Abstract
Among the processes that play essential roles in both genome defense and organism survival are those involved in chromosome comparison. They are acutely active in the meiotic cells of Neurospora crassa, where they evaluate the mutual identity of homologs by a process we call trans-sensing. When nonsymmetrical regions are found, they are silenced. The known molecular components of this meiotic silencing machinery are related to RNA-dependent RNA polymerases, Argonautes and Dicers, suggesting that the mechanisms of how heterologous chromosomal regions are silenced involves, at some stage, the production of small interfering RNAs. Neurospora has two active and clearly distinct RNA interference pathways: quelling (vegetative specific) and meiotic silencing (meiosis specific). Both pathways require a common set of protein types like RNA-dependent RNA polymerases, Argonautes and Dicers. In this work we demonstrate the involvement of quelling defective-2 interacting protein (qip+), a Neurospora gene whose function is essential to silencing by quelling, in meiotic silencing, and normal sexual development. Our observations reinforce the molecular connection between these two silencing pathways.
EUKARYOTIC genomes require the activity of complex mechanisms aimed at maintaining and preserving their molecular integrity. At least four distinct but potentially interrelated such processes are known in Neurospora crassa: DNA methylation, quelling, repeat-induced point mutation (RIP), and meiotic silencing (Galagan et al. 2003; Borkovich et al. 2004).
Quelling, or vegetative RNA silencing, was observed at the very dawn of the RNA silencing field (Cogoni et al. 1996). Like in other organisms, its discovery was born by the need to explain why having two copies of a gene was equivalent to having none (Cogoni et al. 1994). Genetic dissection of this pathway uncovered the genes quelling defective-1 (qde-1+), -2 (qde-2+), -3 (qde-3+), encoding for an RNA-dependent RNA polymerase (QDE-1), an Argonaute (QDE-2), and a RecQ DNA helicase (QDE-3), respectively (Cogoni and Macino 1997, 1999a, 1999b; Catalanotto et al. 2000).
Like the discovery of quelling, meiotic silencing was also born out of the need to explain an odd observation: How could a gene be dominant by being absent? (Aramayo and Metzenberg 1996). Our initial observation was that mutants carrying a deletion of the transcription factor Ascospore maturation-1 (asm-1+) were viable, and yet, when crossed to wild type, the progeny was composed of dead sexual spores (Aramayo and Metzenberg 1996; Aramayo et al. 1996). The connection between this position-effect phenomenon to RNA silencing was made by the characterization of a suppressor of meiotic silencing, Suppressor of ascus dominance-1 (Sad-1) (Shiu et al. 2001), which implicated SAD-1, an RNA-dependent RNA polymerase in the pathway. Further characterization of the pathway revealed the involvement of the Argonaute Suppressor of meiotic silencing-2 (Sms-2) and of dicer-like-1/Suppressor of meiotic silencing-3 (Dcl-1/Sms-3) (Galagan et al. 2003; Lee et al. 2003b; Alexander et al. 2008). Unlike quelling, the molecular properties required to trigger meiotic silencing are well understood (Kutil et al. 2003; Lee et al. 2003b; Lee et al. 2004; Pratt et al. 2004). Exactly how this symmetry-evaluation process works is still a mystery, but we know that it involves the evaluation of identity at the most detailed level (Pratt et al. 2004). A model for meiotic silencing has been described previously (Aramayo and Pratt 2010). It involves the recognition of nonhomology between homologous chromosomes by unknown molecular players. Once identified, heterologous regions are predicted to produce some form of aberrant RNA, which is converted into dsRNA by the action of the RdRP, SAD-1. Once produced, dsRNA triggers the initiation step of the pathway, which is predicted to involve the conversion of the dsRNA trigger into siRNAs. This step is most likely executed by the DCL-1/SMS-3 Dicer. The maintenance of silencing probably involves amplification, also through SAD-1, and degradation of the target mRNAs via an RNA-induced silencing complex (RISC), of which the Argonaute-like protein SMS-2 is predicted to be an essential component. Interestingly, the SAD-2 protein is also required for meiotic silencing, but its role appears to be to anchor or recruit the SAD-1 RdRP to the cytoplasmic face of the nuclear periphery (Shiu et al. 2006) (D. W. Lee and R. Aramayo, unpublished results). This indicates that SAD-1's RdRP activity is required outside of the nucleus, which is never completely dissolved in Neurospora meiosis. RNAs might be processed by SAD-1 as they transit nuclear pores as a sort of quality check. In any case, RdRP activity is likely a downstream effector of meiotic silencing and not a component of the trans-sensing mechanism that must necessarily be nuclear. An “RNA quality” monitoring system, however, implies that deficient pairing triggers production of aberrant transcripts to alert the system, and hence that proficient recognition and pairing suppresses this transcription.
