Abstract
Type IV pili (T4P) are critical to virulence for Vibrio cholerae and other bacterial pathogens. Among their diverse functions, T4P mediate microcolony formation, which protects the bacteria from host defenses and concentrates secreted toxins. The T4P of the two V. cholerae disease biotypes, classical and El Tor, share 81% identity in their TcpA subunits, yet these filaments differ in their interaction patterns as assessed by electron microscopy. To understand the molecular basis for pilus-mediated microcolony formation, we solved a 1.5 Å resolution crystal structure of N-terminally-truncated El Tor TcpA and compared it to that of classical TcpA. Residues that differ between the two pilins are located on surface-exposed regions of the TcpA subunits. By iteratively changing these non-conserved amino acids in classical TcpA to their respective residues in El Tor TcpA, we identified residues that profoundly affect pilus:pilus interaction patterns and bacterial aggregation. These residues lie on either the protruding D-region of the TcpA subunit or in a cavity between pilin subunits in the pilus filament. Our results support a model whereby pili interact via intercalation of surface protrusions on one filament into depressions between subunits on adjacent filaments as a means to hold V. cholerae cells together in microcolonies.
Keywords: Type IV pili, x-ray crystallography, bacterial pathogenesis, colonization, virulence factor, secretion
INTRODUCTION
The surfaces of many Gram-negative bacteria display Type IV pili (T4P), filamentous appendages that have a broad range of functions in colonization and pathogenesis (Mattick, 2002; Pelicic, 2008). T4P are adhesive organelles that in some cases bind specifically to host cell receptors to facilitate bacterial attachment, but more commonly cause bacteria to aggregate in microcolonies, which protect them from host defenses, increase their density and concentrate secreted toxins and hydrolytic enzymes. T4P are required for microcolony formation for many bacterial pathogens including Vibrio cholerae, enteropathogenic Escherichia coli (EPEC), Neisseria gonorrhoeae and Neisseria meningitidis (Taylor et al., 1987; Marceau et al., 1995; Bieber et al., 1998; Park et al., 2001; Higashi et al., 2007). T4P-mediated aggregation also facilitates biofilm formation in some organisms (Klausen et al., 2003; Jurcisek et al., 2007; Luke et al., 2007; Barken et al., 2008; Varga et al., 2008).
T4P-mediated microcolony formation is not well-understood, but appears to require very precise interactions and to be a dynamic and transient process. T4P are polymers of the pilin protein subunit. The amino acid sequence of the N-terminal segment of Type IV pilins, α1N (residues 1-28), is conserved and hydrophobic and acts as the primary polymerization domain (Craig and Li, 2008). α1N, together with α1C (residues 29-53), form a continuous α-helix, α1, in the full length crystal structures of PilE from N. gonorrhoeae (Parge et al., 1995; Craig et al., 2006) and PAK pilin from Pseudomonas aeruginosa (Craig et al., 2003). The α1N segments associate in a helical array in the pilus filament core, anchoring the more variable C-terminal globular domains, which form the external surface of the pilus filaments. Within the globular domain, α1C is embedded in a β-sheet to form a conserved structural core that supports two variable regions, the αβ-loop and the D-region, which form the exposed surface of the protein and define the diverse pilus functions (Craig et al., 2003; Craig et al., 2006; Li et al., 2008). Since each pilus filament is comprised of thousands of pilin subunits, small differences in surface-exposed residues can have a major impact on the overall stereochemistry of the filament surface. Kirn et al. showed that single amino acid substitutions in the D-region of the TcpA subunit of the V. cholerae toxin coregulated pilus (TCP) disrupt pilus:pilus interactions, as indicated by decreased autoagglutination levels (a phenotypic manifestation of microcolony formation), by increased sensitivity of the bacteria to complement-mediated killing, and by reduced colonization of the infant mouse (Kirn et al., 2000). An important finding of this study was that wild type V. cholerae were unable to interact with non-aggregative tcpA deletion mutants to form mixed microcolonies, implying that pilus:pilus interactions rather than pilus:cell interactions are responsible for bacterial aggregation. T4P-mediated aggregation is a prerequisite for intimate adherence of pathogenic Neisseria to host epithelial cells (Todd et al., 1984; Marceau et al., 1995; Park et al., 2001; Higashi et al., 2007). Natural antigenic variation in the N. meningitidis pilin PilE results in bacteria that differ in their ability to aggregate (Nassif et al., 1993), and a single amino acid change in the D-region converted a low-adhesive strain to a high-adhesive one, with a corresponding increase in pilus bundling, visualized by transmission electron microscopy (TEM) (Marceau et al., 1995). In N. meningitidis, pilus-mediated bacterial aggregation involves an additional protein, PilX. PilX is a pilin-like protein that shares N-terminal sequence homology with PilE and other Type IV pilins and is incorporated irregularly along the length of the filament (Helaine et al., 2005; Helaine et al., 2007). A crystal structure of N-terminally-truncated PilX revealed an architecture characteristic of the Type IV pilins, with an N-terminal α-helix (α1C), a conserved structural core, an αβ-loop and a D-region (Helaine et al., 2007). Helaine and colleagues (2007) modeled PilX into the N. gonorrhoeae pilus structure, obtained by fitting the PilE crystal structure into a 12.5 Å native cryoelectron microscopy (cryoEM) reconstruction of the pilus filament (Craig et al., 2006). PilX fits into the N. gonorrhoeae pilus in an orientation similar to that of PilE, with its globular domain and D-region exposed on the filament surface and its putative N-terminal α-helix embedded in the filament core. Disruption of the pilX gene, or of a region of this gene encoding a short surface-exposed α-helix in the D-region of PilX, resulted in a non-aggregative strain without affecting pilus expression levels, implicating PilX, and the D-region in particular, in bacterial aggregation (Helaine et al., 2007). Importantly, PilX-expressing cells could not form aggregates with PilX-deficient cells, providing further evidence that pilus:pilus interactions rather than pilus:cell interactions cause these bacteria to aggregate.
T4P-mediated bacterial aggregation represents an early and transient step in host colonization. Disruption of microcolonies is necessary for progression of colonization and for dissemination of bacterial pathogens into the environment. For some species, disruption of these aggregates appears to be facilitated by retraction of the pili, a process that also mediates twitching motility and uptake of bound DNA and bacteriophage (Mattick, 2002; Burrows, 2005). T4P retraction, which is a reversal of the pilus assembly process, is powered by ATP hydrolysis by a cytosolic inner membrane-associated nucleotide binding protein (Wolfgang et al., 1998; Merz et al., 2000; Skerker and Berg, 2001; Chiang et al., 2005). Pilus retraction is necessary for aggregated Neisseria to establish intimate adherence to host epithelial cells to promote invasion (Park et al., 2001; Nassif et al., 1997; Higashi et al., 2007; Higashi et al., 2009). In EPEC infections, pilus-mediated aggregation facilitates bacterial reproduction, but full virulence requires that these aggregates eventually dissociate, allowing bacteria to colonize other sites in the small intestine and to be released into the environment (Anantha et al., 1998; Bieber et al., 1998). The dispersal process requires functional BfpF, the putative EPEC nucleotide binding protein (Bieber et al., 1998), and thus may involve pilus retraction. The N. gonorrhoeae PilT retraction ATPase is one of the strongest molecular motors known, generating forces greater than 100 piconewtons on single pili (Maier et al., 2002) and 10-fold higher forces for pilus bundles (Biais et al., 2008). Presumably such forces are necessary to pull bacteria along surfaces and also to disrupt intercellular pilus:pilus interactions. These data imply a fine balance for pilus:pilus interaction affinities, which must be strong enough to hold cells together in microcolonies, but not so strong that they can’t be disrupted for the infection to progress for invasion and dissemination.
V. cholerae use TCP to establish microcolonies in the human small intestine (Taylor et al., 1987), a process that can be simulated in an in vitro autoagglutination assay (Kirn et al., 2000). Transmission electron microscopy (TEM) analysis reveals dramatically different pilus:pilus interaction patterns for the two V. cholerae biotypes associated with cholera epidemics, classical and El Tor (DiRita et al., 1996), in spite of their TcpA subunits having 81% amino acid sequence identity. The x-ray crystal structure of the TcpA globular domain from classical V. cholerae (ΔN-TcpACl) has been determined and a computational model for the TCP filament is available (Craig et al., 2003; Li et al., 2008). Thus, T4P are an attractive system to investigate the molecular nature of pilus:pilus interactions. An understanding of the mechanism governing pilus-mediated bacterial aggregation is crucial for understanding V. cholerae pathogenesis and developing antibacterial therapies. Molecular differences between classical and El Tor biotypes are pertinent to cholera epidemiology, particularly given the recent emergence of hybrid V. cholerae strains. Here we exploit differences between classical and El Tor TCP to define the mechanism for pilus:pilus interactions mediating V. cholerae aggregation and microcolony formation. We report a 1.5 Å resolution x-ray crystal structure of N-terminally-truncated El Tor TcpA (ΔN-TcpAET) and combine computational modeling and mutational analysis to identify key residues that define the pilus surface and its ability to interact with pili on adjacent cells. Our results provide important insights into the molecular nature of V. cholerae microcolony formation with implications for T4P-mediated aggregation in other bacterial pathogens.
