Abstract
The versatile coiled-coil protein motif is widely used to induce and control macromolecular interactions in biology and materials science. Yet the types of interaction patterns that can be constructed using known coiled coils are limited. Here we greatly expand the coiled-coil toolkit by measuring the complete pair-wise interactions of 48 synthetic coiled coils and 7 human bZIP coiled coils using peptide microarrays. The resulting 55-member protein ‘interactome’ includes 27 pairs of interacting peptides that preferentially hetero-associate. The 27 pairs can be used in combinations to assemble sets of 3 to 6 proteins that compose networks of varying topologies. Of special interest are heterospecific peptide pairs that participate in mutually orthogonal interactions. Such pairs provide the opportunity to dimerize two separate molecular systems without undesired crosstalk. Solution and structural characterization of two such sets of orthogonal heterodimers provide details of their interaction geometries. The orthogonal pair, along with the many other network motifs discovered in our screen, provide new capabilities for synthetic biology and other applications.
The coiled coil is a fundamental building block for molecular engineering. Its simple structure, which consists of two or more alpha helices twisted into a supercoiled rod-like bundle, is encoded in a seven amino-acid repeat designated [abcdefg]n. Coiled coils have been used to induce and stabilize protein oligomers, to promote protein-protein interactions, to rewire cellular networks, to assemble functional scaffolds, to construct hydrogel materials, and to self-assemble nano-scale fibers and/or recruit ligands to nanopartices1-9. Important early advances in coiled-coil engineering included demonstrating that leucine-zipper peptides, which are short coiled coils of ~40 amino acids, can fold to give stable structures composed of two to four helices, and that coiled coils can be modified using charge patterning to encode heterospecificity and helix orientation10. In particular, peptide “Velcro” is a designed heterospecific coiled-coil dimer with glutamates at all interfacial e and g positions on one helix and lysines at all e and g positions on the other; this heterodimer and variants of it have been widely employed in bio-molecular engineering. Further experiments have illustrated how residues at the hydrophobic interface, particularly those in a positions, can be mutated to modulate interaction affinity and introduce additional specificity11. Prior studies not only generated reagents that have found many uses, but also elucidated structural principles that control interaction selectivity12-14.
Heterodimeric coiled-coil pairs have proven particularly useful for molecular engineering12-18. Exciting recent applications have included using coiled-coil heterodimers to modulate MAP kinase signaling in yeast and inducing ordered structure via coiled coils in nano-scale fibers. Notably, while coiled-coil reagents for inducing homo-oligomerization or hetero-oligomerization of single complexes are widely used, the modern coiled-coil toolkit does not provide access to more complex interaction patterns. Lacking is a large set of coiled coils that participate in specific and defined interactions with one another. Such reagents could be used to construct interaction networks containing multiple associations in a logical manner. For example, when engineering cellular circuits it might be desirable to implement multiple parallel pathways, each using coiled coils to direct assembly of signaling complexes without crosstalk. Likewise, to engineer artificial transcription factors, heterodimers with specified cross-interactions could provide access to combinatorial control of binding to different DNA sites. For complex applications such as these, greater versatility is required than is currently provided by characterized coiled-coil peptides.
Results and Discussion
We recently reported the computational design of synthetic peptides that interact with the coiled-coil regions of human bZIP transcription factors. These designed peptides are 35-54 residues in length and share an amino-acid composition characteristic of bZIP leucine zippers (Figure S1, Table S1). Homodimerization of the designed peptides was disfavored by a variety of strategies, and experiments confirmed that most designs do not form strong self-associations19. Speculating that this set of heterospecific reagents might harbor interesting and useful interactions patterns, we systematically measured all pair-wise interactions involving 48 designed peptides and 7 additional coiled coils from human bZIPs that do not strongly self-associate.
To identify new heterospecific coiled-coil interactions in a high-throughput manner, we used a protein microarray assay. A complete 55 × 55 interaction matrix was generated by spotting small amounts of each peptide onto aldehyde-derivatized slides (Figure S2, Table S2). Each of the 55 proteins in turn was labeled with Cy3 dye and used in solution to probe subarrays printed on the slides. This assay is highly reproducible and shows good reciprocity with respect to which protein is immobilized (Figures S2 and S3). The relative ordering of fluorescence intensities on the arrays has also been shown to agree qualitatively with solution stability measurements19,20.
