Abstract
This study concerns the problem of odor receptor gene choice in the fruit fly Drosophila melanogaster. From a family of 60 Odor receptor genes, only one or a small number are selected for expression by each olfactory receptor neuron. Little is known about how an olfactory receptor neuron selects a receptor, or how the nucleotide sequences flanking a receptor gene dictate its expression in a particular neuron. Previous investigation has primarily concerned the maxillary palp, the simpler of the fly's two olfactory organs. Here we focus on genes encoding four antennal receptors that respond to fly odors in an in vivo expression system. To investigate the logic of odor receptor expression, we carry out a genetic analysis of their upstream regulatory sequences. Deletion analysis reveals that relatively short regulatory regions are sufficient to confer expression in the appropriate neurons, with limited if any misexpression. We find evidence for both positive and negative regulation. Multiple repressive functions restrict expression to the antenna, to a region of the antenna, and to neurons. Through deletion and base substitution mutagenesis we identify GCAATTA elements and find evidence that they act in both positive and negative regulation.
OLFACTORY systems of insects and vertebrates contain many odor receptors and many olfactory receptor neurons (ORNs), yet each ORN expresses only one or a small number of receptors (Mombaerts 2004; Fuss and Ray 2009). A critical problem in this system is the mechanism of receptor gene choice. What factors conspire to express one particular receptor in a particular ORN? Conversely, what factors prevent all the other receptors from being expressed in that ORN? This problem is especially challenging because in many organisms, including Drosophila, the expression of individual receptors is subject to a spatial constraint: each receptor is expressed only in a subset of ORNs that lie in a particular subdomain of the olfactory field.
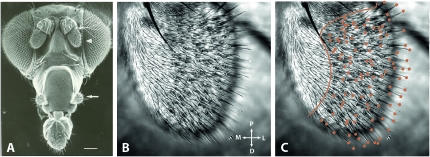
The olfactory system of the fruit fly contains two organs, the third segment of the antenna, referred to henceforth for simplicity as “the antenna,” and the maxillary palp (Figure 1A). Each organ is covered with sensilla, ∼400 in the case of the antenna, and ∼60 in the maxillary palp (Shanbhag et al. 2001). Each sensillum is innervated by up to four ORNs. There are three major morphological types of antennal sensilla: trichoid, basiconic, and coeloconic sensilla. Trichoid sensilla are distributed predominantly in the distolateral region (Figure 1, B and C), basiconic sensilla are most highly concentrated in the proximomedial region of the antenna, and coeloconic sensilla are spread broadly on the antennal surface as well as in a sensory pit known as the sacculus. All of the olfactory sensilla on the maxillary palp are of the basiconic type.
Figure 1.—
The olfactory organs and sensilla of D. melanogaster. (A) Electron micrograph of a Drosophila head indicating the two main olfactory organs, the antenna (arrowhead) and maxillary palp (arrow). Scale bar is 100 μm. (B and C) Transmitted-light confocal image showing the anterior face of a Drosophila third antennal segment. In C, the orange line separates the proximomedial region, which is devoid of trichoid sensilla, from the central and distolateral regions of the antenna, which are covered by basiconic, coeloconic, and trichoid sensilla. Orange dots indicate the tips of visible trichoid sensilla. Abbreviations: P, proximal; D, distal; M, medial; L, lateral. Image in A reproduced from Carlson(1996) with permission from Elsevier.
Trichoid sensilla, examined in this study, have been implicated in the response to fly odors (Clyne et al. 1997; Ejima et al. 2007; Kurtovic et al. 2007; van der Goes van Naters and Carlson 2007; Datta et al. 2008), whereas ORNs of basiconic sensilla respond strongly to fruit odors (de Bruyne et al. 2001; Hallem et al. 2004; Hallem and Carlson 2006). Trichoid sensilla fall into four subtypes, designated at1, at2, at3, and at4 (Shanbhag et al. 1999; Couto et al. 2005; Fishilevich and Vosshall 2005). The at1 sensilla are most highly concentrated in the proximomedial portion of the trichoid zone, while the at4 sensilla are more densely distributed in the distolateral portion. Each at1 sensillum is innervated by a single ORN; an at2 sensillum contains two ORNs; and at3 and at4 sensilla each contain three ORNs. Among the 60 Or (Odor receptor) genes of Drosophila melanogaster (Clyne et al. 1999b; Gao and Chess 1999; Vosshall et al. 1999), 12 map to individual ORNs in trichoid sensilla (Couto et al. 2005; Fishilevich and Vosshall 2005). Of these 12, 4 were found to respond to fly odors in an in vivo expression system, the “empty neuron” system (Dobritsa et al. 2003; van der Goes van Naters and Carlson 2007). Of these four receptors, Or67d is expressed in the ORN of at1, Or47b is expressed in 1 ORN of at4, Or65a is expressed (along with the closely related genes Or65b and Or65c) in a second ORN of at4, and Or88a is expressed in the third ORN of at4 (Couto et al. 2005; Fishilevich and Vosshall 2005).
