyl-2-(4-(trifluoromethyl)phenyl)-5-thiazolyl)methyl)thio)phenoxy)-(GW501516), N-(2-benzoylphenyl)-O-[2-(methyl-2-pyridinylamino)ethyl]-l-tyrosine hydrochloride (GW1929), and 2-[[4-[2-[[(cyclohexylamino) carbonyl](4-cyclohexylbutyl)amino]ethyl]phenyl]thio]-2-methylpropanoic acid (GW7647) were purchased from Sigma-Aldrich (Steinheim, Germany). Rosiglitazone was purchased from BIOMOL Research Laboratories (Plymouth Meeting, PA).
Animals and Treatments.
Animal experiments were approved by the Institutional Animal Care and Use Committee at Pennsylvania State University, which conforms to the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health. For RNA and DNA analysis, male wild-type and Pparβ/δ-null mice (Peters et al., 2000) were administered vehicle (corn oil), GW0742 (10 mg/kg), GSK3787 (10 mg/kg), or GW0742 and GSK3787 by oral gavage 3 h before euthanasia. After euthanasia, colons were carefully dissected. To isolate colon epithelium, colons were flushed with phosphate-buffered saline, and epithelial cells were scraped from mucosa using a razor blade. The isolated tissues were used for RNA isolation. For glucose-tolerance tests, male wild-type and Pparβ/δ-null mice were administered vehicle (corn oil), GW0742 (10 mg/kg), GSK3787 (10 mg/kg), or rosiglitazone (20 mg/kg) by oral gavage once a day for 2 weeks.
RNA Analysis.
Colon samples were immediately homogenized in TRIzol reagent (Invitrogen, Carlsbad, CA), and total RNA was prepared according to the manufacturer's recommended protocol. The mRNA encoding angiopoietin-like protein 4 (Angptl4), adipose differentiation-related protein (Adrp), and glyceraldehyde 3-phosphate dehydrogenase (Gapdh) was measured by quantitative real-time polymerase chain reaction (qPCR) analysis. cDNA was generated from 2.5 μg of total RNA using a MultiScribe Reverse Transciptase kit (Applied Biosystems, Foster City, CA). The real-time primers for Angptl4, Adrp, and Gapdh have been described previously (Hollingshead et al., 2008). qPCR reactions were carried out using SYBR green PCR master mix (Quanta BioSciences, Gaithersburg, MD) in the iCycler and detected using the MyiQ Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA). The following reaction conditions were used for PCR: 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s, repeated for 45 cycles. Each PCR included a no-template reaction to control for contamination, and all PCR reactions had greater than 85% efficiency. The relative mRNA value for each gene was normalized to the relative mRNA value for Gapdh and analyzed for statistical significance using a two-way analysis of variance with Bonferroni's multiple comparison test (Prism 5.0; GraphPad Software Inc., San Diego, CA).
Chromatin Immunoprecipitation.
Male wild-type and Pparβ/δ-null mice were treated with vehicle, GW0742, GSK3787, or GW0742 and GSK3787 by oral gavage 3 h before euthanasia as described above, and colon and liver were carefully dissected. Colon epithelium samples from five mice per group were individually snap-frozen, pooled, and then pulverized using a mortar and pestle. Cross-linking was performed using a 1% formaldehyde saline solution with sample rotation for 10 min, after which the cross-linking was quenched by the addition of glycine to a final concentration of 125 mM, and samples were rotated for 10 min. Cells were washed twice with phosphate-buffered saline before the addition of lysis buffer (50 mM Tris-HCl, pH 8, 1% SDS, 10 mM EDTA, and protease inhibitor cocktail). The lysates from each treatment group were pooled, and the DNA was sheared to obtain sheared chromatin in the range of 500 to 1500 base pairs with the Diagenode Bioruptor (Diagenode, Sparta, NJ). The sheared chromatin was precleared by the addition of protein A agarose (Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h that was blocked previously with bovine serum albumin/salmon sperm DNA (Invitrogen). The precleared chromatin was immunoprecipitated by gentle agitation with specific antibodies for either anti-PPARβ/δ antibody (Girroir et al., 2008b), anti-acetylated histone H4 (Millipore, Billerica, MA) as a positive control or rabbit IgG (Santa Cruz Biotechnology) as a negative control. After 4 h, the immune complexes were captured by the addition of preblocked protein A agarose (Santa Cruz Biotechnology) and incubated overnight. The beads were washed three times with a low-salt wash buffer (20 mM Tris-HCl, pH 8, 2 mM EDTA, 0.1% sodium deoxycholate, 1% Triton-X, 150 mM NaCl, and protease inhibitor cocktail) and once with a high-salt wash buffer (20 mM Tris-HCl, pH 8, 2 mM EDTA, 0.1% sodium deoxycholate, 1% Triton-X, 500 mM NaCl, and protease inhibitor cocktail). The beads were washed once with 10 mM Tris-HCl, pH 8, and 1 mM EDTA, and the immune complexes were released by the addition of elution buffer (100 mM NaHCO3 and 1% SDS). The formaldehyde cross-links were reversed by overnight incubation at 65°C. Immunoprecipitated DNA was purified by phenol/chloroform/isoamylalcohol (25:24:1) extraction and subjected to real-time qPCR analysis for occupancy in the Adrp or Angptl4 peroxisome proliferator response elements (PPREs). The Adrp PPRE (Chawla et al., 2003) and a primer set spanning this region have been described previously (Hollingshead et al., 2008). The primer set for Angptl4 was designed based on the previous identification of PPREs in intron 3 of the mouse Angtpl4 gene (Heinäniemi et al., 2007). The primers for Angptl4 were 5′-CTAGCCAAGTAGAGGAAAGTTCAGAGC-3′ (forward) and 5′-CCAATCCCTCGGGCAGCTAGC-3′ (reverse). qPCR reactions were carried out as described above. The specific values were normalized to treatment inputs and were verified to be greater than rabbit IgG controls. Promoter occupancy was determined based on fold accumulation to normalized vehicle values.
Reporter Assays.
The LexA-mPPARβ/δ, LexA-mPPARα, LexA-mPPARγ, 7L-TATA initiator module, and PPRE-TATA initiator module plasmids have been described previously (Jérôme and Müller, 1998; Fauti et al., 2005; Naruhn et al., 2010). Transfections were performed with polyethylenimine (average molecular weight, 25,000; Sigma-Aldrich). NIH-3T3 cells were transfected on six-well plates at 70 to 80% confluence in Dulbecco's modified Eagle's medium (DMEM) plus 2% fetal calf serum with 5 μg of plasmid DNA and 5 μl of polyethylenimine (1:1000 dilution, adjusted to pH 7.0 and preincubated for 15 min in 200 μl of phosphate-buffered saline for complex formation). Four hours after transfection, the medium was changed, and cells were incubated in normal growth medium for 24 h with and without the presence of the PPARα ligand GW7647 (0.3 μM), the PPARβ/δ ligand GW501516 (0.3 μM), the PPARγ ligand GW1929 (0.3 μM), and/or GSK3787 (1.0 μM). Luciferase assays were performed as described previously (Gehrke et al., 2003). Values from three independent experiments were combined to calculate averages and standard deviations.
Time-Resolved Fluorescence Resonance Energy Transfer Assays In Vitro.
The interaction of coregulator peptides with PPARs in vitro was determined by time-resolved fluorescence resonance energy transfer (TR-FRET) (Stafslien et al., 2007) using the Lanthascreen TR-FRET PPARα, PPARβ/δ, and PPARγ coregulator assays according to the manufacturer's (Invitrogen) instructions with the following peptides: coactivator peptide C33, HVEMH PLLMGLLMESQWGA; coactivator peptide thyroid hormone receptor-associated protein 220/vitamin D receptor interacting protein-1 (TRAP220/DRIP-1), KVSQNPILTSLLQITGNGG; corepressor silencing mediator for retinoid and thyroid hormone receptors interaction domain 2 (SMRT-ID2), HASTNMGLEAIIRKALMGKYDQW; and nuclear receptor corepressor interaction domain 2 (NCoR-ID2), DPASNLGLEDIIRKALMGSFDDK. Incubation times were 15 to 60 min for all assays shown in this study. The assay buffer contained 100 mM KCl, 20 mM Tris, pH 7.9, 0.01% Triton X-100, and 1 μg/μl bovine serum albumin. All assays were validated for their robustness by determining the respective Z′ factors (Zhang et al., 1999). Measurements were performed on a VICTOR3 V Multilabel Counter (PerkinElmer Life and Analytical Sciences, Waltham, MA) with instrument settings as described in the manufacturer's instructions for LanthaScreen assays.
Cell Culture.