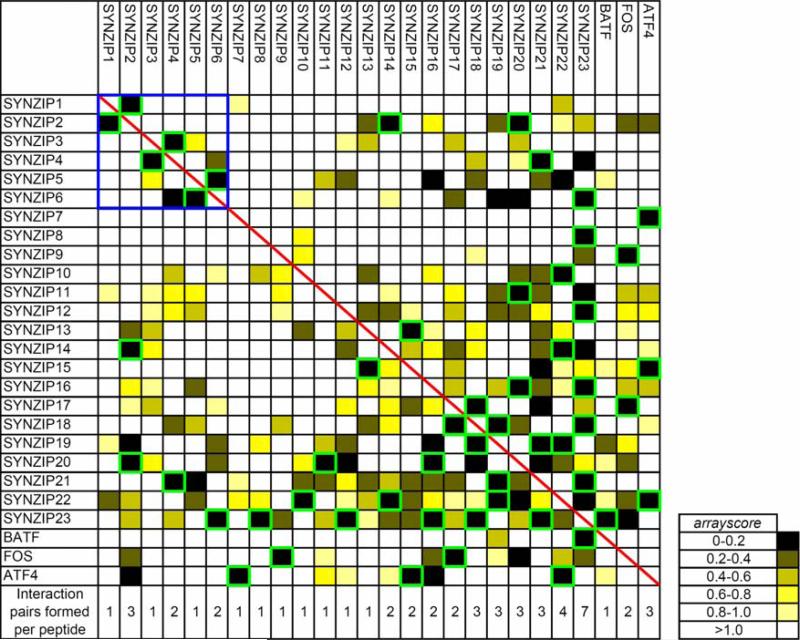
To discover new pairs of hetero-associating coiled coils, the interaction matrix was examined for peptides that: (1) did not show evidence of homo-association and (2) made strong, reciprocal interactions with a partner. Interacting and non-interacting pairs were chosen conservatively based on comparisons of prior array data with solution data. A total of 27 heterospecific pairs involving 23 synthetic peptides (named SYNZIPs 1-23) and 3 human bZIPs were selected for further analysis (Figure 1).
Figure 1.
Array data describing the interactions of 26 peptides that form specific interaction pairs. Peptides printed on the surface are listed in rows, and fluorescently labeled peptides in solution are listed in columns. Color indicates the strength of the array fluorescence signal, given as arrayscore values (see Methods) according to the color bar at right with 0 (black) indicating the strongest signal and >1 (white) indicating the weakest. SYNZIP peptides 1-6, which are further described in Figures 2-4, are in the top left corner, boxed in blue. The red diagonal highlights the absence of homoassociation of peptides on the arrays. Interactions that showed arrayscore ≤ 0.2 in both measurement directions are boxed in green. The number of strong, reciprocal interactions formed by each peptide is listed at bottom of each column.
Coiled coils can vary in their oligomerization state, helix orientation and axial helix alignment21. For the heterospecific pairs uncovered in this assay to be maximally useful, knowledge of their interaction geometry is important. The synthetic coil-coiled peptides were designed to interact with individual human bZIPs as parallel dimers, and we hypothesize that most of the design-design and design-human complexes detected on the arrays also form parallel dimers. Several lines of evidence support this. First is the special role of paired a-position asparagines in leucine zippers. Interaction of an asparagine residue with another asparagine on an opposing helix is common in coiled-coil dimers and is much more favorable than an interaction with a hydrophobic residue (which we term an “Asn mismatch,” unless the Asn occurs very close to the end of the coiled coil)10, 11. Paired asparagines at a favor parallel dimer formation and are strongly conserved in the parallel, dimeric leucine-zipper transcription factors10, 13, 22. Almost all (23 out of 26) peptides analyzed here contain at least one Asn residue at a coiled-coil a position, and of the 27 heterospecific pairs considered, 24 can be aligned such that two a-position Asn residues are paired. All heterospecific pairs can be aligned as parallel dimers without any Asn mismatches11. In addition to the role of Asn residues, half of the 26 peptides also include a charged residue in one or two non-terminal a positions. Lysine in a positions has been reported to favor dimer formation over higher order oligomerization, presumably because a positions in dimers are less buried10, 23; this likely applies for other charged side chains as well, as is supported by the lower frequencies of Lys, Arg and Glu residues in a positions of parallel trimers compared to parallel dimers (K. Gutwin and A. Keating, unpublished data). Additional indirect criteria support parallel dimer formation. For example, when considered as parallel dimers, all pairs can be aligned such that net g-e’ electrostatic interactions are not unfavorable and destabilizing10, 14. Finally, none of the heterospecific interactions encode a motif that has been reported to favor trimer formation24.