RESULTS
Classical and El Tor pili have distinct interaction patterns
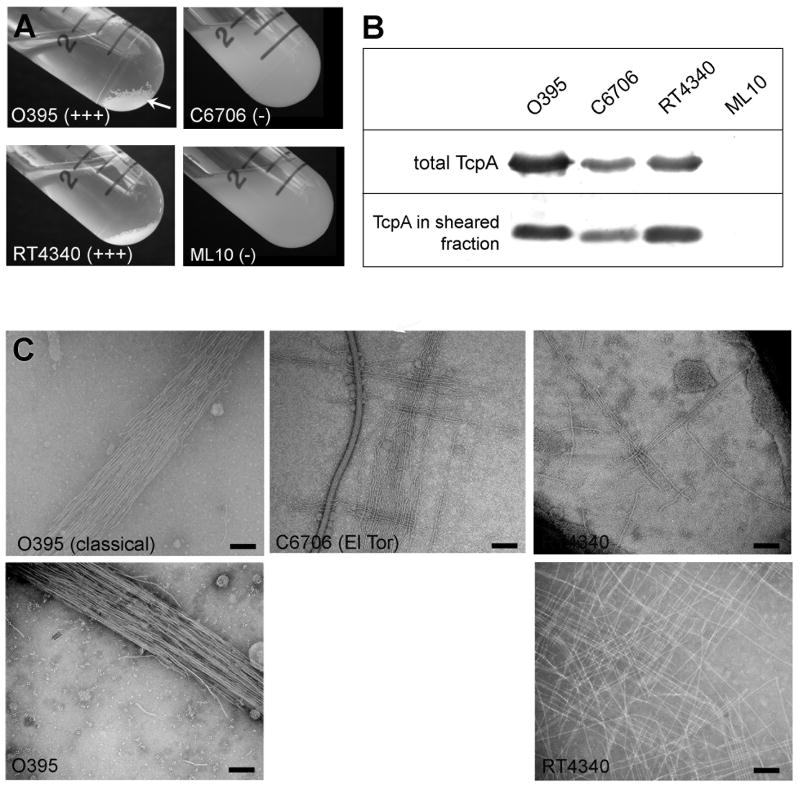
Efficient colonization of the human and mouse gut requires functional TCP for both the classical and El Tor biotypes of the V. cholerae O1 serogroup (Herrington et al., 1988; Thelin and Taylor, 1996; Tacket et al., 1998). Expression of functional pili can be assessed using an in vitro autoagglutination assay where cells are grown in liquid culture overnight in TCP-expressing conditions (Luria Broth [LB], starting pH 6.5, 30 °C), and pilus:pilus interactions cause the cells to aggregate and fall out of suspension (Fig. 1A). El Tor strain C6706 is grown in AKI medium to optimize pilin expression (Iwanaga et al., 1989, Jonson et al., 1990). In general, in vitro autoagglutination correlates well with colonization of the infant mouse small intestine (Kirn et al., 2000). Autoagglutination of classical biotype strain O395 is enhanced by upregulating expression of the tcp operon using plasmid pMT5, which encodes the transcriptional activator ToxT under the control of an inducible promoter (Fig. 1A) (DiRita et al., 1996). In contrast, the El Tor strain C6706 does not autoagglutinate even when ToxT is upregulated. C6706-pMT5 overnight cultures are indistinguishable from those of strain ML10, a non-agglutinating classical strain that has a C120A substitution in the TcpA subunit (Fig. 1A). To determine whether differences in TcpA expression or TCP assembly levels are responsible for differences in autoagglutination levels observed for classical and El Tor V. cholerae, TcpA levels were compared by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting for whole cell cultures and for sheared cell supernatants containing assembled pili (Fig. 1B). TcpA and TCP levels are reduced by ~50% for El Tor C6706 relative to classical O395, which partially explains the inability of C6706 to autoagglutinate. However, when the El Tor C6706 tcpA gene was expressed in classical strain O395 in place of its endogenous tcpA gene, resulting in classical strain RT4340, autoagglutination is comparable to that of O395, as are TcpA and TCP expression levels (Fig. 1A, B). This result indicates that El Tor TCP are fully capable of mediating V. cholerae aggregation and provides a system whereby pilus-mediated interactions could be directly compared for the classical and El Tor pili.
Fig. 1. Comparison of autoagglutination, pilin expression, pilus assembly and TCP:TCP interaction patterns for classical and El Tor V. cholerae biotypes.
(A) Autoagglutination of overnight V. cholerae cultures grown under TCP-inducing conditions: wild type classical strain O395 and O395 expressing El Tor TcpA (RT4340) autoagglutinate well (+++) whereas El Tor strain C6706 and the classical mutant strain ML10 (C120A) do not autoagglutinate (-). (B) Immunoblots of TcpA in whole cell cultures (upper panel) and sheared cell supernatant (lower panel) detected with polyclonal anti-TcpA antibody (TcpA6, residues 174-199, (Sun et al., 1991)) to assess TcpA expression and TCP assembly, respectively. (C) Morphology and bundling characteristics of V. cholerae TCP as assessed by negative stain TEM of whole cell cultures (upper panels) and partially purified pili (lower panels). Scale bars, 100 nm.
To further characterize TCP from the classical and El Tor biotypes, pili were imaged by negative stain TEM in whole cell cultures and after partial purification, and pilus morphology and pilus:pilus interaction characteristics were compared (Fig. 1C). TCP from both biotypes have similar morphologies, being long, relatively straight filaments approx. 80 Å in diameter. However, the pilus:pilus interactions are markedly different for the two biotypes (Fig. 1C). Classical TCP form rope-like bundles, with the filaments aligned along their length, associated via extended lateral interactions. In contrast, El Tor TCP interact in an orthogonal manner, with individual filaments crossing over each other at ~90° angles to form a “crosshatch” pattern. The crosshatch pattern is observed for El Tor TCP in whole cell culture for both the C6706 and RT4340 strains, and is even more evident in TCP samples that were mechanically sheared from RT4340 and partially purified (Fig. 1C, lower panel). While rope-like bundles are also observed for El Tor strains, the crosshatch pattern is dominant and suggests that pili from the two biotypes interact in different ways to form microcolonies.
Atomic structure of El Tor TcpA
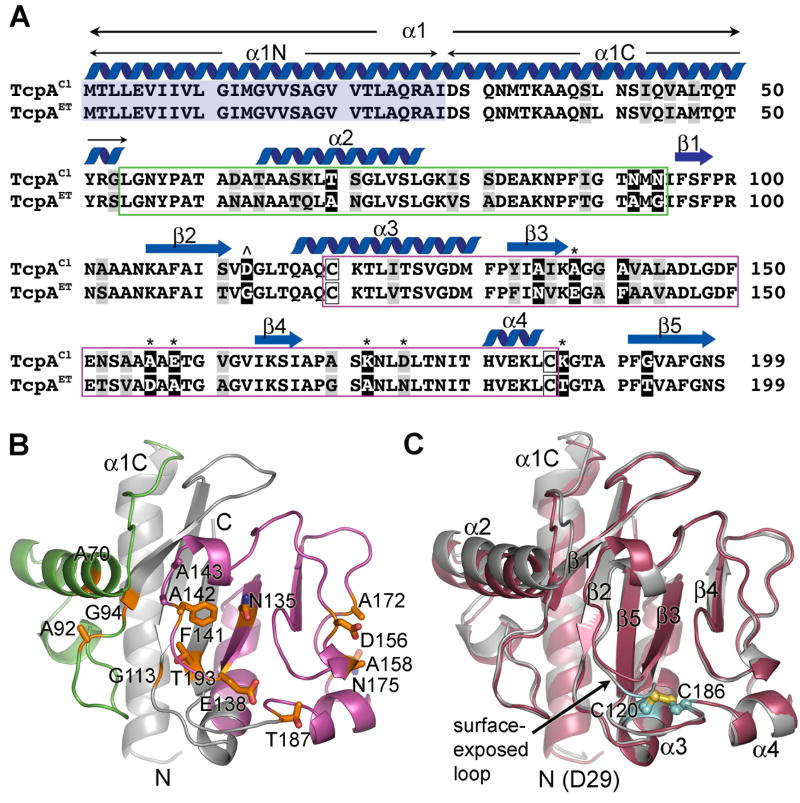
Since TCP filaments are comprised of thousands of copies of the TcpA subunit, we reasoned that differences in pilus:pilus interaction patterns could be explained by differences between the classical and El Tor TcpA subunits. The amino acid sequences of classical and El Tor TcpA are 81% identical, as shown in Fig. 2A. Nine of the 12 non-conserved amino acid differences are located in either the αβ-loop or the D-region of the globular domain. These regions are highly variable among the Type IV pilins (Craig et al., 2004) and are predicted together to form the exposed surface of TcpA in the TCP filament (Craig et al., 2003; Li et al., 2008). To understand the difference between classical and El Tor TCP interaction patterns, we determined the crystal structure of ΔN-TcpAET (residues 29-199, Table 1) and compared this structure to that of ΔN-TcpACl (Craig et al., 2003). Both structures lack the first 28 residues, which are predicted to form a hydrophobic α-helix, α1N, based on amino acid sequence homology with the full length Type IV pilin structures from P. aeruginosa and N. gonorrhoeae (Parge et al., 1995; Craig et al., 2003; Craig et al., 2006).
Fig. 2. Comparison of the amino acid sequences and atomic structures of classical and El Tor TcpA.
(A) Amino acid sequence alignment for mature TcpA proteins from classical V. cholerae O395 (NCBI accession no. ABQ19609) and El Tor strain C6706 (NCBI accession no. AAA85786). The sequences are 81% identical. The secondary structure for TcpACl is indicated above the sequence. The αβ-loop is boxed in green and the D-region is boxed in magenta. α1N (residues 1-28, shaded blue) were replaced by a hexahistidine tag plus a linker region for both pilin crystal structures. α1 is predicted to be a continuous α-helix based on amino acid sequence homology with N. gonorrhoeae and P. aeruginosa pilins, whose full-length structures have been determined (Craig et al., 2003, Craig et al., 2006, Parge et al., 1995). Conserved amino acid differences between classical and El Tor TcpA are shaded in grey and non-conserved differences are shown in black with white letters. Classical-to-El Tor substitutions in the D-region patch are indicated by an asterisk (*) above the corresponding amino acid and the D113A cavity change is indicated by a carat (ˆ). (B)ΔN-TcpAET crystal structure at 1.5 Å resolution, shown as a ribbon diagram. Non-conserved residues between classical and El Tor TcpA are show as stick representations. (C) Superposition of ΔN-TcpAET structure (red) onto ΔN-TcpACl (grey) (Craig et al., 2003). The conserved cysteines are shown in cyan/yellow using ball-and-stick representation.