The human hepatocarcinoma cell lines HepG2 and Huh7, lung adenocarcinoma cell lines A549 and H1838, squamous carcinoma cell line A431, and the breast cancer cell line MCF7 were obtained from the American Type Culture Collection (Manassas, VA). Cells were cultured according to the recommended procedures: HepG2, Huh7, A431, and MCF7 cells were cultured in DMEM; A549 cells were cultured in Ham's F12K medium; and H1838 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. For mRNA analysis, cells were plated in 12-well tissue culture plates and cultured until 80% confluence, at which time they were treated with either DMSO, GW0742, GSK3787, or GW0742 and GSK3787 for 24 h. The concentrations of GW0742 (0.05–1.0 μM) were used because they have been shown previously to specifically activate PPARβ/δ (Shearer and Hoekstra, 2003). The concentrations of GSK3787 (0.1–10 μM) were used because they have been shown previously to antagonize PPARβ/δ in other cell-based models and are probably in the range of concentrations that could be achieved in vivo (Shearer et al., 2010). After this treatment, mRNA was isolated and used for qPCR as described above. It is noteworthy that all of the cell lines have been shown to respond to ligand activation of PPARβ/δ and express PPARβ/δ mRNA (Hollingshead et al., 2007; Girroir et al., 2008a; He et al., 2008; and data not shown). However, relative expression of PPARβ/δ has been noted to be lower in these cancer cell lines compared with normal cells/tissue (data not shown).
Isolation of Mouse Keratinocytes and Fibroblasts.
Keratinocytes and fibroblasts were isolated from newborn mouse skin and were cultured as described previously (Dlugosz et al., 1995).
Cell Proliferation Assays.
Cell proliferation was examined using real-time monitoring of cell proliferation of adherent cells using the xCELLigence System (Roche Applied Science, Indianapolis, IN). In brief, the optimal number of cells required to obtain exponential growth in a single well of an E-Plate 16 was determined by monitoring cell proliferation in real time using an increasing number of cells per well (Supplemental Fig. 1). The number of cells seeded per well to obtain exponential growth curves and the length of time examined for each cell line is shown in Supplemental Fig. 1. Cell proliferation was monitored every 15 min using the RTCES System (ACEA Biosciences, San Diego, CA) for up to 120 h. Cell-sensor impedance is expressed as an arbitrary unit called the Cell Index. The Cell Index at each time point is defined as (Rn-Rb)/15, where Rn is the cell-electrode impedance of the well when it contains cells and Rb is the background impedance of the well with the media alone. Start and end times were selected during the log-growth phase (Supplemental Fig. 1) and used to calculate doubling time with RTCA Software version 1.2 (ACEA Biosciences, Inc., San Diego, CA) from independent triplicate wells per treatment. For examination of the effect of GW0742 or GSK3787 on cell proliferation, the human squamous cell carcinoma cell line A431, human liver cancer cell lines HepG2 and Huh7, the human lung cancer cell lines A549 and H1838, or the human breast cancer cell line MCF7 were seeded as described (Supplemental Fig. 1) and treated with GW0742 (0.1, 1.0, or 10 μM) or GSK3787 (0.1, 1.0, or 10 μM).
Quantitative Western Blotting.
Protein samples were prepared from fibroblasts and keratinocytes using lysis buffer containing protease inhibitors. Seventy-five microgram of protein per sample was resolved using SDS 10% polyacrylamide gels. Proteins were transferred onto a polyvinylidene fluoride membrane. The membrane was blocked with 5% nonfat milk or 0.5% gelatin in Tris-buffered saline/Tween 20 and incubated overnight with primary antibodies against PPARβ/δ (Girroir et al., 2008b) or lactate dehydrogenase. Membranes were washed and incubated with biotinylated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) followed by incubation with 125I-labeled streptavidin. Membranes were exposed to plates, and the level of radioactivity was quantified by filmless autoradiographic analysis. Hybridization signals for PPARβ/δ were normalized to the hybridization signals for the loading control lactate dehydrogenase. Three independent samples were analyzed for each group.
3T3-L1 Preadipocyte Cell Culture and Differentiation.
Mouse 3T3-L1 preadipocytes were cultured in DMEM with 10% fetal calf serum and 1% penicillin/streptomycin. Cells were cultured to confluence and then treated with differentiation medium. The differentiation medium was DMEM with 10% FBS, 10 μg/ml insulin, 200 μM 3-isobutyl-1-methylxanthine, and 250 μM dexamethasone. One day after treatment with the differentiation medium, cells were treated with 1.0 to 10 μM rosiglitazone, 1.0 to 10 μM GSK3787, or both rosiglitazone and GSK3787. For analysis of PPARγ-dependent gene expression, RNA was isolated as described above from cells 24 h after treatment for analysis of the PPARγ target gene ap2 by qPCR as described previously by others (Rockwell et al., 2006). For analysis of adipocyte differentiation, cells were cultured for 4 days in maintenance medium containing DMEM with 10% FBS, 1% penicillin/streptomycin, and insulin (10 μg/ml) beginning 24 h after treatment with rosiglitazone, GSK3787, or both. Cells were fixed in 10% formalin and stained with Oil Red O (0.2% in 60% isopropanol) for 15 min. After washing with 60% isopropanol, Oil Red O stain was extracted with 4% Nonidet P-40 in isopropanol. The intensity of staining was determined by measuring absorbance at 570 nm.
Glucose Tolerance Test.
Male wild-type and Pparβ/δ-null mice were administered GW0742, GSK3787, or rosiglitazone for 2 weeks as described above. After a 6-h fast, mice were injected with glucose (1.5 mg/g body weight) by intraperitoneal injection. After this injection, blood was collected from the mandibular vein every 30 min for 2 h and used for analysis of blood glucose using Accu-Chek Active Glucometer (Roche Diagnostics, Indianapolis, IN).
Results
GSK3787 Antagonizes Ligand-Induced PPARβ/δ-Dependent Gene Expression In Vivo.
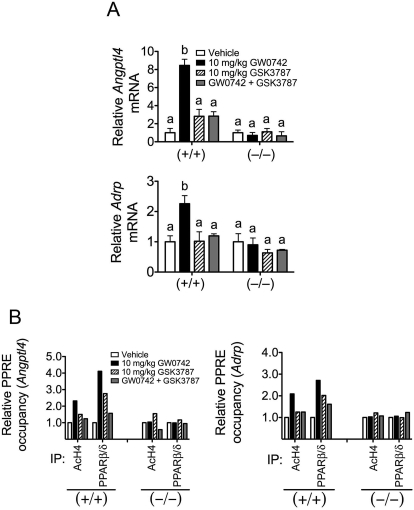
Preliminary characterization of GSK3787 indicated that this antagonist inhibits both basal and ligand-induced expression of pyruvate dehydrogenase kinase 4 (PDK4) and carnitine palmitoyl transferase 1a (CPT1a) in human skeletal muscle cells at concentrations up to 1 μM (Shearer et al., 2010). In addition, this study also demonstrated that oral administration of GSK3787 (10 mg/kg) led to a serum Cmax of 2.2 ± 0.4 μM in C57BL/6 male mice (Shearer et al., 2010). To more definitively examine the effect of GSK3787 in vivo, qPCR analysis of PPARβ/δ target genes and ChIP assays were performed using tissue collected from wild-type and Pparβ/δ-null mice. Oral administration of GW0742 caused an increase in expression of Angptl4 and Adrp mRNA (known PPARβ/δ target genes) in wild-type mouse colon epithelium, and this effect was not found in Pparβ/δ-null mouse colon epithelium (Fig. 2A). Oral administration of GSK3787 had no effect on the expression of Angptl4 and Adrp mRNA in mouse colon epithelium in either genotype (Fig. 2A). Coadministration of GSK3787 with GW0742 effectively prevented the ligand-induced expression of both Angptl4 and Adrp mRNA in wild-type mouse colon epithelium, and this effect was not found in Pparβ/δ-null mouse colon epithelium (Fig. 2A). GSK3787 did not modulate PPARβ/δ-dependent gene expression or antagonize ligand-induced PPARβ/δ-dependent gene expression in liver (data not shown). Because the antagonism of PPARβ/δ by GSK3787 was more evident in colon epithelium, consistent with high expression of PPARβ/δ in this tissue (Girroir et al., 2008b), ChIP assays were performed using colon epithelial DNA obtained from mice treated with either GW0742, GSK3787, or both compounds. Ligand activation of PPARβ/δ with GW0742 caused an increase in acetylated histone H4 (AcH4) associated with the PPRE region of both the Angptl4 and Adrp genes in wild-type mouse colon epithelium, and this effect was not found in similarly treated Pparβ/δ-null mice (Fig. 2B), consistent with past results (Hollingshead et al., 2008). Acetylation of histone H4 is important for chromatin remodeling and recruitment of the transcription initiation complex. Although oral administration of GSK3787 had essentially no effect on promoter occupancy of AcH4 in the PPRE region of both the Angptl4 and Adrp genes, coadministration of GSK3787 with GW0742 resulted in markedly less accumulation of AcH4 in the PPRE region of both the Angptl4 and Adrp genes in wild-type mouse colon epithelium (Fig. 2B). Ligand activation of PPARβ/δ with GW0742 caused an increase in promoter occupancy of PPARβ/δ in the PPRE region of both the Angptl4 and Adrp genes in wild-type mouse colon epithelium, and this effect was not found in similarly treated Pparβ/δ-null mice (Fig. 2B). Oral administration of GSK3787 caused a modest increase in promoter occupancy of PPARβ/δ in the PPRE region of both the Angptl4 and Adrp genes, but coadministration of GSK3787 with GW0742 resulted in markedly less accumulation of PPARβ/δ in the PPRE region of both the Angptl4 and Adrp genes in wild-type mouse colon epithelium (Fig. 2B). These changes observed with coadministration were not found in similarly treated Pparβ/δ-null mouse colon epithelium. Although promoter occupancy of AcH4 on the Angptl4 gene was modestly lower in Pparβ/δ-null mouse colon epithelium after cotreatment with GW0742 and GSK3787, there was no change in promoter occupancy of AcH4 on the Adrp gene in Pparβ/δ-null mouse colon epithelium after cotreatment with GW0742 and GSK3787 (Fig. 2B). It remains possible that the former change could reflect an off-target effect of GSK3787 or a limitation because of the use of technical replicates for the ChIP assay, which precluded assessing measures of variability. Collectively, these results demonstrate that GSK3787 can effectively antagonize ligand-induced effects on PPARβ/δ target genes in vivo and that these effects are due to receptor-dependent mechanisms because they are not found in Pparβ/δ-null mice.
