Abstract
Background
Date palm (Phoenix dactylifera L.), a member of Arecaceae family, is one of the three major economically important woody palms—the two other palms being oil palm and coconut tree—and its fruit is a staple food among Middle East and North African nations, as well as many other tropical and subtropical regions. Here we report a complete sequence of the data palm chloroplast (cp) genome based on pyrosequencing.
Methodology/Principal Findings
After extracting 369,022 cp sequencing reads from our whole-genome-shotgun data, we put together an assembly and validated it with intensive PCR-based verification, coupled with PCR product sequencing. The date palm cp genome is 158,462 bp in length and has a typical quadripartite structure of the large (LSC, 86,198 bp) and small single-copy (SSC, 17,712 bp) regions separated by a pair of inverted repeats (IRs, 27,276 bp). Similar to what has been found among most angiosperms, the date palm cp genome harbors 112 unique genes and 19 duplicated fragments in the IR regions. The junctions between LSC/IRs and SSC/IRs show different features of sequence expansion in evolution. We identified 78 SNPs as major intravarietal polymorphisms within the population of a specific cp genome, most of which were located in genes with vital functions. Based on RNA-sequencing data, we also found 18 polycistronic transcription units and three highly expression-biased genes—atpF, trnA-UGC, and rrn23.
Conclusions
Unlike most monocots, date palm has a typical cp genome similar to that of tobacco—with little rearrangement and gene loss or gain. High-throughput sequencing technology facilitates the identification of intravarietal variations in cp genomes among different cultivars. Moreover, transcriptomic analysis of cp genes provides clues for uncovering regulatory mechanisms of transcription and translation in chloroplasts.
Introduction
The chloroplast (cp), believed to arise from endosymbiosis between a photosynthetic bacterium and a non-photosynthetic host [1], is the photosynthetic organelle that provides essential energy for plants and algae. A given cell, such as that of a plant leaf, often contains 400 to 1,600 copies of cp genome [2]. Other biological activities also take place in chloroplast including the production of starch, certain amino acids and lipids, vitamins, certain pigments in flowers and several key pathways of sulfur and nitrogen metabolism [3]. In angiosperm, most cp genomes are circular DNA molecules ranging from 120 to 160 kb in length and have a quadripartite organization consisting of two copies of inverted repeats (IRs) of ∼20–28 kb in size, which divides the rest of genome into a large-single-copy region (LSC; 80–90 kb) and a small-single-copy (SSC; 16–27 kb) region. Usually, the gene content of angiosperm cp genome is rather conserved, encoding 4 rRNAs, ∼30 tRNAs, and ∼80 unique proteins [4]. Since the completion of the first cp genome of tobacco (Nicotiana tabacum) [5], there have been many discoveries on rearrangements and IR expansions within cp genomes, perhaps the most remarkable one is that of Pelargonium x hortorum, in which numerous rearrangements and large IR expansions were found [4]. Gene losses have been found frequently in angiosperm cp genomes [6], [7], [8].
The date palm (Phoenix dactylifera, L.), an Arecaceae family member, is one of the most economically important woody plant cultivated in Middle East and North Africa [9]. Its fruit provides staple food across Asian (mainly in Arabian Peninsula, Iran, and Pakistan), African, Europe, and American tropics. A recent report suggests that there are more than 340 cultivars in Saudi Arabia and nearly 2,000 cultivars around the world [10]. The total number of date palm trees grown in the world is ∼100 million, producing 15 million metric ton fruit each year [11]. The cultivated hybrids of date palm are mostly diploid (2n = 36) and propagated from offshoots [12]. Because cp genome is maternally inherited [13], deeper knowledge about its structure, sequence variation, and evolution provides useful information for developing propagation technologies, such as cytoplasmic breeding and transgenic insertion.
In the past 10 years, we have witnessed a dramatic increase in the number of complete cp genomes. Up to now, 132 complete land plant cp genomes have been deposited in GenBank Organelle Genome Resources, albeit only 22 of them are monocot cp genomes and most of these genomes were sequenced using capillary sequencers [14]. With the emergence of next-generation sequencers, new approaches for genome sequencing have been gradually proposed due to their high-throughput, time-saving, and low-cost advantages [15], [16], [17]. Here, we report the complete cp genome sequence of date palm, the first member of the Arecaceae family from an elite cultivar Khalas (Al-Hssa Oasis, Saudi Arabia), using one of the next-generation sequencing method – pyrosequencing (Roche GS FLX). We also describe details in genome assembly, annotation, and comparative analysis, as well as information on sequence variations and trancriptomics for the date palm cp genome.
Results and Discussion
1. Genome assembly and validation
Using Roche GS FLX system, we carried out five sequencing runs to generate 4,169,506 raw reads (347 bp in average read length) for the project. After screening the reads through alignment with reference cp genomes and extensive contig extension efforts (see Materials and Methods for details), we collected 369,022 cp-genome-related reads (8.8% of total reads and an average of 384 bp in length), reaching 1,081x coverage on average over the cp genome. After validating the homopolymer regions and the junctions between single-copy and IR regions with PCR-based confirmation, we obtained a complete cp genome sequence of 158,462 bp in length.
Typically, two types of errors are characteristic in pyrosequencing data processing; one is associated with contig ends and the other involves heterogeneous insertion/deletions (Indels) arisen from homopolymeric repeats [17]. The first is basically a sequence quality issue and usually overcome by increasing coverage (here we have 369,022 cp reads) and removing low-quality reads. The second is intrinsic to pyrosequencing and can not be improved by increasing coverage; we therefore performed alternative types of experiments to correct the erroneous homopolymer calls found in the assembly. Based on the homopolymer (n> = 7) distribution in the preliminary cp genome assembly, we designed 151 pairs of PCR primers to validate all homopolymer runs in the entire assembly by using capillary sequencing (Table S1). The result was very satisfactory where we added 117 base pairs in 108 homopolymers, and all previously uncertain or missed nucleotides, either A or T (except for one homopolymer run that has 13 cytosines) were satisfactorily validated. These recovered nucleotides appeared larger in number as compared to what encountered when sequencing Mungbean cp genome, where only 49-bp sequences were lost in the initial assembly (74.6x raw data) [15]. Since we have 161 homopolymers above 7 bp, and 181 equal to 7 bp, the potential errors are too large to not handle them with care and a simple increase of sequence coverage is not likely to be useful [17].
2. Genome features
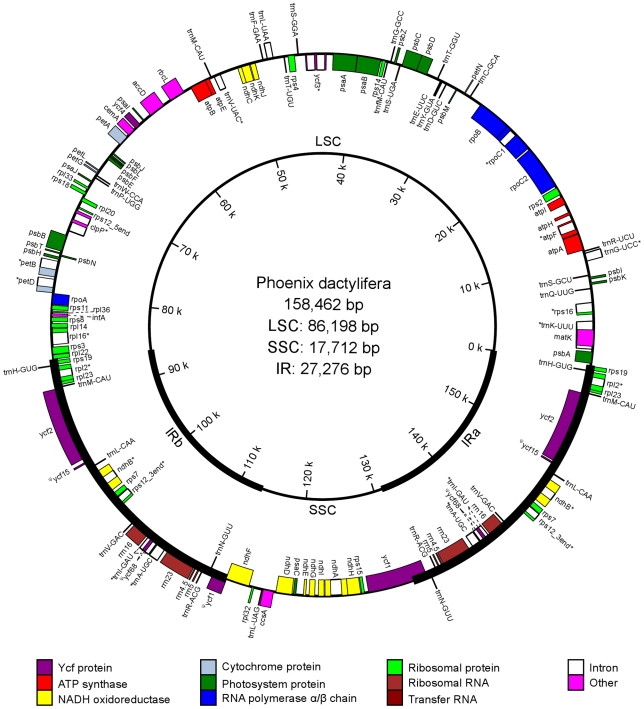
The date palm cp genome is a typical circular double-stranded DNA molecule, and it shares a common quadripartite structure with the vast majority of other angiosperms: a pair of IRs (27,276 bp) separated by the LSC (86,198 bp) and SSC regions (17,712 bp) (Figure 1). It encodes 131 predicted functional genes; 112 are unique and 19 are duplicated in the IR regions. Among the 112 unique genes, we identified 79, 29, and 4 protein-coding, transfer RNA, and ribosomal RNA genes, respectively. 50.93%, 1.79%, and 5.71% of the genome sequence encode proteins, tRNAs, and rRNAs, respectively, whereas the remaining 41.57% are non-coding and filled with introns, intergenic spacers, and pseudogenes. Similar to other cp genomes [18], [19], the date palm cp genome is also AT-rich (62.77%), and the values vary slightly among defined sequences of non-coding, protein-coding, tRNA, and rRNA, where their A+T contents are 66.60%, 61.03%, 57.94%, and 52.19%, respectively.
Figure 1. The map of date palm cp genome sequence.
Genes shown outside the outer circle are transcribed clockwise, whereas those shown inside are transcribed counterclockwise. The thick line indicates IRs. The genome coordinate is shown in the inner circle. Genes belonging to different groups are color-coded (Ψ, pseudogene).