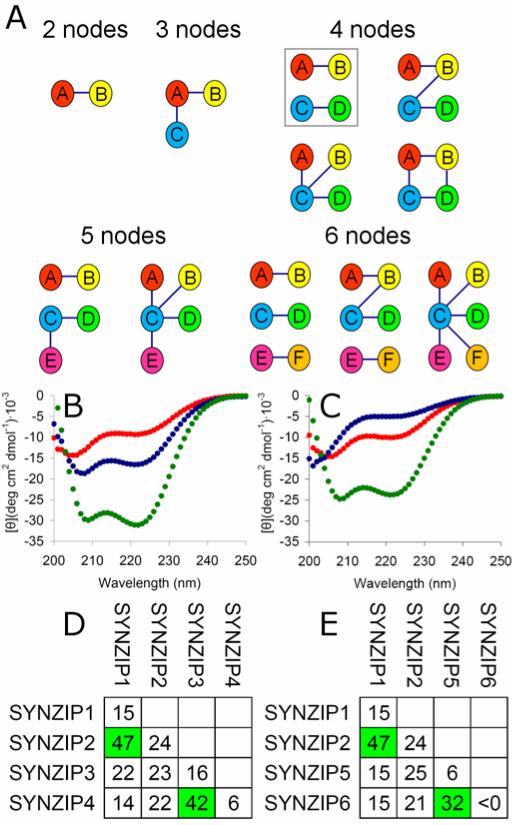
Given 27 heterospecific pairs among 26 peptides that likely form parallel coiled-coil dimers, we analyzed these to identify higher-order patterns of interaction and non-interaction. Each of the 26 peptides participates in 1-7 interactions, suggesting that subnetworks involving more than 2 peptides could be common in our data (Figure 1). We searched exhaustively for all subnetworks containing 3-6 proteins and found examples of the 10 topologies shown in Figure 2A (Table S3)25. In that figure, an edge indicates a high-confidence observation of an interaction on the array and the absence of an edge indicates that an interaction was not observed. Most networks are based on motifs we describe as “pair”, “line”, or “hub” structures. Many networks are composed of smaller networks, such as the 4 node “orthogonal pair” (2 pairs with no cross-interactions), “orthogonal triplet” (3 pairs with no cross-interactions) or the 5 node “pair + line” (similarly with no cross interactions). Interestingly, protein nodes in the networks are sparsely connected. It may be that features engineered to diminish self-association also reduce interaction promiscuity more broadly.
Figure 2.
SYNZIP coiled coils form specific interaction subnetworks. (A) Graphical representation of subnetworks detected in the coiled-coil array data. Edges indicate an interaction and the absence of an edge between nodes indicates no interaction in the microarray screen. The orthogonal pair motif is boxed in grey. (B, C) CD spectra for two pairs of heterospecific coiled coils (4 μM of each protein and 8 μM total for mixtures, 25 °C). (B) SYNZIP2 (blue), SYNZIP1 (red), and SYNZIP2 + SYNZIP1 (green). (C) SYNZIP4 (blue), SYNZIP3 (red), and SYNZIP4 + SYNZIP3 (green). (D, E) Melting temperatures (Tms) derived from fits to thermal melts of peptide mixtures. Tm values for the “on” interacting pair mixtures are highlighted in green.
Because of its immediate utility, e.g. for direct extension of existing applications, we chose the orthogonal-pair motif for further characterization1, 2, 6. Three coiled-coil pairs were selected that participate in two sets of orthogonal interactions. All three pairs were evaluated in solution using circular dichroism (Figure 2B and C, Figure S4). The six individual peptides gave only weak helical signal in isolation. But mixing each peptide with its appropriate partner gave a spectrum characteristic of a coiled coil, confirming heterospecific interaction. The orthogonal sets that can be constructed from these three pairs each consist of four peptides that participate in two interactions (‘on’ states) and eight non-interactions (‘off’ states). We measured the thermal stabilities of the ten possible interactions for each set (Figure 2D and E, Figure S5). The ‘on’ states had melting temperatures between 32 and 47 °C, at 8 μM total peptide concentration. For [SYNZIP6:SYNZIP5, SYNZIP1:SYNZIP2] the difference between the weakest ’on’ state and the strongest ’off’ state was ~8 °C. For [SYNZIP4:SYNZIP3, SYNZIP1:SYNZIP2] the difference was ~18 °C. (See Figure S6 for characterization of an additional orthogonal set.) Previously published orthogonal coiled-coil pairs are either much less stable than this, have the property that at least one “off” interactions is more stable than one “on” interaction, or incorporate non-natural amino acids15-17, 26.
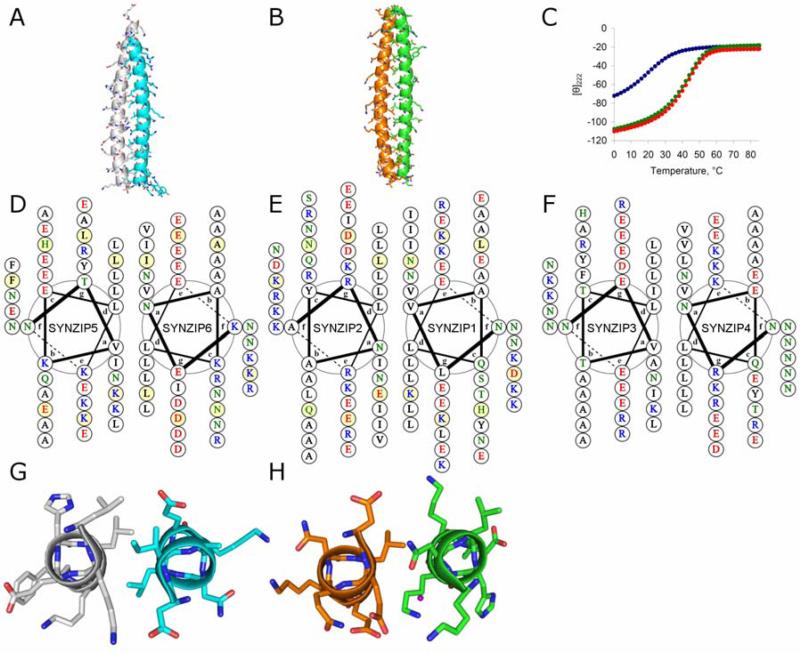
To confirm the interaction geometry of complexes composing the orthogonal pairs, we solved the structures of SYNZIP6:SYNZIP5 and SYNZIP1:SYNZIP2 to 2.5 and 1.8 Å, respectively (Figure S7, Table S4). Both complexes are parallel heterodimers, as anticipated (Figure 3A and B). We were unable to obtain crystals of SYNZIP4:SYNZIP3. While it is likely that this pair forms a parallel dimer (it includes a-position Asn and Lys residues and highly charged e- and g-position residues), SYNZIP3 is shorter than SYNZIP4, and the precise axial alignment of its two helices is uncertain. Either of two Asn residues in SYNZIP4 could be paired with the single a-position Asn in SYNZIP3, while maintaining a similar extent of coiled-coil dimer. To experimentally determine the alignment, two truncated versions of SYNZIP4 were generated. Each was mixed with SYNZIP3, and the thermal stabilities of the resulting complexes were measured by CD. The N-terminal SYNZIP4 truncation had very similar stability to the full-length peptide, while the C-terminal truncation was markedly destabilized (Figure 3C). Thus, the two most N-terminal heptads of SYNZIP4 are dispensable for the interaction. Based on these experiments, helical wheel diagrams were generated for the three heterospecific pairs (Figure 3 D-F).