Fig. 2.
GSK3787 antagonizes ligand-induced changes in PPARβ/δ-dependent gene expression in vivo. Wild-type (+/+) or Pparβ/δ-null (−/−) mice were treated with the PPARβ/δ ligand GW0742 (10 mg/kg), the PPARβ/δ antagonist GSK3787 (10 mg/kg), or both GW0742 and GSK3787 (both at 10 mg/kg) as described under Materials and Methods. A, quantitative real-time PCR was performed using total RNA isolated from colon epithelium to quantify mRNA expression of the PPARβ/δ target genes Angptl4 or Adrp. Values are the average normalized fold change compared with vehicle control and represent the mean ± S.E.M., n = 4 biological replicates. Values with different letters are significantly different (P ≤ 0.05), as determined by ANOVA and Bonferroni's multiple comparison test. B, chromatin immunoprecipitations were carried out as described under Materials and Methods to examine the recruitment of PPARβ/δ and histone acetylation at the regulatory regions of Angptl4 (left) and Adrp (right) in chromatin isolated from colon epithelium from (+/+) and (−/−). Values are the input-normalized average of technical replicates as fold change compared with vehicle control; n = 1 technical replicate of five pooled biological replicates.
GSK3787 Antagonizes Ligand-Induced PPARβ/δ-Dependent Gene Expression In Vitro.
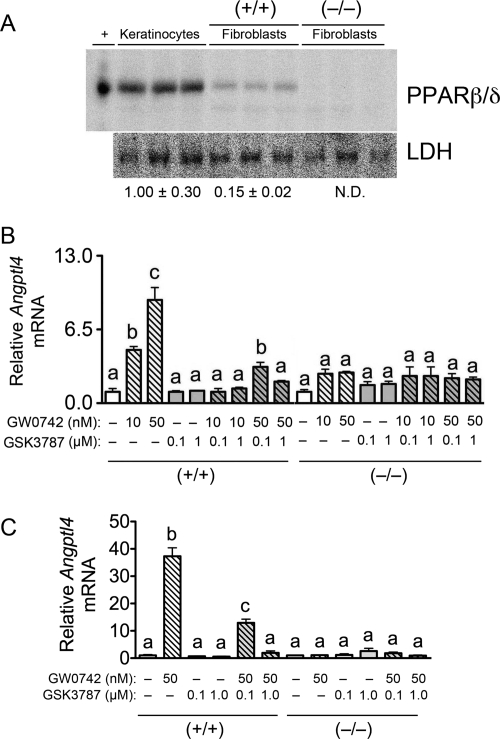
Although preliminary characterization of GSK3787 indicates that this compound can antagonize both basal and ligand-induced expression of PDK4 and CPT1a in human skeletal muscle cells at concentrations up to 1 μM (Shearer et al., 2010), the specificity of this effect on gene expression was not examined. For this reason, the effect of GSK3787 in cells expressing a relatively low level of PPARβ/δ (fibroblasts) and a relatively high level of PPARβ/δ (keratinocytes) was examined using cells isolated from wild-type and Pparβ/δ-null mice. Expression of PPARβ/δ protein is ∼7-fold lower in fibroblasts compared with keratinocytes (Fig. 3A). Keratinocytes are known to express PPARβ/δ at a high level compared with most other mouse tissues/cells (Girroir et al., 2008b). Despite the relatively low level of expression of PPARβ/δ observed in fibroblasts, treatment with 10 or 50 nM GW0742 caused up to ∼10-fold increase of Angptl4 mRNA compared with control; this effect was not found in fibroblasts from Pparβ/δ-null mice (Fig. 3B). The increase in Angptl4 mRNA observed in response to 10 nM GW0742 was not found in wild-type fibroblasts that were cultured with both 10 nM GW0742 and 0.1 or 1.0 μM GSK3787 (Fig. 3B). The increase in Angptl4 mRNA observed in response to 50 nM GW0742 was markedly lower in wild-type fibroblasts that were cultured with 50 nM GW0742 and 0.1 μM GSK3787 and essentially absent in wild-type fibroblasts that were cultured with 50 nM GW0742 and 1.0 μM GSK3787 (Fig. 3B). None of these effects were found in similarly treated fibroblasts from Pparβ/δ-null mice (Fig. 3B). Consistent with the difference in expression of PPARβ/δ protein (Fig. 3A), the change in expression of Angptl4 mRNA in response to GW0742 was greater in keratinocytes compared with fibroblasts, because 50 nM GW0742 caused greater than a 30-fold increase in Angptl4 mRNA compared with control (Fig. 3C). The increase in Angptl4 mRNA observed in response to 50 nM GW0742 was markedly lower in wild-type keratinocytes that were cultured with 50 nM GW0742 and 0.1 μM GSK3787 and not found in wild-type keratinocytes that were cultured with 50 nM GW0742 and 1.0 μM GSK3787 (Fig. 3C). None of these changes were found in similarly treated keratinocytes from Pparβ/δ-null mice (Fig. 3C). Combined, these results establish that GSK3787 can effectively antagonize ligand-induced gene expression mediated by PPARβ/δ in cultured fibroblasts and keratinocytes using concentrations ranging from 0.1 to 1.0 μM in the presence of an agonist with affinity for PPARβ/δ in the nanomolar range.
Fig. 3.
GSK3787 antagonizes ligand-induced changes in PPARβ/δ-dependent gene expression in mouse primary fibroblasts and keratinocytes. A, expression of PPARβ/δ protein in keratinocytes and fibroblasts from wild-type (+/+) or Pparβ/δ-null (−/−) mice. Normalized expression values are fold expression relative to keratinocytes and represent the mean ± S.E.M., n = 3 biological replicates. Expression of PPARβ/δ is ∼7-fold lower in fibroblasts compared with keratinocytes. +, positive control (lysate from COS1 cells transfected with PPARβ/δ expression vector; N.D., not detected. fibroblasts (B) or keratinocytes (C) from (+/+) and (−/−) were cultured in the presence of the PPARβ/δ ligand GW0742 and/or GSK3787 at the indicated concentration as described under Materials and Methods. Quantitative real-time PCR was performed using total RNA isolated from cells to quantify mRNA expression of the PPARβ/δ target gene Angptl4. Values are the average normalized fold change compared with vehicle control and represent the mean ± S.E.M., n = 3 biological replicates. Values with different letters are significantly different (P ≤ 0.05), as determined by ANOVA and Bonferroni's multiple comparison test.
GSK3787 Antagonizes Ligand-Induced PPARβ/δ-Dependent Gene Expression in Human Cancer Cell Lines.