Clearly, the presence of mechanisms that evolved to preserve genome integrity have had profound evolutionary consequences for Neurospora and shaped its genome. Currently, Neurospora has the lowest number of paralogous genes in comparison to other organisms (Galagan et al. 2003; Borkovich et al. 2004), suggesting that if related molecular mechanisms operate at different stages of its life cycle (e.g., RNA silencing operating in vegetative cells vs. meiotic cells), the corresponding protein complexes involved must then share as many molecular components as possible. When the Neurospora genome became available we found that many components involved in quelling had corresponding paralogs, and, as predicted, at least some of them were involved in meiotic silencing (Lee et al. 2003b). The unexpected presence of paralogs was explained to be due to the complexity of meiotic silencing.
A common hallmark of the conserved RNA interference pathways (RNAi) in all organisms is the activation of the RISC by Argonaute proteins, a process that requires the separation of the siRNA duplex into single strands (Paroo et al. 2007). In the Neurospora quelling pathway, QIP, the product of the quelling defective-2 interacting protein (qip+) gene, is essential for this purpose and acts as an exonuclease that cleaves and removes the nicked passenger strand from the siRNA duplex in a QDE-2-dependent manner (Maiti et al. 2007). Here we report the involvement of qip+ in meiotic silencing and sexual development. This establishes QIP as a common component of quelling and meiotic silencing and reinforces the notion that quelling and meiotic silencing are intrinsically connected (Catalanotto et al. 2000; Cogoni 2001; Lee et al. 2004).
MATERIALS AND METHODS
Molecular biology:
Basic procedures for DNA cloning, analysis, sequencing, Southern blot, and other nucleic acid manipulations were performed as described (Pratt and Aramayo 2002; Pratt et al. 2004).
Strain description and construction:
Escherichia coli K12 XL1-Blue MR (Stratagene) was the host for all bacterial manipulations. All N. crassa strains used in this study are described in Table 1. The formulas for the Vogel's medium N, the Westergaard's medium, and the sugar mixture of Brockman and de Serres have been described by Davis and De Serres (1970). Similarly, growth conditions, conidial spheroplast preparation, and fungal transformation were performed as described (Pratt and Aramayo 2002). Homokaryon purification was performed as described (Pratt and Aramayo 2002; Lee et al. 2003a).
TABLE 1.
Fungal strains used in this study
| Namea | Genotypeb | Originac |
|---|---|---|
| 367-4 | QipΔ∷hph+A | From Yi Liu (Maiti et al. 2007) |
| DLNCR517 | fl; Asm-1-Ama-1+∷hph+A | Progeny from DLNCR455 × DLNCR469 |
| DLNCR518 | fl; Asm-1-Ama-1+∷hph+a | Progeny from DLNCR455 × DLNCR469 |
| DLNCR553 | gle-1+-sgfp+∷hph+A | Progeny from RANCR05A × DLNCT545 |
| DLNCR554 | gle-1+-sgfp+∷hph+a | Progeny from RANCR05A × DLNCT545 |
| DLNCR707 | A | Progeny from RANCR05A × RANCR49A |
| DLNCR708 | a | Progeny from RANCR05A × RANCR49A |
| DLNCR750 | QipΔ∷hph+a | Progeny from 367-4 × KBNCR06A |
| DLNCR769 | ridRIP1; qip+-sgfp+∷hph+, mus-53RIPA | Progeny from DLNCT756 × DLNCR708 |
| DLNCR770 | qip+-sgfp+∷hph+, mus-53RIPa | Progeny from DLNCT756 × DLNCR708 |
| DLNCT807 | ridRIP1, his-3+∷ccg-1(p)-qde-2-sgfp+; inl A | Transformation of DLNCR243 with pDLAM384 |
| DLNCT808 | ridRIP2, his-3+∷ccg-1(p)-qde-2-sgfp+; inl a | Transformation of DLNCR244 with pDLAM384 |
| KBNCR05A | his-3, RspRIP93; fl A | Progeny from DLNCR93 × RANCR50A |
| KBNCR06A | RspRIP93; fl a | Progeny from DLNCR93 × RANCR50A |
| RMNCR113 | QipΔ∷hph+; gle-1+-sgfp+∷hph+A | Progeny from 367-4 × DLNCR554 |
| RMNCR114 | QipΔ∷hph+; gle-1+-sgfp+∷hph+a | Progeny from 367-4 × DLNCR554 |
DLNC, KBNC, RANC, and RMNC indicate strains constructed or provided for this study by Dong W. Lee, Kevin Baker, Rodolfo Aramayo and Ryan Millimaki, respectively.
