Abstract
Apurinic/apyrimidinic (AP) endonuclease 1 (Ape1) is an essential DNA repair protein that plays a critical role in repair of AP sites via base excision repair. Ape1 has received attention as a druggable oncotherapeutic target, especially for treating intractable cancers such as glioblastoma. The goal of this study was to identify small-molecule inhibitors of Ape1 AP endonuclease. For this purpose, a fluorescence-based high-throughput assay was used to screen a library of 60,000 small-molecule compounds for ability to inhibit Ape1 AP endonuclease activity. Four compounds with IC50 values less than 10 μM were identified, validated, and characterized. One of the most promising compounds, designated Ape1 repair inhibitor 03 [2,4,9-trimethylbenzo[b][1,8]-naphthyridin-5-amine; AR03), inhibited cleavage of AP sites in vivo in SF767 glioblastoma cells and in vitro in whole cell extracts and inhibited purified human Ape1 in vitro. AR03 has low affinity for double-stranded DNA and weakly inhibits the Escherichia coli endonuclease IV, requiring a 20-fold higher concentration than for inhibition of Ape1. AR03 also potentiates the cytotoxicity of methyl methanesulfonate and temozolomide in SF767 cells. AR03 is chemically distinct from the previously reported small-molecule inhibitors of Ape1. AR03 is a novel small-molecule inhibitor of Ape1, which may have potential as an oncotherapeutic drug for treating glioblastoma and other cancers.
Introduction
DNA repair pathways maintain the integrity of eukaryotic and prokaryotic genomes. The base excision repair (BER) pathway repairs bases damaged by endogenous and exogenous alkylating and oxidizing agents (Christmann et al., 2003). The BER pathway is initiated by a damage-specific DNA glycosylase, which recognizes and excises the damaged base to generate an apurinic/apyrimidinic site (AP) site. Alternatively, AP sites also can be generated by spontaneous depurination (Wilson and Barsky, 2001). AP endonuclease 1 (Ape1) cleaves the phosphodiester backbone 5′ to the AP site, generating 3′-hydroxyl and 5′-deoxy ribose phosphate termini. Polymerase β removes the 5′-deoxy ribose phosphate, fills in the one-base gap, and the nick is ligated by DNA ligase III/X-ray cross-species complementing 1 to complete repair (Evans et al., 2000; Robertson et al., 2009).
Ape1 belongs to the xth family of class II endonucleases, whose prototypical member is the Escherichia coli exonuclease III. The xth-related proteins account for almost 95% of the cellular AP endonuclease activity in most eukaryotic species (Evans et al., 2000). Ape1 exhibits a prominent 5′ hydrolytic AP endonuclease, a weak 3′-diesterase, and a 3′-5′ exonuclease activity (Chou and Cheng, 2003; Parsons et al., 2004). In Saccharomyces cerevisiae (yeast), however, AP endonuclease 1 is the most abundant AP endonuclease (Popoff et al., 1990) and along with E. coli endonuclease IV belongs to the nfo family of the class II endonucleases (Evans et al., 2000). Although the E. coli endonuclease IV and Ape1 proteins have similar biological roles, their protein sequences are dissimilar (Wilson and Barsky, 2001; Garcin et al., 2008).
Ape1 is also a redox factor, which stimulates the DNA binding capacity of several transcription factors [e.g., p53, activator protein-1 (Fos/Jun), hypoxia-inducible factor 1-α, nuclear factor-κB] by reducing cysteine residues in their DNA binding domains (Xanthoudakis et al., 1994; Tell et al., 2009; Luo et al., 2010). Through its DNA repair and redox functions, Ape1 is involved in protein-protein interactions within the BER pathway and several other signaling pathways (Fan and Wilson, 2005). Probably, both these functions of Ape1 contribute to the resistance of cancer cells to chemotherapeutic agents. Although the repair function of Ape1 probably plays a direct role in resistance, the redox function may play an indirect role through its modulation of transcription factors, which control the expression of genes with important roles in cell survival, tumor promotion, and tumor progression (Fishel and Kelley, 2007; Reed et al., 2009; Luo et al., 2010).
Malignant glioma is a common and often fatal brain cancer, with a 2-year survival rate of ∼26%. Treatment options for gliomas include surgical resection followed by chemotherapy with temozolomide (TMZ) or radiation, both of which generate lesions repaired by the BER pathway (Evans et al., 2000; Chaudhry, 2007; Stupp et al., 2007). These treatments rarely result in complete remission and are often accompanied by adverse toxic effects in longer surviving patients (Stupp et al., 2007). High expression levels of Ape1 in cancer cells, including glioblastomas, compared with normal cells have been reported, and altered subcellular localization of Ape1 correlates with cellular resistance to chemotherapeutic agents such as methyl methane sulfonate (MMS), TMZ, hydrogen peroxide, bleomycin, and gemcitabine in several cancer cell lines (Bobola et al., 2001; Kelley et al., 2001; Robertson et al., 2001; Silber et al., 2002). Congruously, treatment with Ape1-targeted small interfering RNA or expression of dominant-negative Ape1 increases cellular sensitivity to ionizing radiation and alkylating and oxidizing agents, suggesting Ape1 as a potential therapeutic target in cancer cells (Evans et al., 2000; Fishel and Kelley, 2007; Fishel et al., 2007; McNeill and Wilson, 2007).
Known inhibitors of the repair activity of Ape1 include methoxyamine (MX) as well as small molecules identified in high-throughput screens. MX reacts with and covalently modifies AP sites to nonspecifically inhibit enzymes that bind to AP sites (Rosa et al., 1991), including Ape1, DNA polymerase β (Horton and Wilson, 2007), and other BER enzymes (Taverna et al., 2001). MX enhances the sensitivity of several cancer cell lines to TMZ and radiation (Fishel et al., 2007). Recent studies identified small-molecule inhibitors of the endonuclease activity of Ape1: 7-nitro-indole-2-carboxylic acid (NCA; CRT0044876) (Madhusudan et al., 2005), arylstibonic acid compounds (Seiple et al., 2007), pharmacophore-based compounds (Zawahir et al., 2009), and Reactive Blue 2,6-hydroxyl-dl-DOPA and myricetin (Simeonov et al., 2009). The arylstibonic compounds effectively inhibit Ape1 in vitro but are ineffective in vivo due to poor cellular uptake (Seiple et al., 2007), whereas the pharmacophore compounds were not tested in cell-based assays (Zawahir et al., 2009). Reactive Blue 2,6-hydroxyl-dl-DOPA and myricetin have numerous other targets besides Ape1 in cells; therefore, they are not very promising agents with selectivity or specificity as Ape1 inhibitors (Simeonov et al., 2009).
Here, we aimed to identify and characterize specific inhibitor(s) of the Ape1 AP endonuclease, using a high-throughput screening (HTS) assay to screen a library of 60,000 small-molecule compounds, which is the largest HTS performed to date. Four compounds with IC50 values less than 10 μM were identified, validated, and characterized. The most promising compound, designated Ape1 repair inhibitor 03 (AR03), inhibits the AP endonuclease activity of purified human Ape1 in vitro and inhibits cleavage of AP sites in SF767 cell extracts in vitro and in SF767 glioblastoma cells in vivo. AR03 also potentiates the cytotoxicity of MMS and TMZ in SF767 cells.
Materials and Methods
Enzymes.