Similar to tobacco cp genome, the date palm cp genome has 18 intron-containing genes among the 112 unique genes; almost all are single-intron except for two genes, ycf3, and clpP, whose exons are separated by two introns. The gene rps12 is trans-spliced; one of its exons is in the LSC region (5′) and the two reside in the IR regions separated by an intron. The introns of all protein-coding genes share the same splicing mechanism as Group II introns [20]. In the IR regions, both ycf15 and ycf68 became pseudogenes duo to internal stop codons identified in their coding sequences (CDS). Specifically, the CDS of ycf15 is interrupted by a stop codon at downstream (57 bp away from the start codon), whereas several stop codons appeared in ycf68 gene. Similar mutations have also been known to happen in the cp genomes of other species [19]. Another pseudogene is ycf1 in the boundary of IRb and SSC because of the incomplete duplication of the normal copy of ycf1 at the IRa and SSC boundary (Figure 1).
A total of 22,950 codons represent the coding capacity of all protein-coding genes of date palm cp genome (Table 1). Among these codons, 2001 (8.72%) encode for isoleucine and 271 (1.18%) for cysteine, which were the most and the least amino acids, respectively. These extremes are the same as what in nuclear genomes. The base compositions at each codon position are slightly biased: 54.4%, 61.8%, and 70.6% for the first to the third, respectively.
Table 1. Codon usage and codon-anticodon recognition pattern for tRNA in date palm chloroplast genome.
| Amino acid | Codon | NO. | RSCU | tRNA | Amino acid | Codon | NO. | RSCU | tRNA |
| Phe | UUU | 836 | 1.29 | Ser | UCU | 498 | 1.69 | ||
| UUC | 462 | 0.71 | trnF-GAA | UCC | 289 | 0.98 | trnS-GGA | ||
| Leu | UUA | 734 | 1.89 | trnL-UAA | UCA | 371 | 1.26 | trnS-UGA | |
| UUG | 500 | 1.29 | trnL-CAA | UCG | 162 | 0.55 | |||
| Tyr | UAU | 700 | 1.6 | Cys | UGU | 207 | 1.53 | trnC-GCA | |
| UAC | 177 | 0.4 | trnY-GUA | UGC | 64 | 0.47 | |||
| Ter | UAA | 37 | 0 | Ter | UGA | 20 | 0 | ||
| Ter | UAG | 22 | 0 | Trp | UGG | 392 | 1 | trnW-CCA | |
| Leu | CUU | 473 | 1.22 | Pro | CCU | 367 | 1.57 | ||
| CUC | 162 | 0.42 | CCC | 185 | 0.79 | ||||
| CUA | 314 | 0.81 | trnL-UAG | CCA | 275 | 1.18 | trnP-UGG | ||
| CUG | 150 | 0.39 | CCG | 107 | 0.46 | ||||
| His | CAU | 423 | 1.54 | Arg | CGU | 322 | 1.39 | trnR-ACG | |
| CAC | 127 | 0.46 | trnH-GUG | CGC | 78 | 0.34 | |||
| Gln | CAA | 608 | 1.5 | trnQ-UUG | CGA | 307 | 1.32 | ||
| CAG | 200 | 0.5 | CGG | 106 | 0.46 | ||||
| Ile | AUU | 959 | 1.44 | Thr | ACU | 465 | 1.56 | ||
| AUC | 415 | 0.62 | trnI-GAU | ACC | 223 | 0.75 | trnT-GGU | ||
| AUA | 627 | 0.94 | ACA | 382 | 1.28 | trnT-UGU | |||
| Met | AUG | 545 | 1 | trn(f)M-CAU | ACG | 125 | 0.42 | ||
| Asn | AAU | 857 | 1.59 | Ser | AGU | 361 | 1.23 | ||
| AAC | 223 | 0.41 | trnN-GUU | AGC | 84 | 0.29 | trnS-GCU | ||
| Lys | AAA | 884 | 1.5 | trnK-UUU | Arg | AGA | 445 | 1.92 | trnR-UCU |
| AAG | 294 | 0.5 | AGG | 135 | 0.58 | ||||
| Val | GUU | 470 | 1.44 | Ala | GCU | 559 | 1.83 | ||
| GUC | 162 | 0.5 | trnV-GAC | GCC | 188 | 0.62 | |||
| GUA | 496 | 1.52 | trnV-UAC | GCA | 347 | 1.14 | trnA-UGC | ||
| GUG | 181 | 0.55 | GCG | 127 | 0.42 | ||||
| Asp | GAU | 741 | 1.6 | Gly | GGU | 550 | 1.4 | ||
| GAC | 184 | 0.4 | trnD-GUC | GGC | 137 | 0.35 | trnG-GCC | ||
| Glu | GAA | 935 | 1.53 | trnE-UUC | GGA | 633 | 1.61 | trnG-UCC | |
| GAG | 290 | 0.47 | GGG | 251 | 0.64 |
There are some exceptional cases in start codons. We identified two ACG as start codons in rpl2 and ndhD and one GUG start codon in rps19. The non-canonical starts have been detected in other angiosperms [19] and even in fern-like plant Alsophila spinulosa, where 20 genes start with ACG [18]. We also found an unconventional start codon in cemA that encodes a heme-binding protein functioning in the inner chloroplast envelope membrane [21]. Multiple alignments of cp genomes show that there is an 8-bp homopolymer (AAAAAAAA) in the downstream of its start codon sequences commonly found among certain monocots, such as rice. However, in date palm (also in oil palm and Yucca), this homopolymer is 10 bp in length, resulting a 2-bp coding frame shift that changes ATG to AAT and GAA (Figure 2). Moreover, a stop codon—TAG—12 bp in the upstream sequence makes the start codon of cemA in date palm rather ambiguous. Our transcriptome analysis demonstrated that its mRNA is in a polycistronic transcription unit, together with psaI, ycf4, and petA (Part 7). However, whether this gene is translatable to protein or not remains unclear due to the obscurity of its start codon. To make further inspection to this gene, we aligned its orthologs from 74 angiosperms and 3 gymnosperms (Ginkgo, Pinus and Welwitschia) and found a start codon among all other species either at the origin or the upstream other than what in date palm, oil palm, Yucca and Lemna (Figure S1). Although ATG is typical the start codon, GUG is also used in 14 species. Most of the 14 species are either basal angiosperms or gymnosperms except two in Acoraceae family (Acorus calamus and Acorus americanus) [22]. These results suggest that the start codon of cemA gene must have endured a strong selective pressure during the early evolution of angiosperms but it is somewhat relaxed among certain plant taxa.
Figure 2. A multiple alignment of cemA genes from representative monocot cp genomes.
The start codon is assigned as origin. The possible stop codons in the upstream regions are marked by ellipse. 663-bp less-relevant aligned sequences were omitted.
3. Gene order and content
Since the tobacco cp genome is often regarded to be unarranged [5], we compared our assembly with it and observed a high degree of synteny between the two cp genomes. Two minor exceptions were found—additional copies of rps19 and trnH-GUG around the junction between LSC and IRs— due to IR expansions in the date palm cp genome. The gene content is also nearly identical with that of tobacco; only a single gene, namely infA common among monocots, appeared degenerated into a pseudogene in tobacco. However, structural rearrangements and gene loss-and-gain events are quite often among other monocot cp genomes, mainly found in Poaceae family, where the canonical order is disrupted by three inversions in their LSC regions and rpl23 are translocated from IR to LSC regions [23]. Indels are also frequently found in Poaceae such as intron-loss in rpoC1 and insertion in rpoC2 [24]. Other variations are gene-loss (deletion or becoming pseudogene) of accD, ycf1, and ycf2 in Poaceae cp genomes [25]. We calculated the vestigial lengths of these three pseudogenes and found that their sequence variations can explain in part the cp genome size differences between Poaceae species (137,851 bp in average) and date palm, since the average size of the Poaceae cp genomes is ∼14 kb shorter than that of date palm. In addition, gene-loss event often occurs in other monocot families (cp genomes of 22 monocot species are available in public databases and 15 of them are Poaceae species). There is only one Typhaceae cp genome published recently, which has the same gene content and order as date palm [26]. Among the remaining cases, Lemna and Dioscorea, each lost a single gene: infA and rps16, respectively; two Acoraceae members lost accD [6], [7], [24] and Phalaenopsis and Oncidium lost most of ndh genes [8], [27]. Rearrangements also occurred in Dioscorea, such as the inversion of SSC. Nevertheless, similar to that of tobacco, date palm cp genome appeared less rearranged and having very limited gene loss-and-gain especially when compared to these monocots.
4. IR expansion and evolutionary implications
Around the borders of JLB and JLA, date palm has the same structure as what in Poaceae cpDNAs; specifically, in JLB, rpl22 and its 5′-end adjacent rps19 are completely fell in LSC and IRb, respectively, whereas in JLA, another copy of rps19 in IRa adjoins its 3′-end to psbA in LSC. However, surrounding JSB and JSA, the gene order of the date palm assembly is similar to those of Amborella and certain dicots (i.e., tobacco, Panax, and Arabidopsis), namely JSB locates between ycf1 pseudogene and ndhF, whereas JSA resides in the 3′ region of the normal ycf1 gene.