Allele numbers or designations are: mating type A, A; mating type a, a; Ascospore maturation-africana-1, Ama-1; Ascospore maturation-1, Asm-1; clock-controlled gene-1 promoter, ccg-1(p); fluffy, fl (P); glycine-leucine-phenylalanine-glycine lethal-1, gle-1; histidine-3, his-3 (1-234-723); hygromycin B phosphotransferase, hph; inositol, inl (89601); mutagen-sensitive-53, mus-53; quelling-deficient-2, qde-2; quelling-deficient-2-interacting protein, qip; repeat-induced-deficient, rid; Roundspore, Rsp; S65T green fluorescent protein, sgfp.
Information regarding the construction of parental strains and/or plasmids used will be provided upon request.
Preparation of perithecial tissues for microscopy:
Perithecia were collected (4 days after fertilization), fixed (90 mm Pipes pH 6.9, 10 mm EGTA pH 7.5, 10 mm MgSO4, 0.3% Triton X-100, and 3.7% formaldehyde for 30 min), washed (twice with 1× PBS), and observed (while suspended in 1× PBS, 15% glycerol, pH 7.0 solution containing 0.75 μg/ml DAPI) at 600× magnification. Pictures were taken with Photometrics CoolSnap HQ2 camera using RSimage software from Roper Scientific.
Assaying meiotic silencing:
All crosses are listed in Table 2. For quantification of Asm-1 silencing, progeny from directional crosses were allowed to shoot onto petri dish lids. At 15 days postfertilization (dpf), spores were collected from the lid with 1 ml of water. An aliquot of this population was placed on a hemocytometer and covered with a glass cover slip. Six pictures were taken from each cross and the number of white and black spores was determined from these pictures. Quantification of Rsp silencing was done as follows: at 15 dpf, ascospores were collected in 1 ml water from the lid of the plate then transferred to a microfuge tube. Round-shaped spores are not ejected as efficiently as wild-type spores and tend to ooze out of the perithecia, so the surface of crossing plate was scraped using a glass rod and 3 ml of water. Collected spores were combined and filtered through Miracloth. The combined spores were centrifuged at 13,500 rpm for 10 min to concentrate. An aliquot of ascospores from each cross was transferred to a hemocytometer and covered with a glass cover slip. Six pictures were taken from each cross and the number of round and spindle ascospores were counted. For both Asm-1- and Rsp-silencing experiments, two or three duplicate crosses were performed per experiment. Multiple duplicate crosses were performed to account for plate-to-plate variation. The percentage of wild-type spores was averaged between the crosses and a standard deviation was determined to provide error bars.
TABLE 2.
