Abstract
The three Drosophila atypical soluble guanylyl cyclases, Gyc-89Da, Gyc-89Db, and Gyc-88E, have been proposed to act as oxygen detectors mediating behavioral responses to hypoxia. Drosophila larvae mutant in any of these subunits were defective in their hypoxia escape response—a rapid cessation of feeding and withdrawal from their food. This response required cGMP and the cyclic nucleotide-gated ion channel, cng, but did not appear to be dependent on either of the cGMP-dependent protein kinases, dg1 and dg2. Specific activation of the Gyc-89Da neurons using channel rhodopsin showed that activation of these neurons was sufficient to trigger the escape behavior. The hypoxia escape response was restored by reintroducing either Gyc-89Da or Gyc-89Db into either Gyc-89Da or Gyc-89Db neurons in either mutation. This suggests that neurons that co-express both Gyc-89Da and Gyc-89Db subunits are primarily responsible for activating this behavior. These include sensory neurons that innervate the terminal sensory cones. Although the roles of Gyc-89Da and Gyc-89Db in the hypoxia escape behavior appeared to be identical, we also showed that changes in larval crawling behavior in response to either hypoxia or hyperoxia differed in their requirements for these two atypical sGCs, with responses to 15% oxygen requiring Gyc-89Da and responses to 19 and 25% requiring Gyc-89Db. For this behavior, the identity of the neurons appeared to be critical in determining the ability to respond appropriately.
SENSORY input is critical for an animal to produce a relevant response to changes in its environment. For animals to survive in hypoxic environments, they need to acquire a wide variety of physiological and behavioral adaptations. Many studies have focused on long-term changes in response to hypoxia, especially those involving the hypoxia inducing factor-1 (HIF-1) and other modifications in gene expression (Semenza 2000; Hochachaka and Rupert 2003). In addition, animals also show short-term responses to hypoxia that occur in a time frame of seconds or minutes and likely do not involve transcription-mediated events. The mammalian carotid body is a peripheral chemoreceptor that senses a number of blood-borne stimuli, including hypoxia and hypercapnia, transducing them into neural discharges that initiate a number of cardiorespiratory reflexes (Gonzalez et al. 1994; López-Barneo et al. 2008). Both long-term and short-term responses to hypoxia critically depend on the ability to detect decreases in O2 supply; however, the molecular bases for O2-sensing systems have only recently begun to be identified (Gray et al. 2004; Morton 2004b; Vermehren et al. 2006). For example, within the carotid body, heme oxygenase-2 has been proposed as the O2 sensor for the sensing of hypoxia in vertebrates (Kemp 2005; Ortega-Sáenz et al. 2006).
Recently, members of a novel class of soluble guanylyl cyclases (sGCs), the atypical sGCs, have been identified as likely molecular O2-sensors in invertebrates. Guanylyl cyclases catalyze the synthesis of the intracellular second messenger cyclic guanosine monophosphate (cGMP), which mediates a wide variety of physiological and developmental events in both invertebrates and vertebrates (Lucas et al. 2000; Morton and Hudson 2002).The nematode Caenorhabditis elegans has been shown to have a strong behavioral preference for 5–10% O2 and mutations in their sGC genes alter this O2 preference (Gray et al. 2004; Chang et al. 2006). Recent studies in C. elegans also showed that URX neurons, which express the sGC subunits gyc-35 and gyc-36, are required for responses to O2 upshifts, while the gyc-31 sGC subunit expressed in BAG neurons mediates responses to O2 downshifts (Zimmer et al. 2009). The Drosophila melanogaster genome contains five sGC subunit-encoding genes. As in mammalian systems, flies express the conventional α- and β-subunits (Gycα-99B and Gycβ-100B) that function as obligate heterodimers and can be activated by nitric oxide (NO) (Morton et al. 2005). In contrast, the remaining three sGC subunits, Gyc-88E, Gyc-89Da, and Gyc-89Db, belong to the atypical sGC family and have been shown to be regulated by oxygen (Morton 2004a).
The Gyc-89Da and Gyc-89Db subunits are up to 80% identical throughout their coding regions and are biochemically indistinguishable from each other, suggesting that they have arisen from a recent duplication (Morton et al. 2005). Transient expression of different combinations of the Drosophila atypical sGCs in COS-7 cells showed that Gyc-89Da and Gyc-89Db form obligate heterodimers with Gyc-88E, and are potently activated in the absence of O2, therefore suggesting a function as O2 sensors (Morton 2004b; Vermehren et al. 2006; Morton and Vermehren 2007). Gyc-88E is also active as a homodimer (Morton 2004b; Huang et al. 2007), directly binds O2 and is also activated by reduced O2 (Huang et al. 2007). In the current study we examine the role of the atypical sGCs in larval behavioral responses to changes in O2 levels.
MATERIALS AND METHODS
Fly stocks and genetics:
Drosophila stocks were raised on standard cornmeal–agar–molasses medium at 25° with a 12 hr light/12 hr dark photocycle. The Gyc-89Da mutant (w1118; PBac{w+mC=RB}Gyc-89Dae01821) and Gyc-89Db mutant (w1118; Mi{3xP3-EGFP.ET1}Gyc-89DbMB03197) lines were obtained from the Bloomington Drosophila Stock Center (stocks 17991, 23519). The Gyc-89Da/Gyc-89Db double mutant was obtained by standard meiotic recombination, selecting animals that expressed both the white+ and green fluorescent protein (GFP). Deficiency lines that covered portions of the genome including Gyc-89Da/Gyc-89Db, Gyc-88E and cyclic nucleotide-gated ion channel (cng) genes were obtained from the Bloomington Drosophila Stock Center (stocks 7987, 24137, 7545, 25442, 23692). The UASGyc-88E and UASCNG RNAi lines were obtained from the Vienna Drosophila RNAi Center (VDRC stocks 21798 for Gyc-88E, and 101745, 29046, 28625, 40964, 100882, 11817, 11816, and 38850 for cng). Generation of the UASbPDE5 (bovine phosphodiesterase 5) overexpression line and the RNAi lines for the cGMP-dependent protein kinases, dg1 and dg2, have previously been described (Broderick et al. 2004; S.-A. Davies, unpublished data). Fly lines containing two Gyc-88E point mutations (V474M and S451F) were obtained from the Seattle Fly tilling project (http://tilling.fhcrc.org; lines Z31083 and Z31071, respectively). All crosses were carried out using standard methods at 25° (Greenspan 2004).
Gyc-89Da, Gyc-89Db, and Gyc-88E rescue fly lines:
Gyc-89Da ORF was amplified by PCR, cloned into pcDNA3.1 (Invitrogen, Carlsbad, CA) as described previously (Morton et al. 2005), and then subcloned into the pUASt vector (https://dgrc.cgb.indiana.edu/) using the KpnI and XbaI restriction sites (UASGyc-89Da). The Gyc-89Db ORF was obtained from the Berkeley Drosophila Genome Project (http://www.fruitfly.org/; clone ID GH09958), subcloned into pcDNA3.1, and then into the pUASt vector using the KpnI and XbaI restriction sites (UASGyc-89Db). Gyc-88E ORF was amplified by PCR and cloned into pcDNA3.1, as described previously (Morton et al. 2005), and then subcloned into the pUASt vector using the KpnI and NotI restriction sites (UASGyc-88E). All constructs were confirmed by sequencing and injected into w1118 embryos (http://www.thebestgene.com/). Four insertion lines were recovered for Gyc-89Da-pUAST, 11 for Gyc-89Db-pUAST, and 6 for Gyc-88E-pUAST. Chromosomal locations of the transposon insertions were determined by standard crossing methods to a balancer line (w1118; CyO/nub1b1nocScoli1stw3; MKRS/TM6B, Tb1, Bloomington stock 3703). Most of the parental rescue lines were viable and healthy, suggesting that the insertion of the constructs is not deleterious on its own. The only exception was UASGyc-89Db in the Gyc-89Da−/− background, which had to be kept over a MKRS balancer, with sufficient double homozygous flies eclosing for the rescue crosses. The transgenic GAL4 driver lines for Gyc-89Da and Gyc-89Db (Gyc-89DaGAL4, Gyc-89DbGAL4) were generated by germline transformation as described earlier (Morton et al. 2008).
Fly lines with the Gyc-89Da and Gyc-89Db rescue constructs (Gyc-89DaGAL4, Gyc-89DbGAL4, UASGyc-89Da, UASGyc-89Db) inserted on the second chromosome were crossed with the single and double mutant fly lines (third chromosome, standard crossing methods) to create double homozygous fly lines, with the GAL4 and UAS insertions on the second and the Gyc-89Da and Gyc-89Db mutations on the third chromosomes. Similarly, Gyc-88E rescue constructs (Gyc-89DaGAL4, Gyc-89DbGAL4, UASGyc-88E) inserted on the second chromosome were crossed with the Gyc-88E point mutations (third chromosome, standard crossing methods) to create double homozygous fly lines, with the GAL4 and UAS insertions on the second and the Gyc-88E point mutations on the third chromosomes. For the rescue experiments, one promoter GAL4 and one UAS line with the same mutant background were crossed with each other, obtaining larvae expressing the Gyc-89Da, Gyc-89Db, or Gyc-88E subunit in either the Gyc-89Da or Gyc-89Db neurons in each mutant background.