Figure 3.
Interaction geometries for three heterospecific SYNZIP pairs. (A, B) Crystal structures of SYNZIP5:SYNZIP6 (A) (grey:teal) and SYNZIP2:SYNZIP1 (B) (orange:green) show that both complexes are parallel coiled-coil heterodimers. (C) Determination of the axial alignment of SYNZIP4:SYNZIP3 using CD thermal melts. SYNZIP41-54: SYNZIP3 (red), SYNZIP41-42: SYNZIP3, (blue), and SYNZIP415-54: SYNZIP3 (green). Each mixture was measured at 8 μM total peptide concentration, 4 μM of each peptide. (D-F) Helical wheel diagrams for SYNZIP5:SYNZIP6 (D), SYNZIP2:SYNZIP1 (E), and SYNZIP3:SYNZIP4 (F). Charged residues are colored red/blue, polar residues are in green, and hydrophobic residues are in black. Residues shaded yellow in (D) and (E) correspond to those shown in panels (G) and (H), respectively. (G) The fourth heptad of SYNZIP5 (residues 23-29):SYNZIP6 (residues 37-43), and (H) the fourth heptad of SYNZIP2 (residues 23-29):SYNZIP1 (residues 23-29) are shown in cross-section, as viewed from the N-terminus. A partially buried water molecule is represented in purple. Crystal structure figures generated using PyMOL (DeLano Scientific, Palo Alto, CA). Helical wheel diagrams created using DrawCoil 1.0. http://www.gevorggrigoryan.com/drawcoil/)
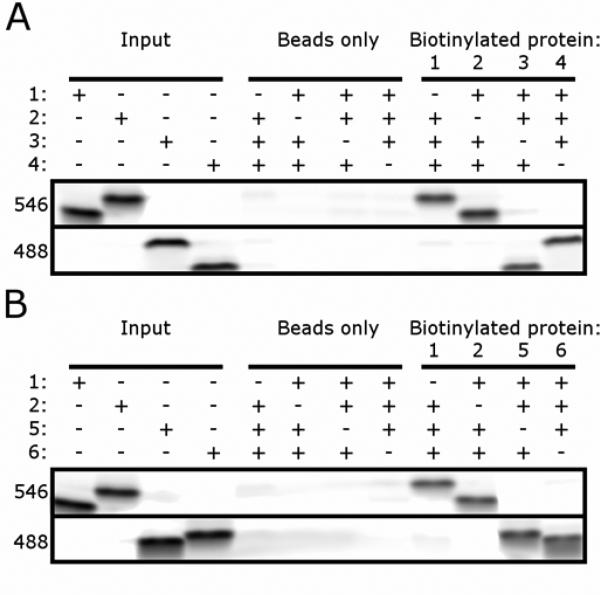
These experiments suggested that portions of each complex were dispensable for the formation of orthogonal pairs. To demonstrate that shorter experimentally determined interaction regions interact specifically, truncated versions of SYNZIPs 1-6 (shown in Figure 3 D-F) were cloned with an N-terminal cysteine. Each protein was labeled with biotin. SYNZIPs 1 and 2 were also labeled with Alexa Fluor 546, and SYNZIPs 3, 4, 5, and 6 were labeled with Alexa Fluor 488. For each orthogonal set, each biotinylated protein was pre-mixed with the three other fluorescent proteins and then incubated with NeutrAvidin coated beads. These pull-down experiments showed that each biotinylated protein interacted specifically with its cognate partner (Figure 4A and B). Thus, the shorter peptides are sufficient to form specific interactions in four-component mixtures.
Figure 4.