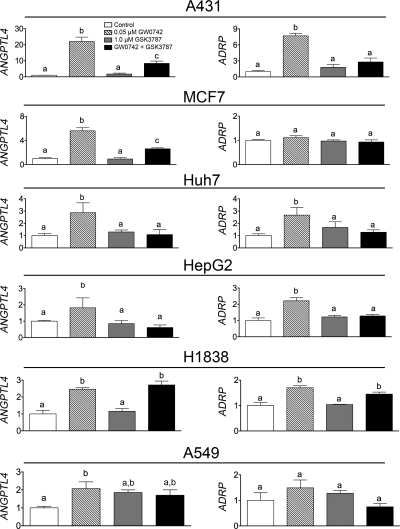
There is considerable interest in the effect of PPARβ/δ in human cancer (Burdick et al., 2006; Peters et al., 2008; Peters and Gonzalez, 2009). Thus, the relative ability of GSK3787 to antagonize PPARβ/δ was examined in human cancer cell lines, including those for skin (A431), liver (HepG2, Huh7), breast (MCF7), and lung cancer (H1838, A549). Expression of ANGPTL4 and ADRP mRNAs in A431 cells were increased by ∼20- and ∼6-fold, respectively, by 50 nM GW0742 (Fig. 4). Cotreatment with 50 nM GW0742 and 1.0 μM GSK3787 largely diminished these GW0742-induced responses in A431 cells (Fig. 4). The magnitude of ligand-induced changes in PPARβ/δ target genes was greater in A431 cells compared with the other human cancer lines. Treatment with 50 nM GW0742 caused an increase in expression of ANGPTL4 mRNA in MCF7, Huh7, HepG2, H1838, and A549 cells ranging from ∼2- to 6-fold (Fig. 4). The magnitude of change in GW0742-induced ANGPTL4 mRNA expression in these cells was markedly lower compared with primary mouse keratinocytes, in which >30-fold increases were noted (Fig. 3C). Cotreatment of 50 nM GW0742 and 1.0 μM GSK3787 antagonized the GW0742-induced increase of ANGPTL4 mRNA in MCF7, Huh7, and HepG2 cells but not in H1838 or A549 cells (Fig. 4). ADRP mRNA increased after treatment with 50 nM GW0742 in Huh7, HepG2, and H1838 cells but not in MCF7 or A549 cells (Fig. 4). Cotreatment of 50 nM GW0742 and 1.0 μM GSK3787 antagonized the GW0742-induced increase of ADRP mRNA in Huh7 and HepG2 cells but not in H1838 cells (Fig. 4). These data show that GSK3787 can antagonize ligand-induced changes in gene expression in most but not all human cancer cell lines examined in this study. This is in contrast to effective antagonism of ligand-induced changes in gene expression observed in mouse primary fibroblasts and keratinocytes using the same concentrations of GW0742 and GSK3787 (Fig. 3, B and C). No decrease in basal expression of either ANGPTL4 or ADRP mRNA was observed after treatment with GSK3787, suggesting that GSK3787 does not antagonize basal expression of either of these two PPARβ/δ target genes in A431, MCF7, Huh7, HepG2, H1838, or A549 human cancer cell lines.
Fig. 4.
GSK3787 antagonizes ligand-induced changes in PPARβ/δ-dependent gene expression in some but not all human cancer cell lines. Human cancer cell lines were cultured in the presence of the PPARβ/δ ligand GW0742 and/or GSK3787 at the indicated concentration as described under Materials and Methods. Quantitative real-time PCR was performed using total RNA isolated from cells to quantify mRNA expression of the PPARβ/δ target genes ANGPTL4 or ADRP. Values are the average normalized fold change compared with vehicle control and represent the mean ± S.E.M., n = 3 biological replicates. Values with different letters are significantly different (P ≤ 0.05), as determined by ANOVA and Bonferroni's multiple comparison test.
Effect of Ligand Activation of PPARβ/δ by GW0742 and Antagonism of PPARβ/δ by GSK3787 on Cell Proliferation.
There is considerable controversy regarding the effects of PPARβ/δ on the proliferation of cultured human cancer cells because there is evidence that PPARβ/δ either increases, decreases, or has no effect on cell growth (Burdick et al., 2006; Peters et al., 2008; Peters and Gonzalez, 2009). Thus, the effect of GW0742 and/or GSK3787 on cell proliferation was examined in the human cancer cell lines A431, HepG2, Huh7, MCF7, H1838, and A549. Neither GW0742 nor GSK3787 had any effect on cell proliferation in MCF7, Huh7, HepG2, A431, A549, or H1838 human cancer cell lines at concentrations ranging from 0.1 to 10 μM (Table 1).
TABLE 1.
Doubling times (hours) for human cancer cell lines after treatment with either a PPARβ/δ agonist (GW0742) or antagonist (GSK3787)
Doubling times were calculated during the exponential growth phase as described under Materials and Methods. Values represent the mean ± S.E.M. There were no significant differences between any of the values.
| Treatment | MCF7 | Huh7 | HepG2 | A431 | A549 | H1838 |
|---|---|---|---|---|---|---|
| DMSO | 9.2 ± 0.2 | 18.5 ± 0.6 | 23.5 ± 1.0 | 22.8 ± 1.0 | 14.8 ± 0.3 | 8.8 ± 0.2 |
| GW0742 | ||||||
| 0.1 μM | 8.4 ± 0.1 | 17.1 ± 0.4 | 22.8 ± 0.6 | 22.0 ± 0.3 | 15.9 ± 0.2 | 8.9 ± 0.3 |
| 1.0 μM | 8.5 ± 0.1 | 19.0 ± 0.2 | 22.8 ± 0.4 | 22.5 ± 0.9 | 15.1 ± 0.3 | 8.4 ± 0.3 |
| 10 μM | 8.6 ± 0.1 | 16.5 ± 0.6 | 22.9 ± 0.8 | 22.0 ± 0.8 | 15.6 ± 0.3 | 9.0 ± 0.2 |
| DMSO | 6.9 ± 0.1 | 19.0 ± 0.5 | 23.6 ± 1.2 | 21.5 ± 1.0 | 15.4 ± 0.3 | 9.0 ± 0.4 |
| GSK3787 | ||||||
| 0.1 μM | 6.8 ± 0.1 | 18.9 ± 0.4 | 21.9 ± 0.5 | 20.7 ± 0.8 | 14.9 ± 0.3 | 8.4 ± 0.3 |
| 1.0 μM | 6.7 ± 0.1 | 18.7 ± 0.4 | 23.4 ± 1.1 | 22.1 ± 1.0 | 14.8 ± 0.3 | 8.1 ± 0.4 |
| 10 μM | 6.6 ± 0.1 | 18.8 ± 0.4 | 22.1 ± 0.5 | 24.6 ± 1.8 | 15.1 ± 0.3 | 7.8 ± 0.4 |
GSK3787 Selectively Antagonizes Ligand-Induced PPARβ/δ Transcription but also Modulates PPARγ Activities.
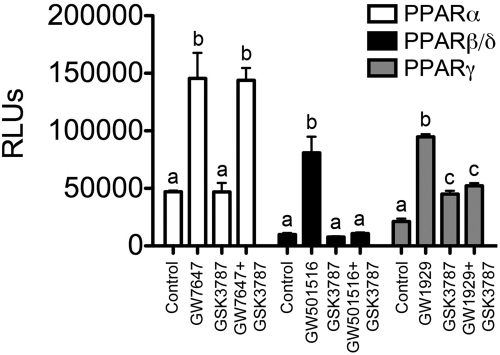
Reporter assays were performed to determine whether GSK3787 could activate the two other members of the PPAR family, PPARα and PPARγ. GSK3787 did not modulate PPARα-dependent transactivation and had no effect on ligand-induced transactivation of PPARα by GW7647 (Fig. 5). In contrast, GW501516-induced PPARβ/δ-dependent transactivation was effectively antagonized by GSK3787 (Fig. 5). It is noteworthy that GSK3787 was able to modestly increase PPARγ-dependent reporter activity compared with the PPARγ agonist GW1929 (Fig. 5). In addition, GSK3787 also antagonized GW1929-induced PPARγ transactivation (Fig. 5). These data revealed that GSK3787 had no influence on PPARα activity, is effective as a PPARβ/δ antagonist, and has weak PPARγ agonistic and antagonistic activities.
Fig. 5.
GSK3787 antagonizes ligand-induced reporter activity modulated by PPARβ/δ but also has weak agonist and antagonist activity for PPARγ. NIH-3T3 cells were transiently transfected with reporter vectors to measure PPARα, PPARβ/δ, or PPARγ-dependent activity as described under Materials and Methods. Cells were cultured for 24 h in the presence of 0.3 μM GW7647, 0.3 μM GW501516, 0.3 μM GW1929, and/or 1.0 μM GSK3787 before analysis of reporter activity. Values represent the average of independent triplicate samples ± S.D., n = 3 biological replicates. Mean values for each PPAR reporter assay with different letters are significantly different (P ≤ 0.05), as determined by ANOVA and Bonferroni's multiple comparison test.