Table 1.
Crystallographic Data Collection and Analysis
| Data collection | Native TcpA |
|---|---|
| Beamline | SSRL 11-1 |
| Space group | I212121 |
| Cell a, b, c (Å) | 62.8, 94.8, 128.4 |
| Cell α, β, γ | 90.0, 90.0, 90.0 |
| Resolution range (Å) | 20 - 1.5 |
| Wavelength (Å) | 0.979 |
| Completeness (%)a | 99.3/100 |
| Wilson B-value (Å2) | 20.5 |
| Observed reflections | 1,334,927 |
| Unique reflections | 61075 |
| Rsym (%)a,b | 6.8/41.3 |
| I/σa | 23.8/5.0 |
| Redundancy | 6.8 |
| Mosaicity | 0.5 |
| Refinement Statistics | |
| Resolution limits (Å) | 20-1.50 |
| Molecules/A.U. | 2 |
| Rcryst (%)c,e | 21.9/31.0 |
| Rfree (%)d,e | 22.4/33.3 |
| Average B-factor (Å2) | 25.5 |
| No. protein atoms | 2964 |
| No. water molecules | 462 |
| RMSD bond angles | 1.90 |
| RMSD bond lengths | 0.017 |
| Ramachandron plot | |
| Most favored (%) | 94.3 |
| Allowed (%) | 5.7 |
Overall/last shell (resolution range for last shell is 1.55-1.50).
Rsym is the unweighted R value on I between symmetry mates.
Rcryst= Σhkl∥Fobs(hkl)∣-∣Fcalc(hkl)∥/Σhkl∣Fobs(hkl)∣.
Rfree is the cross validation R factor for 10% of reflections against which the model was not refined.
Overall/last shell (resolution range for last shell is 1.51-1.50).
ΔN-TcpAET is a globular protein with an N-terminal α-helical spine (α1C, residues 29-53) embedded in a twisted antiparallel β-sheet (Fig. 2B and C and Supplementary Fig. S1). As in ΔN-TcpACl, the αβ-loop and D-region of ΔN-TcpAET together comprise one face of the globular domain as well as its outer edges. The αβ-loop (residues 54-93, colored green in Fig. 2B), which lies between α1C and the β-sheet, begins with an irregular loop followed by a four-turn α-helix, α2, that lies at right angles to α1C. The D-region, which is delineated by the conserved, disulfide-bonded cysteines (C120 and C186), comprises a large portion of the globular domain including part of the β-sheet (colored magenta in Fig. 2B). The secondary structural elements of ΔN-TcpAET are indicated in Fig. 2C and a detailed description of the structure is provided in Fig. S1 of Supplementary Materials.
ΔN-TcpAET is very similar to ΔN-TcpACl, with the two structures having a root mean square deviation of 0.69 Å for all Cα (1.03 Å for all atoms) (Fig. 2C). Most of the non-conserved residues between ΔN-TcpAET and ΔN-TcpACl are exposed on the surface of the globular domain within the αβ-loop and D-region (Fig. 2A, B). Only a few of the non-conserved residues appear to be important for the pilin fold, and changes in these residues that could potentially disrupt the TcpA fold are accompanied by additional “compensatory” changes that maintain the same fold for both biotypes. For instance, an Asp113→Gly change between TcpACl and TcpAET is accompanied by a Gly193→Thr change that maintains the distance and H-bond pattern between β2 and β5 of the β-sheet (Fig. 2B, C). Similarly, an Ala156→Asp change between TcpACl and TcpAET is accompanied by a Glu158→Ala change that prevents repulsion between these negatively-charged side chains in the irregular turn at the D-region edge of the globular domain. The only significant backbone difference is in a surface-exposed loop (138-147) that is shifted by 2-3 Å between ΔN-TcpACl and ΔN-TcpAET when the two pilins are superimposed (Fig. 2B and C). This misalignment is caused by a tighter turn for the ΔN-TcpAET backbone at the point at which the polypeptide chain exits β3 to form the surface-exposed loop (residue 138), followed by a backbone bulge at Ala142-Ala143. The backbone at Ala142-Ala143 bulges in part to accommodate the bulky Phe141 in the ΔN-TcpAET surface-exposed loop, which would otherwise clash with Asn135 on β3. No such clash occurs in ΔN-TcpACl, which has alanines at positions 135 and 141. The Ala142-Ala143 bulge forces the αβ-loop α-helix (α2) of ΔN-TcpAET outward ~2.3 Å relative to its position in ΔN-TcpACl. In spite of the small differences in the backbone structure of the surface-exposed loop, there are no differences in the surface-exposed side chains in this region, with the exception of residue 138, and thus the surfaces of ΔN-TcpACl and ΔN-TcpAET are very similar in this region. In summary, no substantial structural differences are identified between ΔN-TcpACl and ΔN-TcpAET, in part because changes in residues involved in protein fold are accompanied by compensatory changes to maintain the fold, and in part because the majority of their non-conserved differences lie on the surface of the pilin subunits. The similarity in pilin subunit structure supports a common TCP filament architecture for both biotypes.
Computational model of the El Tor TCP filament
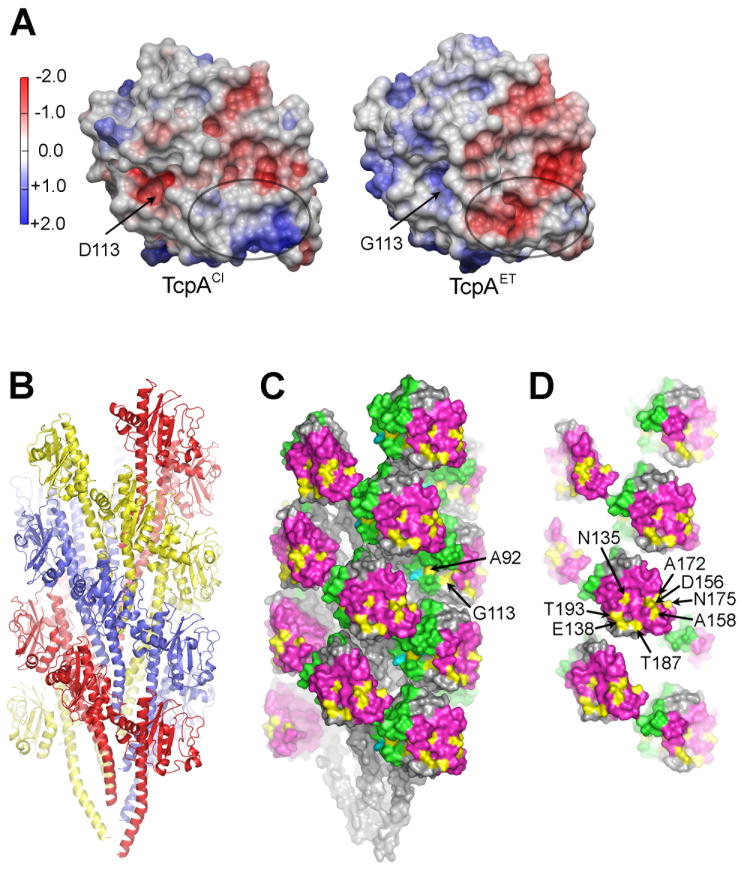
While the ΔN-TcpAET and ΔN-TcpACl structures are very similar, comparison of their electrostatic surfaces reveals a patch in the D-region that differs substantially between the two proteins, having an overall positive charge in ΔN-TcpACl and a negative charge in ΔN-TcpAET (Fig. 3A). Most of the non-conserved amino acids lie on this patch and represent either a loss or gain of charge: A138E, A156D, E158A, K172A, K187T, and D175N (here and throughout the paper the residues in classical TcpA are shown on the left and in El Tor TcpA on the right). Several of these changes are from the small, uncharged alanine to a bulky charged residue, or vice versa, which alters both the shape and the chemistry of the D-region surface. The D-region of ΔN-TcpACl, especially residues ~145-183, is the most exposed region of the protein in the TCP filament according to a hydrogen/deuterium exchange mass spectrometry study (Li et al., 2008). Thus, amino acid differences in the D-region of TcpA would have a profound effect on the chemistry and structural landscape of the TCP surface. We hypothesized that residues within the D-region patch that differ between classical and El Tor TcpA are responsible for the different interaction patterns observed for classical and El Tor TCP filaments. To understand how these amino acid differences might affect pilus:pilus interactions, we fit the ΔN-TcpAET structure into our classical TCP model, which was generated based on solvent accessibility, TCP symmetry parameters and crystal packing of the ΔN-TcpACl subunit (Li et al., 2008) (Fig. 3B, C). The atomic coordinates of the N-terminal 28 residues of the full-length PAK pilin structure were used to position α1N of ΔN-TcpAET. TcpAET fits into the classical TCP assembly model well, with only minor clashes among the N-terminal α-helices. Subunits follow a dominant left-handed three-start helix, shown as red, yellow and blue strands in Fig. 3B. In the TCP model the N-terminal α-helices form a hydrophobic core anchoring the globular domains, which are loosely-packed on the filament surface leaving pronounced gaps or cavities between the subunits (Fig. 3B, C). The αβ-loop edge of the globular domain (green) is turned inward toward the core of the filament and contacts the N-terminal α-helix of a neighboring subunit. The D-region edge of the globular domain (magenta) is turned outward, making minimal contact with neighboring subunits, and protruding slightly from the filament surface. This filament model places the non-conserved residues (yellow) on the exposed face of the pilin subunits. Exposed residues lining the cavities between the subunits are for the most part conserved, with the exception of A92 and G113, which result in a negative charge in the depression around D113 in classical TcpA and a positive one at G113 in El Tor TcpA (Fig. 3A). The symmetry of the classical and El Tor TCP models differs from that of the N. gonorrhoeae Type IV pilus cryoEM structure (Craig et al., 2006), yet the overall architectures are very similar, with subunits arranged in a dominant three-start helix, held together by hydrophobic interactions among the N-terminal α-helices, with the globular domains forming a corrugated filament surface.