The logic of receptor gene expression has been studied in the maxillary palp of Drosophila, but little is known about this process in the antenna. In the maxillary palp, computational analysis was used to identify sequence motifs shared by maxillary palp Or genes, or by Or genes coexpressed in the same ORN. Proper Or expression was found to depend on these sequences, on a combinatorial code of transcription factors, and on the asymmetric segregation of regulatory factors from progenitor cells (Endo et al. 2007; Ray et al. 2008; Ray et al. 2007).
In this study we analyze the logic of odor receptor expression in the antenna using a genetic approach. In particular, the four Or genes shown to encode receptors for fly odors in trichoid sensilla are examined. We construct Or–GAL4 drivers for each gene and then synthesize a series of deletion mutations that remove varying amounts of DNA from each. The effects of these deletions on gene expression are examined. Sequences of particular interest are investigated by base substitution mutation. The simplest interpretation of our analysis is that relatively small regulatory regions are sufficient to confer expression, with limited if any misexpression, and that expression patterns are restricted by multiple repressive functions that limit expression to the antenna, to a particular region of the antenna, and to ORNs. We identify particular sequence motifs that act positively or negatively to dictate expression in the proper subset of ORNs.
MATERIALS AND METHODS
DNA constructs:
We used the GAL4–UAS expression system (Fischer et al. 1988; Brand and Perrimon 1993) to visualize the expression patterns driven by Or promoter fragments. Except for the full-length Or65a–GAL4 construct, which has been described previously (Ejima et al. 2007), all Or–GAL4 reporter constructs were based on the GAL4 expression vector plasmid pG4PN+, which was, in turn, derived from pG4PN (Dobritsa et al. 2003). The original pG4PN plasmid includes a P-element transformation cassette carrying the GAL4 coding region preceded by a multiple cloning site and a mini-w+ transformation marker. The derivative plasmid, pG4PN+, has a modified multiple cloning site with unique recognition sequences including AgeI, BstEII, AvrII, BglII, and NotI (listed from 5′ to 3′).
The initial, long promoter fragments for Or47b, Or67d, and Or88a were amplified from male Canton-S wild-type D. melanogaster using the Expand high-fidelity PCR system (Roche no. 11-732-641-001). To amplify each of these initial promoter fragments we used a unique upstream primer and an anchor primer that bound immediately upstream of the associated Or gene's predicted start codon (supporting information, Table S1). Restriction enzyme recognition sites engineered into the primer tails allowed ligation of the amplified promoter fragments into pG4PN+ subsequent to enzymatic digestion and purification using standard methods. Both junctions between the vector and the promoter fragment were sequenced to confirm the structure of each construct.
Or gene promoter truncation and mutation constructs were prepared in a similar manner. All constructs derived from a given Or gene upstream region were amplified using the same anchor primer and a varying upstream primer (Table S1). We amplified some fragments with PfuTurbo DNA polymerase (Stratagene, La Jolla, CA, no. 600252) instead of Expand hi-fidelity polymerase. Also, we used primers carrying engineered mutations in putative regulatory motifs to create mutated promoter fragments. In some cases we used previously prepared Or–GAL4 plasmids containing a promoter subregion of interest as PCR templates. For example, the synthesis of the plasmid containing the Or88a−486M4 construct was a two-step process. First, we created the Or88a−465M1 construct. Second, we used this plasmid as template for PCR with primers for the Or88a−486 construct to yield the Or88a−486M4 promoter fragment. We confirmed the desired promoter fragment sequence of all truncation and mutation constructs by completely sequencing these inserts and the immediately flanking vector DNA.
Fly stocks:
We injected all Or88a and Or47b constructs, as well as the Or67d−6.1kb GAL4 construct, into a “Cantonized” w1118 background, i.e., w1118 flies that had been back-crossed multiple times to Canton-S wild-type flies. The Or65a−4.7kb construct was injected into w1118 flies (Ejima et al. 2007). The Or65a−4.7kb line analyzed here carries a different chromosomal insertion of the same Or65a–GAL4 construct reported previously. The antennal GFP expression pattern of the insertion examined here appears to be more consistent with the expression pattern of Or65a determined by in situ hybridization (Vosshall et al. 2000; Couto et al. 2005; Fishilevich and Vosshall 2005). However, our analysis of axon projection patterns in the antennal lobe of the brain substantially agrees with that of the previously examined insertion. All Or65a and Or67d truncations were injected into w1118 flies by BestGene, Inc. (Chino Hills, CA, http://www.thebestgene.com/).