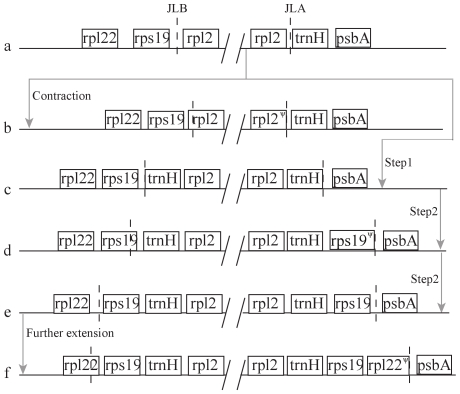
Among other monocots, various degrees of IR to LSC expansions were identified. In Dioscorea and Acorus, only the 5′-end of rps19 are included in IRb to generate a rps19 pseudogene in IRa [28], whereas in Lemna, a contraction was detected as IRb shrunk into the 3′-end of rpl2 [6]. Other than Lemna, all other monocots studied thus far possess a copy of trnH-GUG (in 5′-end of rps19 in IRb) that is absent in tobacco and Amborella. Therefore, based on the two-step hypothesis of IR expansion [6], we deduced that the formation of IR-LSC boundaries among monocots (except Lemna) occurs in the following manner: trnH-GUG is duplicated in IRb as the first step of inclusion of trnH-GUG to IRa; and along with the second expansion step in recruiting rps19 to IRb, another copy of rps19 (or pseudogene) is generated in IRa. However, in Phalaenopsis and Oncidium, the expansion was unusual where IRb extended to the 3′-end of rpl22 gene [8], [27] (Figure 3).
Figure 3. Expansion of IR into the LSC region.
The dashed lines depict the junction boundaries. The arrowed grey lines indicate the process of IR expansions. a, Amborella and Nicotiana; b, Lemna; c, intermediate; d, Acorus and Dioscorea; e, date palm, typha, and all Poaceaes; f, Phalaenopsis and Oncidium.
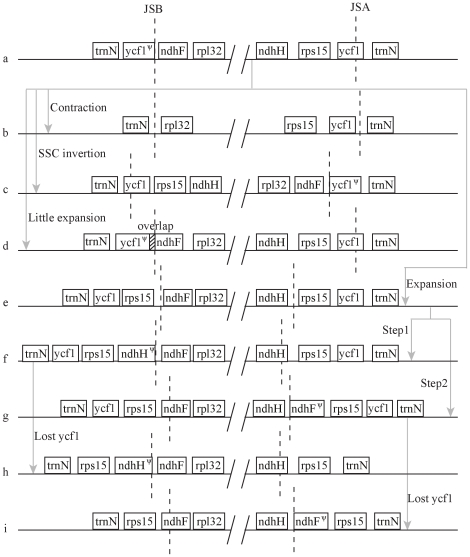
At the two boundaries of SSC, the general structure revealed in Amborella and dicots (i.e., tobacco, Panax and Arabidopsis) is that ycf1 spans JSA and ycf1 pseudogene adjacent to JSB. In monocots, two Acoraceae members share this structure; and similar to Arabidopsis, a small expansion occurred in date palm, which formed an overlap between ndhF and ycf1 pseudogene (55 bp in date palm) in JSB in date palm [28]. In Dioscorea, this structure is inverted—the normal ycf1 in JSB and the corresponding ycf1 pseudogene in JSA—because of the inversion of SSC. In Phalaenopsis, there is a short contraction of IRs where ycf1 is completely included in SSC [8]. A special case in monocots is found in Lemna, its expansion pattern of IRs to SSC is similar to that in Poaceae family if we exclude the complete duplicate of ycf1 [6]. Since we can find the symmetrically degenerated ycf1 in several Poaceae cp genomes (i.e., the position 99,623–100,458 and 114,660–115,495 in IRb and IRa, respectively, in Oryza sativa) in the corresponding position of Lemna, we speculate that in the beginning, the ancestors of Poaceae experienced similar IR expansion into SSC. Therefore, there are two types in IR/SSC junctions in the early evolution of monocots: one is little changed such as the case of date palm and Acoraceae members, which has no obvious expansion to share similar structure with Amborella or tobacco; the other experienced apparent expansion to firstly include the whole duplicates of rps15 and ycf1 in IRb, and thereafter, two possible expansions occur. The first one is the incorporation of the 5′-end of ndhH, resulting in an incomplete copy of ndhH pseudogene in IRb. This expansion happened in Lemna as well as probably among most Poaceaes. The second type is found in Panicoideae subfamily of Poaceae cp genomes and IRb expand into SSC to include the 3′-end of ndhF, accompanying with an incomplete copy of ndhF pseudogene in JSA. After these expansions, ycf1 became nonfunctional and resulted in the present Poaceaes structure (Figure 4).
Figure 4. Expansion of IR into the SSC region.
The dashed lines depict the junction boundaries. The arrowed grey lines indicate the process of IR expansions. a, Amborella, Nicotiana, typha and two Acoraceae members; b, Phalaenopsis; c, Dioscorea; d, date palm and Arabidopsis; e, intermediate; f, Lemna; g, intermediate; h, most Poaceaes; i, Panicoideae subfamily including Coix, Maize, Saccharum and Sorghum.
Gene order in the four junctions has been known to vary among different cp genomes due to the expansion or contraction of the IR regions [8], [19], [28], [29]. The general process of IR changes has also been surveyed for monocots cp genomes [6], [26]. We proposed a most likely evolution rout of IRs and summarized patterns of expansions between junctions of IR/LSC and IR/SSC in comparison to Amborella and tobacco. The basal monocots in our analysis are members of Acoraceae family, whereas the higher monocots belong to Poaceae. However, in the IR/LSC junctions, Lemna has a more contracted IR structure than Acrous and the basal angiosperm— Amborella [6], [22]; and larger expansion occurred in Orchidaceae than in Poaceae. In junctions of IRs/SSC, we observed obvious expansion in Lemna and all Poaceae members, whereas little expansion was noticed in all other monocots, and there is even an IR contraction in Phalaenopsis. Therefore, the expansion rate may not be in accordance with the taxonomic relationship among monocots.
5. Repeat structure and small inversions
Using REPuter, we identified 11 forward and inverted repeats 30 bp or longer with a sequence identity greater than 90% (Table 2). Three pairs of forward repeats were found in the coding region, while seven pairs were identified either in intergenic or intronic regions. The remaining was mostly in the two trnS genes. Compared to other species, the number of repeats in date palm cp genome is fairly low; for example, in Poaceae families, there are 19–37 forward and inverted repeats with the size ranging from 30 bp to 60 bp [30]; and in some dicots, such as Gossypium [31] and Citrus [3], the number of repeats are 54 and 29, respectively. The lengths of repeats in the date palm assembly are also much shorter, and the longest repeat is only 39 bp in length, whereas a much longer 91-bp repeat was found in Poaceae family [30].
Table 2. Repeated sequences in the date palm chloroplast genome.
| Repeat number | Repeat size(bp) | Repeat1 start | Repeat2 start | Repeat type | Repeat1 location | Repeat2 location |
| 1 | 35 | 8334 | 45650 | I | ItrnS-GCU | ItrnS-GGA |
| 2 | 39 | 44165 | 143194 | I | intron (ycf3) | IGS(trnV-GAC - rps12_3end) |
| 3 | 31 | 84427 | 112085 | I | intron (rpl16) | IGS(trnN-GUU - ycf1) |
| 4 | 30 | 126644 | 126677 | I | IGS (rps15 - ycf1) | IGS (rps15 - ycf1) |
| 5 | 30 | 8342 | 35995 | F | trnS-GCU | trnS-UGA |
| 6 | 34 | 39250 | 41474 | F | psaB | psaA |
| 7 | 39 | 44166 | 101428 | F | intron (ycf3) | IGS (rps12_3end - trnV-GAC) |
| 8 | 30 | 69830 | 69863 | F | IGS (rpl33 - rps18) | IGS (rpl33 - rps18) |
| 9 | 30 | 83886 | 120587 | F | intron (rpl16) | IGS (psaC - ndhE) |
| 10 | 31 | 84428 | 132545 | F | intron (rpl16) | IGS (ycf1 - trnN-GUU) |
| 11 | 38 | 93972 | 94020 | F | ycf2 | ycf2 |
partly in the IGS region; I-Inverted, F-forward, IGS-Intergenic spacer region.
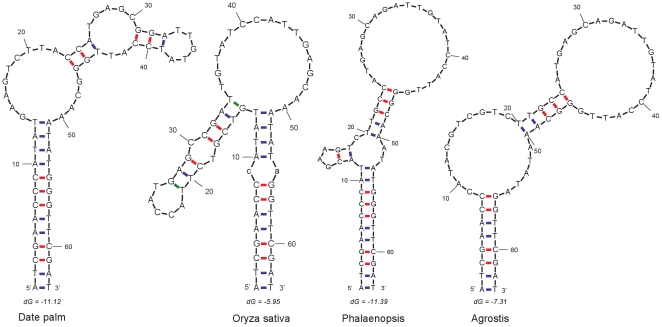
Small inversions or SIs between IRs are quite interesting upon examination. SIs vary in length from 5 to 50 bp, and are flanked by a pair of IRs ranging from 11 to 24 bp in size. SIs can generally be determined through pair-wise comparison between the sequences from closely related taxa. A comprehensive result of 16 SIs has been reported previously for plant chloroplasts [32]. Additional two SIs have been found recently in the intergenic region of psbA-trnH and psbC-trnS [33], [34]. Here, we also identified a new SI in date palm cp genome after performing alignments with its orthologs in other monocots (Table 3). This SI is located in psaB coding region, 63 bp in length, and has the stem and loop being 13 and 37 bp, respectively, in most analyzed cases. The loop regions in the aligned sequences have the same orientation across different taxa and have at least 90% sequence identity to that of data palm. Sequence variations in IR region mostly occurred as a G-to-A single nucleotide polymorphism (SNP), which decrease the stability of the secondary structures. In Lemna and Acrous, there were three SNPs found in the stem region, making the stem-loop structure rather unstable as their free energy values are −1.49 and −1.1, respectively; in tobacco and Panax, numerous variations disrupt the secondary structure seriously. Different loop configurations of the SIs are generated when folding to secondary structure due to their large size and variations in the loop sequences. The alteration is conspicuous in Phalaenopsis and Agrostis, whose loops are even longer (41 and 47 bp, respectively) than most of others (37 bp) due to the variation near the two ends of the loop in the stem region (Figure 5). However, this did not affect the free energy of their secondary structures. Phalaenopsis has almost identical free energy (−11.39) with date palm (−11.12), whereas the free energy of Agrostis (−7.31) is a little lower than that of the Pooideae subfamily (−9.14).