Fig. 3. TcpA electrostatic surface and El Tor TCP filament model.
(A) Comparison of the electrostatic surfaces of ΔN-TcpACl and ΔN-TcpAET. The orientation is similar to that shown in Fig. 2C. The electrostatic surface differs in the D-region patch (circled) where there are six non-conserved amino acid differences between the two proteins, each resulting in a change (gain or loss) of a single charge. A second difference is in the location of an aspartate→glycine change at residue 113. (B) Side view of the El Tor TCP filament model generated by superimposing ΔN-TcpAET onto TcpACl in the classical TCP model (Li et al., 2008). The filament is colored to show the subunits arranged in a left-handed three-start helix. (C) Space-filling representation of El Tor TCP to show the positions of the non-conserved amino acids (yellow) in the cavities of the filament and (D) on the protruding D-region. The αβ-loop is colored green and the D-region is magenta in (C) and (D). D-region residues 138, 156, 158, 172, 175 and 187 were selected for classical-to-El Tor mutations, as was cavity residue 113.
Mutational analysis of the non-conserved surface-exposed residues in classical TcpA
To assess the role of the exposed, non-conserved residues between classical and El Tor TcpA in pilus:pilus interactions, we focused on the six residues in the protruding part of the D-region that either gain or lose a charge: A138E, A156D, E158A, K172A, D175N and K187T. A panel of V. cholerae tcpA mutants was generated whereby each of these six D-region residues in classical TcpA was changed to its corresponding residue in El Tor TcpA, both individually and as composite changes. The phenotypes of the mutant V. cholerae strains were assessed in terms of their ability to synthesize TcpA, to assemble pili and to autoagglutinate. The pili were further examined by TEM to assess their filament morphology and interaction characteristics. The TcpA alterations are listed in Table 2 along with the phenotypic properties of the mutant strains. Pilin expression in all mutant strains was comparable to that of wild type O395, as assessed by SDS-PAGE and immunoblotting of whole cell cultures, indicating that the TcpA alterations did not disrupt the protein stability (Fig. 4A, upper panel). The level of TCP expression was assessed for each strain by shearing the pili from the cells and analyzing the sheared cell supernatant by SDS-PAGE and immunoblotting (Fig. 4A, lower panel). While some variation was evident among the strains, all pilus yields were comparable to or better than O395, suggesting that none of the alterations were deleterious to pilus assembly.
Table 2.
Phenotypes of V. cholerae tcpA mutants
| Strain1 | TcpA alteration | Residues in D-region2 | Pilin | Pili | AI3 | AS4 | Pilus:pilus interactions |
|---|---|---|---|---|---|---|---|
| O395 | Wild type, classical | AAEKDK | + | + | 1.00 | +++ | bundles |
| C6706 | Wild type, El Tor | EDAANT | + | + | 0.02 | - | crosshatch/bundles |
| RT4340 | O395, tcpAET | EDAANT | + | + | 0.53 | +++ | crosshatch/bundles |
| ML7 | RT4524, tcpACl | AAEKDK | + | + | 0.84 | +++ | bundles |
| ML10 | C120A | AAEKDK | -/+ | - | 0.05 | - | - |
| D-region alterations | |||||||
| LC1 | A138E------------------------------------------------- | EAEKDK | + | + | 0.53 | +++ | bundles |
| ML15 | ----------A156D--------------------------------------- | ADEKDK | + | + | 1.27 | +++ | bundles/crosshatch |
| ML17 | --------------------E158A----------------------------- | AAAKDK | + | + | 2.09 | ++++ | bundles |
| ML18 | ------------------------------K172A------------------- | AAEADK | + | + | 2.09 | ++++ | bundles |
| ML12 | ----------------------------------------D175N--------- | AAEKNK | + | + | 0.17 | + | bundles |
| LC6 | --------------------------------------------------K187T | AAEKDT | + | + | 1.9 | ++++ | bundles |
| ML14 | ------------------------------K172A/D175N--------- | AAEANK | + | + | 1.16 | +++ | bundles |
| LC7 | ----------A156D/E158A----------------------------- | ADAKDK | + | + | 0.97 | +++ | bundles/crosshatch |
| LC9 | A138E/A156D/E158A------------------------------ | EDAKDK | + | + | 0.58 | +++ | bundles/crosshatch |
| LC10 | A138E/A156D/E158A/K172A-------------------- | EDAADK | + | + | 0.11 | + | bundles/crosshatch |
| LC11 | A138E/A156D/E158A---------------------K187T | EDAKDT | + | + | 0.11 | + | bundles/crosshatch |
| LC12 | ----------A156D/E158A/K172A/D175N--------- | ADAANK | + | + | 0.72 | +++ | bundles/crosshatch |
| LC13 | A138E/A156D/E158A/K172A/D175N---------- | EDAANK | + | + | 0.04 | - | bundles/crosshatch |
| LC15 | ----------A156D/E158A/K172A/D175N/K187T | ADAANT | + | + | 1.52 | ++++ | bundles/crosshatch |
| ML16 | A138E/A156D/E158A/K172A-----------K187T | EDAADT | + | + | 0.04 | - | bundles/crosshatch |
| LC16 | A138E/A156D/E158A/K172A/D175N/K187T | EDAANT | + | + | 0.04 | - | crosshatch |
| ML25 | A138E-----------E158A/K172A/D175N/K187T | EAAANT | + | + | 0.47 | ++ | bundles |
| Cavity alterations | |||||||
| ML28 | D113G | + | + | 0.04 | - | bundles | |
| ML9 | D113A | + | + | 0.07 | - | bundles/crosshatch | |
| Combined D-region/cavity mutants | |||||||
| DN1 | D113G/A138E/A156D/E158A/K172A/K187T | EDAADT | + | + | 0.08 | - | crosshatch |
| DN2 | D113A/A138E/A156D/E158A/K172A/K187T | EDAADT | + | + | 0.08 | - | crosshatch |
All strains contain pMT5.
D-region residues 138, 156, 158, 172, 175 and 187 are listed; underlined residues are the classical-to-El Tor changes; non-underlined residues are the unchanged classical residues; residue 156 is shown in bold type.
AI, autoagglutination index, reported as at fraction of wild type O395 autoagglutination, calculated as the ratio of the inverse of the test supernatant OD600 over the inverse of O395 supernatant OD600 for n=3.
AS, autoagglutination scale: (-), AI of 0-0.10; (+), 0.11-0.20, detectable autoagglutination; (++), 0.21-0.50, moderate autoagglutination; (+++), 0.51-1.50, good or “wild type” (O395) autoagglutination; (++++), ≥1.51, better than wild type autoagglutination (clear supernatant).
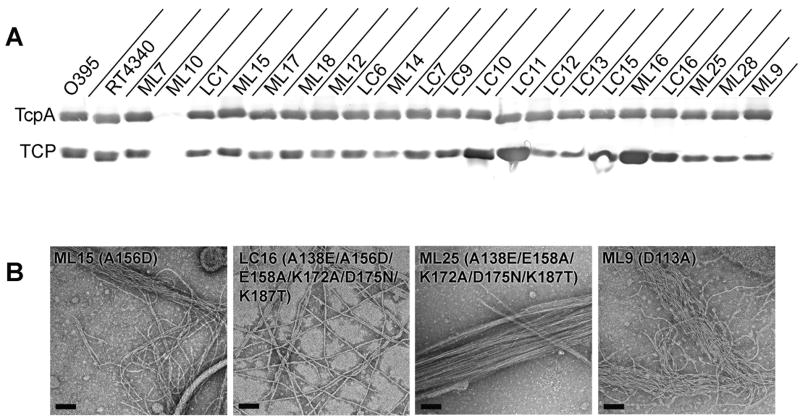
Fig. 4. Analysis of classical-to-El Tor mutants for pilin and pilus expression and pilus morphologies and interaction patterns.
(A) Immunoblots of total TcpA in whole cell cultures (upper panel) and TCP in sheared cell supernatant (lower panel) for tcpA mutants and control strains, detected with polyclonal anti-TcpA antibody (TcpA6). All mutants, with the exception of strain ML10, produced TcpA and TCP at or above wild type levels, indicating that the mutations do not aberrantly affect pilin fold or pilus assembly. (B) TEM images of TCP from whole cell cultures of V. cholerae strains harboring the single A156D change, ML15, the complete classical-to-El Tor D-region mutant LC16, the D156A reversion of LC16, strain ML25, and the D113A mutant, ML9. TCP from strains ML15 and ML9 display a mixed pattern of crosshatches and rope-like bundles, whereas LC16 pili are predominantly crosshatched and ML25 TCP form mostly rope-like bundles. Scale bars, 100 nm.
The presence of functional pili was assessed by autoagglutination assay. The autoagglutination level for each strain is reported in Table 2 as an autoagglutination index (AI) relative to autoagglutination of wild type classical O395 V. cholerae (AI 1.0, see Experimental Procedures). Autoagglutination levels varied substantially for the different mutants despite the fact that comparable levels of pili were expressed, suggesting that the amino acid changes introduced in TcpA affected the strength of interaction between the pili and hence between the cells (Fig. 4A, lower panel). Most of the single D-region classical-to-El Tor mutants autoagglutinated at wild type levels or better. The exception was strain ML12 (D175N), which gave inconsistent results, but typically autoagglutinated poorly. However, several of the composite mutants possessing the D175N change autoagglutinated well, which suggests that additional amino acid changes that make the D-region more like El Tor TcpA can compensate for negative effects of the D175N alteration. The A138E substitution does not have a significant impact on its own (strain LC1, AI 0.53) but 5 of the 7 composite mutants carrying this change autoagglutinate poorly or not at all (LC10, LC11, LC13, ML16, LC16). In addition to the A138E change, these non-agglutinating composite mutants also bear the A156D substitution, which may imply that having negative charges at both these positions disrupts pilus:pilus interactions. Notably, strains LC13, ML16 and LC16 have 5 or 6 classical-to-El Tor alterations, so their D-region stereochemistry should be very similar to that of El Tor TcpA in strain RT4340, yet unlike RT4340, these strains do not autoagglutinate. This implies that the D-region is not solely responsible for cell aggregation. Assuming pilus:pilus interactions rather than pilus:cell interactions hold V. cholerae cells together, our results implicate a second site on TcpA in these interactions. This second site represents the surface with which the D-region interacts and thus must be complementary to the D-region for autoagglutination to occur.