QIP is involved in meiotic silencing
| Cross no. | Linkage group: Relevant genotypea | I rsp | III qip | VI asm-1 | Parents | Total ascospores examined | Wild-type ascospores (%)b | Observations | Suppression of silencing? |
|---|---|---|---|---|---|---|---|---|---|
| 1 | rsp+ | qip+ | asm-1+ | DLNCR707 × DLNCR708 | 5161 | 86.8 ± 0.5 | Control | n/ac | |
| rsp+ | qip+ | asm-1+ | |||||||
| 2 | RspRIP93 | qip+ | asm-1+ | KBNCR05 × KBNCR06 | 2541 | 0 | Control | n/ac | |
| RspRIP93 | qip+ | asm-1+ | |||||||
| 3 | rsp+ | qip+ | asm-1+ | DLNCR707 × KBNCR06 | 2743 | 6.1 ± 0.9 | Meiotic silencing | No | |
| RspRIP93 | qip+ | asm-1+ | |||||||
| 4 | rsp+ | QipΔ | asm-1+ | 367-4 × KBNCR06 | 4139 | 14.1 ± 1.1 | Experimental | Yes | |
| RspRIP93 | qip+ | asm-1+ | |||||||
| 5 | rsp+ | qip+ | Asm-1-Ama-1+ | DLNCR517 × DLNCR518 | 8382 | 73.1 ± 1.3 | Control | n/ac | |
| rsp+ | qip+ | Asm-1-Ama-1+ | |||||||
| 6 | rsp+ | qip+ | Asm-1-Ama-1+ | DLNCR517 × DLNCR708 | 7187 | 9.2 ± 5.4 | Meiotic silencing | No | |
| rsp+ | qip+ | asm-1+ | |||||||
| 7 | rsp+ | QipΔ | asm-1+ | 367-4 × DLNCR518 | 7034 | 36.6 ± 2.1 | Experimental | Yes | |
| rsp+ | qip+ | Asm-1-Ama-1+ |
RESULTS AND DISCUSSION
QIP is involved in meiotic silencing:
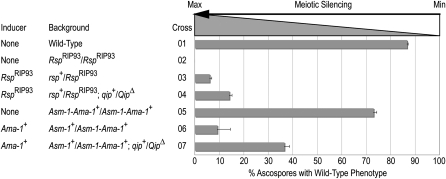
To test the role of QIP in meiotic silencing we used a strain carrying a deletion of the gene or its derivatives (Table 1). Testing its involvement in the homozygous loss-of-function condition proved to be impossible due to the barren phenotype of these crosses (see below). We therefore assayed meiotic silencing in the heterozygous condition following a very-well-established protocol (Lee et al. 2003b, Figure 1, and Table 2). For this we used the best-characterized and sensitive inducers of meiotic silencing, the Roundspore (Rsp) allele RspRIP93 (Pratt et al. 2004) and a chimeric fusion where the promoter of the Ascospore maturation-1 (Asm-1) gene of N. crassa is fused to the coding and trailer region of the Ascospore maturation-1 (Ama-1) gene of N. africana (Asm-1-Ama-1+). Unlike the RspRIP93 allele, which is nonfunctional and methylated, the Asm-1-Ama-1+ allele is both functional and not methylated (data not shown), but induces meiotic silencing due to a series of polymorphisms present in both its coding and 3′-untranslated regions. As expected, the crosses between the “wild-type” strains rsp+ × rsp+ or Asm-1-Ama-1+ × Asm-1-Ama-1+ produced ascospores with the wild-type phenotype (86.8 ± 0.5% and 73.1 ± 1.3%; Figure 1, crosses 1 and 5, respectively), while crosses between the mutant strains RspRIP93 × RspRIP93 produced only round ascospores (Figure 1, cross 2). Unpairing Rsp in an otherwise wild-type background results in the production of ascospores with a mutant round (instead of the wild-type spindle) phenotype (Pratt et al. 2004) (6.1 ± 0.5%; Figure 1, cross 3). Silencing is compromised when RspRIP93 is unpaired in the presence of an unpaired copy of Qip (14.1 ± 1.1%; Figure 1, cross 4). We also induced meiotic silencing by crossing strains carrying the Asm-1-Ama-1+ allele with those carrying the wild-type asm-1+. In heterozygous crosses (i.e., asm-1+ × Asm-1-Ama-1+), Asm-1 unpairing results in efficient silencing as measured by the low production of black, mature ascospores (9.2 ± 5.4%; Figure 1, cross 6). As expected, this silencing is suppressed by unpairing the same reporter gene in the presence of an unpaired copy of Qip (36.6 ± 2.1%; Figure 1, cross 7). Combined, these results suggest that, under these conditions, QIP is involved in meiotic silencing.
Figure 1.—
QIP is required for efficient meiotic silencing. The involvement of qip+ in the silencing induced by homeologous and indel alleles was determined by quantifying the ratio of wild-type to mutant ascospores in each cross. Efficient meiotic silencing results in the production of ascospores that are round instead of spindle shaped for Rsp and white instead of black for Asm-1. The weaker the silencing the higher the percentage of wild-type ascospores produced in the cross. Note that the absence of spindle spores in cross 2 is not due to silencing but to the loss of Rsp function. Similarly, the number of wild-type ascospores observed in cross 5 is due to the nature of the Asm-1–Ama-1+ fusion allele. Crosses are described in Table 2. See text and Pratt et al. (2004) for details.