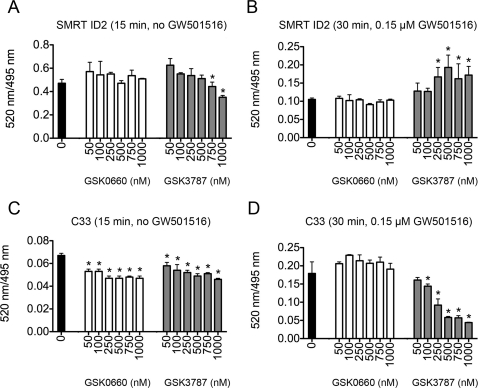
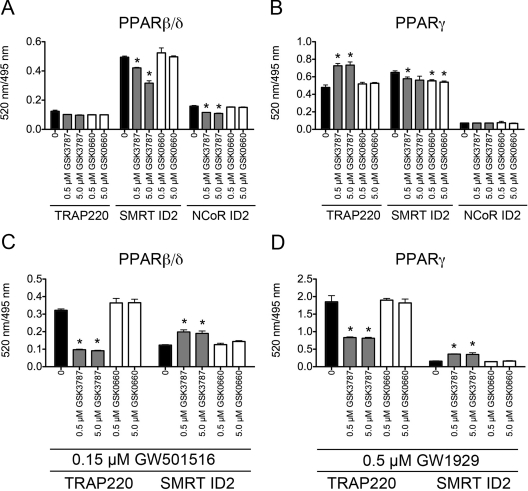
To more closely examine the ability of GSK3787 to antagonize PPAR activity, the interaction between PPARβ/δ, PPARγ, and coactivator or corepressor peptides was determined using TR-FRET. In this assay, the interaction between the PPARβ/δ or PPARγ ligand binding domain (LBD; indirectly labeled with terbium) with either the coactivator peptides C33 or TRAP220/DRIP-1 (labeled with fluorescein) or the corepressor peptides SMRT-ID2 or NCoR-ID2 (labeled with fluorescein) was determined. The in vitro TR-FRET assay measures the intensity of terbium-induced fluorescence emission of the fluorescein moiety of the labeled peptides, expressed as the ratio of fluorescein- and terbium-derived fluorescence. In the absence of ligand (GW501516), GSK3787 decreased corepressor peptide SMRT-ID2 dissociation at higher concentrations (0.75 and 1.0 μM) and dose-dependently decreased recruitment of coactivator peptide C33 to the PPARβ/δ LBD (Fig. 6, A and C). In the absence of ligand, GSK0660 did not influence corepressor peptide SMRT-ID2 dissociation but did decrease recruitment of coactivator peptide C33 to the PPARβ/δ LBD (Fig. 6, A and C). Previous studies established that maximal SMRT-ID2 dissociation from and maximal C33 recruitment with the LBD of PPARβ/δ occurs by 30 min after ligand treatment (data not shown). A dose-dependent prevention of corepressor peptide SMRT-ID2 dissociation (Fig. 6B) and inhibition of coactivator peptide C33 recruitment (Fig. 6D) was observed after cotreatment of 0.15 μM GW501516 with GSK3787. Similar changes in PPARβ/δ LBD/coactivator/corepressor interactions were not observed with cotreatment of GW501516 with GSK0660, another PPARβ/δ antagonist (Fig. 6, B and D). Because the reporter assays (Fig. 5) indicated that GSK3787 also modulates PPARγ activity, TR-FRET assays were performed to compare coregulator peptide recruitment/dissociation between PPARγ and PPARβ/δ. In the absence of ligand (GW501516), GSK3787 caused dissociation of corepressor peptides SMRT-ID2 and NCoR-ID2 from the PPARβ/δ LBD but had no effect on recruitment of the PPARγ coactivator TRAP220/DRIP-1 to the PPARβ/δ LBD (Fig. 7A). GSK0660 had no effect on either dissociation of corepressor peptides SMRT-ID2 or NCoR-ID2 from the PPARβ/δ LBD or recruitment of the PPARγ coactivator TRAP220/DRIP-1 to the PPARβ/δ LBD (Fig. 7A). In the absence of ligand (GW1929), GSK3787 and GSK0660 caused some dissociation of corepressor peptides SMRT-ID2 but not NCoR-ID2 from the PPARγ LBD (Fig. 7B). In the absence of ligand, GSK3787 enhanced recruitment of the PPARγ coactivator TRAP220/DRIP-1 to the PPARγ LBD, whereas GSK0660 had no effect on recruitment of TRAP220/DRIP-1 to the PPARγ LBD (Fig. 7B). GW501516-induced recruitment of TRAP220/DRIP-1 to the PPARβ/δ LBD and dissociation of SMRT-ID2 from the PPARβ/δ LBD were both inhibited by GSK3787, and GSK0660 did not influence either of these effects (Fig. 7C). It is noteworthy that GW1929-induced recruitment of TRAP220/DRIP-1 to the PPARγ LBD and dissociation of SMRT-ID2 from the PPARγ LBD were both inhibited by GSK3787 and GSK0660 did not change either of these effects (Fig. 7D). Combined, these data suggest that although GSK3787 can antagonize PPARβ/δ, this compound can also cause molecular interactions between coactivators and corepressor peptides with the LBD of PPARγ, similar to the effects found with PPARγ agonists and antagonists, consistent with the reporter assays (Fig. 5).
Fig. 6.
GSK3787 antagonizes ligand-induced recruitment of coactivator peptides and dissociation of corepressor peptides from the LBD of PPARβ/δ in vitro. Interaction of fluorescein-labeled coactivator or corepressor peptide and recombinant GST-PPARβ/δ bound by a terbium-labeled anti-GST antibody was determined by TR-FRET. A, interaction of the corepressor peptide SMRT-ID2 with the LBD of PPARβ/δ in the absence of ligand (GW501516). B, interaction of the corepressor peptide SMRT-ID2 with the LBD of PPARβ/δ in the presence of ligand (0.15 μM GW501516). C, interaction of the coactivator peptide C33 with the LBD of PPARβ/δ in the absence of ligand (GW501516). D, interaction of the coactivator peptide C33 with the LBD of PPARβ/δ in the presence of ligand (0.15 μM GW501516). Results are expressed as the ratio of fluorescence intensity at 520 nm (fluorescein emission excited by terbium emission) and 495 nm (terbium emission). Values represent the average of independent triplicate samples ± S.D., n = 3 biological replicates. *, significantly different from control (P ≤ 0.05), as determined by ANOVA and Bonferroni's multiple comparison test.
Fig. 7.
GSK3787 antagonizes ligand-induced recruitment of coactivator peptides and dissociation of corepressor peptides from the LBD of PPARβ/δ and PPARγ in vitro. Interaction of fluorescein-labeled coactivator or corepressor peptide and recombinant GST-PPARγ bound by a terbium-labeled anti-GST antibody was determined by TR-FRET. A, interaction of the coactivator peptide TRAP220 and the corepressor peptides SMRT-ID2 and NCoR-ID2 with the LBD of PPARβ/δ in the absence of ligand (GW501516). B, interaction of the coactivator peptide TRAP220 and the corepressor peptides SMRT-ID2 and NCoR-ID2 with the LBD of PPARγ in the absence of ligand (GW1929). C, interaction of the coactivator peptide TRAP220 and the corepressor peptide SMRT-ID2 with the LBD of PPARβ/δ in the presence of ligand (0.15 μM GW501516). D, interaction of the coactivator peptide TRAP220 and the corepressor peptide SMRT-ID2 with the LBD of PPARγ in the presence of ligand (0.5 μM GW1929). Results are expressed as the ratio of fluorescence intensity at 520 nm (fluorescein emission excited by terbium emission) and 495 nm (terbium emission). Values represent the average of independent triplicate samples ± S.D., n = 3 biological replicates. *, significantly different from control (P ≤ 0.05), as determined by ANOVA and Bonferroni's multiple comparison test.