Reverse transcription PCR (RT–PCR):
Total RNA was isolated from third-instar larvae (n = 15) using Trizol (Invitrogen). Five micrograms of total RNA were used for synthesizing cDNA using Superscript III reverse transcriptase with oligo(dT) primers (Invitrogen) according to the manufacturer's protocol. Two rounds of PCR reactions (50 μl) were performed using 3 μl of cDNA for the first round and 1 μl of PCR product for the second round. Both PCR amplifications rounds used 35 cycles with an annealing temperature of 55°. Primers sequences for the Gyc-89Da and Gyc-89Db subunits as well as the housekeeping gene β-actin were designed using Primer3 online software and synthesized by Invitrogen (supporting information, Table S1). To control for equivalent levels of input RNA, we used primers targeting β-actin.
Real-time PCR:
Total RNA was extracted from adult fly heads (n = 20) using TRIzol following the manufacturer's instructions (Invitrogen, Carlsbad, CA), resuspended in 10 mm Tris pH 8.5, and stored at −80° until use. To make cDNA, 1 μg of total RNA was reverse transcribed (20 μl) using the Tetro cDNA synthesis kit (Bioline, Taunton, MA) with oligo(dT) primers. Real-time PCR was performed using a MX3000P real-time PCR system (Stratagene) in a final volume of 10 μl using Sensimix Plus SYBR master mix (Quantace, Taunton, MA), 2 μl of cDNA (diluted 1:4), and forward and reverse primers (3 μm). Primers sequences for the Gyc-89Da, Gyc-89Db, and Gyc-88E as well as the housekeeping gene β-actin were designed using Primer3 online software and synthesized by Invitrogen (Table S2). The real-time amplification data were collected continuously and analyzed using the quantification software supplied with the MX3000P real-time PCR system.
Larval hypoxia escape responses:
Larval escape responses to hypoxia were performed as previously described (Wingrove and O'farrell 1999) with the following modifications. Five third-instar larvae (∼100 hr after hatching) were rinsed in water and placed on a small pile of fresh yeast paste with the consistency of toothpaste (∼0.8 ml) in a petri dish. Larvae were allowed to burrow into the yeast for 5 min, replacing the ones that failed to burrow. The dish was placed in a plexiglass chamber (12 × 12 × 23 cm) and exposed to a mixture of O2 and N2 flowing through the chamber at 4–5 liters/min, which was constantly monitored with an O2 monitor (model 5120, Ohmeda, Helsinki, Finland). Larvae were monitored for 20 min and scored as “escaping” when they exited their vertical feeding position and began exploratory behaviors. The time at which each individual larvae first exited the food was recorded. Each animal was tested once.
Gyc-88E Point mutation constructs:
To test the enzyme activity of the Gyc-88E point mutations uncovered by the Seattle Tilling Project, we used site-directed mutagenesis to replace single bases in the Gyc-88E pcDNA3.1 expression vector (Langlais et al. 2004). Point mutations were introduced into Gyc-88E using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA), the vectors sequenced and enzyme activity measured as described previously (Langlais et al. 2004).
Channelrhodopsin-2 expression assays:
A modified version of channelrhodopsin-2 (ChR2 H134R) (Pulver et al. 2009) was expressed in Gyc-89Da and Gyc-89Db neurons by crossing the Gyc-89DaGAL4 and Gyc-89DbGAL4 driver lines with a UASH134R-ChR2 line, kindly provided by Dr. L. C. Griffith (Brandeis University, Waltham, MA). Stable lines that expressed ChR2 H134R in either the Gyc-89Da or Gyc-89Db neurons were generated and larvae were raised on a diet containing 1 mm all trans-retinal (Sigma). Control w1118 larvae were also raised on this diet. Third-instar larvae were placed in a small patch of yeast paste (∼1 cm diameter) placed on a layer of 1% agarose in a 10-cm petri dish, which was then inverted. Neurons expressing ChR2 H134R were activated by illuminating the yeast paste with an ultrabright blue LED (Luxeon III Star; 470-nm wavelength, Thor Labs, Newton, NJ). Trains of light pulses were generated by a custom-made digital circuit using three monostables to control train duration, pulse width, and pulse separation. Digital pulses drove a BuckPuck current source (Thor Labs) with an intensity control (LuxDrive) that provided current pulses up to 700 mA to the LED. Rated luminous flux at full power was 23 L. The pulse generator was capable of delivering pulse trains of 5–50 sec in length, consisting of individual pulses 10-420 msec long, separated by 10–420 msec. Maximum intensity illumination from the LED was focused using a 10× stereo microscope eyepiece and focused so that yeast was illuminated with a spot of light approximately 5 mm in diameter. The behavioral responses of the larvae to illumination were recorded on a digital video recorder (Canon, Vixia HG21) fitted with a +4 and a +2 magnifying lens and a red filter to reduce glare. Three third-instar larvae were placed in each patch of yeast paste and the number leaving the food during illumination was recorded.
Effect of hypoxia and hyperoxia on crawling behavior in larvae:
To assess the behavioral response of freely moving larvae to hypoxia and hyperoxia, the number of stops and turns made by larvae crawling on agarose was recorded after they had been transferred to a chamber containing different oxygen concentrations. Third-instar larvae were removed from the food, rinsed with water, placed in a petri dish (85 mm) containing 1.5% agarose, and allowed to acclimate for 10 min. Larvae were moved to a fresh 1.0% agarose dish. The dish was then transferred to a custom assay apparatus (10.5 × 8 × 5.5-in. plexiglass chamber with two holes drilled into it for tubing). Oxygen levels within this chamber were controlled using a ProOx oxygen controller (model 110, BioSpherix, Lacona, NY). This system injected either oxygen or nitrogen into the chamber until a desired concentration had been reached and then maintained this concentration with additional gas as required. The number of stops and turns made by each larva during the first 2 min in the chamber was then recorded.
Scanning electron microscopy (SEM):
The terminal sensory cones were imaged with an FEI Quanta 200 ESEM (FEI, Hillsborough, OR) using the low-vacuum environmental SEM facility that allows fresh biological samples to be imaged with no fixation or coating. Third-instar larvae were killed by placing them in warm water for 20 min followed by transfer to the imaging chamber.
Statistical analysis:
All statistical analyses were carried out with Prism 4 software (GraphPad Software Inc., San Diego CA). Unless otherwise stated the data were analyzed using one-way ANOVA followed by Dunnett's post-hoc test. Where shown the significance levels are as follows: *P < 0.05, **P < 0.01, ***P < 0.001.
RESULTS
Characterization of flies with disrupted expression of the atypical sGCs:
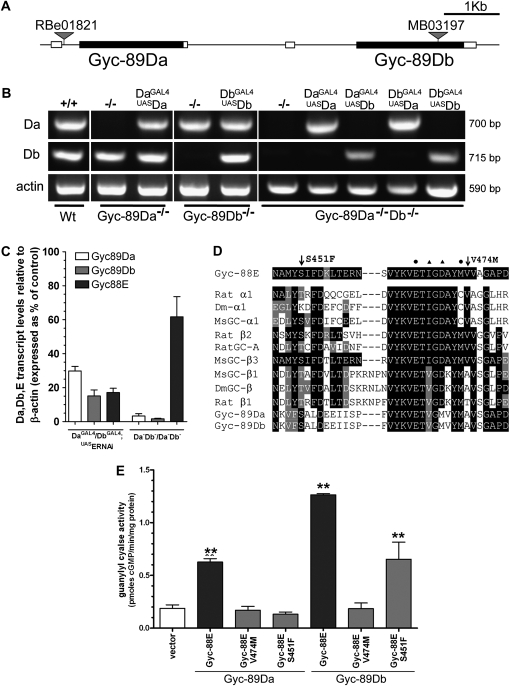
Heterologous expression of the three atypical sGC subunits in COS-7 cells has shown that they are activated by hypoxia and therefore could function as O2 sensors (Morton 2004b). To examine the role of these enzymes in vivo, we identified and characterized fly lines with disrupted atypical sGC genes. Fly lines with transposon insertions 494 bp upstream of the translational initiation codon of Gyc-89Da (Gyc-89Dae01821) and within the Gyc-89Db ORF, 1510 bp downstream of the ATG start site (Gyc-89DbMB03197), were used (Figure 1A).
Figure 1.—
Characterization of Gyc-89Da, Gyc-89Db, and Gyc-88E mutants. (A) Schematic showing the position of Gyc-89Da and Gyc-89Db and transposon insertions. The coding and untranslated regions are represented as solid and open boxes, respectively. (B) Expression levels of Gyc-89Da and Gyc-89Db in the different mutant backgrounds and rescue fly lines. The size of the PCR products is shown on the right. Actin was used as loading control and is shown on the bottom. (C) Real-time PCR of Gyc-89Da, Gyc-89Db, and Gyc-88E transcripts in adult flies expressing Gyc-88E RNAi (UASERNAi) in both Gyc-89Da and Gyc-89Db neurons and in Gyc-89Da/Gyc-89Db double mutant flies. Transcript levels were normalized to the levels of β-actin and expressed as percentages of wild-type controls, mean ± SEM, n = 3. All three transcripts are significantly reduced in flies expressing Gyc-88E RNAi (paired t-test). Only Gyc-89Da and Gyc-89Db were significantly reduced in the Gyc-89Da−/−Db−/− double mutants (paired t-test). (D) Gyc-88E point mutations affect conserved residues in the GTP-binding domain. Multiple sequence alignment of a variety of GCs showing the positions of the two point mutations (arrows) and their proximity to residues that have been modeled to interact with the GTP substrate (Liu et al., 1997). These residues are marked (▴) for α- and (•) for β-subunit residues. Dm-α1: Drosophila Gycα-99B. MsGC-α1, MsGC-β1. MsGC-β3. Manduca sexta α1, β1, and β3 subunits. Rat GC-A is the homodimeric receptor GC-A. (E) Enzyme activity of the Gyc-88E point mutations. Plasmids coding for Gyc-88E with each point mutation were transiently transfected into COS-7 cells together with either Gyc-89Da or Gyc-89Db and guanylyl cyclase activity was measured. Activity levels are expressed as the mean ± SEM of four determinations.