QIP localization is perinuclear in meiosis:
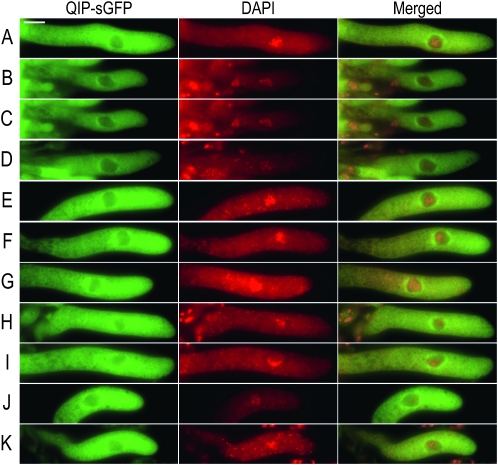
Previous studies have established that components of the meiotic silencing machinery are perinuclearly localized (Shiu et al. 2006; Alexander et al. 2008). Their common localization and function suggests that they work together, possibly through direct interactions (Bardiya et al. 2008). It has been previously established that QIP interacts with QDE-2 (Maiti et al. 2007). To understand how QIP works in meiotic silencing we first determined its subcellular localization in meiosis. We constructed a fusion between qip+ and the reporter gene sgfp+ and inserted the resulting fusion (qip+–sgfp+) at the canonical chromosomal location by gene replacement. Homozygous crosses between these strains were fertile, suggesting the fusion is functional. We observed that QIP–sGFP has a perinuclear localization in meiosis I (Figure 2, A–K). To determine if QDE-2 had a similar localization, we then constructed a qde-2+–sgfp+ fusion gene under the control of the clock controlled gene-1 promoter (ccg-1(p)) and inserted it at the histidine-3 (his-3) chromosomal location (Freitag et al. 2004). In homozygous crosses we were unable to observe perinuclear localization of QDE-2–SGFP in meiosis I (supporting information, Figure S1, A–H). Taken together, these observations are consistent with QIP, an Argonaute-interacting protein, working at the perinuclear region in meiosis, the same region where the Argonaute SMS-2, not QDE-2, is located. While these observations reinforce the notion that QIP–sGFP might be working with SMS-2, they do not establish a direct physical interaction.
Figure 2.—
Localization of QIP–sGFP in meiosis. Pictures show representative asci photographed at the pachytene stage of meiosis I. Columns 1 and 2 show the QIP–sGFP (green) and DAPI (red) signals, respectively. Column 3 shows the corresponding merged images of columns 1 and 2. Tissues were harvested at 4 days postfertilization and processed as described in materials and methods The bar on A, column 1, equals 20 mm.
QIP is essential for early sexual development:
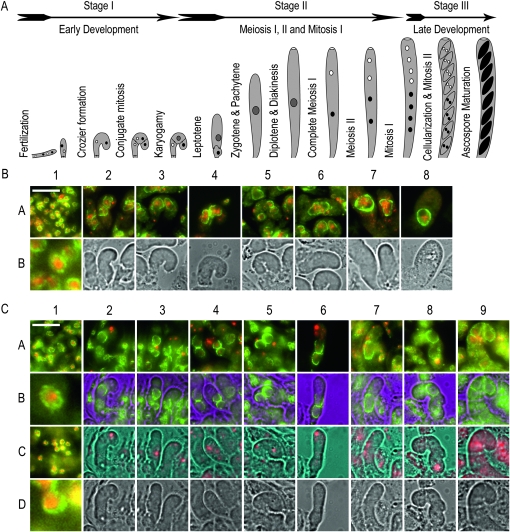
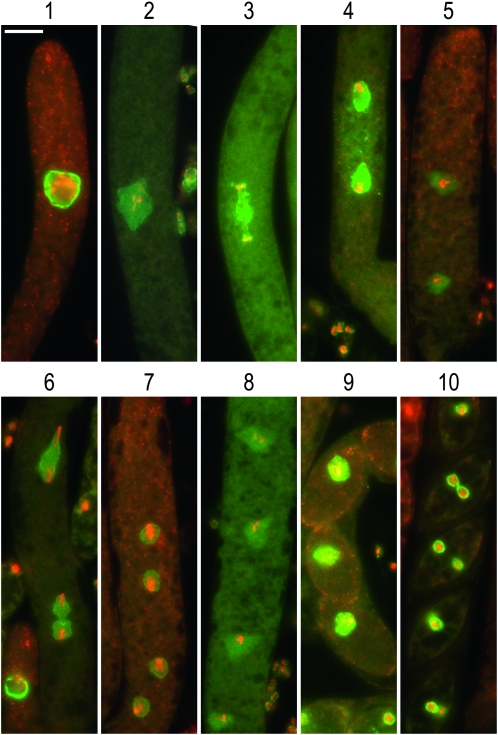
The homozygous barren phenotype of qip loss-of-function mutants was further investigated. Sexual development in Neurospora is complex and can be divided into three stages (Figure 3A). Following fertilization early sexual development is difficult to follow. This is due to the syncytial nature of both the maternal and ascogenous tissue involved and to their colocalization inside the incipient perithecium. To distinguish between these tissues and their corresponding nuclei, we constructed strains carrying canonical fusions of the glycine–leucine–phenylalanine–glycine lethal-1 (gle-1+, NCU01198) nucleopore complex component gene (Murphy and Wente 1996) and sgfp+. GLE-1 is predicted to be localized to the nuclear membrane and this was confirmed in all tissues examined (Figures 3 and 4). Note that during sexual