The only region other than the D-region that showed a marked difference in electrostatic surface potential between classical and El Tor TcpA was around residue 113, which lies in a depression on the TcpA surface that forms a cavity between subunits on the TCP filament (Fig. 3). We hypothesized that TCP interactions might occur by intercalation of the protruding D-regions into the cavities formed between the subunits in adjacent filaments. Residues that line this cavity are conserved, with the exception of positions 92, 94 and 113, which are within ~10 Å of each other (Cα-Cα). These residues have bulky polar side chains Asn92, Asn94 and Asp113 in classical TcpA resulting in a negative electrostatic potential at this site, and small uncharged side chains Ala92, Gly94 and Gly113 in El Tor TcpA, with an overall positive charge (Fig. 3A). To address the role of both charge and bulk at this site, we changed D113 in classical TcpA to alanine (strain ML9), and to its corresponding glycine in El Tor TcpA (strain ML28). Both changes resulted in complete loss of autoagglutination (Table 2), indicating that residue 113 plays a key role in V. cholerae aggregation.
Since classical and El Tor TCP display different interaction patterns (Fig. 1C), we examined the pili from the classical-to-El Tor mutants by TEM to see (i) whether the TcpA surface stereochemistry affected the pilus:pilus interaction pattern, and (ii) if the changes in pilus:pilus interactions corresponded with changes in cell aggregation phenotype. We also wanted to exclude the possibility that the differences in autoagglutination levels were due to gross morphological changes in the pili. Whole cell cultures grown overnight in pilus-inducing conditions were examined by negative stain TEM (Table 2, Fig. 4B and Supplementary Fig. S2). The pilus morphologies of the mutants were similar to those of classical strain O395 TCP, being long, thin, relatively straight filaments. However TCP from strains ML16 and ML9 and to a lesser extent ML15 are wavier than O395 TCP, implying that they are more flexible. For all but one of the single mutants, TCP interacts in rope-like bundles characteristic of the classical strain. However, for one single substitution, A156D (strain ML15), the El Tor crosshatch pattern was observed in addition to the rope-like bundles (Fig. 4B and Supplementary Fig. S2). Notably, the crosshatch pattern was also present in all composite mutants bearing the A156D alteration, and strain LC16, which has all 6 of the classical-to-El Tor D-region changes, was predominantly crosshatched. Strain ML15, which has the single A156D change, autoagglutinates at wild type levels, but several of the composite mutants that contain this substitution autoagglutinate poorly or not at all (Table 2). These results suggest that (i) the pilus:pilus interaction pattern observed by TEM is affected by amino acid changes in the D-region and in particular at position 156, and (ii) the crosshatch pattern may not represent productive interactions that allow cell aggregation. To further test the role of residue 156 in pilus:pilus interactions and autoagglutination, we changed Asp156 back to alanine in the 6-residue composite TcpA mutant strain LC16 (A138E/A156D/E158A/K172A/D175N/K187T), which has predominantly crosshatched pili, but autoagglutinates poorly. Remarkably, this single amino acid change, D156A in LC16, to give strain ML25 (A138E/E158A/K172A/D175N/K187T), restored autoagglutination to near wild type levels, and produced exclusively ropelike interaction pattern characteristic of classical TCP (Table 2 and Fig 4B).
Alterations at residue 113 in the cavity between subunits also affected the pilus interaction pattern. The classical-to-El Tor substitution D113G (strain ML28) produced rope-like bundles characteristic of classical TCP, but the D113A change (ML9) resulted in wavy pili that form disordered bundles with some crosshatch characteristics (Fig. 4B and Supplementary Fig. S2). The change in TCP appearance for ML9, together with the loss of aggregation for both these strains, imply that residue 113 in the filament cavities is directly involved in pilus-mediated V. cholerae aggregation.
Non-agglutinating D-region variants can aggregate with non-agglutinating cavity variants
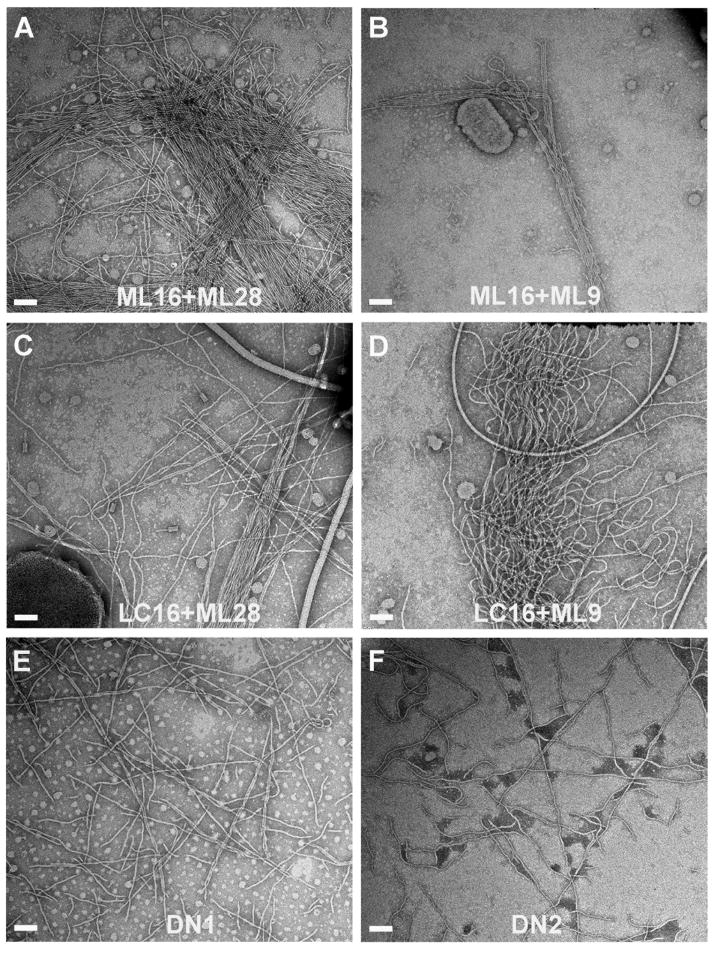
Since changes in either the protruding D-region or the cavity between TcpA subunits profoundly affected pilus:pilus interactions and autoagglutination, we sought to identify a direct interaction between these two regions by demonstrating that non-agglutinating mutants that have an El Tor-like TcpA D-region (strains ML16 and LC16) could aggregate with non-agglutinating mutants that have an El Tor-like cavity (strains ML28 and ML9). LB broth was co-inoculated with ML16 or LC16 cells together with either ML28 or ML9 and the cultures were grown overnight in pilus-inducing conditions. While none of these strains autoagglutinate on its own, strain ML16 in combination with ML28 or ML9 showed weak but reproducible autoagglutination when grown together (Table 3). We further tested cell aggregation by mixing the individual non-agglutinating overnight cultures together to see if these cells could form mixed aggregates after only a short contact time. Autoagglutination was apparent for ML16+ML9 within an hour of mixing the two cultures and considerable aggregation was seen after sitting for 2 hours. In contrast, the individual cultures remained turbid with cells fully suspended for this time period. These results strongly support a direct interaction between the D-regions of one pilus filament and the cavities on adjacent filaments to induce V. cholerae aggregation. Not surprisingly, both rope-like bundles and crosshatched pili were observed by TEM for all combinations of overnight mixed cultures (Fig. 5A-D).
Table 3.
Autoagglutination for mixed cultures and combined D-region/cavity alterations
| Co-inoculates | Cultures mixed after ON growth1 | |||
|---|---|---|---|---|
| Strain | AI2 | AS3 | AI | AS |
| O395 | 1.00 | +++ | 1.00 | +++ |
| Single cultures | ||||
| ML16 | 0.04 | - | 0.04 | - |
| LC16 | 0.04 | - | 0.03 | - |
| ML28 | 0.04 | - | 0.04 | - |
| ML9 | 0.07 | - | 0.09 | - |
| Mixed cultures: D-region mutants + cavity mutants | ||||
| ML16 + ML28 | 0.21 | ++ | 0.09 | - |
| ML16 + ML9 | 0.14 | + | 0.25 | ++ |
| LC16 + ML28 | 0.04 | - | 0.03 | - |
| LC16 + ML9 | 0.04 | - | 0.08 | - |
| Combined D-region/cavity mutants | ||||
| DN1 | 0.08 | - | ||
| DN2 | 0.08 | - | ||
Single cultures and cultures mixed after overnight (ON) growth were left to sit on ice for 2 hrs prior to reading OD600.
AI, autoagglutination index, reported as at fraction of wild type O395 autoagglutination, calculated as the ratio of the inverse of the test supernatant OD600 over the inverse of O395 supernatant OD600 for n=3.
AS, autoagglutination scale: (-), AI of 0-0.10; (+), 0.11-0.20, detectable autoagglutination; (++), 0.21-0.50, moderate autoagglutination; (+++), 0.51-1.50, good or “wild type” (O395).
Fig. 5. TEM images of TCP from overnight mixed cultures and from the combined D-region/cavity mutants.
Negatively stained pili are from whole cell cultures of (A) ML16 co-inoculated with ML28, (B) ML16 co-inoculated with ML9, (C) LC16 co-inoculated with ML28, (D) LC16 co-inoculated with ML9, and from single inocula combined D-region/cavity mutants (E) DN1 and (F) DN2. Scale bars, 100 nm.