Antennal imaging and analysis:
Flies used for microscopy were reared at 25° and, except as noted, were heterozygous for both UAS–GFP and the particular Or–GAL4 construct. All insertions were autosomal and were analyzed in males, except that in the case of −486D, lines 2 and 3 were X-linked; on the basis of Figure 7, B and C, we do not expect their position on the X chromosome to have a sufficiently major effect on expression level as to alter any conclusions. The UAS–GFP was in all cases UAS–gapGFP and was a gift from Akira Chiba (Chiba 1998; Ritzenthaler et al. 2000).
Figure 7.—
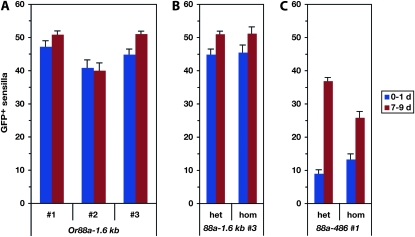
The effect of age and gene dosage on the expression levels of different Or88a–GAL4 constructs. (A) Three independent lines (1–3) of Or88a−1.6kb show similar numbers of labeled sensilla in flies heterozygous for both the Or88a–GAL4 construct and the UAS–GFP reporter. These expression levels are the same at both 0–1 and 7–9 days post-eclosion. (B) Flies either heterozygous (“het”) or homozygous (“hom”) for both Or88a−1.6kb insertion 3 and the UAS–GFP reporter show similar numbers of labeled sensilla at both 0–1d and 7–9 d. (C) Flies either heterozygous (“het”) or homozygous (“hom”) for both an Or88a−486 truncation and the UAS–GFP reporter show an age-dependent increase in number of GFP-labeled sensilla. Bars indicate mean numbers of GFP-labeled sensilla per antenna (±SEM) dissected from male flies. For each bar, 6 ≤ n ≤ 10 antennae.
Flies were anesthetized with CO2, and antennae and maxillary palps were dissected at room temperature into 1× PBS supplemented with 0.05% Tween-20. Tissues were then rinsed, mounted in 1× PBS, and imaged immediately. GFP expression was analyzed with a Bio-Rad MRC-1024 laser scanning confocal microscope and Lasersharp software. Images were collected as confocal stacks with 1 μm spacing between optical sections. Laser power, photomultiplier tube gain, and iris settings were kept constant for all images used in sensillum counts; instrument drift was compensated using black-level adjustments.
Images were processed using ImageJ (Rasband 1997), Photoshop, and Illustrator (Adobe Systems, Inc., San Jose, CA) software. To distinguish GFP signal from cuticle autofluorescence in antennal images, the red channel image, showing only autofluorescence, was merged with the green channel image containing the GFP signal (Manoli et al. 2005). GFP expression appears green and cuticle autofluorescence appears magenta.
For sensillum counts, antennae from male flies of the indicated ages were dissected and imaged as described above. We counted GFP-labeled sensilla on Z-projections of entire antennae and sampled at least five antennae per time point for each line; however, in some cases when three consecutive flies showed no GFP expression, no further data were collected and a value of zero was recorded. Data were collected from only one antenna per fly.
Brain imaging and analysis:
Flies were reared as described above and immunostained as described previously (Python and Stocker 2002). We used primary antibodies nc82 (Developmental Studies Hybridoma Bank, University of Iowa) and rabbit polyclonal anti-GFP (Invitrogen Corp., Carlsbad, CA; no. A-11122) at 1:100 and 1:1000 dilutions, respectively. Secondary antibodies Alexa Fluor 568 goat anti-mouse IgG (Invitrogen, no. A-11004) and Alexa Fluor 488 goat anti-rabbit (Invitrogen, no. A-11008) were used at 1:1000 and 1:500 dilutions, respectively. Images were acquired with a Zeiss LSM510 laser scanning microscope with 0.72 μm spacing between optical sections. Gain and offset were optimized for each image due to the wide range of signal intensities observed. Analysis of brain images to determine glomerular labeling was performed with Imaris 5.0.1 software (Bitplane AG, Zurich, Switzerland); two-dimensional brain images were processed with ImageJ software (Rasband 1997). Glomeruli were identified on the basis of previous descriptions of antennal lobe anatomy (Laissue et al. 1999; Couto et al. 2005).
Cross-species sequence analysis:
We examined the conservation of particular DNA sequences using the whole genome comparative analysis of the D. melanogaster (CAF1) genome visualized with the VISTA Genome Browser, accessed at http://pipeline.lbl.gov/cgi-bin/gateway2?bg=droMel_caf1&selector=vista (Dubchak et al. 2000; Frazer et al. 2004).