We used RT–PCR to determine whether the expression of the Gyc-89Da or Gyc-89Db genes was affected by these transposon insertions. In the Gyc-89Dae01821 mutant (Gyc-89Da−/−) we were unable to detect the Gyc-89Da transcript, while the expression of Gyc-89Db was normal. Similarly, Gyc-89Db was undetectable in the Gyc-89DbMB03197 mutant (Gyc-89Db−/−) larvae and expression of Gyc-89Da was normal (Figure 1B). We then recombined Gyc-89Dae01821 with Gyc-89DbMB03197 flies to obtain a double mutant (Gyc-89Da−/−Db−/−). RT–PCR analysis showed that both Gyc-89Da and Gyc-89Db subunits were undetectable in this fly line (Figure 1B). Occasionally we were able to detect a faint, smaller band for the Gyc-89Db gene in the double mutant. The sequence of this transcript showed that both the transposon and part of Gyc-89Db were spliced out, resulting in a loss of 39 amino acids (position 500–539) in the catalytic domain, which probably resulted in a subunit that was enzymatically inactive.
Gyc-88E is the obligatory partner of Gyc-89Da and Gyc-89Db (Langlais et al. 2004; Morton et al. 2005). Since no lines with transposons in the Gyc-88E gene are currently available, we used RNA interference (RNAi) (Montgomery and Fire 1988) to reduce the levels of Gyc-88E in specific subsets of cells. When we expressed the Gyc-88E RNAi in both the Gyc-89Da and Gyc-89Db neurons, real-time PCR showed that the levels of Gyc-88E transcript were reduced to about 20% of that seen in controls. Interestingly the levels of Gyc-89Da and Gyc-89Db transcripts were also reduced to about 20% of the control values (Figure 1C). This suggests that transcript levels are regulated in part by the levels of the active heterodimeric enzyme. To determine whether there was a reciprocal relationship between the subunits, we measured the levels of Gyc-88E transcripts in the Gyc-89Da−/−Db−/− double mutants and found that they were also reduced, in this case to 62% of controls (Figure 1C).
We utilized the Seattle Tilling project to identify point mutations in Gyc-88E and uncovered two point mutations, S451F and V474M, in Gyc-88E that resulted in the substitution of residues in the portion of the catalytic domain predicted to determine substrate specificity of GTP binding. Multiple sequence alignment showed that these residues are conserved in several classes of guanylyl cyclases across several species (Figure 1D). Both Gyc-88E mutant lines are homozygous lethal, but as the lethality was complemented by deficiencies that covered Gyc-88E it is likely that the lethality is due to second-site mutations. To determine the effect that these point mutations had on the enzyme activity of Gyc-88E, we made constructs in which we introduced the same mutations in the native Gyc-88E subunit and expressed them in COS-7 cells. Wild-type Gyc-88E is active as a homodimer when expressed in COS-7 cells, although it shows higher levels of activity when co-expressed with either Gyc-89Da or Gyc-89Db (Langlais et al. 2004). No detectable activity was measured when Gyc-88EV474M or Gyc-88ES451F were transiently expressed in the absence of additional subunits (data not shown). Similarly, when either of these mutated subunits was co-expressed with Gyc-89Da, no activity was detected (Figure 1E). By contrast when co-expressed with Gyc-89Db, Gyc-88EV474M showed no activity, whereas Gyc-88ES451F showed about 50% of the activity compared to the wild-type subunit (Figure 1E). These results suggest that Gyc-88EV474M acts as a null mutation whereas Gyc-88ES451F is hypomorphic.
We used the GAL4-UAS system (Brand and Perrimon 1993) to rescue these mutant lines, by subcloning the Gyc-88E, Gyc-89Da, and Gyc-89Db cDNA constructs (Langlais et al. 2004; Morton et al. 2005) into the pUASt vector. We then used these fly lines in combination with the previously described Gyc-89Da and Gyc-89Db promoter-GAL4 lines (Morton et al. 2008) to express either subunit in either subset of cells in all the different mutant backgrounds. To test the expression of Gyc-89Da and Gyc-89Db driven by the GAL4-UAS system, we performed an RT–PCR analysis, which showed that the expression of both genes was restored in the different mutant backgrounds (Figure1B and Figure S1).
Gyc-89Da and Gyc-89Db are required for a normal larval hypoxia escape response:
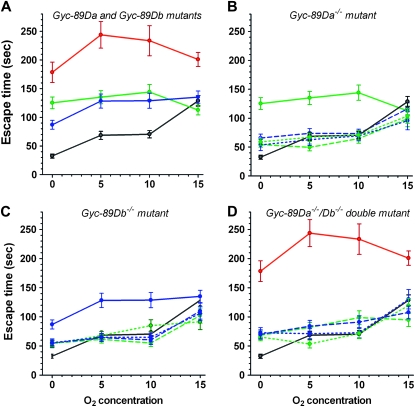
Drosophila larvae feed while buried in their food, with their posterior spiracles protruding above the surface. In response to reduced O2 levels they rapidly withdraw from the food and begin exploratory movements (Wingrove and O'farrell 1999). To study the role of the atypical sGCs in this hypoxia escape response, we placed third-instar larvae onto a pile of fresh yeast in normal atmospheric air (21% O2) and allowed them to burrow into the food and commence feeding. We then placed the dish in a chamber with low O2 concentrations (0, 5, 10, and 15%), and monitored the time it took them to completely crawl out of the yeast and start the exploratory behavior. Wild-type larvae responded rapidly to low O2 conditions by withdrawing from the food (Figure 2A). The time taken to leave the food was shortest for the lowest O2 concentration tested and gradually increased with increasing O2 concentration. Larvae mutant for either Gyc-89Da or Gyc-89Db showed a significant increase in the escape time in response to hypoxia when compared to wild-type controls for 0, 5, and 10% O2 concentrations and showed no difference in response at 15% O2 (Figure 2A). Larvae lacking both Gyc-89Da and Gyc-89Db subunits took significantly longer than either of the single mutants to leave the food and took significantly longer than wild-type larvae for all four O2 concentrations (Figure 2A).
Figure 2.—
Gyc-89Da and Gyc-89Db are required for a normal hypoxia escape response. The time taken for third-instar larvae to exit yeast paste when exposed to four different O2 concentrations, 0, 5, 10, and 15%, was measured. (A) Gyc-89Da and Gyc-89Db single and double mutants. Single Gyc-89Da−/− and Gyc-89Db−/− mutant larvae (green and blue, respectively), as well as double Gyc-89Da−/−Db−/− mutant larvae (red), took significantly longer to withdraw from the food compared to control wild-type larvae (black) at 0, 5, and 10% O2 (P < 0.0001). At 15% O2 only the Gyc-89Da−/−Db−/− double mutant larvae showed a significant delay compared to control larvae (P < 0.01). (B) Rescue of Gyc-89Da single mutants. The increased response time in Gyc-89Da−/− mutant larvae (green solid line) was rescued by expressing either the Gyc-89Da (green dashed/dotted lines) or Gyc-89Db cDNA (blue dashed/dotted lines) in either the Gyc-89Da (dashed lines) or Gyc-89Db (dotted lines) neurons in the Gyc-89Da−/− mutant background at 0, 5, and 10% O2. (C) Rescue of Gyc-89Db single mutants. Similarly, the increased response time in Gyc-89Db−/− mutant larvae (blue solid line) was rescued by expressing either the Gyc-89Db (blue dashed/dotted lines) or Gyc-89Da cDNA (green dashed/dotted lines) in the Gyc-89Db (dashed lines) or Gyc-89Da (dotted lines) neurons in the Gyc-89Db−/− background at 0, 5, and 10% O2. (D) Rescue of Gyc-89Da−/−Db−/− double mutants. Expression of either Gyc-89Da (green dashed/dotted lines) or Gyc-89Db (blue dashed/dotted lines) subunits into either Gyc-89Da or Gyc-89Db neurons (dashed lines for the correct expression, dotted lines for the reversed rescue) in the double Gyc-89Da−/−Db−/− mutant larvae (red solid line) significantly decreased the time of response concentration compared to wild-type control larvae (P < 0.001). All values show mean ± SEM. n = 40-60 for mutant fly lines; N=20-25 for rescue fly lines.
We also varied the number of copies of either Gyc-89Da or Gyc-89Db subunits using a combination of the single and double mutants and small deficiencies to generate larvae that contained 0, 1, 2, 3, or 4 wild-type copies of these two genes. These larvae were all tested for their response to hypoxia and not surprisingly, there was a gradual increase in the delay to leave the food as the number of mutant copies increased. Interestingly, the relationship fit an exponential curve better than a linear regression (Figure S2) suggesting that there might be some synergism between these proteins.
To ensure that the increased response times of the mutant lines was not due to defects in their locomotion, we measured the distance third-instar larvae traveled during 5 min on 1% agarose plates in 21% O2 (Osborne et al. 1997). We found no significant differences in the distance traveled in 5 min for the different genotypes (wild type, 3.0 ± 0.4 cm; Gyc-89Da−/−, 2.6 ± 0.3 cm; Gyc-89Db−/−, 2.7 ± 0.5 cm; and Gyc-89Da−/−Db−/−, 3.16 ± 0.29 cm, n = 10 for each, mean ± SEM).