development the size of the nucleus undergoes dramatic changes; from the small nuclei observed in maternal tissue (Figure 3A, stage I, and Figure 3B, 1A and 1B), to the large nuclei observed in ascogenous tissue (Figure 3A, stages I to III, and Figure 3B[2A−8A]). The nuclei reach their maximal size at the pachytene stage of meiosis I (Figure 4[1] ). Following meiosis I and II (Figure 4[2–9]), nuclei regain “normal” GLE-1 signal after mitosis I (Figure 4[10]). In contrast to the wild-type condition, crosses homozygous for strains carrying GLE-1–sGFP in a qipΔ mutant background (Table 1) were severely developmentally compromised. In no circumstances did we observe the formation of large asci or ascospores. Although the formation of maternal tissue appeared to be normal (Figure 3C[1]), the development of ascogenous tissue was compromised at an early stage (Figure 3C[2–9]). A hallmark of ascogenous tissue is the formation of the crozier, a three-celled apparatus consisting of a basal, a tip, and a penultimate cell (see Figure 3A for details). Our observations suggest that while being formed, crozier development seems to be compromised in qip loss-of-function mutants. In summary, our results show that QIP is essential for early sexual development and suggest that it is also required for meiotic silencing.
Figure 3.—
QIP is essential for the completion of early sexual development. (A) Sexual development in N. crassa. The cartoon diagram represents the three main stages (early development, middle development, and late development), from fertilization to ascospore maturation observed during sexual development. Drawings are not to scale. Cartoon was inspired on the work of Raju (1980). (B). Early wild-type development. Maternal and ascogenous tissue corresponding to a wild-type cross (DLNCR553 × DLNCR554; Table 2) was examined. In 1A, 1B, and 2A−8A, GLE-1–sGFP signal is green, whereas DAPI signal is red. 1A shows representative maternal tissue. 1B shows a 10× enlargement of one of the nuclei present in 1A. 2A−8A show merge images of developing croziers as they go through formation, conjugate mitosis, karyogamy, and early meiosis I. 2B−8B show light images. (C). Early QipΔ development. Maternal and ascogenous tissue corresponding to a mutant Qip loss-of-function cross (RMNCR113 × RMNCR114; Table 1) was examined. In 1A, 1B, 1C, and 1D and 2A, 2B−9A, 9B, GLE-1–sGFP signal is green. Whereas in 1A, 1B, 1C, and 1D and 2A, 2C−9A, 9C, DAPI signal is red. 1A and 1C show representative maternal tissue from days 3 and 5 postfertilization, respectively. 1B and 1D show a 10× enlargement of one of the nuclei present in 1A and 1C, respectively. 2A to 9A show merge images of developing croziers as they go through development. 2B to 9B show GLE-1–sGFP localization. 2C−9C show DAPI signal. 2D−9D show light images. All tissues were harvested at 3 and 4 days postfertilization, unless otherwise noted, and processed as described in materials and methods. Pictures were taken at 600×. The bar on 1A equals 20 μm.