Biotin pull-down assay demonstrating specific interactions in each orthogonal set. (A, B) SYNZIPs 1 and 2 were labeled with Alexa Fluor 546 and SYNZIPS 3, 4, 5, and 6 were labeled with Alexa Fluor 488. Input lanes show each protein run individually. The beads-only lanes shows mixtures of the indicated fluorescent proteins incubated with NeutrAvidin beads. The biotinylated-protein lanes show mixtures of the 3 indicated fluorescent proteins (4 μM each) mixed with the indicated biotinylated protein at 4 μM, then incubated with NeutrAvidin beads. The two fluorescent channels 546 nm (top) and 488 nm (bottom) are indicated. (A) SYNZIP pairs 1-2 and 3-4. (B) SYNZIP pairs 1-2 and 5-6.
The crystal structures of SYNZIP6:SYNZIP5 (PDB ID 3HE4) and SYNZIP1:SYNZIP2 (PDB ID 3HE5) reveal interactions involving polar and charged residues that likely play a role in encoding specificity. Both structures include paired asparagines at a-a’ positions that adopt conformations seen frequently in other parallel coiled-coil dimers. Neither structure contains any asparagine mismatches at non-terminal heptad positions, although both have mismatches at the extreme N-terminal heptad. At that position, asparagine is paired with valine but remains largely solvent exposed due to its location at the end of the helix. In the SYNZIP6:SYNZIP5 complex, in both the fourth and fifth heptads, Lys at a across from Ile interacts with an aspartate at the proceeding g’ position (Figure 3G). In the SYNZIP1:SYNZIP2 complex, the fourth heptad contains a complex polar network involving a partly buried water molecule. The water is coordinated by SYNZIP1 residues Asn 24 at a and Lys 27 at d, as well as by SYNZIP2 residue Glu 24 at a’. In the 3 copies of the heterodimer in the asymmetric unit, Lys 23 at g on SYNZIP1, as well as Gln 25 at b’ and Glu 28 at e’ on SYNZIP2, are involved to varying degrees in this extended network (Figure 3H). These interactions suggest that charged residues in coiled-coil core positions can contribute specificity in parallel dimers, although such residues may be accommodated in ways that are difficult to anticipate, as illustrated here by incorporation of a water molecule.
It is interesting to speculate about how specificity in the orthogonal sets is determined. The simple ACID-BASE charge repulsion strategy used in peptide “Velcro” is not sufficient to encode complex interaction patterns in coiled coils only ~40 amino acids long. How are so many different ‘off’ states disfavored? Using a simple model, 5 of the 14 ‘off’ pairs among the two orthogonal pair sets have net repulsive electrostatic interactions at g-e’ positions, when considered as parallel dimers. Unavoidable Asn mismatches appear in an additional 2 pairs. In the remainder, charged residues at a and d positions appear important, with a-position Lys and Glu residues disfavoring homodimerization, and repulsive charges at g-a’ and d-e’ pairs disfavoring both homo- and heterodimers11. All of these interactions are implicated as useful and important negative design features. In terms of improving specificity, if this is required, we stress that the undesired complexes that form are weak and are not necessarily parallel dimers.
The orthogonal pairs introduced here dramatically increase the number of small, heterospecific protein-protein interaction partners that can be used as modular components for molecular engineering27. The peptides can be over-expressed in Escherichia coli, contain aromatic amino acids for quantification using spectrometry and lack cysteines. While most of these peptides do partner with human bZIPs, they are likely to be effective for applications in yeast or bacteria, where human orthologs are absent, as well as in vitro and for materials applications. These reagents, or molecular parts, are also likely to be useful when paired with other types of synthetic or native interaction domains such as zinc fingers28. It is reasonable to consider using them to design novel transcription factors that do not cross-interact, or to elaborate molecular scaffolds1, 6. Finally, the large number of interactions measured in the course of characterizing these peptides will be useful for testing computational models and further understanding the interaction specificity of “simple” coiled coils.
Methods and Materials
Plasmid construction, protein expression and purification
Proteins used in the array experiments were cloned, expressed and purified as published previously19. For solution studies and crystallography, genes were cloned into pSV282 (Vanderbilt University Medical Center, Center for Structural Biology) using BamHI and XhoI restriction enzymes (NEB). For the pull-down assays, synthetic genes for truncated peptides including an N-terminal cysteine and a short linker (GSCGS) were cloned based on experimentally determined alignments. SYNZIP6 was mutated at a c-position lysine to include a tyrosine for concentration determination. Each plasmid was transformed into RP3098 cells and 1 L cultures in LB were grown to 0.4-0.6 OD and induced at 37 °C for 3-4 hours with the addition of 1mM IPTG. MBP fusion proteins with a His6 tag were purified under native conditions by binding to Ni-NTA resin (Qiagen) and eluting with 8 ml elution buffer (300 mM imidazole, 20 mM Tris, 500 mM NaCl, 1mM DTT, pH 7.9). Fusion proteins were then dialyzed overnight at 4 °C in TEV cleavage buffer (50 mM Tris, 50 mM NaCl, 1 mM DTT, 0.5 mM EDTA, pH 7.5). Peptides were cleaved from MBP by incubating with 100 μl TEV protease (1mg/ml) for 3 hours at room temp. After cleavage, the mixture was added to Ni-NTA resin and the flow through was collected. In the case of SYNZIP2, the peptide bound the Ni-NTA resin after cleavage. SYNZIP2 was eluted from the resin with 6 M guanidine-HCl and the eluate was then dialyzed into water. Peptides were additionally purified using reverse-phase HPLC and lyophilized. The molecular weights of the peptides were confirmed by mass spectrometry. Protein concentrations were determined using the Edelhoch method29 of measuring UV absorbance at 280 in 6 M guanidine-HCl/100 mM sodium phosphate pH 7.4. Protein and DNA sequences are listed in Table S1.