To confirm that the increased time to respond to hypoxia was due to the loss of Gyc-89Da and Gyc-89Db we crossed GAL4 driver lines that drive expression in either the Gyc-89Da or Gyc-89Db cells (Langlais et al. 2004; Morton et al. 2008) with the UASGyc-89Da and UASGyc-89Db cDNA rescue lines into each of the three mutant backgrounds (see Table S3 for hypoxia escape response values of parental lines). On the basis of our previous data, the promoter regions used in the Gyc-89DaGAL4 and Gyc-89DbGAL4 constructs drive expression in populations of neurons that are broadly nonoverlapping, with only a few sensory neurons observed that co-express both Gyc-89Da and Gyc-89Db (Morton et al. 2008). The delayed hypoxia escape response seen in the single mutants was fully rescued for 0, 5, and 10% O2 concentration by expressing either Gyc-89Da or Gyc-89Db in either the Gyc-89Da or the Gyc-89Db neurons (Figure 2, B and C). Interestingly, expressing Gyc-89Da or Gyc-89Db in the Gyc-89Db neurons in a Gyc-89Da−/− background also rescued the escape response time (Figure 2B), as did expressing Gyc-89Da or Gyc-89Db in the Gyc-89Da neurons in a Gyc-89Db−/− background (Figure 2C). This suggests that both subunits and both expression patterns are functionally equivalent to the other. Similarly, the longer delay seen in the double mutants was also rescued by expressing either Gyc-89Da or Gyc-89Db in either set of neurons (Figure 2D). Although expressing a single copy of either Gyc-89Da or Gyc-89Db in either set of neurons significantly reduced the time taken to leave the food, it only partially rescued the response at 0% O2, as the time taken to leave the food was still significantly longer than in wild-type larvae (Figure 2D).
Downregulation of Gyc-88E levels in Gyc-89Da or Gyc-89Db expressing neurons reduces the hypoxia escape response:
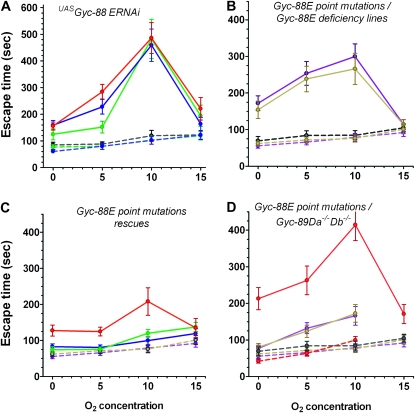
To test the effects of reducing the levels of Gyc-88E, the obligatory partner of Gyc-89Da and Gyc-89Db, we expressed Gyc-88E RNAi in either the Gyc-89Da or the Gyc-89Db neurons. We made stable double homozygous fly lines (Gyc-89DaGAL4; UASGyc-88E RNAi and Gyc-89DbGAL4; UASGyc-88E RNAi) and tested the larvae for their hypoxia escape response. There was no apparent effect on locomotion, development, or survival and the adults eclosed at the same time as their parental controls (data not shown). In contrast, expression of Gyc-88E dsRNA in either Gyc-89Da or Gyc-89Db neurons significantly delayed the response to hypoxia at 0, 5, and 10% O2 concentrations, but no difference was seen at 15% O2. When Gyc-88E RNAi was expressed in both Gyc-89Da and Gyc-89Db neurons, the response was not significantly different from the expression of Gyc-88E RNAi in either population of neurons alone (Figure 3A).
Figure 3.—
Gyc-88E subunit is required for a normal hypoxia escape response. (A) Gyc-88E RNAi. Reducing Gyc-88E expression in Gyc-89Da (green), Gyc-89Db (blue), or both sets of neurons (red) using RNA interference significantly increased the escape response at 0, 5, and 10% O2 concentrations (P < 0.0001) but had no effect at 15% O2 compared to controls (Gyc-89DaGAL4, green dashed line; Gyc-89DbGAL4, blue dashed line; and UASGyc-88E-ERNAi, black dashed line). (B) Gyc-88E point mutations over deficiencies. Larvae with one copy of the point mutation (Gyc-88EV474M/Df in brown and Gyc-88ES451F/Df in purple, continuous lines) took significantly longer to withdraw from the food compared to larvae with one wild-type copy of Gyc-88E (Gyc-88E Df/+ in gray; Gyc-88EV474M/+ in brown; and Gyc-88ES451F/+ in purple; dashed lines) at 0, 5, and 10% O2 (P < 0.0001). (C) Rescue of Gyc-88E point mutations. The Gyc-88EV474M/S451F trans-heterozygote (red) shows a significantly delayed response at 0, 5, and 10% O2 (P < 0.0001) compared to either heterozygote Gyc-88EV474M/+ (brown dashed line) and Gyc-88E S452F/+ (purple dashed line) control larvae that is restored by expressing wild-type Gyc-88E in either Gyc-89Da or Gyc-89Db neurons (solid green and blue lines, respectively) neurons. (D) Gyc-88E point mutations and deficiencies over Gyc-89Da−/−Db−/− double mutant. Larvae trans-heterozygous for the Gyc-88E deficiency and Gyc-89Da−Db− double mutant (red continuous line) took significantly longer to withdraw from the food compared to larvae with only one Gyc-89Da and Gyc-89Db subunits (Gyc-89Da−Db−/+, red dashed line) and larvae with one copy of Gyc-88E (same controls as in 3B) at 0, 5, and 10% O2 (P < 0.0001). Similarly, larvae trans-heterozygous for the Gyc-88E point mutations and the Gyc-89Da−Db− double mutant (Gyc-88EV474M/Gyc-89Da−Db− in brown and Gyc-88ES451F/Gyc-89Da−Db− in purple) also showed significantly increased escape times. All values show mean ± SEM. N = 20–25.
We also utilized fly lines that carried point mutations in Gyc-88E. Because the two point mutations are homozygous lethal (see materials and methods), we generated larvae that had each point mutation over a deficiency that covered the Gyc-88E gene. These larvae (Gyc-88ES451F/Df or Gyc-88EV474M/Df) also showed a significant delay in their hypoxia escape response at 0, 5, and 10% O2, compared to larvae that were heterozygous for the deficiency or for either point mutation (Figure 3B). Similarly, larvae that were trans-heterozygous for the two point mutations (Gyc-88ES451F/Gyc-88EV474M) also showed significantly longer delays in leaving the food at 0, 5, and 10% O2 compared to their heterozygous controls (Figure 3C). As with the Gyc-88E RNAi-expressing larvae, no differences in the escape response were detected at 15% O2. To confirm that this delay was due to the loss of Gyc-88E function, we expressed wild-type Gyc-88E in either Gyc-89Da or Gyc-89Db neurons in the trans-heterozygote background (Figure 3C). This fully rescued the hypoxia escape response. We also generated larvae that had either the Gyc-88E deficiency or each Gyc-88E point mutation trans-heterozygous to the Gyc-89Da−/−Db−/− double mutation (Gyc-88E Df/Gyc-89Da− Gyc-89Db−, Gyc-88ES451F/Gyc-89Da− Gyc-89Db−, Gyc-88EV474M/Gyc-89Da− Gyc-89Db−). All these larvae showed a significant delay in their hypoxia escape response at 0, 5, and 10% O2 compared to controls (Figure 3D).
The hypoxia escape response is mediated via cGMP and cGMP-gated ion channels but not via the cGMP-dependent kinases:
Since guanylyl cyclases catalyze the synthesis of cGMP, we examined whether reduced levels of cGMP in the Gyc-89Da and the Gyc-89Db neurons account for the reduced ability of these larvae to respond to hypoxia. We reduced cGMP levels by expressing bovine phosphodiesterase 5 (bPDE5, a cGMP-specific PDE) in either or both of these populations of neurons. Previous studies using this construct to express bPDE5 in Malpighian tubules, the osmoregulatory and detoxifying organs of flies (Dow and Davies 2003), resulted in reduced cGMP levels and activity, similar to the effects of sGC inhibitors (Broderick et al. 2004).
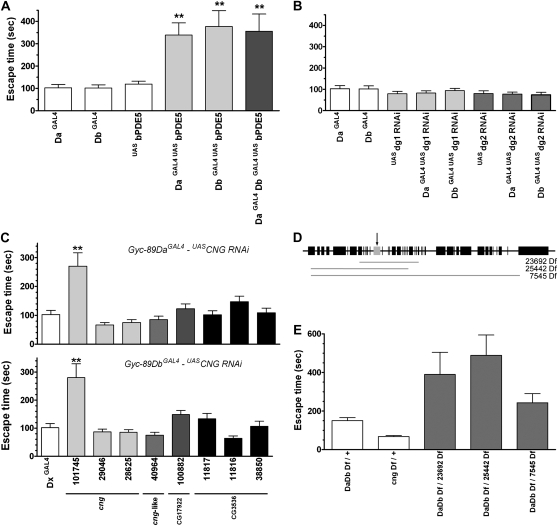
We made stable double homozygous fly lines (Gyc-89DaGAL4; UASbPDE5 and Gyc-89DbGAL4; UASbPDE5) and examined the effect of hypoxia on these larvae. The expression of bPDE5 in Gyc-89Da or Gyc-89Db neurons had no apparent effect on locomotion, fly development, or survival, and the adults eclosed at the same time as their parental controls (data not shown). Larvae expressing bPDE5 in either Gyc-89Da or Gyc-89Db neurons took significantly longer to respond to hypoxia at 0, 5 (data not shown), and 10% (Figure 4A) O2 concentrations compared to the parental control GAL4 and UAS larvae. In addition, there were no significant differences in the escape time between the larvae expressing bPDE5 in either Gyc-89Da or Gyc-89Db neurons or both populations of neurons (Figure 4A).