Figure 4.—
Localization of GLE-1–sGFP in normal meiosis. The cellular localization of GLE-1–sGFP was followed during meiosis in a cross between strains (DLNCR553 × DLNCR554; Table 1), representing cells in (1) pachytene, (2) metaphase, (3) telophase, (4) interphase, (5) prophase II, (6) anaphase II through telophase II, (7) early interphase II, (8) late interphase II, (9) metaphase IV, and (10) postmitosis II. Stages were assigned following Raju (1980). All stages show the merge image of GLE-1–sGFP (green) and DAPI (red) signals. Tissues were harvested at 5 and 7 days postfertilization and processed as described in materials and methods. The bar in l 1 equals 20 μm. Pictures were taken at 600×.
A substantial component of all eukaryotic genomes studied to date is formed of repeated sequences (Hsieh and Fire 2000). The genome of N. crassa is intriguing in this regard because it has mechanisms, like quelling and meiotic silencing, that arguably have evolved to cope with, and to respond to, the presence of genomic-repeated elements. This is despite the presence of RIP, an efficient repeat-inactivating mechanism. This redundancy suggests that both quelling and meiotic silencing might have biological functions that go beyond simple repeat recognition and silencing. In evolutionary terms, it is safe to think that meiosis is a “late” evolutionary invention (i.e., meiosis vs. mitosis). As such, it is likely that the meiotic silencing machinery was built upon an already existing RNA silencing apparatus. While the exact evolutionary order of emergence between the primitive RNA silencing machineries and RIP is not known, the evolutionary pressures for function imposed by RIP must have selected against the presence of proteins specific for each pathway while favoring sharing of proteins between both pathways. Our finding of an additional common component reinforces this hypothesis.
Acknowledgments
We thank Michelle Yeoman for technical help, Yi Liu for providing strains, and our other lab colleagues for support. This work was supported by U.S. Public Health Service Grants GM58770 to R.A.
Note added in proof: See Xiao et al. in this issue (pp. 119–126) for a related work.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.118422/DC1.
This work is dedicated to the memory of David Dexter Perkins (1919–2007), whose leadership and spirit guided and inspired our work. David, your light shall illuminate our work forever.
References
- Alexander, W. G., N. B. Raju, H. Xiao, T. M. Hammond, T. D. Perdue et al., 2008. DCL-1 colocalizes with other components of the MSUD machinery and is required for silencing. Fungal Genet. Biol. 45 719–727. [DOI] [PubMed] [Google Scholar]
- Aramayo, R., and R. L. Metzenberg, 1996. Meiotic transvection in fungi. Cell 86 103–113. [DOI] [PubMed] [Google Scholar]
- Aramayo, R., and R. J. Pratt, 2010. Meiotic trans-sensing and silencing in Neurospora, pp. 132–144 in Cellular and Molecular Biology of Filamentous Fungi, edited by K. A. Borkovich and D. J. Ebbole. ASM Press, Washington, DC.