Table 3. The location and sequences of small inversion in psaB.
| Taxa | Genome coordination | aSequence alignment | Free energy (ΔG) |
| Phoenix | 38,667–38,729 | ATCGAACCCATATGAAGTCTTACCATGAGCGGATTGTATCCATTGGGCAAATATGGGTTCGAT | −11.12 |
| bPhalaenopsis | 39,524–39,586 | ATCGAACCCATACGAAGTCTTGCCATGAGCAGATTGTATCCATTGGGCAAATATGGGTTCGAT | −11.39 |
| bOncidium | 37,640–37,702 | ATCGAACCCATACGAAGTCTTGCCATGAGCAGATTGTATCCATTGGGCAAATATGGGTTCGAT | −11.39 |
| Typha | 40,779–40,839 | ATCGAACCCATATGAAGTTTTACCATGAGCAGATTGTATCCATTGGGCAAATATGGGTTCGAT | −10.05 |
| Brachypodium | 37,341–37,403 | ATCGAACCCATATGTCGTCTTGCCATGAGCAGATTGTATCCATTGGGCAAATATaGGTTCGAT | −9.14 |
| Festuca | 37,657–37,719 | ATCGAACCCATATGTCGTCTTGCCATGAGCAGATTGTATCCATTGGGCAAATATaGGTTCGAT | −9.14 |
| Hordeum | 38,178–38,240 | ATCGAACCCATATGTCGTCTTGCCATGAGCAGATTGTATCCATTGGGCAAATATaGGTTCGAT | −9.14 |
| Lolium | 37,293–37,355 | ATCGAACCCATATGTCGTCTTGCCATGAGCAGATTGTATCCATTGGGCAAATATaGGTTCGAT | −9.14 |
| Triticum | 38,014–38,076 | ATCGAACCCATATGTCGTCTTGCCATGAGCAGATTGTATCCATTGGGCAAATATaGGTTCGAT | −9.14 |
| bAgrostis | 37,863–37,925 | ATCGAACCCATACGTCGTCTTGCCATGAGCAGATTGTATCCATTGGGCAAATATAGGTTCGAT | −7.31 |
| Dioscorea | 37,740–37,802 | ATCGAACCCATATGAAGTCTTACCATGAGCAGATTGTATCCATTGGGCAAATATGGGTTCaAT | −7.06 |
| Anomochloa | 39,677–39,739 | ATCGAACCcATATGTCGTCTTACCATGAGCGGATTGTATCCATTGGGCAAATATaGGTTCGAT | −6.12 |
| Oryza.niv | 37,482–37,544 | ATCGAACCCATATGTCGTCTTACCATGAGCCGATTGTATCCATTGAGCAAATATaGGTTCGAT | −5.95 |
| Oryza.sat | 37,548–37,610 | ATCGAACCCATATGTCGTCTTACCATGAGCCGATTGTATCCATTGAGCAAATATaGGTTCGAT | −5.95 |
| Bambusa | 39,617–39,679 | ATCGAACCCATATGTCGTCTTACCATGAGCAGATTGTATCCATTGGGCAAATATaGGTTCGAT | −5.2 |
| Coix | 39,617–39,679 | ATCGAACCCATATGTCGTCTTACCATGAGCAGATTGTATCCATTGGGCAAATATaGGTTCGAT | −5.2 |
| Dendrocalamus | 39,782–39,844 | ATCGAACCCATATGTCGTCTTACCATGAGCAGATTGTATCCATTGGGCAAATATaGGTTCGAT | −5.2 |
| Saccharum | 40,453–40,515 | ATCGAACCCATATGTCGTCTTACCATGAGCAGATTGTATCCATTGGGCAAATATaGGTTCGAT | −5.2 |
| Sorghum | 40,672–40,734 | ATCGAACCCATATGTCGTCTTACCATGAGCAGATTGTATCCATTGGGCAAATATaGGTTCGAT | −5.2 |
| Zea | 39,899–39,961 | ATCGAACCCATATGTCGTCTTACCATGAGCAGATTGTATCCATTGGGCAAATATaGGTTCaAT | −2.06 |
| Lemna | 41,245–41,307 | ATCaAACCCATATGAAGTCTTCCCATGAGCAGATTGTATCCATTGGGCAAATATaGGTTCaAT | −1.49 |
| Acorus.ame | 38,253–38,314 | ATCaAACCCATATAAAGTCTTACCATGAGCGGATTGTATCCACTGGGCAAATATaGGTTCaAT | −1.1 |
| Acorus.cal | 38,245–38,307 | ATCaAACCCATATAAAGTCTTACCATGAGCGGATTGTATCCACTGGGCAAATATaGGTTCaAT | −1.1 |
| cPanax | 40,104–40,166 | ATCGAAaCCATATGAAGTTTTACCATGTGCGGATTGTATCCATTGAGCAAATATaGgTTCGAT | |
| cNicotiana | 39,789–39,851 | ATCGAAaCCATATGAAGTTTTACCATGAGCGGATTGTATCCATTGAGCAAATATaGgTTCaAT |
The small inversions and SNPs in stem are marked with underlined bold capitals and bold lowercases, respectively.
The length of small inversion is different from others.
The sequence can not fold into stem-loop structure.
Figure 5. Folded stem-loop structures of the new SI from four representative species.
Free energy values (dG) are shown for SI in each species.
In addition, we detected 29 SIs when searching for IRs; nine SIs were confirmed and the rest were all putative because we could not find homologous sequences among other monocots (Table S2). They were mostly located in intergenic or intron regions and stem-loop forming. Excluding the previously-mentioned SI in psaB, we found three putative SIs in the coding region; one was known in ccsA [32]. If these putative SIs are proven common in the genome, they may provide phylogenetic information or may even play functional roles in stabilizing their corresponding mRNAs [32], [34].
6. Intravarietal single nucleotide polymorphisms (intraSNPs)
A plant cell often contains multiple clones or copies of cp genomes, and chloroplast can be regarded as a population with high genetic heterogeneity. Therefore, when thousands of high-quality cp sequence reads are aligned, polymorphic sites can be detected readily with software tools and confirmed experimentally. Similar phenomenon is also observed in mitochondrial genomes. The sequence variations within a variety (or cultivar and subspecies), often discovered by high-coverage sequencing, can be separated into major and minor intravarietal genotypes within a chloroplast or mitochondrial genome assembly based on sequence counts. Variations can be further defined based on experimentation over large population sampling from different varieties and subspecies of the same species as intervarietal or intersubspecific genotypes when one of the alleles become unique to certain faction of the samples [35], [36].
High throughput next-generation sequence technologies provide us a great opportunity to collect a huge number of raw reads for surveying intravarietal polymorphism within cp genomes. Using the 369,022 raw 454 cp reads, we surveyed each locus in date palm cp genome for possible intravarietal SNPs. A total of 113 SNPs were originally detected, and after an exclusion of homopolymer runs, the remaining 78 SNPs were considered to be stable SNPs. Among them, 16 fell into intergenic or intronic regions (Table S3) and the remaining were all located in protein-coding regions spreading over 23 genes (Table 4). There are 26 transitions and 52 transversions and more transversions are seen in both non-coding (93.8%) and coding (59.7%) regions. In the CDS regions, 29 and 31 mutations were synonymous and nonsynonymous substitutions, respectively. Major or minor genotypes occurred simultaneously in two SNPs of rpoC2 at positions 21,213 and 21,215 in genome. In addition to these intravarietal SNPs, we also detected a 4-bp intravarietal Indel in intergenic region of accD and psaI from the position 61,482 to 61,485. This Indel, characterized as “TAGA”, however, is defined as a minor genotype, because in the total of 255 original reads (quality value greater than 20) in these four sites, only 75 displayed as “TAGA” insertion.