The human Δ40 Ape1 protein was purified as described previously (Georgiadis et al., 2008). In brief, the Δ40Ape1 protein in the pET15b vector with an N-terminal hexa-His tag was expressed in the Rosetta E. coli strain. The cell pellets were resuspended in buffer A (50 mM phosphate, pH 7.8, 0.3 M NaCl, and 10 mM immidazol) and lysed using a French press. The supernatant was first eluted from a nickel column with buffer B (50 mM phosphate, pH 7.8, 0.3 M NaCl, and 250 mM immidazol), and the pooled Ape1 fractions were diluted five times with buffer C (50 mM MES, pH 6.5, and 1 mM DTT) to a salt concentration of 50 mM, which was eluted a second time from an S-Sepharose column with buffer D (50 mM MES, pH 6.5, 1 M NaCl, and 1 mM DTT). The Ape1 fractions digested overnight with thrombin (2 U) to remove the hexa-His tag were diluted eight times to 50 mM NaCl using buffer E (50 mM MES, pH 6.0, and 1 mM DTT) and gradient-eluted from an S-Sepharose column with buffer F (50 mM MES, pH 6.0, 1 M NaCl, and 1 mM DTT). Ape1 fractions were concentrated using a 10,000 Da-cut-off protein concentrator, and protein concentration and activity of the protein were determined.
The E. coli endonuclease IV protein (100 units) used in the gel-based AP endonuclease assays was purchased from Trevigen (Gaithersburg, MD).
Cell extracts from SF767 glioblastoma cells were prepared as described previously (Kreklau et al., 2001). The protein concentration of the SF767 cell extracts was determined using the Bio-Rad Bradford assay (Bio-Rad Laboratories, Hercules, CA; Bradford, 1976).
Oligonucleotides.
The pair of oligonucleotides used in the HTS assay were 30 base pairs (5′-6-FAM-GCCCCC*GGGGACGTACGATATCCCGCTCC-3′ and 3′-Q-CGGGGGCCCCCTGCATGCTATAGGGCGAGG-5′) and were synthesized via custom order from Eurogentec Ltd. (San Diego, CA) (Madhusudan et al., 2005). Of the pair, one of the oligonucleotides has a fluorescein label (6-FAM) at the 5′ end and contains an AP site mimic called tetrahydrofuran (THF, represented as * in the oligonucleotide). The complimentary strand has a quenching moiety (dabcyl-Q in the oligonucleotide) at its 3′ end. A stretch of G and C base pairs was used before and after the THF moiety in the HTS assay to prevent spontaneous dissociation of the short labeled fragment before cleavage by Ape1. The 26-base pair oligonucleotides used in the gel-based AP endonuclease assay were obtained from the Midland Certified Reagent (Midland, TX). The oligonucleotides comprise one strand (5′-HEX-AATTCACCGGTACC*CCTAGAATTCG-3′) with a hexa-chloro phosphoramidite fluorescein (HEX) label and tetrahydrofuran (THF, represented as *) molecule, and its opposite strand (3′-TTAAGTGGCCATGGTGGATCTTAAGC-5′) (Kreklau et al., 2001).
For both the HTS and the gel-based AP endonuclease assay, the single-stranded oligonucleotides were dissolved and annealed in 1× TEN buffer (25 mM Tris, pH 7.5, 1 mM EDTA, and 50 mM NaCl) at 95°C for 5 min at a 1:1 ratio at a concentration of 10 μM and allowed to cool to room temperature overnight. The DNA was appropriately diluted, aliquoted, and stored at –20°C.
HTS Assay.
The HTS assay used here was modified for our use from that described by Madhusudan et al. (2005). A library of 60,000 diverse compounds adhering to Lipinski's rules (Lipinski and Hopkins, 2004) from Chemical Diversity Ltd. Inc. (San Diego, CA) was tested at the Chemical Genomics Core Facility at Indiana University School of Medicine (Indianapolis, IN). The library was aliquoted and screened at a concentration of 10 μM in black 384-well plates in a 50-μl reaction volume including 0.8% DMSO. The Tecan Genesis Workstation 150 TeMo (Tecan, Durham NC) with a 96-channel pipetting head was used to first add 20 μl of a reaction mix containing the annealed fluorescein-dabcyl-containing oligonucleotide substrate (100 nM) and the assay buffer (50 mM Tris, pH 8.0, 1 mM MgCl2, 50 mM NaCl, and 2 mM DTT) to the plates containing the compound library (10 μM) in a 20-μl volume. Then, 10 μl of the Ape1 protein at a final concentration of 0.35 nM was added to the reaction mix in the assay plates to initiate the reaction, and changes in fluorescence were measured at 37°C over 5 min using an Ultra384 plate reader (Tecan) in the kinetic mode with an excitation frequency of 495 nm and an emission frequency of 530 nm. The presence of Ape1 results in cleavage of the AP site mimic and a subsequent release of the short fluorescein-labeled fragment, resulting in a proportionate increase in fluorescence. The rates of reaction were used to determine percentage of inhibition, and the rate of reaction Ape1 protein without inhibitors was considered as the 100% control; subsequent inhibition by the compounds was considered relative to that of the control.
Calculation of IC50 Values of Compounds.
After two rounds of screening, IC50 values of the compounds selected for validation were determined. The assay used to determine the IC50 values was similar to the HTS assay, where a wide range of concentrations (0.1–100 μM) of the compounds were tested with 0.35 nM Ape1, 100 nM annealed substrate, and the assay buffer (50 mM Tris, pH 8.0, 1 mM MgCl2, 50 mM NaCl, and 2 mM DTT) in a 50-μl reaction volume, with a final DMSO concentration of 0.8%. Fluorescence readings were taken at 37°C for 5 min, and percentage of inhibition for each compound concentration was determined compared with the control with no inhibitor. IC50 values were calculated using the SigmaPlot graphing software (Systat Software, Inc., San Jose, CA) using the four-parameter logistic equation as follows:
Each data value is an average of three independent experiments with standard errors. Statistical analysis of the data was done using the student's t test with p values representing the comparison between lanes with inhibitor and without inhibitor.
Gel-Based AP Endonuclease Assay.
An annealed 26-base pair oligonucleotide substrate containing a THF moiety at position 13 is used in this assay, such that cleavage by Ape1 results in two 13 mer fragments of DNA: one fragment with the fluorescent HEX label and one fragment unlabeled. Ape1 (0.175 nM) was added to the reaction mix also containing assay buffer (50 mM HEPES, pH 7.5, 50 mM KCl, 1 mM MgCl2, and 2 mM DTT), 25 nM HEX-labeled DNA substrate, and the inhibitor compounds in a range of concentrations in a total reaction volume of 20 μl. The reaction mixture was then incubated at 37°C for 15 min, and the reaction was stopped by the addition of 10 μl of formamide without dyes. Then, 15 μl of the resultant reaction mixture was resolved on a 20% denaturing (7 M urea) polyacrylamide gel in 1× Tris-borate EDTA at 300 V for 35 min to reveal two bands: the longer full-length labeled strand and the shorter cleaved fragment with the HEX label.
For the endonuclease IV protein (100 units), the assay was performed as described above with 6.25 units of endonuclease IV protein and different concentrations of the inhibitor compounds.
The assay with SF767 cell extracts also was performed as described above. We incubated 3.75 ng of the SF767 cell extract along with the assay buffer, DNA substrate, and inhibitor compounds at 37°C for 30 min, and the reaction was stopped by the addition of 10 μl of formamide without dyes. For the experiments where pure Ape1 protein was added back to the SF767 cell extracts treated with the inhibitor compounds, 3.75 ng of the SF767 cell extracts was first incubated with the compounds for 30 min at 37°C. The DNA substrate and pure Ape1 (0.7–5.6 nM) was added to the reaction mixture, and the reaction was allowed to proceed at 37°C for 30 min, after which it was terminated by the addition of 10 μl of formamide without dyes. Similarly, for SF767 whole cell extracts immunodepleted of Ape1, the reaction was carried out as described above (Kreklau et al., 2001).