Figure 4.—
A normal hypoxia escape response depends on cGMP, cGMP-gated ion channels but not cGMP-dependent protein kinases. The time taken for third-instar larvae to escape from yeast paste when exposed to 10% O2 was recorded. Because previous experiments showed the largest effect at 10% O2, all escape response times are shown only at this concentration. (A) Expression of a bovine phosphodiesterase 5 (bPDE5) in either the Gyc-89Da or Gyc-89Db neurons increased the hypoxia escape response. A significant (P < 0.01) increased latency was measured when bPDE5 was expressed in either the Gyc-89Da or Gyc-89Db (light shading) or both sets of neurons (dark shading) when compared to parental Gyc-89DaGAL4, Gyc-89DbGAL4, and UASbPDE5 controls. (B) Reduced levels of the cGMP-dependent protein kinases dg1 and dg2 had no effect on the hypoxia escape response. dsRNA complementary to dg1 and dg2 was expressed in either Gyc-89Da or Gyc-89Db neurons and the hypoxia escape response compared to parental Gyc-89DaGAL4, Gyc-89DbGAL4, UASdg1, and UASdg2 controls. (C–E) The CNG channel is required for a normal hypoxia escape response. (C) RNAi for a variety of cyclic nucleotide-gated ion channels was expressed in either the Gyc-89Da (top) or Gyc-89Db (bottom) neurons. Several lines for each of the four predicted CNG channels were tested: GAL4 control (open), cng (light shading), cng-like (medium light shading), CG17992 (medium dark shading), and CG3536 (solid bars). The escape response was significantly delayed when the cGMP-specific cng subunit was downregulated using one of the UASRNAi lines (line CNG101745, P < 0.01). P values compared to Gyc-89DaGAL4 (top) and Gyc-89DbGAL4 (bottom) controls. (D) Schematic showing the position of known genes (solid boxes), cng (arrow marking the shaded box), and the relative sizes and positions of the cng deficiencies (shaded lines). (E) Genetic interaction between Gyc-89Da/Db and cng. Larvae that were trans-heterozygous for the Gyc-89Da, Gyc-89Db, and a variety of deficiencies that covered the cng gene (shaded) took significantly longer to withdraw from the food compared to larvae that were heterozygous for either gene at 10% O2 (P < 0.05). The x-axis indicates the genotype of the larvae, and the y-axis the time (seconds) that the larvae took to completely exit the yeast. All values show mean ± SEM. n = 20–25.
Downstream cellular targets of cGMP include the cGMP-dependent protein kinases (cGKs) (Hofmann 2005) and cGMP-gated ion channels (Bradley et al. 2005). To investigate components of the cGMP-signaling cascade in this behavioral response, we used RNAi to reduce the expression of these downstream effectors. When we expressed RNAi targeting two of the cGKs in Drosophila, dg1 or dg2, in either the Gyc-89Da or Gyc-89Db neurons, no significant effect on the hypoxia escape response was observed (Figure 4B) suggesting that these cGKs are not involved in this pathway.
Cyclic nucleotide-gated (CNG) channels bind cyclic nucleotides to allow cations (mainly Ca2+), to flow into the cell and thus modulate signaling networks (Bradley et al. 2005; Davies 2006; Davies and Terhzaz 2009). Drosophila contains at least four genes that appear to code for CNG channels. Two of these have been cloned and characterized (cng and cng-like), while two additional genes (CG3536 and CG17922) have been identified from sequencing the Drosophila genome (Morton and Hudson 2002). cng encodes a cGMP-sensitive homomeric ion channel, similar to the vertebrate CNG3, that is 50-fold more sensitive to cGMP than to cAMP (Baumann et al. 1994). The cng-like gene, by contrast, is similar to the mammalian CNG channel β-subunit; it does not form functional homomeric channels and is predicted to form a heteromeric channel specific to cAMP (Miyazu et al. 2000). Sequence analysis suggests that CG3536 and CG17922 have little selectivity toward cGMP or cAMP (Morton and Hudson 2002).
There are three available RNAi lines for cng, one for cng-like, one for CG3536, and three for CG17922. When we expressed these in either the Gyc-89Da or Gyc-89Db neurons we found no apparent effect on locomotion, fly development, or survival (data not shown). Expression of one of the cng dsRNA sequence (RNAi line 101745) in either Gyc-89Da or Gyc-89Db neurons significantly delayed the response to hypoxia at 10% O2. Downregulation of any of the other three (cng-like, CG3536, and CG17922) CNG channels had no significant effect on the hypoxia escape response (Figure 4C). If cng acts in the same pathway as the atypical sGCs, we would expect that there would be a genetic interaction between these genes. To test this we generated larvae that were trans-heterozygous for the Gyc-89Da/Db deficiency with a deficiency covering the cng gene (Figure 4D). These larvae also showed significant delays in their hypoxia escape response at 10% O2 (Figure 4E).
Activation of the Gyc-89Da neurons is sufficient to trigger the hypoxia escape response:
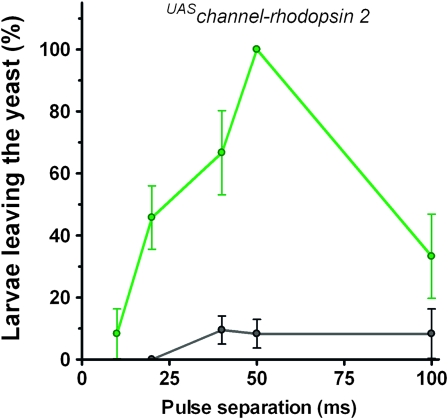
Channel-rhodopsin 2 (ChR2), a light-activated cation channel, has been used to study the roles of neurons in their native circuitry (Nagel et al. 2005). To determine whether we could trigger the hypoxia escape response by directly activating the neurons that express the atypical sGCs, we expressed a mutated form of ChR2 (ChR2-H134R) using the Gyc-89Da–GAL4 driver. ChR2-H134R shows enhanced responses to blue light pulses and less spike frequency adaptation than neurons expressing ChR2 (Pulver et al. 2009). Larvae expressing ChR2-H134R in the Gyc-89Da neurons were placed in yeast paste and illuminated with pulses of blue light. They quickly responded to the blue light by crawling out of yeast (see movie in File S1), similar to the behavior seen at low O2 concentrations. We varied the light pulse length and time between pulses and found that optimal stimulation was achieved with a pulse of 10 msec and a pulse separation of 50 msec (Figure 5). This corresponds to a stimulation frequency of 17 Hz and triggered 100% of the larvae to leave the yeast within the 50-sec stimulation period. When larvae expressing ChR2-H134R in the Gyc-89Db neurons were stimulated in a similar fashion, they did not exit the food at a higher frequency than control animals. Whether this was due to insufficient expression of ChR2 or whether activation of all the Gyc-89Db neurons is insufficient to activate the behavior is not clear at this time.
Figure 5.—
Activation of the Gyc-89Da neurons is sufficient to trigger the hypoxia escape response. Channel-rhodopsin 2 (ChR-2-H134R) was expressed in Gyc-89Da neurons and third-instar larvae allowed to feed in yeast paste. They were then exposed to pulses of blue light (470 nm, 10 msec long) for 50 sec delivered at 0- to 100-msec pulse separation and the number of larvae leaving the food during the stimulus time was recorded. Up to 100% of the larvae expressing ChR-2 in the Gyc-89Da neurons (green) could be driven out from the food whereas control (w1118) larvae (black) rarely exited the food during light stimulation. Values plotted as the mean ± SEM, n = 12–42.
Abrupt changes in O2 levels reduce stopping and turning frequencies in Drosophila larvae and require the atypical sCGs:
During most of the larval phase, Drosophila larvae remain immersed within the food source and feed constantly (Sokolowski et al. 1984). Food-associated taxis behaviors induced by temperature (Ainsley et al. 2008), odorants (Fishilevich et al. 2005), and light (Busto et al. 1999) require that the animal integrates sensory inputs from its current environment for comparison with previously established environmental set points associated with a food source.
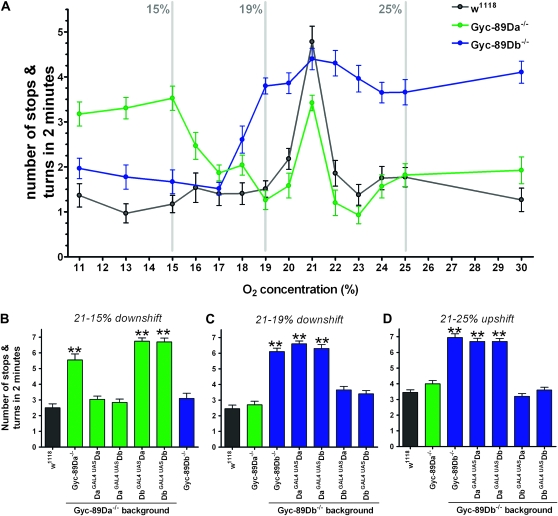
One foraging strategy used by larvae to fine tune their spatial position in order to reorient in the direction of a food source is to perform changes in their stopping and turning behaviors (Yang et al. 2000). By measuring the number of stops and turns that a larva makes when exposed to a change in environment, we conducted a quantitative assessment of exploratory behavior in response to a uniform O2 shift (Ainsley et al. 2008). In the absence of food and O2 shift, wild-type larvae showed an area-restricted search (ARS) behavior with a high number of stops and turns. This number significantly decreased when exposed to an O2 downshift or upshift from 21% O2 (Figure 6A). A change of only 1% (either up or down) was sufficient to trigger this change and shifts of larger magnitude had little additional effect.