Coiled-coil array assay
All array experiments were carried out as previously published19, with the exception that only two spots for each protein were printed per subarray, for a total of 8 measurements of each heteromeric interaction. Briefly, lyophilized proteins were resuspended to a concentration of 40 μM in 6 M guanidine-HCl/100 mM sodium phosphate pH 7.5/0.04% Triton X-100/10 μM Alexa Fluor 633 hydrazide. Proteins were printed on aldehyde-derivatized glass slides and 12 identical subarrays per slide were physically divided by drawing a hydrophobic boundary. Slides were blocked, and then each subarray was probed with Cy3-labeled proteins diluted six-fold from 6 M guanidine-HCl/100 mM sodium phosphate pH 7.5/6 mM TCEP to a concentration of ~160 nM in 1.2X buffer (1.2% BSA, 1.2X PBS, 0.12% Tween-20). Slides were then washed, dried, and scanned to obtain fluorescence values for each spot. Average background-corrected fluorescence values are listed in Table S2.
Data analysis
For each peptide pair, fluorescence intensities for the 4 replicate spots corresponding to the same surface/solution arrangement were corrected for background and then averaged. Averages were corrected further by subtracting the median signal for all proteins on the surface interacting with the same solution probe; this gave a value F. The quantity arrayscore was calculated by taking –log(F/Fmax) where Fmax was the maximum F value for a given solution probe. To identify heterospecific pairs, a strict criterion was employed by comparing arrayscore values to Tm measurements of previously published data19. Non-interactions were required to have arrayscore > 1, which corresponds to an average Tm of 14 °C (based on 13 comparisons). Interactions were required to have arrayscore < 0.2, which corresponds to an average Tm of 43 °C (based on 7 comparisons). These same criteria for interactions and non-interactions were employed to identify subnetworks when using Fanmod 25 to search for all possible 3-6 node networks. Motifs are listed in Table S3.
Circular dichroism
Circular dichroism spectra were measured on an AVIV 400 spectrometer in 12.5 mM potassium phosphate (pH 7.4)/150 mM KCl. Individual measurements were made at 4 μM peptide or 4 μM of each peptide (8 μM total peptide) for mixtures. All measurements were made in a 1 cm cuvette. Mixtures of peptides were incubated for several hours at room temperature before measurement. Spectra were measured at 25 °C. Wavelength scans were monitored from 280 nm to 195 nm in 1 nm steps, averaging for 5 seconds at each wavelength. Three scans for each sample were averaged. Thermal unfolding curves were performed at 4 μM peptide for individual measurements or 4 μM of each peptide (8 μM total peptide) for mixtures and measured in a 1 cm cuvette with stirring. Melting curves were determined by monitoring ellipticity at 222 nm with an averaging time of 30 seconds, an equilibration time of 1.5 minutes, and a scan rate of 2 °C/min. All samples were measured from 0 to 85 °C. Tm values were estimated as reported previously19. All thermal denaturations were reversible, with differences in Tm values upon folding vs. unfolding of < 2°C for all but 2 weak complexes, and < 5 °C in all cases.
For a third orthogonal set of coiled-coil heterodimers, a slightly modified CD protocol was employed. The CD spectra in Figure S6 were measured on an Aviv Model 202 spectrometer in 12.5 mM potassium phosphate (pH 7.4)/150 mM KCl. Individual measurements were made at 40 μM peptide and mixtures at 20 μM of each peptide, 40 μM total peptide. Mixtures of peptides were incubated for several hours at room temperature before measurement. Spectra were measured at 25 °C. Wavelength scans were performed in a 0.1 cm cuvette and were monitored from 260 nm to 195 nm in 1 nm steps averaging for 5 seconds at each wavelength.