For each of the above-mentioned gel-based assays (with pure Ape1 protein, endonuclease IV protein, and SF767 cell extracts), assays were performed three times, and the data are presented as the average of three individual experiments and standard error. The Student's t test was used to calculate p values. For the gel assays with pure Ape1 and SF767 cell extracts alone, lanes with inhibitor are compared with the lane without inhibitors. For the experiments showing restoration of AP endonuclease activity of the cell extracts, lanes that contain pure Ape1 protein are compared with the lane with the cell extract and inhibitor alone.
Fluorescence Intercalation Displacement Assay to Determine DNA Binding.
All validated Ape1 inhibitors were tested for DNA binding using a fluorescence intercalation displacement (FID) assay that takes advantage of the greatly increased fluorescence observed for ethidium bromide (EtBr) when bound to DNA (Fox, 1997; Glass et al., 2010), in this case using calf thymus DNA to present all possible DNA binding sites. Compounds that effectively compete with EtBr for binding to DNA will result in a decrease in the fluorescence measured for EtBr. Triplicate measurements were made using an Envision 2102 multilabel plate reader (PerkinElmer Life and Analytical Sciences, Boston, MA) for 1, 10, and 100 μM compound (final concentration of 0.8% DMSO) in a total of 50 μl including 21 μM calf thymus DNA, 6.5 μM ethidium bromide, 100 mM NaCl, and 10 mM Tris, pH 7.4, in 384-well black Nunc plates (Nalge Nunc International, Rochester, NY). The fluorescence was measured for each well at 530 nm excitation and 615 nm emission and compared with the negative control (no added compound) to calculate percentage of fluorescence decrease for each well and standard deviations. Positive controls included known DNA binding agents, the minor groove binder netropsin, and the intercalator actinomycin D.
Immunodepletion of Ape1 from SF767 Whole Cell Extracts.
SF767 cell extracts were immunodepleted of the Ape1 protein using a polyclonal Ape1 antibody (Novus Biologicals, Inc., Littleton CO). SF767 cell extracts (250 μg) in 1× PBS were precleared by adding 50 μl of washed (beads were washed with 450 μl of PBS twice and resuspended in 50 μl of PBS) protein A/G Plus-agarose beads (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) to the cell extracts and gently rocking them at 4°C for 1 h. The extracts were centrifuged at 10,000g for 1 min, and the supernatants were collected. The supernatants were incubated with 10 μg of the polyclonal Ape1 antibody (Novus Biologicals, Inc.) or normal rabbit IgG (Santa Cruz Biotechnology, Inc.) at 4°C with gentle shaking for 2 h. Then, 50 μl of washed beads was added to the cell extracts, and the extracts were incubated for another 2 h with gentle rocking at 4°C. The cell extracts were centrifuged at 10,000g for 5 min, and the supernatants were collected. The protein concentration of the immunodepleted cell extracts was measured with the Bradford assay (Bio-Rad Laboratories), and aliquots were stored at −80°C.
Western Blot Analysis.
To determine Ape1 levels in SF767 cell extracts after immunodepletion, appropriate samples were mixed with equal amounts of 2× protein loading dye and boiled in boiling water bath for 5 min. The samples were loaded onto a 12% Tris-HCl precast gel (Bio-Rad Laboratories) and allowed to separate at 150 V for 40 min. The gel was transferred onto a nitrocellulose membrane at 90V for 2 h at room temperature. After the transfer of proteins onto the membrane, the membrane was blocked in 5% blocking solution made from blotting grade blocker, nonfat dry milk (Bio-Rad Laboratories) dissolved in 1× TBS for 2 to 4 h. Ape1 monoclonal antibody at a dilution of 1:1000 in 1% blocking solution was added to the membrane and allowed to rotate overnight at 4°C. The next day, the membranes were washed with 1× TBS + 0.1% Tween 20, and a secondary anti-mouse horseradish peroxidase-labeled antibody was added to the blots at a dilution of 1:1000 and allowed to rotate for 1 to 2 h at room temperature. After washing the membranes with 1× TBS + 0.1% Tween 20, the SuperSignal West Femto Maximum Sensitivity Substrate (Pierce Chemical, Rockford, IL) was added to the blots, and the blots were developed using the Bio-Rad Chemi Doc XRS system in the chemiluminescence hi-sensitivity mode. Actin was used as the internal loading control, and the actin antibody (NeoMarkers, Fremont, CA) was used at a dilution of 1:1000, and actin was detected using the ECL Western Blotting Detection reagent (GE Healthcare, Little Chalfont, Buckinghamshire, UK). The experiment was performed three times, and the data are an average of three individual experiments and standard error. For both the gel assays with the immunodepleted extracts and the Western blot analysis, comparisons were made between corresponding lanes in the IgG-treated and the immunodepleted samples.
Cell Culture-Based Survival Assay.
SF767 glioblastoma cells were cultured in Dulbecco's modified Eagle's medium with high glucose supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. The effects of the inhibitor compounds on the growth and survival of these SF767 glioblastoma cells were determined using the xCELLigence DP System (Roche Applied Science, Indianapolis IN) (Solly et al., 2004). SF767 glioblastoma cells (3000) were plated in each well of the 16-well plates in a 100-μl volume, and before plating the cells, a background reading of the wells with 90 μl of appropriate media was recorded. After adding the cells to the wells, the plates were kept at room temperature for 30 min after which they were inserted into the cradles. The cells were allowed to grow for 20 to 24 h before the cytotoxic agents were added. MMS (100 μM), TMZ (2.25 mM), and AR03 (0.5 and 0.75 μM) in a 10-μl volume at a concentration 20× the final concentration were added to the wells, alone or in combination. Continuous impedance measurements were then monitored over 72 h. The assay was performed in triplicate.
AP Site Determination Assay.
To determine number of AP sites formed, SF767 cells were treated with 275 μM MMS and 3 μM AR03 alone or in combination for 24 h. Cells were collected after 24 h, and the genomic DNA was isolated using the Get-Pure DNA isolation kit (Dojindo Molecular Technologies, Rockville, MD). The ARP reagent specifically reacts with the aldehyde group of AP sites in the open conformation, converting them to biotin-tagged AP sites. The amount of biotin can then be quantified by an enzyme-linked immunosorbent assay-like assay (Kow and Dare, 2000). The number of AP sites formed was determined using the AP Site Quantitation kit (Dojindo Molecular Technologies) as per the kit instructions. The experiments were repeated in triplicate, and the data presented are averages of four independent experiments with standard error. An n = 12 was considered while calculating the p values for this experiment, and p values were obtained using the Student's t test where the combined treatment group was compared with AR03 inhibitor- and MMS-treated group alone.
Redox Electrophoretic Mobility Shift Assay.
Redox electrophoretic mobility shift assays were performed as described previously (Georgiadis et al., 2008).
Results
Optimization of the Fluorescence-Based Assay for Ape1 AP Endonuclease Activity.