Figure 6.—
Gyc-89Da and Gyc-89Db mutants respond differently to abrupt changes in O2 levels. A quantitative assessment of larval exploratory behavior in response to a uniform O2 shift was measured as the number of stops and turns in the first 2 min after the shift. (A) Number of stops/turns at different O2 shifts (21% to the O2 concentration indicated on the x-axis) for wild-type larvae (black), Gyc-89Da−/− mutants (green), and Gyc-89Db−/− mutants (blue). Gray lines indicate the shifts used in B, C, and D. (B) Number of stops and turns for larvae downshifted from 21 to 15% O2. Wild-type and Gyc-89Db−/− mutant larvae showed a reduced number of stops and turns. Gyc-89Da−/− mutant larvae, by contrast, maintained a significantly higher number of stops and turns compared to wild-type controls, which were rescued by expressing Gyc-89Da or Gyc-89Db in the Gyc-89Da neurons (DaGAL4, UASDa and DaGAL4, UASDb) but not in the Gyc-89Db neurons (DbGAL4, UASDa and DbGAL4, UASDb) (P < 0.0001). (C and D) number of stops and turns when larvae were downshifted from 21 to 19% O2 (C) or upshifted from 21% to 25% (D). Wild-type and Gyc-89Da−/− mutant larvae showed a reduced number of stops and turns. Gyc-89Db−/− mutant larvae, by contrast, maintained a significantly higher number of stops and turns compared to wild-type controls. As in B the wild-type response was restored by expressing Gyc-89Db or Gyc-89Da in the correct neurons (Gyc-89Db neurons in this case) (DbGAL4, UASDb and DbGAL4, UASDa), but not if expressed in the incorrect neurons (DaGAL4, UASDb and DaGAL4, UASDa) (P < 0.0001). Wild-type controls shown in black, Gyc-89Da−/− mutants green, and Gyc-89Db−/− mutants blue. Data represented as mean ± SEM, n = 20–24.
We then tested the responses of Gyc-89Da−/− and Gyc-89Db−/− mutant larvae to shifts in O2 concentration. When Gyc-89Da−/− mutant larvae were shifted to mildly hypoxic (17–20% O2) or hyperoxic (22–30% O2) conditions, they responded in the same manner as wild-type animals, with a significant reduction in the number of stops and turns. By contrast, when they were shifted to a more severe hypoxic environment (11–15% O2), they showed no behavioral response and maintained their ARS behavior (Figure 6A). When these manipulations were carried out on Gyc-89Db−/− mutant larvae, a complementary pattern was observed. No behavioral response was detected at mildly hypoxic (17–20% O2) or hyperoxic (22–30% O2) conditions, whereas they responded to the more severe hypoxic environment (11–15% O2) with a similar response to wild-type larvae, reducing their stops and turns (Figure 6A). These results suggest that Gyc-89Da is responsible for detecting changes in oxygen concentration between 11 and 15% O2, whereas Gyc-89Db signals changes between 17 and 30% O2.
To confirm these results, we focused on the behavioral responses seen for three different shifts in oxygen concentration: downshifts from 21% to either 15 or 19% O2 and an upshift from 21 to 25% O2 (Figures 6, B–D) and recorded the numbers of stops and turns performed by the larvae. With a downshift from 21 to 15% O2, Gyc-89Da−/− mutant larvae failed to show a reduction in the number of stops and turns, whereas Gyc-89Db−/− mutant larvae responded in the same way as wild-type larvae (Figure 6B). When Gyc-89Da was expressed in the Gyc-89Da neurons of Gyc-89Da−/− mutant larvae, the behavioral response was restored (Figure 6B). Similarly, the behavioral response was restored when Gyc-89Db was expressed in Gyc-89Da neurons (wrong subunit in the correct neuron). However, the response was not restored by expressing either Gyc-89Da or Gyc-89Db in the Gyc-89Db neurons (either subunit in the wrong neuron) (Figure 6B). For the milder downshift from 21 to 19% O2 (Figure 6C) and for the upshift from 21 to 25% O2 (Figure 6D), Gyc-89Db−/− mutants showed no response, which was rescued by expressing Gyc-89Db in the Gyc-89Db neurons. By contrast, the Gyc-89Da−/− mutants showed normal responses to both stimuli (Figure 6, C and D). As with the more severe downshift, when the wrong subunit was expressed in the correct neurons (Gyc-89Da in the Gyc-89Db neurons) the behavioral response was restored. Similarly, when either subunit was expressed in the wrong neuron (Gyc-89Da or Gyc-89Db in the Gyc-89Da neurons) the behavioral response was not restored (Figure 6, C and D).
DISCUSSION
The atypical sGCs are required for a normal hypoxia escape response:
In a wide variety of eukaryotic and prokaryotic cells, heme-containing proteins are used to detect varying levels of O2, CO, and NO (Gilles-González and González 2005). Conventional sGCs are heme proteins that bind, and are regulated by NO and CO, but are unable to bind O2 (Boon et al. 2005). Recent studies have identified the atypical sGCs in C. elegans and Drosophila as likely molecular O2 sensors that can bind and are regulated by O2 (Gray et al. 2004; Morton 2004b; Vermehren et al. 2006; Huang et al. 2007; Zimmer et al. 2009). Results described here show that in Drosophila, like C. elegans, the atypical sGCs are required for behavioral responses to hypoxia. Larvae with disrupted expression of Gyc-88E, Gyc-89Da, or Gyc-89Db subunits showed a significant delay in their hypoxia escape response when compared to wild-type larvae, and this delay was further increased by combining the mutated genes for each subunit. Gyc-89Da and Gyc-89Db were disrupted with transposon insertions either within the coding region (Gyc-89Db) or within an intron (Gyc-89Da) of the gene. Both mutant lines showed a substantial reduction in the levels of transcript for the disrupted gene. We also recombined these two insertions to generate a Gyc-89Da−/−Db−/− double mutant, which showed a significant reduction in the levels of Gyc-89Da and Gyc-89Db.
To reduce the expression of Gyc-88E, we expressed Gyc-88E RNAi in either or both of the neurons that express Gyc-89Da and Gyc-89Db. We also obtained point mutations in the highly conserved catalytic domain of Gyc-88E and showed that the mutated residues were required for enzyme activity. Modeling the structure of sGC catalytic domains has shown that residues from both subunits interact with the GTP substrate, with specific residues being provided by the α-subunit and others provided by the β-subunit (Liu et al. 1997). Although Gyc-88E can form active homodimers, its catalytic activity is substantially lower compared to the heterodimers, where it functions as an α-subunit (Morton and Anderson 2003; Morton 2004b). Both mutated residues are located very close to residues that are required for GTP binding and specificity (Liu et al. 1997; Morton and Hudson 2002).
We showed that the increased time taken to respond to hypoxia in each of the mutant lines was rescued by expressing the subunit that was disrupted. In addition, for the Gyc-89Da and the Gyc-89Db mutants, we could rescue the mutant phenotypes by expressing the complementary subunit, suggesting that they are functionally equivalent. Biochemical experiments using transiently transfected subunits in heterologous cells have also demonstrated that Gyc-89Da and Gyc-89Db are functionally equivalent (Langlais et al. 2004; Morton et al. 2005).
The hypoxia escape response also required an increase in cGMP in the Gyc-89Da and the Gyc-89Db neurons and was likely mediated by the cGMP-gated ion channel cng. Furthermore, our results showed this pathway likely does not involve the cGKs DG1 or DG2, as we observed no change in the hypoxia escape response when dg1 or dg2 RNAi was expressed in either Gyc-89Da or Gyc-89Db neurons. It is possible that another cGK might be involved, since the Drosophila genome contains an additional gene predicted to code for a cGK (Morton and Hudson 2002).
We hypothesize that cGMP-dependent activation of cng in response to reduced oxygen levels activates specific sensory neurons that activate the escape response. By using channel rhodopsin-2 to selectively activate neurons that express Gyc-89Da, we could trigger a behavioral response that mimicked the effect of reduced oxygen. The above experiments show that the atypical sGCs are required for the hypoxia escape response and that activation of the Gyc-89Da neurons are sufficient to activate the response.