Table 4. The intravarietal SNPs found in coding region of date palm chloroplast genome.
| NO. | Gene name | Major(minor) SNP type | Major(minor) Codon | Major(minor) amino acid | Major(minor) Reads number | Major(minor) reads percentage | Strand | Position in CDS | Position in genome |
| 1 | psbAa | C(T) | acC(acT) | Thr(Thr) | 863(124) | 87.4(12.6) | − | 858 | 352 |
| 2 | psbAa | G(A) | gGa(gAa) | Gly(Glu) | 863(206) | 80.7(19.3) | − | 728 | 482 |
| 3 | psbAa | A(G) | gcA(gcG) | Ala(Ala) | 1167(296) | 79.8(20.2) | − | 564 | 646 |
| 4 | psbA | T(G) | cTa(cGa) | Leu(Arg) | 1117(250) | 81.7(18.3) | − | 350 | 860 |
| 5 | psbAa | C(T) | tcC(tcT) | Ser(Ser) | 1061(190) | 84.8(15.2) | − | 303 | 907 |
| 6 | atpFa | A(G) | gaA(gaG) | Glu(Glu) | 928(132) | 87.5(12.5) | − | 282 | 12699 |
| 7 | atpI | A(T) | Agt(Tgt) | Ser(Cys) | 774(93) | 89.3(10.7) | − | 559 | 15572 |
| 8 | rps2a | G(A) | Gcg(Acg) | Ala(Thr) | 558(62) | 90.0(10.0) | − | 475 | 16632 |
| 9 | rpoC2 | A(C) | aaA(aaC) | Lys(Asn) | 564(211) | 72.8(27.2) | − | 711 | 20742 |
| 10 | rpoC2 | C(A) | caC(caA) | His(Gln) | 988(202) | 83.0(17.0) | − | 636 | 20817 |
| 11 | rpoC2a | T(C) | caT(caC) | His(His) | 898(134) | 87.0(13.0) | − | 432 | 21021 |
| 12 | rpoC2 | A(C) | Aat(Cat) | Asn(His) | 828(101) | 89.1(10.9) | − | 352 | 21101 |
| 13 | rpoC2 | T(G) | tTa(tGa) | Leu(−) | 789(99) | 88.9(11.1) | − | 290 | 21163 |
| 14 | rpoC2 | T(C) | Ttt(GtC) | Phe(Val) | 398(94) | 80.9(19.1) | − | 240 | 21213 |
| 15 | rpoC2 | T(G) | 238 | 21215 | |||||
| 16 | rpoC1a | A(G) | aAt(aGt) | Asn(Ser) | 624(231) | 73.0(27.0) | − | 1931 | 21750 |
| 17 | rpoC1 | A(C) | ttA(ttC) | Leu(Phe) | 938(235) | 80.0(20.0) | − | 1503 | 22178 |
| 18 | rpoC1 | A(C) | ttA(ttC) | Leu(Phe) | 702(225) | 75.7(24.3) | − | 1230 | 22451 |
| 19 | rpoC1a | A(G) | ctA(ctG) | Leu(Leu) | 952(223) | 81.0(19.0) | − | 1113 | 22568 |
| 20 | rpoC1a | G(A) | gaG(gaA) | Glu(Glu) | 762(181) | 80.8(19.2) | − | 1092 | 22589 |
| 21 | rpoC1a | T(C) | ctT(ctC) | Leu(Leu) | 674(231) | 74.5(25.5) | − | 885 | 22796 |
| 22 | rpoC1 | T(G) | ttT(ttG) | Phe(Leu) | 256(53) | 82.8(17.2) | − | 537 | 23144 |
| 23 | rpoC1 | A(C) | Aga(Cga) | Arg(Arg) | 529(171) | 75.6(24.4) | − | 199 | 24221 |
| 24 | rpoB | T(G) | tTt(tGt) | Phe(Cys) | 661(165) | 80.0(20.0) | − | 2744 | 24909 |
| 25 | rpoBa | C(T) | tgC(tgT) | Cys(Cys) | 748(137) | 84.5(15.5) | − | 2601 | 25052 |
| 26 | rpoB | A(C) | tcA(tcC) | Ser(Ser) | 319(114) | 73.7(26.3) | − | 2487 | 25166 |
| 27 | rpoB | G(T) | Gat(Tat) | Asp(Tyr) | 963(132) | 87.9(12.1) | − | 2035 | 25618 |
| 28 | psaBa | T(C) | gaT(gaC) | Asp(Asp) | 789(106) | 88.2(11.8) | − | 1092 | 39000 |
| 29 | psaBa | G(T) | ggG(ggT) | Gly(Gly) | 874(100) | 89.7(10.3) | − | 864 | 39228 |
| 30 | psaB | G(T) | Gat(Tat) | Asp(Tyr) | 868(106) | 89.1(10.9) | − | 814 | 39278 |
| 31 | psaBa | A(G) | acA(acG) | Thr(Thr) | 647(87) | 88.1(11.9) | − | 795 | 39297 |
| 32 | psaBa | C(T) | agC(agT) | Ser(Ser) | 861(109) | 88.8(11.2) | − | 720 | 39372 |
| 33 | psaA | A(C) | ttA(ttC) | Leu(Phe) | 521(210) | 71.3(28.7) | − | 2118 | 40252 |
| 34 | psaA | A(C) | tcA(tcC) | Ser(Ser) | 858(145) | 85.5(14.5) | − | 1800 | 40570 |
| 35 | rps4a | A(C) | atA(atC) | Ile(Ile) | 836(132) | 86.4(15.6) | − | 282 | 46306 |
| 36 | ndhJa | T(C) | ttT(ttC) | Phe(Phe) | 407(113) | 78.3(21.7) | − | 456 | 49697 |
| 37 | atpB | A(C) | gtA(gtC) | Val(Val) | 1041(257) | 80.2(19.8) | − | 696 | 55445 |
| 38 | accD | G(A) | gaG(gaA) | Glu(Glu) | 708(88) | 88.9(11.1) | + | 48 | 59201 |
| 39 | accD | C(T) | tcC(tcT) | Ser(Ser) | 680(94) | 87.9(12.1) | + | 276 | 59429 |
| 40 | accD | C(A) | Cat(Aat) | His(Asn) | 863(108) | 88.9(11.1) | + | 403 | 59556 |
| 41 | accD | T(G) | Tac(Gac) | Tyr(Asp) | 852(106) | 88.9(11.1) | + | 406 | 59559 |
| 42 | accD | T(G) | ttT(ttG) | Phe(Leu) | 618(106) | 85.4(14.6) | + | 987 | 60140 |
| 43 | accD | G(A) | gGa(gAa) | Gly(Glu) | 877(114) | 88.5(11.5) | + | 1097 | 60250 |
| 44 | accD | C(T) | tcC(tcT) | Ser(Ser) | 848(102) | 89.3(10.7) | + | 1152 | 60305 |
| 45 | accD | A(C) | Aaa(Caa) | Lys(Gln) | 354(102) | 77.6(22.4) | + | 1177 | 60330 |
| 46 | cemA | G(C) | gtG(gtC) | Val(Val) | 1181(175) | 87.1(12.9) | + | 672 | 63407 |
| 47 | petA | A(G) | gcA(gcG) | Ala(Ala) | 844(113) | 88.2(11.8) | + | 828 | 64480 |
| 48 | psbEa | G(A) | ccG(ccA) | Pro(Pro) | 951(130) | 88.0(12.0) | − | 192 | 66208 |
| 49 | psbB | A(C) | ttA(ttC) | Leu(Phe) | 640(184) | 77.7(22.3) | + | 927 | 75429 |
| 50 | psbT | A(C) | atA(atC) | Ile(Ile) | 695(153) | 82.0(18.0) | + | 48 | 76250 |
| 51 | psbT | A(C) | Aaa(Caa) | Lys(Gln) | 233(151) | 60.7(39.3) | + | 100 | 76302 |
| 52 | psbH | A(G) | acA(acG) | Thr(Thr) | 782(115) | 87.2(12.8) | + | 120 | 76734 |
| 53 | psbH | G(A) | ttG(ttA) | Leu(Leu) | 651(89) | 88.0(12.0) | + | 174 | 76788 |
| 54 | psbH | T(G) | atT(atG) | Ile(Met) | 468(85) | 84.6(15.4) | + | 180 | 76794 |
| 55 | psbH | T(G) | Tat(Gat) | Tyr(Asp) | 469(85) | 84.7(15.3) | + | 181 | 76795 |
| 56 | petB | T(A) | aTa(aAa) | Ile(Lys) | 715(108) | 86.9(13.1) | + | 101 | 77843 |
| 57 | petB | T(G) | Tat(Gat) | Tyr(Asp) | 520(123) | 80.9(19.1) | + | 106 | 77848 |
| 58 | petB | T(G) | Tac(Gac) | Tyr(Asp) | 493(152) | 76.4(23.6) | + | 175 | 77917 |
| 59 | petB | A(C) | gtA(gtC) | Val(Val) | 576(219) | 72.5(27.5) | + | 450 | 78192 |
| 60 | rps11 | A(C) | Att(Ctt) | Ile(Leu) | 1007(120) | 89.4(10.6) | − | 118 | 81436 |
| 61 | ycf2 | T(G) | ttT(ttG) | Phe(Leu) | 917(121) | 88.3(11.7) | + | 3765 | 92696 |
| 62 | ndhA | T(G) | tTa(tGa) | Leu(−) | 793(100) | 88.8(11.2) | − | 1010 | 122904 |
the minor genotype in date palm is major genotype in oil palm, or vice versa.