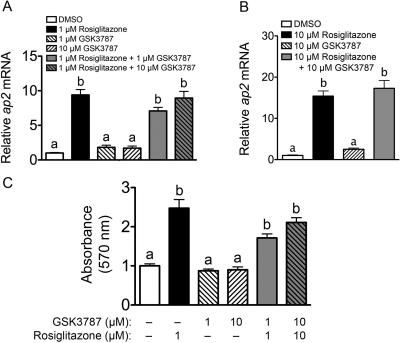
To begin to determine the relative efficacy of GSK3787 to modulate PPARγ activity, the effect of GSK3787 to regulate PPARγ-dependent gene expression was examined in an adipocyte cell-based model. 3T3-L1 preadipocytes were cultured with adipocyte differentiation medium to enhance PPARγ activity and then were treated with either a PPARγ agonist (rosiglitazone), GSK3787, or rosiglitazone and GSK3787. Expression of the PPARγ target gene ap2 mRNA was markedly increased by treatment with 1 or 10 μM rosiglitazone (Fig. 8). GSK3787 (1 or 10 μM) had no effect on the expression of ap2 mRNA in 3T3-L1 cells and did not antagonize rosiglitazone-induced expression of ap2 mRNA (Fig. 8). Likewise, rosiglitazone caused an increase in lipid accumulation associated with adipocyte differentiation as shown by Oil Red O staining, whereas GSK3787 did not significantly influence Oil Red O staining or modulate the increase in adipocyte differentiation caused by rosiglitazone (Fig. 8C and Supplemental Fig. 2). These data suggest that the efficacy of GSK3787 to activate PPARγ is markedly lower compared with rosiglitazone. These data also suggest that the efficacy of GSK3787 to antagonize PPARγ-dependent gene expression is markedly less compared with its ability to antagonize PPARβ/δ-dependent gene expression.
Fig. 8.
Effect of GSK3787 on expression of a PPARγ target gene and adipocyte differentiation. NIH 3T3-L1 cells were differentiated into adipocytes and then cultured in the presence of rosiglitazone, GSK3787, or rosiglitazone and GSK3787 as described under Materials and Methods. A and B, expression of ap2 mRNA was quantified by qPCR. Values are the average normalized fold change compared with vehicle control and represent the mean ± S.E.M. C, cells were stained with Oil Red O and relative staining intensity quantified by measuring absorbance at 570 nm as described under Materials and Methods. Values represent the mean ± S.E.M., n = 3 biological replicates. Values with different letters are significantly different (P ≤ 0.05), as determined by ANOVA and Bonferroni's multiple comparison test.
Effect of GSK3787 on Glucose Tolerance.
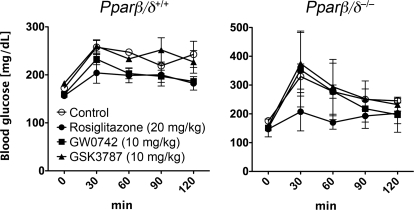
Ligand activation of PPARβ/δ or PPARγ can both improve insulin sensitivity and glucose tolerance (Reilly and Lee, 2008; Quinn et al., 2008). Because GSK3787 may modulate both PPARβ/δ and/or PPARγ activities, the effect of GSK3787 on glucose tolerance was examined. Higher blood glucose, consistent with glucose intolerance, was observed in Pparβ/δ-null mice compared with wild-type mice, similar with past results (Lee et al., 2006). Treatment with GW0742 for 2 weeks markedly improved glucose tolerance in wild-type mice, and this effect was not found in GW0742-treated Pparβ/δ-null mice (Fig. 9). In contrast, treatment with rosiglitazone for 2 weeks improved glucose tolerance in both wild-type and Pparβ/δ-null mice (Fig. 9). Administration of GSK3787 had no effect on glucose tolerance in either genotype (Fig. 9).
Fig. 9.
Effect of GSK3787 on glucose tolerance. Wild-type (+/+) or Pparβ/δ-null (−/−) mice were treated with GW0742 (10 mg/kg), GSK3787 (10 mg/kg), or rosiglitazone (20 mg/kg) as described under Materials and Methods. Glucose tolerance tests were performed to determine the effect of PPARβ/δ agonism, PPARβ/δ antagonism, and PPARγ agonism on glucose tolerance in wild-type (+/+) or null (−/−) mice, n = 3 biological replicates. The P values for the areas under the curve for GW0742-, rosiglitazone-, or GSK3787-treated wild-type mice are 0.03, 0.07, 076, and the P values for GW0742-, rosiglitazone-, or GSK3787-treated Pparβ/δ-null mice are 0.90, 0.08, and 0.87.
Discussion
These studies are the first to demonstrate that GSK3787 can effectively antagonize ligand-induced effects on PPARβ/δ target genes in vivo through receptor-dependent mechanisms because they are not found in Pparβ/δ-null mice. This conclusion is supported by qPCR analysis demonstrating PPARβ/δ-dependent antagonism of ligand-induced changes in gene expression in colon epithelium and ChIP assays demonstrating PPARβ/δ-dependent antagonism of ligand-induced promoter occupancy of PPARβ/δ on target genes in colon epithelium. Further confirmation of receptor specificity for at least some GSK3787 activity was provided by results showing that GSK3787 can antagonize GW0742-induced changes in gene expression in wild-type mouse fibroblasts and keratinocytes but not in Pparβ/δ-null cells. It is noteworthy that GSK3787 caused no overt signs of toxicity as assessed by relative cell proliferation. The specificity of GSK3787 for PPARβ/δ could be due in part to the fact that it is an irreversible antagonist that forms a covalent bond within the ligand binding domain of PPARβ/δ (Shearer et al., 2010). GSK3787 also antagonized ligand-induced changes in gene expression in most but not all human cancer cell lines examined in this study. This is in contrast to PPARβ/δ-dependent antagonism by GSK3787 of ligand-induced changes in gene expression observed in mouse fibroblasts and keratinocytes using the same concentrations of GW0742 and GSK3787. Determining the mechanisms for the resistance of the human lung cancer cell lines H1838 and A549 to PPARβ/δ antagonism by GSK3787 requires further study.
It is worth noting that potency and efficacy of ligand activation of PPARβ/δ on target gene expression in fibroblasts and keratinocytes were markedly greater compared with MCF7, Huh7, HepG2, H1838, and A549 cells. This suggests that the ability of a ligand to activate PPARβ/δ is relatively less in these human cancer cell lines compared with mouse fibroblasts and keratinocytes. Although the mechanism for this difference is not known, the ligand may be more rapidly degraded in the latter cell lines compared with fibroblasts and keratinocytes. Furthermore, because GSK3787 antagonized ligand-induced gene expression in mouse fibroblasts and keratinocytes that express relatively low and relatively high levels of PPARβ/δ protein, respectively, this demonstrates that GSK3787 is effective for in vitro models at concentrations ranging from 0.1 to 1.0 μM in cells with varying levels of receptor expression. This is important to point out because these concentrations are comparable with concentrations achievable in vivo because the Cmax in serum observed in mice administered GSK3787 (10 mg/kg) is 2.2 ± 0.4 μM with a half-life of 2.5 ± 1.1 h (Shearer et al., 2010). In model systems that lack the presence of high-affinity ligands but are dependent on endogenous lower-affinity ligands (e.g., fatty acids, fatty-acid derivatives), concentrations of GSK3787 between 0.1 and 1.0 μM should be capable of antagonizing constitutive PPARβ/δ function. Thus, it is surprising that GSK3787 has no effect on the basal expression of PPARβ/δ target genes examined in these studies in fibroblasts, keratinocytes, or human cancer cell lines. This is in contrast to reduced basal expression of CPT1a and PDK4 in human skeletal muscle cells observed after treatment with GSK3787 (Shearer et al., 2010). This could be due to differences in regulatory elements in the promoters between CPT1a or PDK4 and ANGPTL4 or ADRP. Future in vivo and in vitro studies should consider this possibility.
Despite demonstrating that GSK3787 specifically antagonizes ligand-induced activity of PPARβ/δ based on analysis of cells and mice lacking expression of PPARβ/δ, evidence was also obtained demonstrating that GSK3787 has weak PPARγ agonist and antagonist activities. However, comparison of PPARγ function in 3T3-L1 cells demonstrated negligible PPARγ agonistic or antagonistic activities by GSK3787 as shown by both the lack of change in expression of a known PPARγ target gene and analysis of adipocyte differentiation. Because the concentrations required to specifically antagonize PPARβ/δ are less than or equal to 1 μM, and this concentration of GSK3787 did not cause significant agonism or antagonism of PPARγ activity in 3T3-L1 adipocytes, this suggests that GSK3787 can be used to study the effects of PPARβ/δ antagonism in vitro without considering confounding changes due to PPARγ-dependent activity, as suggested by the FRET analysis and reporter assays that may exhibit increased sensitivity. In vivo examination of glucose tolerance also demonstrated negligible PPARγ activity of GSK3787 compared with a known PPARγ agonist using a dose of GSK3787 that effectively antagonized ligand-induced PPARβ/δ transcriptional activity in colonic epithelium. However, it is important to emphasize that experiments should control for this potential PPARγ agonism/antagonism by GSK3787, because it remains possible that other PPARγ-dependent activities could influence interpretation. Furthermore, whether GSK3787 can be used as an antagonist to study ligand-induced improvement of insulin sensitivity mediated by PPARβ/δ requires further examination.