Gyc-89Da and Gyc-89Db have different effects on oxygen-stimulated changes in crawling and searching behavior:
As described above, Gyc-89Da and Gyc-89Db appear to be functionally redundant, despite having a broadly nonoverlapping expression pattern. It was therefore surprising to observe slightly different effects of mutations in these genes when we examined the responses of crawling larvae to changes in oxygen. In the absence of food, Drosophila larvae exhibit food searching behavior during which bouts of crawling forward are interrupted by frequent stops, turns, and moving the head from side to side (Ainsley et al. 2003). When a larva experiences a potentially deleterious environment, such as low oxygen, it responds by reducing the number of stops and turns that it makes. Both hypoxic and hyperoxic environments elicit this response in wild-type larvae. In larvae that are mutant for either Gyc-89Da or Gyc-89Db, this response is defective, but the concentrations of oxygen at which the response is defective differ depending on the sGC subunit that is affected. Gyc-89Da is required for responses in the range of 11–16% oxygen, whereas Gyc-89Db is required for responses to both mild hypoxia (18–20% oxygen) and hyperoxia (22–30% oxygen). As with the hypoxia escape response, these defective responses could be rescued by expressing the correct subunit in the correct cells. Similarly, expressing the incorrect subunit in the correct cell rescued the response. This latter result is somewhat surprising and contrasts to findings from C. elegans. Preferences for different oxygen concentrations and responses to oxygen upshifts and downshifts are mediated by specific atypical sGCs and sensory neurons (Gray et al. 2004; Zimmer et al. 2009). In nematodes, however, the identity of the subunit appeared to be critical for the correct response to either upshifts or downshifts (Zimmer et al. 2009). In Drosophila larvae Gyc-89Da mutants failed to respond to a 21–15% downshift, which could be rescued by either Gyc-89Da or Gyc-89Db. Gyc-89Db mutants failed to respond to 21–25% upshift, which could also be rescued by either Gyc-89Da or Gyc-89Db. In both cases restoration of the response was not seen if the subunits were expressed in the incorrect cells. This is also in contrast to the molecular basis for odor coding in the antennae of Drosophila where the odor receptor determines the odor specificity of a neuron and the response characteristics of a neuron can be changed by changing the receptors expressed in that cell (Hallem et al. 2004).
Sensory cones are strong candidates for the O2 sensors mediating the hypoxia escape response:
Gyc-89Da and Gyc-89Db subunits are expressed extensively in the Drosophila nervous system, in both central and peripheral neurons (Langlais et al. 2004; Morton et al. 2008) (for a schematic see Figure 7A). There are relatively few cells that co-express both subunits. Because of this broadly nonoverlapping expression pattern of Gyc-89Da and Gyc-89Db, it was possible to express either subunit in both the correct and incorrect cells. All possible combinations fully rescued the hypoxia escape response. For example, in the Gyc-89Db mutant, expression of either Gyc-89Da or Gyc-89Db in either the Gyc-89Da or the Gyc-89Db neurons rescued the response. Similarly, we could rescue the Gyc-88E point mutations by expressing Gyc-88E in either Gyc-89Da or Gyc-89Db neurons. In addition, manipulations of the cGMP levels and the levels of the Gyc-88E and cng had the same effects whether these manipulations were to Gyc-89Da, Gyc-89Db neurons, or both populations. This suggests that both populations of neurons are functionally equivalent to each other. These results contrasted to those where we attempted to rescue the response to oxygen-stimulated changes in crawling and searching behaviors. For this behavior, as described above, the Gyc-89Da and Gyc-89Db subunits were equivalent to each other; however, the cells in which they were expressed were not (see Table 1 for summary).
Figure 7.—
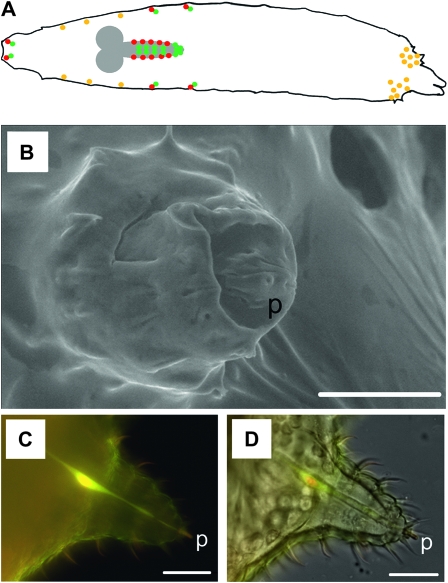
Neurons innervating the larval sensory cones co-express Gyc-89Da and Gyc-89Db. (A) Schematic of a dorsal view of a Drosophila larva showing central and peripheral neurons expressing Gyc-88E/Gyc-89Da (red), Gyc-88E/Gyc-89Db (green), and those co-expressing all three subunits (yellow). Anterior end is on the left and posterior end on the right. Neurons that express Gyc-88E plus only Gyc-89Da or Gyc-89Db are found in the gustatory and olfactory ganglia, abdominal segments of the lateral body wall, and the CNS. The only neurons that express all three atypical sGC subunits are sensory neurons on the thoracic body wall and the terminal sensory cones. (B) Scanning electron micrograph of a larval terminal sensory cone showing what appears to be a terminal pore in the peg (p). (C) Confocal fluorescent micrograph showing the colocalization of Gyc-89Da and Gyc-89Db in the neuron innervating a larval terminal sensory cone. Overlay of two confocal stacks (Gyc-89Da-GFP and Gyc-89DbGAL4, UAS dsRED). (D) Light micrograph of the same chemosensory cone shown in B. Bars measure 20 μm in C and D, and 25 μm in B.
TABLE 1.
Summary of the role of Gyc-89Da and Gyc-89Db and the neurons in which they are expressed during the hypoxia escape response and changes in ARS behavior responses to hypoxia and hyperoxia
| Gyc-89Da−/− mutant |
Gyc-89Db−/− mutant |
|||||||
|---|---|---|---|---|---|---|---|---|
| DaGAL4 UASDa | DaGAL4 UASDb | DbGAL4 UASDa | DbGAL4 UASDb | DaGAL4 UASDa | DaGAL4 UASDb | DbGAL4 UASDa | DbGAL4 UASDb | |
| Hypoxia escape response rescued | ||||||||
| Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | |
| ARS behavior rescued | ||||||||
| 21%-15% | Yes | Yes | No | No | ND | ND | ND | ND |
| 21%-19% | ND | ND | ND | ND | No | No | Yes | Yes |
| 21%-24% | ND | ND | ND | ND | No | No | Yes | Yes |
The hypoxia escape response was rescued at all oxygen concentrations when either subunit was expressed in either Gyc-89Da or Gyc-89Db neurons in both mutants. By contrast the area restricted search (ARS) behavior was rescued by either subunit but only if it was expressed in the correct neurons for that mutation. ND, not determined.
The simplest explanation for these results is that the cells that co-express both Gyc-89Da and Gyc-89Db subunits are the ones that mediate the hypoxia escape response, whereas the cells that express only Gyc-89Da are involved in dispersal responses to more severe hypoxia, while those that express only Gyc-89Db are involved in dispersal behaviors to mild hypoxia and hyperoxia. Our previous studies identified a small subset of neurons that co-expressed both Gyc-89Da and Gyc-89Db subunits (Morton et al. 2008) (see schematic in Figure 7A). These include two sensory neurons on either side of the thoracic segments innervating basoconical sensilla and single sensory neurons innervating each of the seven pairs of terminal sensory cones on the posterior two abdominal segments (Morton et al. 2008). These terminal sensory cones stick out of the food when the larvae are feeding in their vertical position, putting them in an ideal location to sense changes in O2 concentrations in their surrounding environment. Insect chemoreceptors are afferent neurons that are contained within cuticular structures called sensilla, small hollow hairs or pegs bearing one or more pores through their interior, and each containing 2–50 chemosensory neurons (Rogers and Newland 2003). Scanning electron microscopy of these terminal sensory cones showed the existence of a small pore at the tip of the peg located at the top of the sensory cone, suggesting that it could function as a chemosensor (Figure 7B). Confocal imaging using larvae expressing Gyc-89Da-GFP and Gyc-89DbGAL4-UASdsRED showed colocalization of Gyc-89Da and Gyc-89Db in a single neuron innervating each terminal sensory cones (Figure 7, C and D). Due to their location and the co-expression of all three subunits in these cells (Langlais et al. 2004; Morton et al. 2008), we hypothesize that these are the neurons that are responsible for detecting the reduced O2 concentrations and then signaling to the CNS to initiate the escape response.
In summary, we have shown that the atypical sGC subunits are equivalent for a variety of behavioral responses to changes in oxygen concentration. In all cases, the Gyc-89Da and Gyc-89Db subunits appear to be functionally equivalent to each other. By contrast, different subsets of neurons that express these subunits are required to respond to the different stimuli.
Acknowledgments
The authors thank Dennis Hazelett, Philip Copenhaver, and Judy Stewart for helpful discussions and comments on the manuscript. We also thank L. C. Griffith (Brandeis University, MA) for the UASH134R-ChR2 line, Stefan Pulver for help and advice with the channel-rhodopsin activation experiments, Charles Scudder for designing and building the LED stimulation controller, and John Mitchell for taking the scanning EM images of the sensory cones. This study would not have been possible without the resources available from FlyBase and the Bloomington Stock Center. This research was supported by a National Institute of Neurological Disorders and Stroke grant NS29740 to D.B.M.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.118166/DC1.