In general, SNPs in cp genome occurred at a rate of 1 in 1,400 bases. However, the 78 SNPs were not randomly distributed across the whole genome—a majority of these SNPs were clustered in LSC region. In IRs, we only found one SNP in ycf2, harboring a T–G mutation at position 92,696. In SSC region, we detected only two SNP sites: one in an intergenic region at 122,758 (T–G mutation) and the other in ndhA is similar to one locus in rpoC2 (genome position 21163) with T–G mutation causing leucine (TTA) to a stop codon (TGA). The percentage of both minor genotypes in these two genes was about 11% with around 100 original supporting reads. These mutations occurred at 5′-end of rpoC2 and 3′-end of ndhA could terminate the transcription prematurely and therefore make these two genes nonfunctional.
The intraSNPs in LSC region could be classified primarily into four functional gene categories: transcription (ropB, rpoC1, and rpoC2), photosynthesis (psa and psb proteins), energy metabolism (atp, pet and ndh proteins), and translation (rps proteins). Since chloroplasts can be considered a population within the cell, major-allelic variations that change amino acid sequences and translation efficiencies may affect or regulate gene functions. Furthermore, it has been proven in rice that the intravarietal major genotypes in one subspecies are often the minor genotypes in other subspecies, or vice versa [36]. Since subspecies among date palm cultivars are not yet well-characterized, we used the 81 available CDSs in oil palm cp genome for a brief comparative analysis [22]. We have adopted a procedure to avoid read contamination from nuclear or mitochondrial genomes and sequence artifacts (Materials and Methods); therefore, these intravarietal variations are unquestionably characteristic of the Khalas cultivar. Although date palm and oil palm belong to two different genera, we indeed found 19 sites, whose cp minor genotypes in date palm became major genotypes in oil palm, or vice versa (Table 4). This result confirms that some of these genotypes (major in one and minor in another) between the two species are actually real and these intravarietal cp genome variations may also be proven as real intervarietal variations among date palm cultivars if a large survey is to be carried out in the future.
Our study is the first one to quantify intravarietal polymorphisms using the next-generation sequencing technology. A recent analysis has reported a similar discovery of 40 SNP sites in Loloum perenne [37]. The intravarietal polymorphisms are indicators for the heterogeneous nature of chloroplast population in a given species and we have demonstrated in rice that the minor genotypes are also chloroplast in origin [36]. Since the effort of classifying date palm cultivars is early in its way, these intravarietal polymorphisms provide us useful markers for the evaluation of different subspecies. We are definitely able to validate some of these variations in the future genetic studies of date palm.
7. Gene expression analysis based on transcriptome data
We generated transcriptomic data, including 1,076,222, 833,875, and 465,456 raw reads for leaf, root, and bud, respectively (date not shown), for the cp transcriptome analysis. The genes identified from leaf were most abundant: 19,052 and 306,154 for protein-coding and RNA genes, respectively. Most of the genes (99) transcribed in at least two tissues, although we failed to detect transcripts for four genes (trnQ-UUG, rpl32, ndhG, and ndhE) in any tissues due to possible sampling biases (see discussion below). More than half of the 112 unique genes were rather scarcely detected in buds. The transcription-translation apparatus of chloroplasts has a number of prokaryote-like features; for instance, cp genes are often co-transcribed [38]. In our assembly, we found 18 polycistronic transcripts including genes for 63 proteins, 5 tRNAs, and all rRNAs (Table 5).
Table 5. Expression information of date palm cp genome in leaf, root, and bud.
| Gene | Strand | Leaf | Root | Bud | Gene | Strand | Leaf | Root | Bud |
| psbA | N | 44(100) | - | 2(71.1) | apsaJ | P | 76(100) | 16(100) | 1(88.9) |
| matK | N | 107(100) | 57(100) | 2(100) | arpl33 | P | 41(100) | 24(100) | - |
| rps16 | N | 122(100) | 120(100) | 105(100) | arps18 | P | 10(100) | 2(100) | - |
| apsbK | P | 6(100) | - | - | brpl20 | N | 35(100) | 34(100) | - |
| apsbI | P | 13(100) | - | - | brps12_5end | N | 20(100) | 15(100) | 2(100) |
| batpA | N | 143(100) | 28(100) | 6(100) | bclpP | N | 151(100) | 174(100) | 18(100) |
| batpF | N | 15121(100) | 3030(100) | 91(100) | apsbB | P | 34(100) | 5(100) | 6(100) |
| batpH | N | 96(100) | 20(100) | 7(100) | apsbT | P | 14(100) | - | - |
| batpI | N | 67(100) | 10(100) | 6(100) | apsbN | N | 37(100) | - | - |
| brps2 | N | 14(100) | 6(100) | 2(64.7) | apsbH | P | 73(100) | 2(100) | - |
| arpoC2 | N | 30(100) | 29(99.5) | 4(91.1) | apetB | P | 19(100) | 2(77.5) | 2(62.4) |
| arpoC1 | N | 84(96.5) | 63(100) | 9(98.3) | apetD | P | 12(100) | 5(98.7) | - |
| arpoB | N | 59(100) | 47(100) | 5(64) | brpoA | N | 61(100) | 21(100) | 3(100) |
| petN | P | 5(100) | 5(100) | 2(100) | brps11 | N | 121(100) | 15(100) | - |
| psbM | N | 5(100) | 2(100) | 2(100) | brpl36 | N | 8(100) | 2(100) | 1(100) |
| bpsbD | P | 165(100) | 5(72.5) | 2(90.7) | binfA | N | 30(100) | 10(100) | - |
| bpsbC | P | 140(100) | 5(78.2) | - | brps8 | N | 24(100) | 8(100) | - |
| bpsbZ | P | 233(100) | 19(100) | 2(100) | brpl14 | N | 43(100) | 30(100) | 3(100) |
| arps14 | N | 39(100) | - | - | brpl16 | N | 14(100) | 8(97.8) | 3(100) |
| apsaB | N | 150(100) | 16(100) | - | brps3 | N | 40(100) | 24(100) | 4(100) |
| apsaA | N | 29(100) | 7(100) | 2(68.7) | brpl22 | N | 40(100) | 18(100) | 4(100) |
| ycf3 | N | 268(100) | 177(100) | 4(78.9) | brps19 | N | 19(90.3) | 8(90.3) | 5(90.3) |
| rps4 | N | 5(84.8) | 1(84.7) | - | brpl2 | N | 238(100) | 122(100) | 7(100) |
| bndhJ | N | 19(100) | 8(100) | 3(100) | brpl23 | N | 19(100) | 3(97.9) | 1(100) |
| bndhK | N | 9(100) | 4(97.7) | 2(74.9) | ycf2 | P/N | 17(100) | 35(84.3) | 16(97.8) |
| bndhC | N | 13(100) | 4(100) | - | ndhB | N/P | 25(88.5) | 14(70.7) | - |
| aatpE | N | 144(100) | 20(100) | 4(100) | arps7 | N/P | 317(100) | 85(100) | 11(100) |
| aatpB | N | 114(81.7) | 14(87.4) | 7(100) | arps12_3end | N/P | 126(100) | 32(100) | 9(100) |
| rbcL | P | 158(100) | 12(100) | 9(95.4) | ndhF | N | 2(66.1) | 5(60.9) | - |
| accD | P | 25(100) | 47(100) | 12(99.9) | rpl32 | P | - | - | - |
| bpsaI | P | 1(82) | 1(82) | - | ccsA | P | 2(98) | 4(97.7) | 2(91.5) |
| bycf4 | P | 64(100) | 12(100) | 3(100) | bndhD | N | 17(72.5) | - | 5(100) |
| bcemA | P | 7(97.6) | 7(97.6) | - | bpsaC | N | 5(100) | 3(100) | - |
| bpetA | P | 50(100) | 17(100) | 2(50.8) | ndhE | N | - | - | - |
| apsbJ | N | 8(100) | 3(100) | 1(97.6) | ndhG | N | - | - | - |
| apsbL | N | 10(100) | 4(100) | 1(100) | andhI | N | 8(100) | 4(98.7) | - |
| apsbF | N | 8(100) | 1(100) | - | andhA | N | 21(100) | 6(100) | - |
| apsbE | N | 11(100) | 1(100) | - | andhH | N | 18(100) | 6(100) | - |
| bpetL | P | 1(76) | 1(100) | - | arps15 | N | 4(87.9) | 1(95.2) | - |
| bpetG | P | 7(100) | 3(100) | - | aycf1 | N | 33(100) | 32(100) | 14(100) |
1. the numbers outside the parenthesis mean the number of original reads and inside mean the percentage of the gene covered by all related reads (-, no supporting reads).
2. Each continuous bold and italics “a” or “b” superscript at the outset of genes means one polycistronic transcription units.