To further test the interaction between protrusions and cavities on adjacent pili, we generated two additional mutants having both D-region and cavity classical-to-El Tor substitutions within the same strain: DN1 has the D113G substitution from strain ML28 together with the multiple D-region changes (A138E/A156D/E158A/K172A/K187T) of strain ML16; DN2 has the D113A substitution from strain ML9 together with the ML16 D-region changes. Although both strains expressed pilin and pili, neither showed detectable autoagglutination (Tables 2 and 3). These results are somewhat surprising given that the pili for these combined mutants have surfaces that are very similar to El Tor pili, which do induce autoagglutination. Interestingly, pili from both strains appear as crosshatches (Fig. 5E, F).
DISCUSSION
T4P-mediated bacterial aggregation occurs for many bacterial pathogens as an essential step in colonization. Epidemic strains of V. cholerae from both classical and El Tor biotypes use T4P to form microcolonies, yet the pili appear by TEM to interact differently. To understand these different interaction patterns we determined the atomic structure of El Tor ΔN-TcpAfor comparison with the classical ΔN-TcpA structure (Craig et al., 2003), and built an El Tor TCP model based on the published classical TCP model (Li et al., 2008). By comparing these structures we identified two surface-exposed patches, one in the D-region and one in the cavity between the subunits, that differ in charge. We showed that key residues in these regions profoundly affect V. cholerae aggregation: residues 138, 156 and 175 in the D-region and residue 113 in the cavity. Single classical-to-El Tor substitutions at positions 175 (D175N) and 113 (D113G or D113A) resulted in a loss in autoagglutination for these V. cholerae mutants. The A138E and A156D D-region substitutions had no effect on autoagglutination as single changes, but appear to disrupt autoagglutination when present together, as all but one of the strains having both substitutions autoagglutinate poorly. Furthermore, the non-agglutinating composite mutant LC16, which has 6 D-region classical-to-El Tor substitutions including A138E and A156D, was converted to an agglutinating strain by changing residue 156 back to alanine (strain ML25). Although residues 138 and 156 are ~16 Å apart on the TcpA surface, it seems that a negative charge at both positions in an otherwise classical TcpA background disrupts pilus:pilus interactions.
The A156D substitution is particularly interesting as it does not disrupt autoagglutination on its own, but does shift the TCP interaction pattern to the El Tor-like crosshatches, both as a single amino acid change and in combination with other D-region changes. Consistent with this observation, the non-agglutinating composite mutant LC16 displays mostly crosshatched pili, but the reversal of the A156D substitution in this background not only restores autoagglutination, but also restores the classical ropelike bundle phenotype for the pili. These results implicate residue 156 as a key mediator of pilus:pilus interactions, both in terms of their affinity and their interaction pattern. The results also imply that crosshatched pili represent non-productive interactions in that they are not capable of holding cells together. In fact, the three V. cholerae mutants that have the most extensive classical-to-El Tor substitutions, strains LC16, DN1 and DN2, all have predominantly crosshatched pili and none of these strains autoagglutinate. These results can be interpreted in two ways. One explanation is that pili that interact in the crosshatch pattern are not held together tightly enough, or are not in the correct orientation to aggregate cells, and that although this pattern is observed for El Tor TCP in classical strain RT4340, it is the ropelike bundles that are also present that actually hold cells together. An alternative explanation and one we favor is that the crosshatch pattern does represent productive interactions among the pili, provided that pilus surfaces are perfectly complementary, as in strain RT4340, the classical strain expressing El Tor pili. Our non-agglutinating strains LC16, DN1 and DN2 have multiple classical-to-El Tor substitutions but residues remain on the TcpA surface that differ from El Tor TcpA and thus may not be complementary in the context of the crosshatch interaction pattern. The crosshatch pattern is very distinctive and well-ordered in the autoagglutinating strain RT4340 and in El Tor C6706, with pili crossing over each other at almost perfect right angles. In contrast, the crosshatch patterns in the non-agglutinating strains LC16, DN1 and DN2 are irregular, which may suggest sub-optimal complementarity between their pilus surfaces.
We show here that the non-agglutinating D-region mutant ML16 is able to autoagglutinate with the non-agglutinating cavity mutants ML28 and ML9, thereby supporting a direct interaction between the D-regions and the cavities of adjacent pili. However, when these D-region and cavity amino acid changes were combined in the same strain, these mutants failed to autoagglutinate, despite their TCP being highly similar to El Tor pili. Importantly, the pilus:pilus interaction pattern for these combined D-region/cavity mutants, DN1 and DN2, was distinctly different, being crosshatched, as compared to the mixed bundle/crosshatch pattern seen for the mixed cultures. These results suggest that the combined classical-to-El Tor changes on the TCP surface in these mutants cause the pili to reorient themselves from the lateral interactions seen in the ropelike bundles to orthogonal interactions that produce the crosshatch pattern. However, because the TCP surfaces are not perfectly complementary, these interactions are too weak to induce bacterial aggregation. The autoagglutination observed for the mixed cultures may have occurred via ropelike bundle interactions among the pili, which are apparently not favored in the combined D-region/cavity mutants. Thus, although we identified several residues in the D-regions and cavities that are critical for pilus:pilus interactions and provide evidence that these two regions interact in adjacent filaments, our results suggest that other TcpA residues also profoundly affect these interactions.
It should be noted that the appearance of ropelike bundles in TEM images does not necessarily correlate with autoagglutination for a given strain. For instance, ropelike bundle were seen for strain LC10 (Supplementary Fig. S2), which autoagglutinates poorly. These observations suggest that autoagglutination requires a threshold affinity between the pili beyond what is required for TEM-observed interactions. TEM images most likely capture non-productive interactions among pili originating from the same cell in additional to productive interactions between cells that cause them to autoagglutinate. Whether these interactions are productive or not (i.e. are capable of aggregating cells) would require that (i) the interacting pili are attached to separate cells, and (ii) that the overall interaction affinity is high enough to hold cells together. Neither of these requirements can be assessed by TEM, in part because we rarely see pili attached to cells. Thus, the autoagglutination results, which generally correlate with microcolony formation (Kirn et al., 2000), are more reliable as an indicator of pilus-induced bacterial aggregation.
We propose that pilus:pilus interactions must achieve a fine balance, being sufficiently high-affinity to allow initial aggregation of the bacteria, but not so strong that they cannot eventually be disrupted to allow the bacteria to disperse. Each individual D-region:cavity interaction is likely to be relatively weak, as single amino acid changes can disrupt it. But because pili are polymers of repeating subunits, each filament has many interaction sites, regardless of whether the interactions between the pili occur laterally in ropelike bundles or orthogonally in crosshatches. These multiple interaction interfaces would be very effective in holding cells together, each individual interaction (ie. each D-region/cavity interaction) is easily disrupted to release the bacteria from microcolonies and allow invasion of host cells or to disperse the bacteria for dissemination to other sites or into the environment. In Neisseria, pilus retraction would provide the disruption force to release the bacteria from microcolonies and establish intimate adhesion to host cells (Higashi et al., 2009). In animal models, both EPEC and V. cholerae have been shown to disperse from microcolonies as colonization progresses (Bieber et al., 1998, Nielsen et al., 2006). It is not clear whether pilus retraction or some other mechanism is responsible for disruption of microcolonies for these enteric pathogens, but the dispersal process would presumably be facilitated by relatively weak interactions among the pili.
Differences in the primary TcpA sequence, and subsequently in the way TCP interact, may have factored into the emergence of El Tor biotype as the primary cause of modern-day cholera epidemics. Immune pressure is known to drive changes in surface-exposed pilin residues for N. gonorrhoeae and N. meningitidis T4P (Hagblom et al., 1985). Immune pressure may also influence the emergence of new V. cholerae strains with different TcpA sequences. TCP are immunogenic in humans (Chowdhury et al., 2008), and antibodies to TcpA are protective (Rollenhagen et al., 2006). However, the classical and El Tor biotypes are thought to have evolved from separate lineages (Kaper et al., 1982, Karaolis et al., 1995), so the amino acid differences in the TcpA subunits cannot be explained by immunological pressure alone.
An outstanding question regarding pilus-pilus interactions is why El Tor strain C6706 does not autoagglutinate in vitro in spite of the fact that its pili can mediate autoagglutination when expressed in a classical V. cholerae strain (RT4340). TcpA levels are lower in C6706, but this does not account for their complete failure to autoagglutinate. Classical strains may possess a factor that mediates pilus:pilus interactions similar to PilX in N. meningitidis (Helaine et al., 2005). Alternatively, C6706 may possess a factor that disrupts pilus:pilus interactions. However, since both classical and El Tor strains are capable of colonizing the mouse and human intestines in a TCP-dependent manner, the differences we observe in autoagglutination in vitro may not pertain to in vivo growth.
In summary, our structural analysis of El Tor and classical TcpA subunits and the TCP filaments revealed sites critical to pilus:pilus interactions and support a model whereby the protruding D-regions intercalate into the cavities between subunits in adjacent filaments to hold V. cholerae cells together in microcolonies. Stereochemical complementarity between these sites may be optimized when the pili align laterally in ropelike bundles for classical strains and when the pili align orthogonally for El Tor strains. While a comprehensive view of pilus-mediated bacterial interactions requires further study, these results provide valuable insights into pilus:pilus interactions and their role in V. cholerae aggregation, with implications for other bacterial pathogens that rely on T4P for colonization.
EXPERIMENTAL PROCEDURES
Plasmids and Bacterial Strains
Bacterial strains and plasmids used in this study are shown in Supplementary Table S1. Primers are listed in Table S2. E. coli strains were grown in Luria-Bertani (LB) broth at 37°C. V. cholerae strains grown under TCP-expressing conditions were grown in LB broth, starting pH 6.5, 30°C for 16 hours. Antibiotics were used at a final concentration of 100 μg ml-1 streptomycin (Sm), 30 μg ml-1 gentamicin (Gm), and 100 μg ml-1 ampicillin (Ap). Expression of the ToxT transcriptional regulator on the pMT5 plasmid was induced with 1 mM isopropyl thiogalactoside (IPTG).