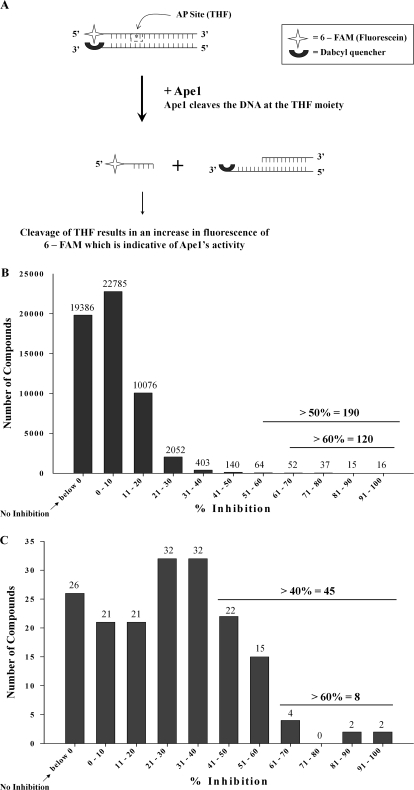
A fluorescence-based AP endonuclease assay described by Madhusudan et al. (2005) was adapted in this study for HTS of a library of 60,000 small-molecule compounds (see Materials and Methods). In brief, the assay uses two complementary 30-bp synthetic oligonucleotides, one oligonucleotide of which carries 5′-fluorescein and a THF AP site mimic (Wilson and Barsky, 2001), whereas the complementary oligonucleotide carries a 3′-dabcyl fluorescence-quenching moiety (Fig. 1A). After cleavage at the THF moiety, a short single-stranded DNA fragment 5′-tagged with fluorescein is released from the duplex DNA substrate, resulting in increased fluorescence. In the linear range of this assay, the increase in fluorescence over time is proportional to the amount of AP endonuclease activity in the reaction (Madhusudan et al., 2005; Fig. 1A). To optimize the HTS, increasing concentrations of Ape1 were added to the reaction mixture, and a concentration of 0.35 nM Ape1 was selected for screening at 37°C for 5 min (data not shown). Under these conditions, the Z′ factor was 0.78, which indicates that the assay provides a statistically valid measure of inhibition of the Ape1 AP endonuclease (Zhang et al., 1999).
Fig. 1.
HTS assay to identify inhibitors of Ape1. A, principle of the HTS Assay. This figure represents the principle of the modified fluorescent HTS assay used for our screening purposes. The fluorescence of 6-FAM is diminished by Q because of their proximity to each other. On addition of Ape1, it cleaves the THF residue releasing the short 6-FAM-labeled fragment that results in a proportionate increase in fluorescence. This increase in fluorescence can be considered to be an indirect measure of cleavage activity of Ape1. B, results of the initial screen of the 60,000-compound library. After the screen of the entire library, we identified 190 compounds that inhibited the activity of Ape1 by 50% or more. C, results of a secondary screen of all the hits from the initial screen showing ≥50% inhibition of the activity of Ape1. Of the 190 compounds identified, 174 were retested in this same assay to weed out false positives, and 41 compounds with ≥40% inhibition of the DNA repair activity of Ape1 were validated. For both B and C, the graphs are representations of the numbers of compounds plotted with their corresponding percentage of inhibition of the activity of Ape1.
Identification and Validation of Ape1 Inhibitors from a 60,000-Compound Library.
The HTS protocol used in this study is described in detail under Materials and Methods (see Supplemental Fig. 1). In the first HTS cycle, 190 compounds were found to inhibit ≥50% of the Ape1 endonuclease activity. In a parallel effort, the same 60,000 compound library was screened for DNA binding activity using a high-throughput fluorescence intercalator displacement assay (Glass et al., 2010). Compounds identified as Ape1 endonuclease inhibitors that also demonstrated strong DNA binding were not further evaluated; these compounds would not be expected to be specific inhibitors of Ape1 endonuclease activity. After two rounds of rescreening, 45 compounds were identified that achieved ≥40% inhibition of Ape1 endonuclease activity (Fig. 1, B and C). Forty-one of these compounds were available from ChemDiv (http://us.chemdiv.com/) at that time and were further validated as described below.
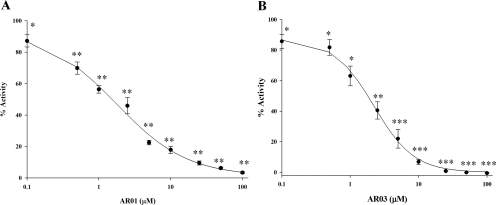
The concentration dependence for inhibition of AP endonuclease activity was determined for the 41 compounds that emerged as hits from the secondary HTS. NCA (Madhusudan et al., 2005) was used as a positive control, and IC50 values were calculated using SigmaPlot software. Eighteen compounds had an IC50 value ≤50 μM, and nine compounds had an IC50 value ≤10 μM (Supplemental Table 1; data not shown in text). Representative data are shown for AR01 and AR03, including four-parameter logistic curves for determining IC50 values, in Fig. 2, A and B, respectively. From the 41 compounds, seven chemically distinct classes of compounds were selected. Table 1 shows the most potent inhibitor of each chemical class, representing the chemical diversity of inhibitors identified in the HTS.
Fig. 2.
Calculation of the IC50 values of the AR01 and AR03 compounds. IC50 values of the top hit compounds were determined using the HTS assay. A range of concentrations of each of the compounds were tested with 0.35 nM Ape1. The percentage of inhibition of Ape1's activity was calculated using the rates of reaction for each of the concentrations of the compounds compared against the percentage of activity of Ape1 alone. The assays for each compound were performed three individual times. SigmaPlot software (see equation described under Materials and Methods) was used to calculate the values, and they are presented here is a semilog plot of the IC50 value determination. A,IC50 value curve for AR01. B, IC50 value curve for AR03. The p values were calculated using the Student's t test comparing lanes with inhibitor with lane with no inhibitor (DMSO), *, p ≤ 0.03; **, p ≤ 0.0005; and ***, p ≤ 0.0001.
TABLE 1.
Inhibitors identified in the HTS
| Small Molecule | Mol. Wt. | cLog P | No. Relative Compounds | First HTS Assay | Second HTS Assay | IC50 | ΔFID, DNA binding |
|---|---|---|---|---|---|---|---|
| μM | % | ||||||
| AR01 | 245.274 | 3.7 | 1 | 74 | 70 | 1.7 ± 0.3 | 8 |
| AR02 | 278.348 | 5.3 | 10 | 54 | 40 | 6.4 ± 1.1 | 9.60 |
| AR03 | 237.3 | 3.7 | 60 | 58 | 2.1 ± 0.1 | 12.80 | |
| AR04 | 350.414 | 4.8 | 1 | 58 | 45 | 14.9 ± 8.4 | <15 |
| AR05 | 351.379 | 2.4 | 75 | 85 | 6.6 ± 0.9 | <15 | |
| AR06 | 262.692 | 2.7 | 1 | 52 | 44 | 1.6 ± 0.1 | 3.10 |
| AR07 | 400.534 | 5.3 | 12 | 80 | 41 | ∼100 | <15 |
For comparison, a similar IC50 analysis was carried out for previously characterized inhibitors of Ape1 AP endonuclease: these compounds included two arylstibonic acid compounds (13755 and 13793) reported by Seiple et al. (2007) and the pharmacophore compound 18 (A1NI2-A3NI1) reported by Zawahir et al. (2009). Previously reported inhibitory effects on Ape1 activity of NCA (Madhusudan et al., 2005), resveratrol (Yang et al., 2005), and arylstilbonic acids 13755 and 13793 (Seiple et al., 2007) were not reproducible or were quantitatively different in the present study than in previous studies (Supplemental Table 2; data not shown in text). Although the precise reason for these discrepancies is not known, they may reflect differences in the AP endonuclease assay conditions used in each study.
The seven chemically distinct AP endonuclease inhibitors were retested for DNA binding using an FID assay (Table 1; Supplemental Fig. 2); Glass et al., 2010). In the FID assay, 10 μM netropsin or 10 μM actinomycin D (positive controls) caused a 32 or 63% decrease in fluorescence, respectively. A decrease in fluorescence of <20% indicated no significant DNA binding activity. The FID data confirm that inhibitors AR01–AR07 do not exhibit significant DNA binding (Table 1).