Results from these studies also illustrate differences in functional roles of PPARβ/δ in cell proliferation. There is considerable controversy regarding the role of PPARβ/δ in cell proliferation because some studies suggest that activating this receptor increases cell proliferation, whereas others indicate that activating PPARβ/δ has either no effect or inhibits cell proliferation in association with the induction of terminal differentiation (Peters et al., 2008; Peters and Gonzalez, 2009). This is largely due to the lack of stringency in approaches used to examine cell proliferation, in particular the limited published time course and concentration-dependent studies. Ligand activation of PPARβ/δ with GW0742 had no effect on cell proliferation in A431, MCF7, Huh7, HepG2, H1838, or A549 human cancer cell lines, results that are consistent with some but not all studies showing that GW0742 or GW501516 has little influence on cell proliferation at concentrations less than 10 μM (Peters et al., 2008; Peters and Gonzalez, 2009). The present studies examined cell proliferation by determining doubling time using real-time analysis, and many published studies typically limit the analysis of cell proliferation to a single time point (Peters et al., 2008; Peters and Gonzalez, 2009). Thus, it remains possible that the approach used to detect cell growth could affect these findings. It is noteworthy that real-time analysis of cell proliferation provides an outstanding approach yielding accurate assessment of doubling time during the linear growth phase over a broad concentration range of compound. Because ligand activation of PPARβ/δ had no influence on cell proliferation, it is not surprising that GSK3787 had no effect on cell proliferation in A431, MCF7, Huh7, HepG2, H1838, or A549 human cancer cell lines, despite the fact that GSK3787 antagonized ligand-induced changes in PPARβ/δ-dependent gene expression in these cells. This is consistent with a recent study in which GSK3787 had no effect on cell proliferation in SW480, HCT116, DLD1, RKO, A549, or human embryonic kidney 293 cancer cell lines with concentrations of GSK3787 up to 10 μM (Shearer et al., 2010). This could be due to the fact that expression of PPARβ/δ is low in tumors and cancer cell lines compared with “normal” cells. For example, expression of PPARβ/δ in the C20 mammary gland cancer cell line is less than 25% of that found in keratinocytes (Foreman et al., 2010). Furthermore, although it has been suggested that PPARβ/δ expression is up-regulated by the APC/β-CATENIN signaling pathway that is often enhanced in cancer, recent findings indicate that this idea is incorrect (Foreman et al., 2009), and many studies show that PPARβ/δ expression is either lower or unchanged in tumors compared with control tissue (Uhlén et al., 2005; Berglund et al., 2008; Peters et al., 2008; Peters and Gonzalez, 2009). Thus, although activation of PPARβ/δ with a high-affinity ligand modulates changes in gene expression in cancer cell lines, the effect of either an agonist or an antagonist on cell proliferation can be negligible in human cancer cell lines.
Results from these studies are in contrast to a recent report suggesting that antagonism of PPARβ/δ with 10 μM SR13904 inhibited cell proliferation of A549 cells (Zaveri et al., 2009). However, data from the present study and recent work by others (Shearer et al., 2010) show that antagonism of PPARβ/δ in the same human lung cancer cell line has no effect on cell proliferation at concentrations that specifically antagonize PPARβ/δ. It is important to note that the study by Zaveri et al. (2009) had limitations that preclude definitive conclusions regarding the specificity of the response observed with SR13904 because they did not demonstrate an increase in A549 cell growth by ligand activation of PPARβ/δ that was prevented by cotreatment with SR13904. This is a concern because no effect on cell proliferation is found in response to ligand activation of PPARβ/δ in A549 cells as shown by the present study and another recent report (He et al., 2008). Moreover, Zaveri et al. (2009) did not demonstrate specific antagonism of ligand-induced PPARβ/δ target gene(s) or altered cell proliferation using knockout or knockdown approaches. This raises the possibility that the observed inhibition of cell proliferation in A549 cells by high concentration SR13904 is due to off target effects rather than antagonism of PPARβ/δ, in particular because SR13904 was also shown to antagonize PPARγ (Zaveri et al., 2009).
Although it has been suggested that antagonism of PPARβ/δ may be a useful approach for chemoprevention (Zuo et al., 2009), this idea is not supported by results from the present study showing no influence on cell proliferation of human cancer cell lines and the fact that the effect of ligand activation of PPARβ/δ on tumorigenesis is entirely unclear (Peters et al., 2008; Peters and Gonzalez, 2009). Furthermore, because ligand activation of PPARβ/δ improves insulin sensitivity, increases skeletal muscle fatty acid catabolism, and has potent anti-inflammatory activities, therapeutic antagonism of PPARβ/δ could probably lead to negative effects on these essential functions. Nevertheless, results from these studies demonstrate that GSK3787 can be used to antagonize PPARβ/δ in vivo and in vitro, providing a new strategy to delineate the functional role of a receptor with great potential as a therapeutic target for the treatment and prevention of diseases, including dyslipidemias, obesity, and cancer. Given the potential for GSK3787 to interact with PPARγ, receptor specificity must be controlled for in future studies.
Supplementary Material
Acknowledgments
We gratefully acknowledge Andrew Billin and Timothy Willson for providing GW0742, GSK0660, and GSK3787.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported by the National Institutes of Health National Cancer Institute [Grants CA124533, CA126826, CA141029]; the Deutsche Forschungsgemeinschaft [Grant SFB-TR17/A3]; and in part by the Intramural Research Program of the National Institutes of Health National Cancer Institute.
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.110.065508.
- PPAR
- peroxisome proliferator-activated receptor
- Adrp
- adipocyte differentiation-related protein
- ANOVA
- analysis of variance
- Angptl4
- angiopoietin-like protein 4
- ChIP
- chromatin immunoprecipitation
- DMSO
- dimethyl sulfoxide
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- PPRE
- peroxisome proliferator response element
- PCR
- polymerase chain reaction
- TR-FRET
- time-resolved fluorescence resonance energy transfer
- TRAP220/DRIP-1
- thyroid hormone receptor-associated protein 220/vitamin D receptor interacting protein-1
- SMRT-ID2
- silencing mediator for retinoid and thyroid hormone receptors interaction domain 2
- NCoR-ID2
- nuclear receptor corepressor interaction domain 2
- AcH4
- acetylated histone H4
- LBD
- ligand binding domain
- qPCR
- quantitative real-time polymerase chain reaction
- PDK4
- pyruvate dehydrogenase kinase 4
- CPT1a
- carnitine palmitoyl transferase 1a
- GST
- glutathione transferase
- GSK3787
- 4-chloro-N-(2-{[5-trifluoromethyl)-2-pyridyl]sulfonyl}ethyl)benzamide
- GW0742
- 4-[2-(3-fluoro-4-trifluoromethyl-phenyl)-4-methyl-thiazol-5-ylmethylsulfanyl]-2-methyl-phenoxy}-acetic acid
- GW501516
- (2-methyl-4-(((4-methyl-2-(4-(trifluoromethyl) phenyl)-5-thiazolyl)methyl)thio)phenoxy)-acetic acid
- GSK0660
- 3-[[[2-methoxy-4-(phenylamino)phenyl]amino]sulfonyl]-2-thiophenecarboxylic acid methyl ester
- GW1929
- N-(2-benzoylphenyl)-O-[2-(methyl-2-pyridinylamino)ethyl]-l-tyrosine hydrochloride
- GW7647
- 2-[[4-[2-[[(cyclohexylamino)carbonyl](4-cyclohexylbutyl)amino]ethyl]phenyl]thio]-2-methylpropanoic acid.