Crystallization of TcpAET
DNA encoding TcpAET residues 29-199 was PCR-amplified from V. cholerae El Tor strain C6706. TcpAET was cloned into the pET-15b vector (Novagen), expressed and purified as described (Rollenhagen et al., 2006). TcpAET contains an N-terminal hexahistidine tag and linker sequence (MGSSHHHHHHSSGLVPRGSHM). TcpAET crystals were grown from Hampton Crystal Screen Reagent #16 (0.1 M Na-HEPES, pH 7.5, 1.5 M LiSO4) by sitting drop vapor diffusion by mixing 2 μl TcpA with 2 μl reservoir solution at room temperature. X-ray diffraction data were collected at beamline 11-1 at Stanford Synchrotron Radiation Laboratories. TcpAET crystallized in the I212121 space group. Data were processed using DENZO and SCALEPAK (Otwinowski, 1993). The TcpAET structure was solved to 1.5 Å by molecular replacement using AMoRe (Navaza, 2001), with the TcpACl (PDB entry 1OQV) as the search model. The sequence of TcpA was fit manually using XtalView/Xfit (McRee, 1999) and the resulting model was refined using SHELX (Sheldrick, 1990) and CNS (Brunger et al., 1998) in combination with Xfit. The TcpAET structure contains two molecules in the asymmetric unit and 462 well ordered water molecules plus. The his-tag/linker segment was disordered in both chains. Electron density consistent with three glycerol molecules and three sulfates was present. PROCHECK analysis (Laskowski et al., 1993) revealed that 94.3% of all residues are in the most favored regions of the Ramachandron plot and the remaining 5.7% are in allowed regions.
Generation and isolation of tcpA mutants
Complementary mutagenic primers (Supplementary Table S2) were designed to generate missense mutations in the tcpA gene within the pTK1 vector (Kirn et al., 2000) using the Quikchange PCR-based mutagenesis system (Stratagene). pTK1 is derived from the pKAS32 suicide vector (Skorupski and Taylor, 1996). Mutations were introduced into the parental V. cholerae strain RT4524 by allelic exchange as described previously (Skorupski and Taylor, 1996). Strain RT4524, derived from classical biotype O395, has the tcpA gene disrupted by a lacZ gene insertion in the 3’ end of the tcpA gene for screening purposes. This strain also carries plasmid pMin1, which encodes for Gm-resistance. Briefly, E. coli S17 cells carrying pTK1-tcpA* (i.e. the mutated tcpA gene) were mated with V. cholerae strain RT4524, and transconjugants were selected on LB-Ap-Gm plates. Transconjugants were passed on LB-Gm plates to select for cells that excised the pTK1. Colonies that grew on LB-Gm plates but not LB-Ap plates were screened by PCR to identify exconjugants that replaced the tcpA-lacZ gene with tcpA*. All mutations were confirmed by sequencing the entire tcpA gene in both directions. Strain ML7 is a positive control that has the tcpA-lacZ gene fusion of RT4524 replaced with the wild type classical tcpA gene. The pMin1 plasmid (Nye et al., 2000) was cured from the mutant strains and pMT5 (DiRita et al., 1996) was introduced to maximize pilus expression, resulting in more readily-identifiable phenotypes. Plasmid pMT5 was mobilized from E. coli X90 into V. cholerae tcpA mutants by triparental mating with E. coli strain MM294 carrying pRK2013 (Figurski and Helinski, 1979).
Autoagglutination assays
V. cholerae cells were grown overnight in 2-ml cultures in LB-Sm-Ap. The overnight culture was diluted 1/100 in fresh LB-Sm and grown to OD600 ~0.8. Fresh LB-Sm-Ap, 1 mM IPTG, starting pH 6.5 was inoculated with a ~1/10,000 dilution of the overnight culture. The dilution factor varied slightly from strain to strain according to their OD600 reading in order to normalize the inocula. Cultures were grown overnight at 30 °C rotating in a ferris wheel rotator). El Tor strain C6706 was grown for four hours at 37°C in stationary tubes containing AKI media (1.5% Bacto-Peptone, 0.4% yeast extract, 0.3% NaHCO3, 1 mM IPTG), followed by 12 hours at 30°C with rotation (Iwanaga et al., 1989). After overnight growth, cultures were placed on the bench for 15 minutes to allow cell aggregates to settle, and the OD600 of the supernatant, drawn from the top of the culture so as to not disturb the settled cells, was measured. Cultures were diluted in LB to obtain OD600 readings between 0.1 and 1.0. Autoagglutination levels are reported as autoagglutination index (AI), which is the ratio of the inverse of the OD600 value of the test strain over the inverse of the OD600 value of V. cholerae O395:
Thus, strains with an AI greater than 1 autoagglutinate better than O395 and those with AI values lower than 1 do not autoagglutinate as well as O395. AI values are also reported in Table 2 as pluses and minuses based on the autoagglutination scale (AS), for facile comparison with O395: (-), AI ≤0.10, no autoagglutination; 0.11-0.20 (+), detectable autoagglutination; 0.21-0.50 (++), moderate autoagglutination; 0.51-1.50 (+++), good or “wild type” (O395) autoagglutination; ≥1.51 (++++), better than wild type autoagglutination (clear supernatant). For the co-inoculations, cell cultures were normalized as described above, and two strains were used to inoculate the growth media. For the post-mixed cultures, individual cells were grown overnight under pilus-inducing conditions, and then 0.8 ml of each culture was mixed in pairwise combinations. Culture mixtures were held stationary at room temperature for 2 hours before measuring OD600.
Pilin and TCP production assays
V. cholerae strains from overnight cultures were grown to mid-log phase in non-pilus-inducing conditions (LB, pH 8.5, 37 °C), and then cultures were normalized by optical density measurement (OD600) and diluted 1/20,000 into LB, pH 6.5, 1 mM IPTG (or AKI, 1 mM IPTG for C6706). Cells were grown for 16 hr at 30 °C. Eight μL aliquots of whole cell culture were analyzed by 15% SDS-PAGE and immunoblotting to assess TcpA expression. Pilus assembly was assessed by physically shearing the pili from the cells using an Ultra-Turrax T8.01 disperser and analyzing the cleared cell homogenate by SDS-PAGE and immunoblotting. Cells from 9 ml overnight cultures were pelleted by centrifugation and resuspended in 1 ml PBS-EDTA (20 mM phosphate, 150 mM NaCl, 20 mM EDTA, pH 7.4). Pili were sheared from the cells by homogenization for 20 seconds using the Ultra-Turrax disperser, the cell debris was removed by centrifugation and 16 μl of the sheared cell supernatant was analyzed by SDS-PAGE. Pilin protein was detected using an antibody against a C-terminal peptide (TcpA-6, residues 174-199 (Sun et al., 1991).
Transmission electron microscopy to assess TCP morphology, interaction phenotype
Five μL aliquots of overnight bacterial culture were applied to untreated carbon-coated copper grids (Electron Microscopy Science) and stained with either 1% phosphotungstic acid (PTA) or 1% ammonium molybdate, pH 7.4. Grids were imaged in the FEI Tecnai F20 at 200 keV. For the partially-purified TCP shown in Fig. 1C, cells from overnight cultures of V. cholerae O395 and RT4340 were pelleted by centrifugation and resuspended in PBS-EDTA. Pili were sheared from cells using a 25-Gauge needle and the mixture was centrifuged at 3000X g to pellet the cells. Pili were removed from the top phase of the supernatant, resuspended in 125 mM ethanolamine, pH 10.5, and applied to grids as described above.
Supplementary Material
Acknowledgments
We thank Magnar Bjørås for technical advice, Francisca Cerda-Maira for production of ΔN-TcpAET for crystallization, Juliana Li for model building, Sumaiya Islam for technical assistance, Subramaniapillai Kolappan for model refinement and the beamline staff at the Stanford Synchrotron Radiation Laboratory. This work was supported by the Canadian Institutes of Health Research (CIHR) grant RGPIN312152 to LC and the National Institutes of Health grants AI022160 to JAT and AI025096 to RKT. LC is supported by salary awards from CIHR and the Michael Smith Foundation for Health Research.
Footnotes
Figure preparation
Molecule structure figures were prepared using PyMOL (http://www.pymol.org).
Accession numbers
The TcpAET crystal structure has been deposited in the Protein Data Bank under accession number 3HRV.
Supplementary material is available online.