At this point in the study, five inhibitors—AR01, 02, 03, 05, and 06—appeared to be the most promising of the chemically distinct inhibitors, with IC50 values less than 10 μM. However, when diluted in aqueous solutions required for cell-based assays, AR05 was found to be completely insoluble. Thus, four chemically distinct inhibitors, AR01, AR02, AR03, and AR06 (Table 1), were further characterized.
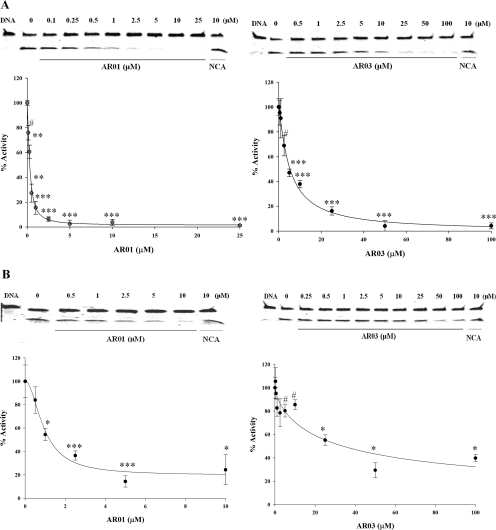
Further validation of AR01, AR02, AR03, and AR06 included inhibition of Ape1 endonuclease activity using a gel-based AP endonuclease assay (see Materials and Methods for details; Kreklau et al., 2001). The results of the gel-based assay confirm that all four inhibitors effectively inhibit Ape1 AP endonuclease activity. Representative results for AR01 and AR03 are shown in Fig. 3A. The inhibitors were also tested for ability to inhibit E. coli endonuclease IV, which is structurally unrelated to Ape1 (Wilson and Barsky, 2001; Garcin et al., 2008). The results of gel-based AP endonuclease assays show that in this assay, AR03 is approximately 8-fold less effective in inhibiting endonuclease IV than Ape1 (40 μM for 50% inhibition versus 5 μM), whereas AR01 inhibited endonuclease IV and Ape1 at similar concentrations (0.6 μM for 50% inhibition versus 1.4 μM; Fig. 3B). AR06 and AR02 inhibited endonuclease IV by 50% at 10 and 50 μM, respectively (Supplemental Fig. 3).
Fig. 3.
Effect of AR01 and AR03 on the activity of purified Ape1 and endonuclease IV proteins in the gel-based AP endonuclease assay. The ability of the top hit compounds to inhibit the activity of Ape1 in a distinct gel-based AP endonuclease assay was tested, as was their effect against the E. coli endonuclease IV protein. The assay was performed as described under Materials and Methods. Effect of AR01 and AR03 on purified Ape1 protein (A) and on endonuclease IV (B). Representative gels are shown. Each assay was performed three individual times and is shown here as the average with standard error (B, AR01 and D, AR03). The p values were calculated using the Student's t test comparing lanes with inhibitor with lane with no inhibitor (DMSO). #, p ≤ 0.05; *, p ≤ 0.005; **, p ≤ 0.001; and ***, p ≤ 0.0001.
Inhibition of Ape1 in SF767 Whole Cell Extracts.
The ability of AR01, AR02, AR03, and AR06 to inhibit AP site cleavage in SF767 glioblastoma whole cell extracts (Kreklau et al., 2001) was examined using the gel-based AP endonuclease assay. AR01 and AR03 inhibited purified Ape1 and AP endonuclease activity in SF767 extracts with similar efficiency (Fig. 4A). However, higher concentrations of AR02 and AR06 (∼93 and 26 μM, respectively, for 50% inhibition) were required to inhibit the AP endonuclease activity in SF767 cell extracts than to inhibit purified Ape1 (Supplemental Fig. 4). It is important to note that addition of purified Ape1 (0.7–5.6 nM) reversed inhibition of AP site cleavage in SF767 cell extracts treated with 50 μM AR03 (Fig. 4B) or 10 μM AR01 in a dose-dependent manner. Immunodepletion of Ape1 from SF767 cell extracts with a polyclonal Ape1 antibody also strongly reduced AP site cleavage activity in the cell extracts (Fig. 4, C and D). These data suggest that Ape1 is the primary AP endonuclease in SF767 extracts and that AR01 and AR03 specifically target the endonuclease activity of Ape1, thereby reducing AP site cleavage within the context of glioblastoma whole cell lysate. The higher concentrations of AR02 and AR06 required to inhibit AP endonuclease activity in SF767 cell extracts suggest that they do not selectively target Ape1 in cell extracts; therefore, they were not further characterized.
Fig. 4.
Ability of the top hits to inhibit the activity of Ape1 in cell extracts made from SF767 glioblastoma cells. A, ability of the top hit compounds to inhibit the AP endonuclease activity of SF767 cell extracts was tested. The effect of the compounds on SF767 whole cell extract was determined using the gel-based AP endonuclease assay (as described under Materials and Methods), and both compounds inhibited the repair activity of Ape1 similarly to that of purified Ape1 protein in the gel assay. The p values were calculated using the Student's t test comparing lanes with inhibitor with lane with no inhibitor (DMSO). B, to further confirm the inhibition of Ape1 in the SF767 cell extracts by AR03, increasing amounts (0.7–5.6 nM) of purified Ape1 protein were added to SF767 cell extracts treated with 10 μM AR01 or 50 μM AR03 (described under Materials and Methods). Addition of purified Ape1 protein results in a linear rescue of the AP endonuclease activity of the SF767 cell extracts compared with those without inhibitors. Lane 1, DNA alone (no extract); lane 2, SF767 cell extract + DMSO; lane 3, SF767 cell extract + inhibitor, 10 μM AR01, or 50 μM AR03; lanes 4 to 7, SF767 cell extract + inhibitor + increasing amounts of purified Ape1 protein (0.7–5.6 nM), and lanes 8 to 11, SF767 cell extract + increasing amounts of purified Ape1 protein (0.7–5.6 nM) without inhibitors. To calculate the p values, lanes 4 to 7 were compared with lane 3. To confirm that Ape1 is the only protein from the cell extracts that can act in the gel-based AP endonuclease assay, SF767 cell extracts were immunodepleted of Ape1 using a polyclonal Ape1 antibody (10 μg), and the AP endonuclease activity of these extracts was determined. C, Western blot analysis to show reduced levels of the Ape1 protein in the immunodepleted SF767 cell extracts (0.75–3 μg of protein loaded). Actin was used as a loading control. D, AP endonuclease activity of the immunodepleted SF767 cell extracts is significantly less than the IgG-treated control extracts. Representative gels are shown. Each assay was performed three individual times and is shown here as the average with standard error. For C and D, IgG control lanes were compared with the lanes with the immunodepleted cell extracts. #, p ≤ 0.08; ##, p ≤ 0.01; *, p ≤ 0.005; **, p ≤ 0.001; and ***, p ≤ 0.0001.
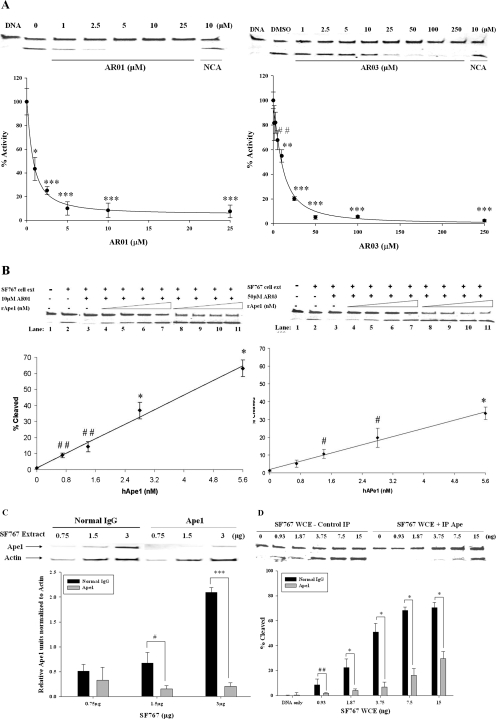
Effect of AR03 on SF767 Cells.