References
- Ainsley, J., J. Pettus, D. Bosenko, C. Gerstein, N. Zinkevich et al., 2003. Enhanced locomotion caused by loss of the Drosophila DEG/ENaC protein Pickpocket1. Curr. Biol. 13 1557–1563. [DOI] [PubMed] [Google Scholar]
- Ainsley, J., M. Kim, L. Wegman, J. Pettus and W. A. Johnson, 2008. Sensory mechanisms controlling the timing of larval developmental and behavioral transitions require the Drosophila DEG/ENaC subunit, Pickpocket1. Dev. Biol. 322 46–55. [DOI] [PubMed] [Google Scholar]
- Baumann, A., S. Frings, M. Godde, R. Seifert and U. Kaupp, 1994. Primary structure and functional expression of a Drosophila cyclic nucleotide-gated channel present in eyes and antennae. EMBO J. 13 5040–5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon, E., S. Huang and M. Marletta, 2005. A molecular basis for NO selectivity in soluble guanylate cyclase. Nature 1 53–59. [DOI] [PubMed] [Google Scholar]
- Bradley, J., J. Reisert and S. Frings, 2005. Regulation of cyclic nucleotide-gated channels. Curr. Opin. Neurobiol. 15 343–349. [DOI] [PubMed] [Google Scholar]
- Brand, A., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 401–415. [DOI] [PubMed] [Google Scholar]
- Broderick, K., L. Kean, J. Dow, N. Pyne and S. Davies, 2004. Ectopic expression of bovine type 5 phosphodiesterase confers a renal phenotype in Drosophila. J. Biol. Chem. 279 8159–8168. [DOI] [PubMed] [Google Scholar]
- Busto, M., B. Iyengar and A. Campos, 1999. Genetic dissection of behavior: modulation of locomotion by light in the Drosophila melanogaster larva requires genetically distinct visual system functions. J. Neurosci. 19 3337–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, A., N. Chronis, D. Karow, M. Marletta and C. Bargmann, 2006. A distributed chemosensory circuit for oxygen preference in C. elegans. PLoS Biol. 4 e274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, S., 2006. Signalling via cGMP: lessons from Drosophila. Cell Signal 18 409–421. [DOI] [PubMed] [Google Scholar]
- Davies, S., and S. Terhzaz, 2009. Organellar calcium signalling mechanisms in Drosophila epithelial function. J. Exp. Biol. 212 387–400. [DOI] [PubMed] [Google Scholar]
- Dow, J., and S. Davies, 2003. Integrative physiology and functional genomics of epithelial function in a genetic model organism. Physiol. Rev. 83 687–729. [DOI] [PubMed] [Google Scholar]
- Fishilevich, E., A. Domingos, K. Asahina, F. Naef, L. Vosshall et al., 2005. Chemotaxis behavior mediated by single larval olfactory neurons in Drosophila. Curr. Biol. 15 2086–2096. [DOI] [PubMed] [Google Scholar]
- Gilles-González, A., and G. González, 2005. Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J. Inorg. Biochem. 99 1–22. [DOI] [PubMed] [Google Scholar]
- Gonzalez, C., L. Almaraz, A. Obeso and R. Rigual, 1994. Carotid body chemoreceptors: from natural stimuli to sensory discharges. Physiol. Rev. 74 829–898. [DOI] [PubMed] [Google Scholar]
- Gray, J., D. Karow, H. Lu, A. Chang, J. Chang et al., 2004. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature 430 317–322. [DOI] [PubMed] [Google Scholar]
- Greenspan, R. J., 2004. Fly Pushing. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Hallem, E.A., M.G. Ho and J.R. Carlson, 2004. The molecular basis of odor coding in the Drosophila antenna. Cell 117 965–979. [DOI] [PubMed] [Google Scholar]
- Hochachaka, P., and J. Rupert, 2003. Fine tuning the HIF-1 ‘global’ O2 sensor for hypobaric hypoxia in Andean high-altitude natives. BioEssays 25 515–519. [DOI] [PubMed] [Google Scholar]
- Hofmann, F., 2005. The Biology of Cyclic GMP-dependent Protein Kinases. J. Biol. Chem. 208 1–4. [DOI] [PubMed] [Google Scholar]
- Huang, S., D. Rio and M. Marletta, 2007. Ligand binding and inhibition of an oxygen-sensitive soluble guanylate cyclase, Gyc-88E, from Drosophila. Biochemistry 46 15115–15122. [DOI] [PubMed] [Google Scholar]
- Kemp, P., 2005. Hemeoxygenase-2 as an O2 sensor in K+ channel-dependent chemotransduction. Biochem. Biophys. Res. Commun. 338 648–652. [DOI] [PubMed] [Google Scholar]
- Langlais, K. K., J. A. Stewart and D. B. Morton, 2004. Preliminary characterization of two atypical soluble guanylyl cyclases in the central and peripheral nervous system of Drosophila melanogaster. J. Exp. Biol. 207 2323–2338. [DOI] [PubMed] [Google Scholar]
- Liu, Y., A. Ruoho, V. Rao and J. Hurley, 1997. Catalytic mechanism of the adenylyl and guanylyl cyclases: modeling and mutational analysis. Proc. Natl. Acad. Sci. USA 94 13414–13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Barneo, J., P. Ortega-Sáenz, R. Pardal, A. Pascual and J. Piruat, 2008. Carotid body oxygen sensing. Eur. Respir. J. 32 1386–1398. [DOI] [PubMed] [Google Scholar]
- Lucas, K., G. Pitari, S. Kazerounian, I. Ruiz-Stewart, J. Park et al., 2000. Guanylyl cyclases and signaling by cGMP. Pharmacol. Rev. 52 375–413. [PubMed] [Google Scholar]
- Miyazu, M., T. Tanimura and M. Sokabe, 2000. Molecular cloning and characterization of a putative cyclic nucleotide-gated channel from Drosophila melanogaster. Insect Mol. Biol. 9 283–292. [DOI] [PubMed] [Google Scholar]
- Montgomery, M., and A. Fire, 1988. Double-stranded RNA as a mediator in sequence-specific genetic silencing and co-suppression. Trends Genet. 14 255–258. [DOI] [PubMed] [Google Scholar]
- Morton, D. B., 2004. a Invertebrates yield a plethora of atypical guanylyl cyclases. Mol. Neurobiol. 29 97–115. [DOI] [PubMed] [Google Scholar]
- Morton, D. B., 2004. b Atypical soluble guanylyl cyclases in Drosophila can function as molecular oxygen sensors. J. Biol. Chem. 279 50651–50653. [DOI] [PubMed] [Google Scholar]
- Morton, D. B., and E. A. Anderson, 2003. MsGC-b3 forms active homodimers and inactive heterodimers with NO-sensitive soluble guanylyl cyclase subunits. J. Exp. Biol. 206 937–947. [DOI] [PubMed] [Google Scholar]
- Morton, D. B., and M. L. Hudson, 2002. Cyclic GMP regulation and function in insects. Adv. Insect Physiol. 29 1–54. [Google Scholar]
- Morton, D. B., and A. Vermehren, 2007. Soluble guanylyl cyclases in invertebrates: targets for NO and O2, pp. 65–82 in Advances in Experimental Biology on Nitric Oxide, edited by B. A. Trimmer and B. Tota. Elsevier Press, New York. [DOI] [PMC free article] [PubMed]
- Morton, D. B., K. K. Langlais, J. A. Stewart and A. Vermehren, 2005. Comparison of the properties of the five soluble guanylyl cyclase subunits in Drosophila melanogaster. J. Insect Sci. 5 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton, D. B., J. A. Stewart, K. K. Langlais, R. A. Clemens-Grisham and A. Vermehren, 2008. Synaptic transmission in neurons that express the Drosophila atypical soluble guanylyl cyclases, Gyc-89Da and Gyc-89Db, is necessary for the successful completion of larval and adult ecdysis. J. Exp. Biol. 211 1645–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel, G., M. Brauner, J. Liewald, N. Adeishvili, E. Bamberg et al., 2005. Light activation of Channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr. Biol. 15 2279–2284. [DOI] [PubMed] [Google Scholar]
- Ortega-Sáenz, P., A. Pascual, R. Gómez-Díaz and J. López-Barneo, 2006. Acute oxygen sensing in heme oxygenase-2 null mice. J. Gen. Physiol. 128 405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne, K., A. Robichon, E. Burgess, S. Butland, R. Shaw et al., 1997. Natural behavior polymorphism due to a cGMP-dependent protein kinase of Drosophila. Science 277 834–836. [DOI] [PubMed] [Google Scholar]
- Pulver, S., S. Pashkovski, N. Hornstein, P. Garrity and L. Griffith, 2009. Temporal dynamics of neuronal activation by channelrhodopsin-2 and TRPA1 determine behavioral output in Drosophila larvae. J. Neurophysiol. 101 3075–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S., and P. Newland, 2003. The neurobiology of taste in insects. Adv. Insect Physiol. 31 141–204. [Google Scholar]
- Semenza, G., 2000. HIF-1: mediator of physiological and pathophysiological responses to hypoxia. J. Appl. Physiol. 88 1474–1480. [DOI] [PubMed] [Google Scholar]
- Sokolowski, M., C. Kent and J. Wong, 1984. Drosophila larval foraging behavior: developmental stages. Anim. Behav. 32 645–651. [Google Scholar]
- Vermehren, A., K. K. Langlais and D. B. Morton, 2006. Oxygen-sensitive guanylyl cyclases in insects and their potential roles in oxygen detection and in feeding behaviors. J. Insect Physiol. 52 340–348. [DOI] [PubMed] [Google Scholar]
- Wingrove, J., and P. O'Farrell, 1999. Nitric oxide contributes to behavioral, cellular, and developmental responses to low oxygen in Drosophila. Cell 98 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P., S. Shaver, A. Hilliker and M. Sokolowski, 2000. Abnormal turning behavior in Drosophila larvae: identification and molecular analysis of scribbler (sbb). Genetics 155 1161–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer, M., J. Gray, N. Pokala, A. Chang, D. Karow et al., 2009. Neurons detect increases and decreases in oxygen levels using distinct guanylate cyclases. Neuron 61 865–879. [DOI] [PMC free article] [PubMed] [Google Scholar]