References
- Anantha RP, Stone KD, Donnenberg MS. Role of BfpF, a member of the PilT family of putative nucleotide-binding proteins, in type IV pilus biogenesis and in interactions between enteropathogenic Escherichia coli and host cells. Infect Immun. 1998;66:122–131. doi: 10.1128/iai.66.1.122-131.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barken KB, Pamp SJ, Yang L, Gjermansen M, Bertrand JJ, Klausen M, et al. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ Microbiol. 2008;10:2331–2343. doi: 10.1111/j.1462-2920.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- Biais N, Ladoux B, Higashi D, So M, Sheetz M. Cooperative retraction of bundled type IV pili enables nanonewton force generation. PLoS Biol. 2008;6:e87. doi: 10.1371/journal.pbio.0060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieber D, Ramer SW, Wu CY, Murray WJ, Tobe T, Fernandez R, Schoolnik GK. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Burrows LL. Weapons of mass retraction. Mol Microbiol. 2005;57:878–888. doi: 10.1111/j.1365-2958.2005.04703.x. [DOI] [PubMed] [Google Scholar]
- Chiang P, Habash M, Burrows LL. Disparate subcellular localization patterns of Pseudomonas aeruginosa Type IV pilus ATPases involved in twitching motility. J Bacteriol. 2005;187:829–839. doi: 10.1128/JB.187.3.829-839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury F, Khan AI, Harris JB, LaRocque RC, Chowdhury MI, Ryan ET, et al. A comparison of clinical and immunologic features in children and older patients hospitalized with severe cholera in Bangladesh. Pediatr Infect Dis J. 2008;27:986–992. doi: 10.1097/INF.0b013e3181783adf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig L, Li J. Type IV pili: paradoxes in form and function. Curr Opin Struct Biol. 2008;18:267–277. doi: 10.1016/j.sbi.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- Craig L, Taylor RK, Pique ME, Adair BD, Arvai AS, Singh M, et al. Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol Cell. 2003;11:1139–1150. doi: 10.1016/s1097-2765(03)00170-9. [DOI] [PubMed] [Google Scholar]
- Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Molecular Cell. 2006;23:651–662. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- DiRita VJ, Neely M, Taylor RK, Bruss PM. Differential expression of the ToxR regulon in classical and E1 Tor biotypes of Vibrio cholerae is due to biotype-specific control over toxT expression. Proc Natl Acad Sci U S A. 1996;93:7991–7995. doi: 10.1073/pnas.93.15.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagblom P, Segal E, Billyard E, So M. Intragenic recombination leads to pilus antigenic variation in Neisseria gonorrhoeae. Nature. 1985;315:156–158. doi: 10.1038/315156a0. [DOI] [PubMed] [Google Scholar]
- Helaine S, Carbonnelle E, Prouvensier L, Beretti JL, Nassif X, Pelicic V. PilX, a pilus-associated protein essential for bacterial aggregation, is a key to pilus-facilitated attachment of Neisseria meningitidis to human cells. Mol Microbiol. 2005;55:65–77. doi: 10.1111/j.1365-2958.2004.04372.x. [DOI] [PubMed] [Google Scholar]
- Helaine S, Dyer DH, Nassif X, Pelicic V, Forest KT. 3D structure/function analysis of PilX reveals how minor pilins can modulate the virulence properties of type IV pili. Proc Natl Acad Sci U S A. 2007;104:15888–15893. doi: 10.1073/pnas.0707581104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrington DA, Hall RH, Losonsky G, Mekalanos JJ, Taylor RK, Levine MM. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J Exp Med. 1988;168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi DL, Lee SW, Snyder A, Weyand NJ, Bakke A, So M. Dynamics of Neisseria gonorrhoeae attachment: microcolony development, cortical plaque formation, and cytoprotection. Infect Immun. 2007;75:4743–4753. doi: 10.1128/IAI.00687-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi DL, Zhang GH, Biais N, Myers LR, Weyand NJ, Elliott DA, So M. Influence of type IV pilus retraction on the architecture of the Neisseria gonorrhoeae-infected cell cortex. Microbiology. 2009;155:4084–4092. doi: 10.1099/mic.0.032656-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanaga M, Nakasone N, Ehara M. Pili of Vibrio cholerae O1 biotype E1 Tor: a comparative study on adhesive and non-adhesive strains. Microbiol Immunol. 1989;33:1–9. doi: 10.1111/j.1348-0421.1989.tb01492.x. [DOI] [PubMed] [Google Scholar]
- Jonson G, Svennerholm AM, Holmgren J. Expression of virulence factors by classical and El Tor Vibrio cholerae in vivo and in vitro. FEMS Microbiol Ecol. 1990;74:221–228. [Google Scholar]
- Jurcisek JA, Bookwalter JE, Baker BD, Fernandez S, Novotny LA, Munson RS, Jr, Bakaletz LO. The PilA protein of non-typeable Haemophilus influenzae plays a role in biofilm formation, adherence to epithelial cells and colonization of the mammalian upper respiratory tract. Mol Microbiol. 2007;65:1288–1299. doi: 10.1111/j.1365-2958.2007.05864.x. [DOI] [PubMed] [Google Scholar]
- Kaper JB, Bradford HB, Roberts NC, Falkow S. Molecular epidemiology of Vibrio cholerae in the U.S. Gulf Coast. J Clin Microbiol. 1982;16:129–134. doi: 10.1128/jcm.16.1.129-134.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaolis DK, Lan R, Reeves PR. The sixth and seventh cholera pandemics are due to independent clones separately derived from environmental, nontoxigenic, non-O1 Vibrio cholerae. J Bacteriol. 1995;177:3191–3198. doi: 10.1128/jb.177.11.3191-3198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirn TJ, Lafferty MJ, Sandoe CM, Taylor RK. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol Microbiol. 2000;35:896–910. doi: 10.1046/j.1365-2958.2000.01764.x. [DOI] [PubMed] [Google Scholar]
- Klausen M, Heydorn A, Ragas P, Lambertsen L, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol. 2003;48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, McArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Applied Crystallography. 1993;26:283–291. [Google Scholar]
- Li J, Lim MS, Li S, Brock M, Pique ME, Woods VL, Jr, Craig L. Vibrio cholerae toxin-coregulated pilus structure analyzed by hydrogen/deuterium exchange mass spectrometry. Structure. 2008;16:137–148. doi: 10.1016/j.str.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke NR, Jurcisek JA, Bakaletz LO, Campagnari AA. Contribution of Moraxella catarrhalis type IV pili to nasopharyngeal colonization and biofilm formation. Infect Immun. 2007;75:5559–5564. doi: 10.1128/IAI.00946-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B, Potter L, So M, Seifert HS, Sheetz MP. Single pilus motor forces exceed 100 pN. Proc Natl Acad Sci U S A. 2002;99:16012–16017. doi: 10.1073/pnas.242523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceau M, Beretti JL, Nassif X. High adhesiveness of encapsulated Neisseria meningitidis to epithelial cells is associated with the formation of bundles of pili. Mol Microbiol. 1995;17:855–863. doi: 10.1111/j.1365-2958.1995.mmi_17050855.x. [DOI] [PubMed] [Google Scholar]
- Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- McRee DE. XtalView/Xfit -A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- Merz AJ, So M, Sheetz MP. Pilus retraction powers bacterial twitching motility. Nature. 2000;407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- Nassif X, Lowy J, Stenberg P, O’Gaora P, Ganji A, So M. Antigenic variation of pilin regulates adhesion of Neisseria meningitidis to human epithelial cells. Mol Microbiol. 1993;8:719–725. doi: 10.1111/j.1365-2958.1993.tb01615.x. [DOI] [PubMed] [Google Scholar]
- Nassif X, Marceau M, Pujol C, Pron B, Beretti JL, Taha MK. Type-4 pili and meningococcal adhesiveness. Gene. 1997;192:149–153. doi: 10.1016/s0378-1119(96)00802-5. [DOI] [PubMed] [Google Scholar]
- Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr D Biol Crystallogr. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- Nielsen AT, Dolganov NA, Otto G, Miller MC, Wu CY, Schoolnik GK. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2006;2:e109. doi: 10.1371/journal.ppat.0020109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nye MB, Pfau JD, Skorupski K, Taylor RK. Vibrio cholerae H-NS silences virulence gene expression at multiple steps in the ToxR regulatory cascade. J Bacteriol. 2000;182:4295–4303. doi: 10.1128/jb.182.15.4295-4303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. Data collection and processing. Science and Engineering Research Council; 1993. [Google Scholar]
- Parge HE, Forest KT, Hickey MJ, Christensen DA, Getzoff ED, Tainer JA. Structure of the fibre-forming protein pilin at 2.6 A resolution. Nature. 1995;378:32–38. doi: 10.1038/378032a0. [DOI] [PubMed] [Google Scholar]
- Park HS, Wolfgang M, van Putten JP, Dorward D, Hayes SF, Koomey M. Structural alterations in a type IV pilus subunit protein result in concurrent defects in multicellular behaviour and adherence to host tissue. Mol Microbiol. 2001;42:293–307. doi: 10.1046/j.1365-2958.2001.02629.x. [DOI] [PubMed] [Google Scholar]
- Pelicic V. Type IV pili: e pluribus unum? Mol Microbiol. 2008;68:827–837. doi: 10.1111/j.1365-2958.2008.06197.x. [DOI] [PubMed] [Google Scholar]
- Rollenhagen JE, Kalsy A, Cerda F, John M, Harris JB, Larocque RC, et al. Transcutaneous immunization with toxin-coregulated pilin A induces protective immunity against Vibrio cholerae O1 El Tor challenge in mice. Infect Immun. 2006;74:5834–5839. doi: 10.1128/IAI.00438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM. Phase annealing in SHELX-90: direct methods for larger structures. Acta Crystallogr. 1990;A46:467–473. [Google Scholar]
- Skerker JM, Berg HC. Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci U S A. 2001;98:6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorupski K, Taylor RK. Positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- Sun D, Seyer JM, Kovari I, Sumrada RA, Taylor RK. Localization of protective epitopes within the pilin subunit of the Vibrio cholerae toxin-coregulated pilus. Infect Immun. 1991;59:114–118. doi: 10.1128/iai.59.1.114-118.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacket CO, Taylor RK, Losonsky G, Lim Y, Nataro JP, Kaper JB, Levine MM. Investigation of the roles of toxin-coregulated pili and mannose-sensitive hemagglutinin pili in the pathogenesis of Vibrio cholerae O139 infection. Infect Immun. 1998;66:692–695. doi: 10.1128/iai.66.2.692-695.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci U S A. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelin KH, Taylor RK. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect Immun. 1996;64:2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd WJ, Wray GP, Hitchcock PJ. Arrangement of pili in colonies of Neisseria gonorrhoeae. J Bacteriol. 1984;159:312–320. doi: 10.1128/jb.159.1.312-320.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga JJ, Therit B, Melville SB. Type IV pili and the CcpA protein are needed for maximal biofilm formation by the gram-positive anaerobic pathogen Clostridium perfringens. Infect Immun. 2008;76:4944–4951. doi: 10.1128/IAI.00692-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang M, Lauer P, Park HS, Brossay L, Hebert J, Koomey M. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol Microbiol. 1998;29:321–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.