- Aramayo, R., Y. Peleg, R. Addison and R. Metzenberg, 1996. Asm-1+, a Neurospora crassa gene related to transcriptional regulators of fungal development. Genetics 144 991–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardiya, N., W. G. Alexander, T. D. Perdue, E. G. Barry, R. L. Metzenberg et al., 2008. Characterization of interactions between and among components of the meiotic silencing by unpaired DNA machinery in Neurospora crassa using bimolecular fluorescence complementation. Genetics 178 593–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich, K. A., L. A. Alex, O. Yarden, M. Freitag, G. E. Turner et al., 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68 1–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto, C., G. Azzalin, G. Macino and C. Cogoni, 2000. Gene silencing in worms and fungi. Nature 404 245. [DOI] [PubMed] [Google Scholar]
- Cogoni, C., 2001. Homology-dependent gene silencing mechanisms in fungi. Annu. Rev. Microbiol. 55 381–406. [DOI] [PubMed] [Google Scholar]
- Cogoni, C., and G. Macino, 1997. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc. Natl. Acad. Sci. USA 94 10233–10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni, C., and G. Macino, 1999. a Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399 166–169. [DOI] [PubMed] [Google Scholar]
- Cogoni, C., and G. Macino, 1999. b Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science 286 2342–2344. [DOI] [PubMed] [Google Scholar]
- Cogoni, C., N. Romano and G. Macino, 1994. Suppression of gene expression by homologous transgenes. Antonie Van Leeuwenhoek 65 205–209. [DOI] [PubMed] [Google Scholar]
- Cogoni, C., J. T. Irelan, M. Schumacher, T. J. Schmidhauser, E. U. Selker et al., 1996. Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. EMBO J. 15 3153–3163. [PMC free article] [PubMed] [Google Scholar]
- Davis, R. H., and F. J. de Serres, 1970. Genetic and microbiological research techniques for Neurospora crassa, pp. 79–143 in Metabolism of Amino Acids and Amines, edited by S. P. Colowick and N. O. Kaplan. Academic Press, New York.
- Freitag, M., P. C. Hickey, N. B. Raju, E. U. Selker and N. D. Read, 2004. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 41 897–910. [DOI] [PubMed] [Google Scholar]
- Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read et al., 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422 859–868. [DOI] [PubMed] [Google Scholar]
- Hsieh, J., and A. Fire, 2000. Recognition and silencing of repeated DNA. Annu. Rev. Genet. 34 187–204. [DOI] [PubMed] [Google Scholar]
- Kutil, B. L., K. Y. Seong and R. Aramayo, 2003. Unpaired genes do not silence their paired neighbors. Curr. Genet. 43 425–432. [DOI] [PubMed] [Google Scholar]
- Lee, D. W., J. R. Haag and R. Aramayo, 2003. a Construction of strains for rapid homokaryon purification after integration of constructs at the histidine-3 (his-3) locus of Neurospora crassa. Curr. Genet. 43 17–23. [DOI] [PubMed] [Google Scholar]
- Lee, D. W., R. J. Pratt, M. McLaughlin and R. Aramayo, 2003. b An argonaute-like protein is required for meiotic silencing. Genetics 164 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D. W., K.-Y. Seong, R. J. Pratt, K. Baker and R. Aramayo, 2004. Properties of unpaired DNA required for efficient silencing in Neurospora crassa. Genetics 167 131–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti, M., H. C. Lee and Y. Liu, 2007. QIP, a putative exonuclease, interacts with the Neurospora argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev. 21 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, R., and S. R. Wente, 1996. An RNA-export mediator with an essential nuclear export signal. Nature 383 357–360. [DOI] [PubMed] [Google Scholar]
- Paroo, Z., Q. Liu and X. Wang, 2007. Biochemical mechanisms of the RNA-induced silencing complex. Cell Res. 17 187–194. [DOI] [PubMed] [Google Scholar]
- Pratt, R. J., and R. Aramayo, 2002. Improving the efficiency of gene replacements in Neurospora crassa: a first step towards a large-scale functional genomics project. Fungal Genet. Biol. 37 56–71. [DOI] [PubMed] [Google Scholar]
- Pratt, R. J., D. W. Lee and R. Aramayo, 2004. DNA methylation affects meiotic trans-sensing, not meiotic silencing, in Neurospora. Genetics 168 1925–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju, N. B., 1980. Meiosis and ascospore genesis in Neurospora. Eur. J. Cell Biol. 23 208–223. [PubMed] [Google Scholar]
- Shiu, P. K. T., B. N. Raju, D. Zickler and R. Metzenberg, 2001. Meiotic silencing by unpaired DNA. Cell 107 905–916. [DOI] [PubMed] [Google Scholar]
- Shiu, P. K. T., D. Zickler, N. B. Raju, G. Ruprich-Robert and R. L. Metzenberg, 2006. SAD-2 is required for meiotic silencing by unpaired DNA and perinuclear localization of SAD-1 RNA-directed RNA polymerase. Proc. Natl. Acad. Sci. USA 103 2243–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, H., W. G. Alexander, T. M. Hammond, E. C. Boone, T. D. Perdue et al. 2010. QIP, a protein that converts duplex siRNA into single strands, is required for meiotic silencing by unpaired DNA. Genetics 186 119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]