Crystallography
Purified lyophilized protein was re-suspended in water to a concentration of 20 mg/ml and mixed to give 20 mg/ml of each complex. Crystals were grown by the hanging drop method at room temperature by mixing 1 μl protein solution with 1 μl of reservoir solution. SYNZIP1:SYNZIP2 was grown in 45% MPD, 100 mM Tris pH 8.0, and 160 mM ammonium acetate. SYNZIP6:SYNZIP5 was grown in 100 mM Tris pH 8.2 and 20% MPD. Crystals were frozen in LN2 without addition of any cryoprotectant. Diffraction data were collected at 100K on a Rigaku MicroMax007-HF with VariMax-HR optics and a RAXIS-IV detector (SYNZIP1: SYNZIP2) or at the NE-CAT 24ID-E beam line of the Advanced Photon Source (SYNZIP6:SYNZIP5) and processed using HKL200030. Both structures were solved by molecular replacement using PHASER4. In each case the search model was derived from a single energy-minimized theoretical model selected from an ensemble of models spanning the space of parameters of native parallel dimeric coiled-coil structures. The ensemble was generated as previously described5. The search models had no overhangs and the side chains at all non-interfacial positions (b, c, and f) were truncated to alanine. Model building was done using COOT31, 32 using twin law corrections for both structures (Table S4). Non-crystallographic symmetry (NCS) restraints between the four copies of the heterodimer in the asymmetric unit (ASU) of the SYNZIP6:SYNZIP5 crystals were used to aid in the refinement of that structure. Geometry was checked using MOLPROBITY33 and no outliers were identified (Table S4). Figures of structures were generated using PyMol (DeLano Scientific, Palo Alto, CA).
Pull down assay
Proteins containing a unique N-terminal cysteine were labeled by mixing 100 μM protein with 0.5 mM Alexa Fluor 488 or 546 maleimide (Invitrogen) or 2 mM maleimide-PEG11-biotin (Thermo Scientific) in 100 mM potassium phosphate pH 7.0/150 mM KCl/1 mM TCEP. Solutions were incubated for three hours at 18-22 °C. Free dye or biotin was removed using desalting spin columns (Thermo Scientific). Biotinylated proteins were concentrated using centrifugal filter units (Millipore). The concentration of unlabeled and biotinylated proteins was determined using the Edelhoch method. The concentration of dye labeled proteins was estimated by assuming a 50% recovery after desalting. Each dye labeled protein was mixed with the unlabeled version (at known concentration) in a 1:10 ratio. 400 pmoles of each protein indicated in Figure 4 were mixed in 75 μl binding buffer (12.5 mM potassium phosphate pH 7.4, 150 mM KCl, 1 mM DTT, 1% BSA, 0.1% Tween-20). Protein mixtures were incubated for 1 hour at 18-22 °C and then 50 μl of a 50% slurry of NeutrAvidin beads (Thermo Scientific) in binding buffer was added. Mixtures were incubated for 2 hours at 18-22 °C with rotation. Beads were then washed 3 times with 1 ml binding buffer at 4 °C and mixed with 100 μl of loading buffer (10 % glycerol, 2% SDS, 100 mM DTT, 0.01% bromophenol blue, 100 mM Tris pH 6.8). Following heating at 65 °C for 15 minutes, 10 μl of each sample was loaded onto an 18% Tris-glycine gel (Invitrogen). Gels were imaged on a Typhoon 9400 imager. Fluorsep software (Amersham Biosciences) was used to remove background fluorescent overlap.
Sequence analysis
Positions a-g in the coiled-coil heptad repeat were assigned manually, as designed previously19, based on conserved Leu residues and overall hydrophobic/polar patterning. Each peptide contains 5-7 full heptads. The following criteria were applied for sequence analysis. To predict the most probable alignments of coiled-coil dimers, all possible helix alignments that overlapped by at least 5 full heptads and did not contain an asparagine mismatch were considered. Asparagine mismatches were defined as an Asn residue at a non-terminal a position across from isoleucine, valine or leucine at a non-terminal a position. A terminal a position was defined as an a position ≤ 3 residues from the end of the coiled coil. For assessing g-e’ electrostatics, the least repulsive alignment of ≥ 5 heptads that did not contain an asparagine mismatch was used. For this purpose, each attractive g-e’ interaction was scored as + 0.5 and each repulsive g-e’ interaction was scored as -0.5. Negatively charged glutamate and aspartate, and positively charged lysine and arginine were considered during scoring. Note that Glu, Lys, Arg and – to a lesser extent – Asp overwhelmingly predominate at g and e positions of the 26 peptides considered (Figure S1).
Supplementary Material
Acknowledgements
This work was supported by NIH award GM067681. This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source, supported by award RR-15301 from the National Center for Research Resources at the National Institute of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357 We thank the MIT BioMicro center for arraying instrumentation, J.R. Apgar for generating models used for structure determination and G. Grigoryan for computational design of synthetic peptides not described elsewhere. We thank members of the Keating laboratory for comments on the manuscript.