3. N, negative strand; P, positive strand.
Since the cDNA identification for chloroplast genome is not as quantitative as it should be due to the lack of polyA tail that provide a handle for mRNA purification, the interpretation of cp gene transcription is rather qualitative. What we observed here is the transcripts whose RNAs are co-purified with mRNA preparation, similar to rRNAs, as they both are so abundant. It is also the unbiased co-purification that provides us useful information about chloroplast transcripts. We indeed observed interesting expressions of the date palm cp transcripts. First, one protein-coding gene—atpF—have 15,121, 3,030, and 91 corresponding cDNA reads, accounting for 79.4%, 66.8%, and 21.9% of the total cp related reads in leaf, root, and bud, respectively. Although the fractions may reflect the real proportions of the gene abundance in three different tissues, its unusual abundance is obvious by examining the assembly—its sequence has a polyA-like track that is 22 bp long and situated at the 57-bp upstream of atpF gene. Since our reverse transcription was primed by using a polyT adaptor, atpF transcripts are thus enriched in the cDNA libraries. However, due to the prokaryote-like features of chloroplast genome, there is no obvious polyA structure at the downstream of other genes. Furthermore, we found that polycistronic transcript rps2-atpI-atpH-atpF-atpA-trnR-trnG had only 50 transcripts containing atpF and atpA and it is obvious a result of enrichment related to the polyA-like track (Figure S2). In fact, other than atpF, the fraction of total transcripts with adaptor sequences account for about 72% of all CDS reads, and it indicated that our transcripts for CDS are indeed non-specifically co-purified with polyA mRNAs rather than enriched due to polyT affinity purification. For example, in the downstream of atpA, there is an 11-bp (10,718-10,728 in genome) rather degenerated polyA unit, whereas only 23 reads with adaptors for atpA were detected. Besides, the extremely high number of atpF transcripts suggested that this gene may not be simply co-transcribed with other genes as a polycistronic transcript but may also be transcribed independently. Second, there are two rRNA genes—rrn23 and rrn16—also display high abundance. In leaf, the number for rrn23 and rrn16 are 280,106 and 28,721, respectively (Table 5). There is no polyA structure downstream of these RNA genes, and therefore their transcripts may be just co-purified with mRNA but such enrichment may reflect their real abundance in leaf. Third, the number of trnA-UGC transcript is also special, accounting for 90.9% of all tRNA transcripts. It is curious why this gene had so high a copy number in the cell, since alanine only accounts for 5.32% amino acids in date palm cp genome. The reason remains to be illuminated.
In summary, we present here the first complete cp genome from Arecaceae family obtained based on pyrosequencing. The genome is A+T rich and we had to design primers to verify the most of the homopolymer runs. As a rather regular cp genome, its gene content and structure are similar to those of tobacco, and its IRs expansion occurred in LSC but not SSC regions. Its number of repeated sequences appeared lower than that of Poaceae cp genomes and certain dicots. The high coverage of raw cp reads demonstrated that the intravarietal SNPs exist not only in intergenic region, but also in coding genes, where these variations may have been functionally selected. Some of the major intervarietal SNPs may serve as genetic markers for classifying subspecies when appropriately developed as simple genotyping assays. Most protein-coding genes of cp genome are transcribed as polycistronic transcripts and transcriptome analysis is to be designed in particular to avoid cross-contamination between the nuclear poyA-containing and organellar polyA-lacking transcripts. Our data are useful for the study of date palm biology and pave a way for further molecular investigations.
Materials and Methods
DNA sequencing, genome assembly, and PCR-based assembly validation
We collected fresh green leaves from an adult plant of Khalas (a common cultivar of date palm) grown in Al-Hass Oasis, Kingdom of Saudi Arabia, and extracted genomic DNA from 50 g leaves according to a CTAB-based protocol [39]. We used 5 µg purified DNA for constructing libraries according to the manufacturer's manual for GS FLX Titanium.
Since our original sequence reads are a mixture of DNA from nucleus and organelles, we separated the cp reads from the total reads based on the known cp genome sequences. We assembled the filtered sequence reads into non-redundant contigs using Newbler, a de novo sequence assembly software, and aligned the contigs to the references, including those of tobacco [5], rice [40], orchid [8], and the coding sequences from oil palm [22]. We obtained 42 cp genome contigs with a collective length of 115 kb. Using a perl script, we made an iterative elongation for both ends of each contig against the total raw reads until the 42 contigs emerged as preliminary cp sequence scaffolds.
To avoid assembly errors from homopolymer runs (characteristics of the pyrosequencing technology) and to acquire a high-quality complete cp genome sequence, we designed 151 pairs of primers covering the preliminary cp genome assembly. Based on a published procedure [41], we sequenced PCR products using BigDyeV3.1 Terminator Kit for 3730XL (Life Technologies) and assembled the high-quality sequences into the complete cp genome using the Phred-Phrap-Consed package [42], [43]. The four junctions between the single-copy segments and inverted repeats were also confirmed based on PCR product sequencing. The final date palm chloroplast sequence has been deposited to GenBank (accession number GU811709).
Genome features, gene content and codon usage
The genome was annotated by using DOGMA [44], coupled with manual corrections for start and stop codons. Intron positions were determined based on those of the tobacco cp genome [20]. The transfer RNA genes were identified by using DOGMA and tRNAscan-SE (version 1.23) [45]. Certain intron-containing genes (i.e., trnK-UUU, petB, and petD), in which exons are too short to be detected with software tools, were identified based on comparisons to corresponding exons in tobacco and other cp genomes. The functional classification of cp genes was referred to CpBase (http://chloroplast.ocean.washington.edu/). The cemA sequences of various species were downloaded from Genbank and aligned by using MUSCLE [46]
Repeat structure and small inversion
We used REPuter [47] to assess both direct (forward) and inverted (palindrome) repeats within the date palm cp assembly. The identity and the size of the repeats were limited to no less than 90% (hamming distance equal to 3) and 30 bp in unit length, respectively. Verification of the identified repeats was performed manually based on intragenomic comparisons.
To obtain possible small inversions, we first searched IRs in the length from 11 to 24 bp with REPtuter, and then collected those repeats whose distance is shorter than 50 bp as candidate SIs. Next, we evaluated the likely secondary structure of these SIs using MFOLD (version3.2) [48], and discarded those do not form obvious step-loop structure. Finally, we run Blast program using the remaining putative SIs against all monocot cp genomes, and only collected those qualified hits into the final SI list.
Intravarietal single nucleotide polymorphisms
We used the total 369,022 cp reads for intravarietal SNP discovery. Based on the Consed graphical interface, we were able to identify genome loci that have more than one nucleotide type as candidates for intravarietal SNPs and to assign major and minor alleles. We only considered those loci with nucleotide sequencing quality value greater than 20.
To make our results more convincing and to eliminate false positives caused by contamination from nuclear and mitochondrial sequences, we adopted the following measures: (1) the number of aligned reads for each selected locus must be above 50; (2) the percentage of the maximal minor genotype (if there are two or more minor genotypes) must be above 10%; (3) if there were gaps in the reads of one aligned locus, we did not count (it may be a one-basepair Indel), and after removing these reads, the number of the remaining reads must be more than 50; and (4) we excluded the sites overlapping with homopolymer runs. These errors often occurred at two ends of a homopolymer run due to high coverage of the original reads; for example, a sequence “AAAAAAAAAC” often displays a minor genotype for C in position 10, but this C is often an artifact due to sequence errors intrinsic to pyrosequencing.
Transcriptome analysis
We purified total RNAs from leaf, root, and flower bud of cultivar Khalas and sequenced their cDNAs using Roche GS FLX system according to the manufacture's protocol. The total RNA was reverse-transcribed by using an oligo-dT adaptor. The adaptor sequences are:
5′-ATTCTAGAGACCGAGGCGGCCGACATGTTTTGTCTTTTGTTCTGTTTCTTTT-3′
After reverse-transcription, the cDNAs were sheared into small fragment to construct sequencing libraries followed by emulsion PCR and sequencing. We mapped all raw reads to the date palm cp genome assembly using both Newbler and Blast programs. We acquired non-redundant contigs and singlets by assembling the mapped reads, and collected only those with over 80% coverage and 99% identity of a gene. If different genes or a single gene are assembled into one contig or singlet, we assign the genes as polycistronic or monocistronic transcription unit, respectively. The poly- or monocistronic transcripts were estimated using the assembling results from leaf where chloroplasts are most abundant. These transcription units are carefully inspected manually using Consed graphical interface to avoid assembling mistakes and interrupting contigs by intron-containing genes.
Supporting Information
A whole list of all pairs of primers used in date palm chloroplast
(1.08 MB PDF)
The location and sequences of all putative small inversions in date palm chloroplast genome
(0.01 MB PDF)
The intravarietal SNPs found in non-coding regions
(0.01 MB PDF)
Multiple alignments of cemA genes from 77 different cp genomes.
(0.02 MB PDF)
Partial 454 transcription reads alignments with date palm cp genome at downstream of atpF and upstream of atpA. Comments are marked with thick blue line and blue color fonts.
(0.25 MB TIF)
Acknowledgments
We thank Dr. Yongjun Fang as well as Mr. Xiaoguang Yu and Guangyu Zhang for their useful suggestions and excellent technical supports in sequencing experiments.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The project is supported by a project grant from KACST 428-29, King Abdulaziz city for science and technology, Riyadh, Saudi Arabia. The funder had no role in study design and analysis.