References
- Barak Y, Liao D, He W, Ong ES, Nelson MC, Olefsky JM, Boland R, Evans RM. (2002) Effects of peroxisome proliferator-activated receptor δ on placentation, adiposity, and colorectal cancer. Proc Natl Acad Sci USA 99:303–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund L, Björling E, Oksvold P, Fagerberg L, Asplund A, Szigyarto CA, Persson A, Ottosson J, Wernérus H, Nilsson P, et al. (2008) A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol Cell Proteomics 7:2019–2027 [DOI] [PubMed] [Google Scholar]
- Billin AN. (2008) PPAR-β/δ agonists for type 2 diabetes and dyslipidemia: an adopted orphan still looking for a home. Expert Opin Investig Drugs 17:1465–1471 [DOI] [PubMed] [Google Scholar]
- Burdick AD, Kim DJ, Peraza MA, Gonzalez FJ, Peters JM. (2006) The role of peroxisome proliferator-activated receptor-β/δ in epithelial cell growth and differentiation. Cell Signal 18:9–20 [DOI] [PubMed] [Google Scholar]
- Chawla A, Lee CH, Barak Y, He W, Rosenfeld J, Liao D, Han J, Kang H, Evans RM. (2003) PPARδ is a very low-density lipoprotein sensor in macrophages. Proc Natl Acad Sci USA 100:1268–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlugosz AA, Glick AB, Tennenbaum T, Weinberg WC, Yuspa SH. (1995) Isolation and utilization of epidermal keratinocytes for oncogene research. Methods Enzymol 254:3–20 [DOI] [PubMed] [Google Scholar]
- Fauti T, Müller-Brüsselbach S, Kreutzer M, Rieck M, Meissner W, Rapp U, Schweer H, Kömhoff M, Müller R. (2005) Induction of PPARβ and prostacyclin (PGI2) synthesis by Raf signaling: failure of PGI2 to activate PPARβ. FEBS J 273:170–179 [DOI] [PubMed] [Google Scholar]
- Foreman JE, Sharma AK, Amin S, Gonzalez FJ, Peters JM. (2010) Ligand activation of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) inhibits cell growth in a mouse mammary gland cancer cell line. Cancer Lett 288:219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman JE, Sorg JM, McGinnis KS, Rigas B, Williams JL, Clapper ML, Gonzalez FJ, Peters JM. (2009) Regulation of peroxisome proliferator-activated receptor-β/δ by the APC/β-CATENIN pathway and nonsteroidal antiinflammatory drugs. Mol Carcinog 48:942–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke S, Jérôme V, Müller R. (2003) Chimeric transcriptional control units for improved liver-specific transgene expression. Gene 322:137–143 [DOI] [PubMed] [Google Scholar]
- Girroir EE, Hollingshead HE, Billin AN, Willson TM, Robertson GP, Sharma AK, Amin S, Gonzalez FJ, Peters JM. (2008a) Peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) ligands inhibit growth of UACC903 and MCF7 human cancer cell lines. Toxicology 243:236–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girroir EE, Hollingshead HE, He P, Zhu B, Perdew GH, Peters JM. (2008b) Quantitative expression patterns of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) protein in mice. Biochem Biophys Res Commun 371:456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross B, Staels B. (2007) PPAR agonists: multimodal drugs for the treatment of type-2 diabetes. Best Pract Res Clin Endocrinol Metab 21:687–710 [DOI] [PubMed] [Google Scholar]
- He P, Borland MG, Zhu B, Sharma AK, Amin S, El-Bayoumy K, Gonzalez FJ, Peters JM. (2008) Effect of ligand activation of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) in human lung cancer cell lines. Toxicology 254:112–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinäniemi M, Uski JO, Degenhardt T, Carlberg C. (2007) Meta-analysis of primary target genes of peroxisome proliferator-activated receptors. Genome Biol 8:R147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingshead HE, Borland MG, Billin AN, Willson TM, Gonzalez FJ, Peters JM. (2008) Ligand activation of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) and inhibition of cyclooxygenase 2 (COX2) attenuate colon carcinogenesis through independent signaling mechanisms. Carcinogenesis 29:169–176 [DOI] [PubMed] [Google Scholar]
- Hollingshead HE, Killins RL, Borland MG, Girroir EE, Billin AN, Willson TM, Sharma AK, Amin S, Gonzalez FJ, Peters JM. (2007) Peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) ligands do not potentiate growth of human cancer cell lines. Carcinogenesis 28:2641–2649 [DOI] [PubMed] [Google Scholar]
- Jérôme V, Müller R. (1998) Tissue-specific, cell cycle-regulated chimeric transcription factors for the targeting of gene expression to tumor cells. Hum Gene Ther 9:2653–2659 [DOI] [PubMed] [Google Scholar]
- Lee CH, Olson P, Hevener A, Mehl I, Chong LW, Olefsky JM, Gonzalez FJ, Ham J, Kang H, Peters JM, et al. (2006) PPARδ regulates glucose metabolism and insulin sensitivity. Proc Natl Acad Sci USA 103:3444–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadra K, Anghel SI, Joye E, Tan NS, Basu-Modak S, Trono D, Wahli W, Desvergne B. (2006) Differentiation of trophoblast giant cells and their metabolic functions are dependent on peroxisome proliferator-activated receptor β/δ. Mol Cell Biol 26:3266–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruhn S, Meissner W, Adhikary T, Kaddatz K, Klein T, Watzer B, Müller-Brüsselbach S, Müller R. (2010) 15-Hydroxyeicosatetraenoic acid is a preferential peroxisome proliferator-activated receptor β/δ agonist. Mol Pharmacol 77:171–184 [DOI] [PubMed] [Google Scholar]
- Peters JM, Gonzalez FJ. (2009) Sorting out the functional role(s) of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) in cell proliferation and cancer. Biochim Biophys Acta 1796:230–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Hollingshead HE, Gonzalez FJ. (2008) Role of peroxisome-proliferator-activated receptor β/δ (PPARβ/δ) in gastrointestinal tract function and disease. Clin Sci 115:107–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Lee SS, Li W, Ward JM, Gavrilova O, Everett C, Reitman ML, Hudson LD, Gonzalez FJ. (2000) Growth, adipose, brain and skin alterations resulting from targeted disruption of the mouse peroxisome proliferator-activated receptor β(δ). Mol Cell Biol 20:5119–5128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn CE, Hamilton PK, Lockhart CJ, McVeigh GE. (2008) Thiazolidinediones: effects on insulin resistance and the cardiovascular system. Br J Pharmacol 153:636–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly SM, Lee CH. (2008) PPARδ as a therapeutic target in metabolic disease. FEBS Lett 582:26–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell CE, Snider NT, Thompson JT, Vanden Heuvel JP, Kaminski NE. (2006) Interleukin-2 suppression by 2-arachidonyl glycerol is mediated through peroxisome proliferator-activated receptor gamma independently of cannabinoid receptors 1 and 2. Mol Pharmacol 70:101–111 [DOI] [PubMed] [Google Scholar]
- Shearer BG, Hoekstra WJ. (2003) Recent advances in peroxisome proliferator-activated receptor science. Curr Med Chem 10:267–280 [DOI] [PubMed] [Google Scholar]
- Shearer BG, Steger DJ, Way JM, Stanley TB, Lobe DC, Grillot DA, Iannone MA, Lazar MA, Willson TM, Billin AN. (2008) Identification and characterization of a selective peroxisome proliferator-activated receptor β/δ (NR1C2) antagonist. Mol Endocrinol 22:523–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer BG, Wiethe RW, Ashe A, Billin AN, Way JM, Stanley TB, Wagner CD, Xu RX, Leesnitzer LM, Merrihew RV, et al. (2010) Identification and characterization of 4-chloro-N-(2-{[5-trifluoromethyl)-2-pyridyl]sulfonyl}ethyl)benzamide (GSK3787), a selective and irreversible peroxisome proliferator-activated receptor delta (PPARdelta) antagonist. J Med Chem 53:1857–1861 [DOI] [PubMed] [Google Scholar]
- Staels B, Dallongeville J, Auwerx J, Schoonjans K, Leitersdorf E, Fruchart JC. (1998) Mechanism of action of fibrates on lipid and lipoprotein metabolism. Circulation 98:2088–2093 [DOI] [PubMed] [Google Scholar]
- Stafslien DK, Vedvik KL, De Rosier T, Ozers MS. (2007) Analysis of ligand-dependent recruitment of coactivator peptides to RXRβ in a time-resolved fluorescence resonance energy transfer assay. Mol Cell Endocrinol 264:82–89 [DOI] [PubMed] [Google Scholar]
- Sznaidman ML, Haffner CD, Maloney PR, Fivush A, Chao E, Goreham D, Sierra ML, LeGrumelec C, Xu HE, Montana VG, et al. (2003) Novel selective small molecule agonists for peroxisome proliferator-activated receptor δ (PPARδ)–synthesis and biological activity. Bioorg Med Chem Lett 13:1517–1521 [DOI] [PubMed] [Google Scholar]
- Uh M, Björling E, Agaton C, Szigyarto CA, Amini B, Andersen E, Andersson AC, Angelidou P, Asplund A, Asplund C, et al. (2005) A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics 4:1920–1932 [DOI] [PubMed] [Google Scholar]
- Zaveri NT, Sato BG, Jiang F, Calaoagan J, Laderoute KR, Murphy BJ. (2009) A novel peroxisome proliferator-activated receptor δ antagonist, SR13904, has anti-proliferative activity in human cancer cells. Cancer Biol Ther 8:1252–1261 [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 4:67–73 [DOI] [PubMed] [Google Scholar]
- Zuo X, Peng Z, Moussalli MJ, Morris JS, Broaddus RR, Fischer SM, Shureiqi I. (2009) Targeted genetic disruption of peroxisome proliferator-activated receptor-δ and colonic tumorigenesis. J Natl Cancer Inst 101:762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.