Footnotes
Supporting Information Available: Sequences and data for all 55 peptides measured on the protein arrays, additional CD data, crystallographic data and refinement statistics and electron density maps of crystal structures. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Bashor CJ, Helman NC, Yan S, Lim WA. Science. 2008;319:1539–1543. doi: 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- 2.Diehl MR, Zhang K, Lee HJ, Tirrell DA. Science. 2006;311:1468–1471. doi: 10.1126/science.1122125. [DOI] [PubMed] [Google Scholar]
- 3.Eckert DM, Malashkevich VN, Hong LH, Carr PA, Kim PS. Cell. 1999;99:103–115. doi: 10.1016/s0092-8674(00)80066-5. [DOI] [PubMed] [Google Scholar]
- 4.Papapostolou D, Smith AM, Atkins EDT, Oliver SJ, Ryadnov MG, Serpell LC, Woolfson DN. Proceedings of the National Academy of Sciences. 2007;104:10853–10858. doi: 10.1073/pnas.0700801104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takagi J, Erickson HP, Springer TA. Nat Struct Mol Biol. 2001;8:412–416. doi: 10.1038/87569. [DOI] [PubMed] [Google Scholar]
- 6.Wolfe SA, Grant RA, Pabo CO. Biochemistry. 2003;42:13401–13409. doi: 10.1021/bi034830b. [DOI] [PubMed] [Google Scholar]
- 7.Petka WA, Harden JL, McGrath KP, Wirtz D, Tirrell DA. Science. 1998;281:389–392. doi: 10.1126/science.281.5375.389. [DOI] [PubMed] [Google Scholar]
- 8.McAllister KA, Zou H, Cochran FV, Bender GM, Senes A, Fry HC, Nanda V, Keenan PA, Lear JD, Saven JG, Therien MJ, Blasie JK, DeGrado WF. Journal of the American Chemical Society. 2008;130:11921. doi: 10.1021/ja800697g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mapp AK, Ansari AZ, Ptashne M, Dervan PB. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3930–3935. doi: 10.1073/pnas.97.8.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mason JM, Muller KM, Arndt KM. Methods Mol Biol. 2007;352:35–70. doi: 10.1385/1-59745-187-8:35. [DOI] [PubMed] [Google Scholar]
- 11.Acharya A, Rishi V, Vinson C. Biochemistry. 2006;45:11324–11332. doi: 10.1021/bi060822u. [DOI] [PubMed] [Google Scholar]
- 12.Arndt KM, Pelletier JN, Müller KM, Plückthun A, Alber T. Structure. 2002;10:1235–1248. doi: 10.1016/s0969-2126(02)00838-9. [DOI] [PubMed] [Google Scholar]
- 13.Moll JR, Ruvinov SB, Pastan I, Vinson C. Protein Sci. 2001;10:649–655. doi: 10.1110/ps.39401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Shea EK, Lumb KJ, Kim PS. Current Biology. 1993;3:658–667. doi: 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- 15.Lai JR, Fisk JD, Weisblum B, Gellman SH. Journal of the American Chemical Society. 2004;126:10514–10515. doi: 10.1021/ja047710f. [DOI] [PubMed] [Google Scholar]
- 16.Diss ML, Kennan AJ. Journal of the American Chemical Society. 2008;130:1321–1327. doi: 10.1021/ja076265w. [DOI] [PubMed] [Google Scholar]
- 17.Bromley EHC, Sessions RB, Thomson AR, Woolfson DN. Journal of the American Chemical Society. 2009;131:928–930. doi: 10.1021/ja804231a. [DOI] [PubMed] [Google Scholar]
- 18.Mason JM, Schmitz MA, Müller KM, Arndt KM. Proceedings of the National Academy of Sciences. 2006;103:8989–8994. doi: 10.1073/pnas.0509880103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grigoryan G, Reinke AW, Keating AE. Nature. 2009;458:859–864. doi: 10.1038/nature07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman JRS, Keating AE. Science. 2003;300:2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- 21.Grigoryan G, Keating AE. Current Opinion in Structural Biology. 2008;18:477–483. doi: 10.1016/j.sbi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harbury PB, Zhang T, Kim PS, Alber T. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 23.Campbell KM, Sholders AJ, Lumb KJ. Biochemistry. 2002;41:4866–4871. doi: 10.1021/bi0159276. [DOI] [PubMed] [Google Scholar]
- 24.Kammerer RA, Kostrewa D, Progias P, Honnappa S, Avila D, Lustig A, Winkler FK, Pieters J, Steinmetz MO. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13891–13896. doi: 10.1073/pnas.0502390102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wernicke S, Rasche F. Bioinformatics. 2006;22:1152–1153. doi: 10.1093/bioinformatics/btl038. [DOI] [PubMed] [Google Scholar]
- 26.Diss ML, Kennan AJ. Org Lett. 2008;10:3797–800. doi: 10.1021/ol801461a. [DOI] [PubMed] [Google Scholar]
- 27.Bromley EHC, Channon K, Moutevelis E, Woolfson DN. ACS Chemical Biology. 2008;3:38–50. doi: 10.1021/cb700249v. [DOI] [PubMed] [Google Scholar]
- 28.Giesecke AV, Fang R, Joung JK. Mol Syst Biol. 2006;2:2006.0011. doi: 10.1038/msb4100053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edelhoch H. Biochemistry. 1967;6:1948–54. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 30.Otwinowski Z, Minor W, Carter Charles W. Jr. Methods in Enzymology. Vol. 276. Academic Press; 1997. [20] Processing of X-ray diffraction data collected in oscillation mode. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. Acta Crystallographica Section D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. Acta Crystallographica Section D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 33.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, III, Snoeyink J, Richardson JS, Richardson DC. Nucl. Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.