References
- 1.Howe CJ, Barbrook AC, Koumandou VL, Nisbet RE, Symington HA, et al. Evolution of the chloroplast genome. Philos Trans R Soc Lond B Biol Sci. 2003;358:99–106; discussion 106-107. doi: 10.1098/rstb.2002.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pyke KA. Plastid division and development. Plant Cell. 1999;11:549–556. doi: 10.1105/tpc.11.4.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bausher MG, Singh ND, Lee SB, Jansen RK, Daniell H. The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var ‘Ridge Pineapple’: organization and phylogenetic relationships to other angiosperms. BMC Plant Biol. 2006;6:21. doi: 10.1186/1471-2229-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chumley TW, Palmer JD, Mower JP, Fourcade HM, Calie PJ, et al. The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 2006;23:2175–2190. doi: 10.1093/molbev/msl089. [DOI] [PubMed] [Google Scholar]
- 5.Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, et al. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mardanov AV, Ravin NV, Kuznetsov BB, Samigullin TH, Antonov AS, et al. Complete sequence of the duckweed (Lemna minor) chloroplast genome: structural organization and phylogenetic relationships to other angiosperms. J Mol Evol. 2008;66:555–564. doi: 10.1007/s00239-008-9091-7. [DOI] [PubMed] [Google Scholar]
- 7.Hansen DR, Dastidar SG, Cai Z, Penaflor C, Kuehl JV, et al. Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms: Buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae). Mol Phylogenet Evol. 2007;45:547–563. doi: 10.1016/j.ympev.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Chang CC, Lin HC, Lin IP, Chow TY, Chen HH, et al. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol Biol Evol. 2006;23:279–291. doi: 10.1093/molbev/msj029. [DOI] [PubMed] [Google Scholar]
- 9.Sghaier-Hammami B, Valledor L, Drira N, Jorrin-Novo JV. Proteomic analysis of the development and germination of date palm (Phoenix dactylifera L.) zygotic embryos. Proteomics. 2009;9:2543–2554. doi: 10.1002/pmic.200800523. [DOI] [PubMed] [Google Scholar]
- 10.Al-Mssallem IS. Date palm. Arabian Global Encyclopedia. 1996;7:182–187. [Google Scholar]
- 11.Caroebter J.B RCL. Date palm breeding, a review. Date Grower's Inst Rep. 1976;53 [Google Scholar]
- 12.Djamila Chabane AA, Bouguedoura Nadia, Haûcour Robert, Ducreux Georges. Induction of callus formation from difficile date palm protoplasts by means of nurse culture. C R Biologies. 2007;330 doi: 10.1016/j.crvi.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 13.CORRIVEAU JL, W.COLEMAN A. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. Amer J Bot. 1988;75:1443–1458. [Google Scholar]
- 14.Jansen RK, Raubeson LA, Boore JL, dePamphilis CW, Chumley TW, et al. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol. 2005;395:348–384. doi: 10.1016/S0076-6879(05)95020-9. [DOI] [PubMed] [Google Scholar]
- 15.Tangphatsornruang S, Sangsrakru D, Chanprasert J, Uthaipaisanwong P, Yoocha T, et al. The chloroplast genome sequence of mungbean (Vigna radiata) determined by high-throughput pyrosequencing: structural organization and phylogenetic relationships. DNA Res. 2010;17:11–22. doi: 10.1093/dnares/dsp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cronn R, Liston A, Parks M, Gernandt DS, Shen R, et al. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res. 2008;36:e122. doi: 10.1093/nar/gkn502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore MJ, Dhingra A, Soltis PS, Shaw R, Farmerie WG, et al. Rapid and accurate pyrosequencing of angiosperm plastid genomes. BMC Plant Biol. 2006;6:17. doi: 10.1186/1471-2229-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao L, Yi X, Yang YX, Su YJ, Wang T. Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: insights into evolutionary changes in fern chloroplast genomes. BMC Evol Biol. 2009;9:130. doi: 10.1186/1471-2148-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raubeson LA, Peery R, Chumley TW, Dziubek C, Fourcade HM, et al. Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics. 2007;8:174. doi: 10.1186/1471-2164-8-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugita* M, Sugiura M. Regulation of gene expression in chloroplast of higher plants. Plant Mol Biol. 1996;32:315–326. doi: 10.1007/BF00039388. [DOI] [PubMed] [Google Scholar]
- 21.Sasaki Y, Sekiguchi K, Nagano Y, Matsuno R. Chloroplast envelope protein encoded by chloroplast genome. FEBS Lett. 1993;316:93–98. doi: 10.1016/0014-5793(93)81743-j. [DOI] [PubMed] [Google Scholar]
- 22.Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doyle JJ, Davis JI, Soreng RJ, Garvin D, Anderson MJ. Chloroplast DNA inversions and the origin of the grass family (Poaceae). Proc Natl Acad Sci U S A. 1992;89:7722–7726. doi: 10.1073/pnas.89.16.7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goremykin VV, Holland B, Hirsch-Ernst KI, Hellwig FH. Analysis of Acorus calamus Chloroplast Genome and Its Phylogenetic Implications. Mol Biol Evol. 2005;22:1813–1822. doi: 10.1093/molbev/msi173. [DOI] [PubMed] [Google Scholar]
- 25.Maier RM, Neckermann K, Igloi GL, Kossel H. Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J Mol Biol. 1995;251:614–628. doi: 10.1006/jmbi.1995.0460. [DOI] [PubMed] [Google Scholar]
- 26.Guisinger MaryM, Chumley TW, Kuehl JenniferV, Boore JeffreyL, Jansen RobertK. Implications of the Plastid Genome Sequence of Typha(Typhaceae, Poales) for Understanding Genome Evolution in Poaceae. J Mol Evol. 2010;70:149–166. doi: 10.1007/s00239-009-9317-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu FH, Chan MT, Liao DC, Hsu CT, Lee YW, et al. Complete chloroplast genome of Oncidium Gower Ramsey and evaluation of molecular markers for identification and breeding in Oncidiinae. BMC Plant Biol. 2010;10:68. doi: 10.1186/1471-2229-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim KJ, Lee HL. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004;11:247–261. doi: 10.1093/dnares/11.4.247. [DOI] [PubMed] [Google Scholar]
- 29.Saski C, Lee SB, Daniell H, Wood TC, Tomkins J, et al. Complete chloroplast genome sequence of Gycine max and comparative analyses with other legume genomes. Plant Mol Biol. 2005;59:309–322. doi: 10.1007/s11103-005-8882-0. [DOI] [PubMed] [Google Scholar]
- 30.Saski C, Lee SB, Fjellheim S, Guda C, Jansen RK, et al. Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Theor Appl Genet. 2007;115:571–590. doi: 10.1007/s00122-007-0567-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee SB, Kaittanis C, Jansen RK, Hostetler JB, Tallon LJ, et al. The complete chloroplast genome sequence of Gossypium hirsutum: organization and phylogenetic relationships to other angiosperms. BMC Genomics. 2006;7:61. doi: 10.1186/1471-2164-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim KJ, Lee HL. Widespread occurrence of small inversions in the chloroplast genomes of land plants. Mol Cells. 2005;19:104–113. [PubMed] [Google Scholar]
- 33.Catalano SAs, Saidman BO, Vilardi JCs. Evolution of small inversions in chloroplast genome: a case study from a recurrent inversion in angiosperms. Cladistics. 2008;25:93–104. doi: 10.1111/j.1096-0031.2008.00236.x. [DOI] [PubMed] [Google Scholar]
- 34.BaRK JansenJF. A chloroplast DNA hairpin structure provides useful phylogenetic data within tribe Senecioneae (Asteraceae). Can J Bot. 2006;84:862–868. [Google Scholar]
- 35.Tian X, Zheng J, Hu S, Yu J. The rice mitochondrial genomes and their variations. Plant Physiol. 2006;140:401–410. doi: 10.1104/pp.105.070060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang J, Xia H, Cao M, Zhang X, Zeng W, et al. A comparison of rice chloroplast genomes. Plant Physiol. 2004;135:412–420. doi: 10.1104/pp.103.031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diekmann K, Hodkinson TR, Wolfe KH, van den Bekerom R, Dix PJ, et al. Complete chloroplast genome sequence of a major allogamous forage species, perennial ryegrass (Lolium perenne L.). DNA Res. 2009;16:165–176. doi: 10.1093/dnares/dsp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugita M, Sugiura M. Regulation of gene expression in chloroplast of higher plants. Plant Mol Biol. 1996;32:315–326. doi: 10.1007/BF00039388. [DOI] [PubMed] [Google Scholar]
- 39.N.J. Gawel RLJ. A Modified CTAB DNA Extraction Procedure for Musa and Ipomoea. Plant Molecular Biology Reporter. 1991;9:262–266. [Google Scholar]
- 40.Hiratsuka J, Shimada H, Whittier R, Ishibashi T, Sakamoto M, et al. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet. 1989;217:185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- 41.S. Tangphatsornruang* DS, Chanprasert J, Uthaipaisanwong P, Yoocha T, Jomchai N, et al. The Chloroplast Genome Sequence of Mungbean (Vigna radiata) Determined by High-throughput Pyrosequencing: Structural Organization and Phylogenetic Relationships. DNA Res. 2009. pp. 1–12. [DOI] [PMC free article] [PubMed]
- 42.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 43.Ewing B GP. Base-calling of automated sequencer traces using phred. II, Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 44.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 45.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, et al. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A whole list of all pairs of primers used in date palm chloroplast
(1.08 MB PDF)
The location and sequences of all putative small inversions in date palm chloroplast genome
(0.01 MB PDF)
The intravarietal SNPs found in non-coding regions
(0.01 MB PDF)
Multiple alignments of cemA genes from 77 different cp genomes.
(0.02 MB PDF)
Partial 454 transcription reads alignments with date palm cp genome at downstream of atpF and upstream of atpA. Comments are marked with thick blue line and blue color fonts.
(0.25 MB TIF)