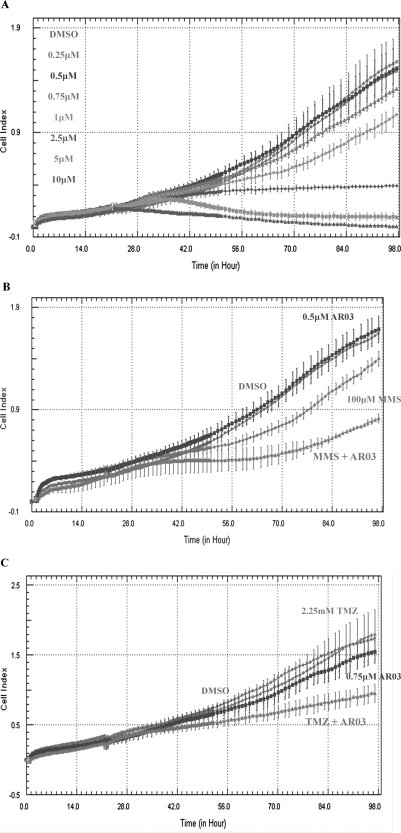
Initially, both AR01 and AR03 were tested in an 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium cell proliferation assays using SF767 glioblastoma cells. However, no effect on cell proliferation was observed for AR01 at concentrations as high as 200 μM, suggesting that it may not be cell-permeable, whereas AR03 was found to effectively kill cells at low micromolar concentrations (LD50 value of ∼1 μM; Supplemental Fig. 5). Therefore, the ability of AR03 to inhibit proliferation, reduce survival of SF767 glioblastoma cells, or both was further characterized using the xCELLigence DP system (Roche Applied Science). This system measures a dimensionless parameter called the cell index, which evaluates the ionic environment at an electrode/solution interface and integrates information on cell viability, number, morphology, and adhesion (Xing et al., 2005; Kirstein et al., 2006). AR03 reduced the cell index of SF767 cells efficiently and in a dose-dependent manner, suggesting that it inhibits growth and/or kills SF767 cells (Fig. 5A). AR03 also enhanced the cytotoxicity of MMS and TMZ (Fig. 5, B and C). These data are consistent with the idea that AR03 inhibits AP site cleavage by Ape1 and that MMS and TMZ increase the load of AP sites, leading to increased cytotoxicity.
Fig. 5.
Cell survival analysis of SF767 glioblastoma cells with AR03 alone and in combination with MMS and TMZ. The xCELLigence DP system was used to determine cell survival and growth after treatment with AR03 alone and in combination with MMS and TMZ. A, AR03 can act as a single agent to kill SF767 glioblastoma cells. Combined treatment of the SF767 cells with AR03 and MMS (B) or TMZ (C) results in greater cell killing compared with either agent alone. The assay was performed in triplicate, three individual times, and a representative experiment is shown.
Quantification of AP Sites in AR03-Treated SF767 Cells.
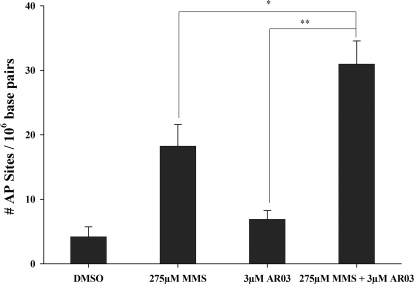
The above-mentioned data predict that treatment with AR03 will increase the number of AP sites in MMS-treated SF767 cells. This idea was tested by treating SF767 cells with MMS and AR03 and quantifying AP sites using the ARP assay (Kow and Dare, 2000). As predicted, more AP sites were detected in cells treated with AR03 and MMS than in cells treated with either agent alone (Fig. 6). These data further support the conclusion that AR03 inhibits Ape1-catalyzed repair of AP sites in SF767 cells.
Fig. 6.
AP Site determination in SF767 cells after treatment with MMS and AR03 alone and in combination. To directly determine inhibition of Ape1 by AR03 in cells, AP site formation was measured using the ARP assay. A significant increase in the number of AP sites was seen after treatment of the SF767 cells with AR03 and MMS in combination compared with either agent alone. The assay was done in triplicate, four individual times, and the data presented here are an average of four individual experiments with standard errors. The p values were determined for an n = 12 using the Student's t test where MMS + AR03 is compared with AR03 alone (**, p ≤ 0.0001) and MMS alone (*, p ≤ 0.01).
Effects of AR Inhibitors on Ape1 Redox Activity.
The effect of AR01, AR02, AR03, and AR06 on Ape1 redox activity (Tell et al., 2009) was investigated using an electrophoretic mobility shift assay (Georgiadis et al., 2008). In this assay, reduction of c-Jun/c-Fos by Ape1 increases its DNA binding affinity for DNA containing an AP-1 binding site and is measured by quantitation of the shifted band. AR02, AR03, and AR06 had no effect on the redox activity of Ape1 or the ability of c-Jun/c-Fos to bind DNA (data not shown). This result confirms the low binding affinity of these compounds for duplex DNA. However, addition of AR01 reduced the ability of c-Jun to bind DNA in this assay (50% inhibition at 15 μM). To determine whether this reduction was dependent on the presence of Ape1, the assay was repeated using reduced c-Jun/c-Fos in the absence of Ape1 protein, and a similar result (50% inhibition at 15 μM) was obtained, suggesting that AR01 affects the ability of c-Jun/c-Fos to bind DNA in the absence of Ape1 and is therefore not a redox inhibitor (data not shown).
Discussion
This study reports the results of a high-throughput screen of 60,000 small-molecule compounds, leading to the identification and characterization of several small-molecule inhibitors of Ape1 AP endonuclease activity. Seven chemically distinct inhibitors were initially selected of which four emerged from initial validation studies as promising inhibitors and were further characterized. AR01 is negatively charged like many of the Ape1 inhibitors reported previously (Madhusudan et al., 2005; Seiple et al., 2007; Simeonov et al., 2009; Zawahir et al., 2009), but it is otherwise unrelated to the previously described inhibitors. AR02, AR03, and AR06 are novel inhibitors of Ape1. Having established that AR02 and AR06 may have significant off-target interactions (Supplemental Fig. 2), the focus of this study was to characterize the specificity of AR01 and AR03 as Ape1 inhibitors in vitro and in SF767 glioblastoma cells.
The most potent inhibitors identified in this screen have low affinity for duplex DNA (Table 1), a critical characteristic for a specific inhibitor of Ape1. The ability of AR01, AR02, AR03, and AR06 to inhibit AP site cleavage by endonuclease IV was also examined as a measure of specificity, due to functional but not structural similarity to Ape1. AR01 inhibits Ape1 and endonuclease IV at similar concentrations; thus, it is possible that AR01 binds to an active site metal (i.e., through its carboxylate moiety) or another positively charged protein region, a site that normally binds to the DNA phosphate backbone. The ability of AR01 to inhibit binding of c-Jun/c-Fos to DNA is consistent with the latter mechanism of inhibition. Significantly higher concentrations of AR02, AR03, and AR06 are required to inhibit endonuclease IV than Ape1; these compounds may interact directly with Ape1 or the Ape1-DNA substrate complex, which is structurally distinct from the endonuclease IV-DNA substrate complex.
An important result of this study is that AR01 and AR03 inhibit the predominant AP endonuclease activity in SF767 cell extracts (Fig. 4A) and do so as effectively as they inhibit pure Ape1 in vitro. Furthermore, this effect is reversed in a linear- and dose-dependent manner by addition of purified Ape1 to the AR03-treated cell extract (Fig. 4B). Immunodepletion of Ape1 from the SF767 cell extract confirms the conclusion, based on other published studies, that Ape1 is the predominant AP endonuclease in SF767 cells (Fig. 4, C and D). Together, these results suggest that AR03 inhibits the AP endonuclease activity of Ape1 protein in cellular extracts and that Ape1 is the primary target of AR03 in SF767 cells; thus, we conclude that AR03 selectively targets Ape1. Although AR01 also efficiently inhibits AP site cleavage in SF767 cell extracts, it inhibits purified Ape1 and endonuclease IV at similar concentrations and inhibits the binding of c-Jun/c-Fos to DNA. Therefore, the specificity of AR01 for Ape1 is less well defined and warrants further study.
Glioblastoma is a relatively intractable cancer that is often treated with alkylating agents or radiation therapy (Stupp et al., 2007). Here, SF767 glioblastoma cells were used as a representative cancer cell line to explore the potential efficacy of AR03 as an oncotherapeutic agent alone or in combination with other cancer drugs. AR03 inhibits growth and viability of SF767 cells using the xCELLigence and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium-based proliferation assays (Fig. 5A; data not shown). In the present study using SF767 cells, the most likely mechanism for AR03-induced loss of cell viability is the accumulation of cytotoxic AP sites (Evans et al., 2000; Wilson and Barsky, 2001), a mechanism that is strongly supported by direct demonstration of increased accumulation of AP sites in these cells (Fig. 6). Further supporting the inhibition of Ape1 by AR03, xCELLigence assays show that AR03 enhances the cytotoxicity of MMS and TMZ (Fig. 5, B and C). Both of these DNA-damaging agents create lesions acted upon by BER and Ape1, and we expected potentiation of MMS and TMZ under conditions in which Ape1 was not functioning. The combination of AR03 and traditional chemotherapeutic agents could collaboratively block and overload the BER pathway, leading to cancer cell death. In contrast, AR01 is not toxic to SF767 cells (data not shown), which may be due to poor uptake of this negatively charged compound by these cells.
In conclusion, this study describes several inhibitors of Ape1 AP endonuclease, the most promising of which is AR03. AR03 appears to be a specific inhibitor of Ape1 AP endonuclease activity, with low micromolar efficacy as an inhibitor of purified Ape1 or Ape1 in cell extracts, low affinity for duplex DNA and much lower ability to inhibit E. coli endonuclease IV than human Ape1 protein. AR03 reduces proliferation and viability of cultured SF767 glioblastoma cells and enhances the cytotoxicity of MMS and TMZ, suggesting that it may have potential therapeutic applications. Additional studies are needed to determine the effect of AR03 in combination with other cancer chemotherapeutic agents, in combination with other Ape1 inhibitors, or with other BER targets such as poly(ADP-ribose) polymerase (Reed et al., 2009) and on other cancer cell lines and in the context of other in vitro or in vivo model systems. Additional analyses also are needed to determine whether there are any off-target effects of AR03, such as effects on mitotic and postmitotic normal cells. Furthermore, some structure-activity studies and pharmacokinetic studies to determine the mechanism of inhibition of AR03 need to be performed to assess the therapeutic potential of this class of small-molecule inhibitor of Ape1 endonuclease activity.
Supplementary Material
Acknowledgments
We thank Lan Chen (Chemical Genomics Core Facility) for invaluable assistance with the HTS.
This work was supported by the National Institutes of Health National Cancer Institute [Grants CA114571 (to M.M.G.), CA94025, CA106298, CA114571, CA121168 (to M.R.K.)], Indiana University Melvin and Bren Simon Cancer Center Translational Research Acceleration Collaboration pilot funding (to M.M.G. and M.R.K.)], and the Riley Children's Foundation (to M.R.K.).
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.110.169128.
The online version of this article (available at http://jpet.aspetjournals.org) contains supplemental material.
- BER
- base excision repair
- AP
- apurinic/apyrimidinic
- Ape1
- apurinic/apyrimidinic endonuclease 1
- TMZ
- temozolomide
- MMS
- methyl methane sulfonate
- MX
- methoxyamine
- NCA
- 7-nitro indole 2-carboxylic acid
- HTS
- high-throughput screen
- AR
- Ape1 repair inhibitor
- AR02
- 4-(2,6,8-trimethylquinolin-4-ylamino)phenol
- MES
- 2-(N-morpholino)ethanesulfonic acid
- DTT
- dithiothreitol
- FAM
- 5-carboxyfluorescein
- THF
- tetrahydrofuran
- HEX
- hexa-chloro phosphoramidite fluorescein
- DMSO
- dimethyl sulfoxide
- FID
- fluorescence intercalator displacement
- EtBr
- ethidium bromide
- PBS
- phosphate-buffered saline
- TBS
- Tris-buffered saline
- ARP
- aldehyde reactive probe
- AR01
- 2-(4-(2,5-dimethyl-1H-prryol-1-yl)phenoxy acetic acid
- AR03
- 2,4,9-trimethylbenzo[b][1,8]-naphthyridin-5-amine
- AR06
- N-(3-chlorophenyl)-5,6-dihyro-4H-cyclopenta[d] isoxazole-3-carboxamide
- AR07
- C22H28N2O3S
- AR04
- C20H22N4O2
- AR05
- C18H13N3O3S
- Compound 18 (A1NI2-A3NI1)
- 4-((2-carboxyphenoxy)methyl)-2,5-dimethylfuran-3-carboxylic acid
- 13755
- C7H6NO7Sb
- 13793
- C12H17O5Sb.
References
- Bobola MS, Blank A, Berger MS, Stevens BA, Silber JR. (2001) Apurinic/apyrimidinic endonuclease activity is elevated in human adult gliomas. Clin Cancer Res 7:3510–3518 [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254 [DOI] [PubMed] [Google Scholar]
- Chaudhry MA. (2007) Base excision repair of ionizing radiation-induced DNA damage in G1 and G2 cell cycle phases. Cancer Cell Int 7:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou KM, Cheng YC. (2003) The exonuclease activity of human apurinic/apyrimidinic endonuclease (APE1). Biochemical properties and inhibition by the natural dinucleotide Gp4G. J Biol Chem 278:18289–18296 [DOI] [PubMed] [Google Scholar]
- Christmann M, Tomicic MT, Roos WP, Kaina B. (2003) Mechanisms of human DNA repair: an update. Toxicology 193:3–34 [DOI] [PubMed] [Google Scholar]
- Evans AR, Limp-Foster M, Kelley MR. (2000) Going APE over ref-1. Mutat Res 461:83–108 [DOI] [PubMed] [Google Scholar]
- Fan J, Wilson DM., 3rd (2005) Protein-protein interactions and posttranslational modifications in mammalian base excision repair. Free Radic Biol Med 38:1121–1138 [DOI] [PubMed] [Google Scholar]
- Fishel ML, He Y, Smith ML, Kelley MR. (2007) Manipulation of base excision repair to sensitize ovarian cancer cells to alkylating agent temozolomide. Clin Cancer Res 13:260–267 [DOI] [PubMed] [Google Scholar]
- Fishel ML, Kelley MR. (2007) The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target. Mol Aspects Med 28:375–395 [DOI] [PubMed] [Google Scholar]
- Fox KR. (1997) Methods in Molecular Biology: Drug DNA Interaction Protocols, Vol. 90, Humana Press, Inc. Totowa, NJ [Google Scholar]
- Garcin ED, Hosfield DJ, Desai SA, Haas BJ, Björas M, Cunningham RP, Tainer JA. (2008) DNA apurinic-apyrimidinic site binding and excision by endonuclease IV. Nat Struct Mol Biol 15:515–522 [DOI] [PubMed] [Google Scholar]
- Georgiadis MM, Luo M, Gaur RK, Delaplane S, Li X, Kelley MR. (2008) Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease. Mutat Res 643:54–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass LS, Bapat A, Kelley MR, Georgiadis MM, Long EC. (2010) Semi-automated high-throughput fluorescent intercalator displacement-based discovery of cytotoxic DNA binding agents from a large compound library. Bioorg Med Chem Lett 20:1685–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JK, Wilson SH. (2007) Hypersensitivity phenotypes associated with genetic and synthetic inhibitor-induced base excision repair deficiency. DNA Repair (Amst) 6:530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley MR, Cheng L, Foster R, Tritt R, Jiang J, Broshears J, Koch M. (2001) Elevated and altered expression of the multifunctional DNA base excision repair and redox enzyme Ape1/ref-1 in prostate cancer. Clin Cancer Res 7:824–830 [PubMed] [Google Scholar]
- Kirstein SL, Atienza JM, Xi B, Zhu J, Yu N, Wang X, Xu X, Abassi YA. (2006) Live cell quality control and utility of real-time cell electronic sensing for assay development. Assay Drug Dev Technol 4:545–553 [DOI] [PubMed] [Google Scholar]
- Kow YW, Dare A. (2000) Detection of abasic sites and oxidative DNA base damage using an ELISA-like assay. Methods 22:164–169 [DOI] [PubMed] [Google Scholar]
- Kreklau EL, Limp-Foster M, Liu N, Xu Y, Kelley MR, Erickson LC. (2001) A novel fluorometric oligonucleotide assay to measure O(6)-methylguanine DNA methyltransferase, methylpurine DNA glycosylase, 8-oxoguanine DNA glycosylase and abasic endonuclease activities: DNA repair status in human breast carcinoma cells overexpressing methylpurine DNA glycosylase. Nucleic Acids Res 29:2558–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski C, Hopkins A. (2004) Navigating chemical space for biology and medicine. Nature 432:855–861 [DOI] [PubMed] [Google Scholar]
- Luo M, He H, Kelley MR, Georgiadis M. (2010) Redox regulation of DNA repair: implications for human health and cancer therapeutic development. Antioxid Redox Signal 12:1247–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhusudan S, Smart F, Shrimpton P, Parsons JL, Gardiner L, Houlbrook S, Talbot DC, Hammonds T, Freemont PA, Sternberg MJ, et al. (2005) Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res 33:4711–4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill DR, Wilson DM., 3rd (2007) A dominant-negative form of the major human abasic endonuclease enhances cellular sensitivity to laboratory and clinical DNA-damaging agents. Mol Cancer Res 5:61–70 [DOI] [PubMed] [Google Scholar]
- Parsons JL, Dianova II, Dianov GL. (2004) APE1 is the major 3′-phosphoglycolate activity in human cell extracts. Nucleic Acids Res 32:3531–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popoff SC, Spira AI, Johnson AW, Demple B. (1990) Yeast structural gene (APN1) for the major apurinic endonuclease: homology to Escherichia coli endonuclease IV. Proc Natl Acad Sci USA 87:4193–4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed AM, Fishel ML, Kelley MR. (2009) Small-molecule inhibitors of proteins involved in base excision repair potentiate the anti-tumorigenic effect of existing chemotherapeutics and irradiation. Future Oncol 5:713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson AB, Klungland A, Rognes T, Leiros I. (2009) DNA repair in mammalian cells: base excision repair: the long and short of it. Cell Mol Life Sci 66:981–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KA, Bullock HA, Xu Y, Tritt R, Zimmerman E, Ulbright TM, Foster RS, Einhorn LH, Kelley MR. (2001) Altered expression of Ape1/ref-1 in germ cell tumors and overexpression in NT2 cells confers resistance to bleomycin and radiation. Cancer Res 61:2220–2225 [PubMed] [Google Scholar]
- Rosa S, Fortini P, Karran P, Bignami M, Dogliotti E. (1991) Processing in vitro of an abasic site reacted with methoxyamine: a new assay for the detection of abasic sites formed in vivo. Nucleic Acids Res 19:5569–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiple LA, Cardellina JH, Akee R, Stivers JT. (2007) Potent inhibition of human apurinic/apyrimidinic endonuclease 1 by arylstibonic acids. Mol Pharmacol 73:669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber JR, Bobola MS, Blank A, Schoeler KD, Haroldson PD, Huynh MB, Kolstoe DD. (2002) The apurinic/apyrimixinic endonuclease activity of Ape1/Ref-1 contributes to human glioma cell resistance to alkylating agents and is elevated by oxidative stress. Clin Cancer Res 8:3008–3018 [PubMed] [Google Scholar]
- Simeonov A, Kulkarni A, Dorjsuren D, Jadhav A, Shen M, McNeill DR, Austin CP, Wilson DM., 3rd (2009) Identification and characterization of inhibitors of human apurinic/apyrimidinic endonuclease APE1. PLoS ONE 4:e5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solly K, Wang X, Xu X, Strulovici B, Zheng W. (2004) Application of real-time cell electronic sensing (RT-CES) technology to cell-based assays. Assay Drug Dev Technol 2:363–372 [DOI] [PubMed] [Google Scholar]
- Stupp R, Hegi ME, Gilbert MR, Chakravarti A. (2007) Chemoradiotherapy in malignant glioma: standard of care and future directions. J Clin Oncol 25:4127–4136 [DOI] [PubMed] [Google Scholar]
- Taverna P, Liu L, Hwang HS, Hanson AJ, Kinsella TJ, Gerson SL. (2001) Methoxyamine potentiates DNA single strand breaks and double strand breaks induced by temozolomide in colon cancer cells. Mutat Res 485:269–281 [DOI] [PubMed] [Google Scholar]
- Tell G, Quadrifoglio F, Tiribelli C, Kelley MR. (2009) The many functions of APE1/Ref-1: not only a DNA repair enzyme. Antioxid Redox Signal 11:601–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DM, 3rd, Barsky D. (2001) The major human abasic endonuclease: formation, consequences and repair of abasic lesions in DNA. Mutat Res 485:283–307 [DOI] [PubMed] [Google Scholar]
- Xanthoudakis S, Miao GG, Curran T. (1994) The redox and DNA-repair activities of Ref-1 are encoded by nonoverlapping domains. Proc Natl Acad Sci USA 91: 23–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing JZ, Zhu L, Jackson JA, Gabos S, Sun XJ, Wang XB, Xu X. (2005) Dynamic monitoring of cytotoxicity on microelectronic sensors. Chem Res Toxicol 18:154–161 [DOI] [PubMed] [Google Scholar]
- Yang S, Irani K, Heffron SE, Jurnak F, Meyskens FL., Jr (2005) Alterations in the expression of the apurinic/apyrimidinic endonuclease-1/redox factor-1 (APE/Ref-1) in human melanoma and identification of the therapeutic potential of resveratrol as an APE/Ref-1 inhibitor. Mol Cancer Ther 4:1923–1935 [DOI] [PubMed] [Google Scholar]
- Zawahir Z, Dayam R, Deng J, Pereira C, Neamati N. (2009) Pharmacophore guided discovery of small-molecule human apurinic/apyrimidinic endonuclease 1 inhibitors. J Med Chem 52:20–32 [DOI] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. (1999) A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen 4:67–73 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.