RESULTS
Initial reporter constructs for trichoid Or genes:
To examine the cis-regulation of Or genes in trichoid sensilla, we first constructed Or promoter–GAL4 fusions of Or47b, Or88a, Or65a, and Or67d. The promoters comprise nearly all the intervening DNA between the start codon of the Or gene and the coding region of the next upstream annotated gene, except in the case of Or65a, in which the next annotated gene is ∼57 kb upstream. Specifically, the extent of upstream DNA is 7.9 kb for Or47b, 1.6 kb for Or88a, 4.7 kb for Or65a, and 6.1 kb for Or67d. We examined the expression of the Or–GAL4 constructs by using them to drive a UAS–GFP reporter construct (Chiba 1998; Ritzenthaler et al. 2000).
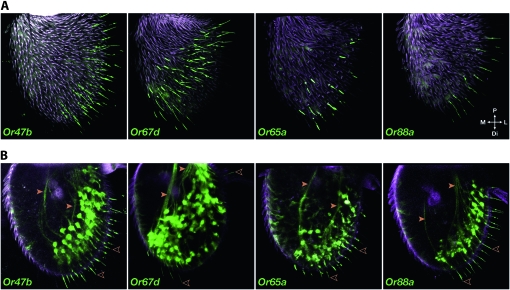
All four Or–GAL4 constructs drive GFP expression in subsets of antennal ORNs (Figure 2). These neurons project dendrites into olfactory sensilla on the antennal surface (Figures 2A and 2B, open arrowheads), and extend axons through the antenna (Figure 2B, solid arrowheads) and into the antennal lobe (AL) of the brain (Figures 3E, 9M, and Figure S1).
Figure 2.—
Antennal expression patterns of initial Or promoter–GAL4 constructs: Or47b−7.9kb, Or67d−6.1kb, Or65a−4.7kb, Or88a−1.6kb. Cuticular autofluorescence appears as magenta. (A) Confocal images of Drosophila antennae homozygous for both the UAS–GFP and the Or–GAL4 construct. Images in A are Z-projected cuticle-level confocal sections of the antennae showing patterns of GFP-labeled olfactory sensilla in the distolateral region (Or47b, Or65a, and Or88a) and midregion (Or67d) of the antenna. (B) Z-projections of interior sections reveal ORN dendrites (open arrowheads) and axons (solid arrowheads). Abbreviations: P, proximal; M, medial; L, lateral; Di, distal.
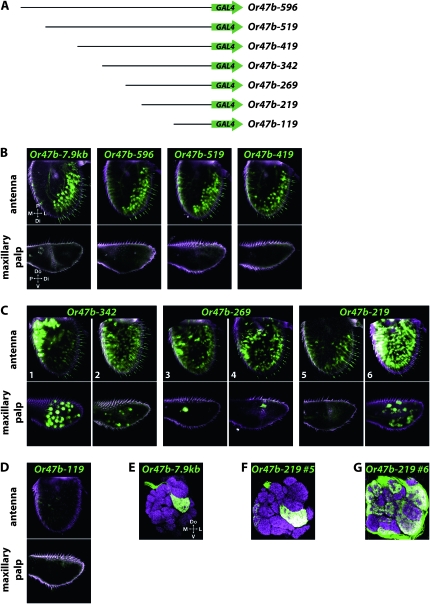
Figure 3.—
Expression of Or47b–GAL4 truncation constructs. (A) Scheme of Or47b–GAL4 truncation constructs. Black lines represent Or47b sequences upstream of the predicted translation start site, and green arrows represent vector and GAL4 coding sequences. (B–D) Images are interior sections of antennae (top row) or maxillary palps (bottom row) heterozygous for the indicated construct and the UAS–GFP reporter, except Or47b−7.9kb, which is homozygous for both. (B) Constructs with 419 bp or more of Or47b upstream DNA show similar patterns of expression. (C) Variable expression patterns of truncated Or47b–GAL4 constructs. For each construct, confocal Z-projections from two independent insertion lines are shown. All lines of these three constructs exhibited some degree of maxillary palp GFP expression. (D) No expression was observed in antennae or maxillary palps of flies carrying Or47b−119. (E–G) Antennal lobes showing neuropil (nc82 antibody) in magenta and GFP expression (anti-GFP antibody) in green. (E) Antennal lobe from a 0- to 1-day-old fly homozygous for Or47b−7.9kb–GAL4 and UAS–GFP. (F and G) Antennal lobes from 7- to 8-day-old flies heterozygous for the indicated Or47b−219 insertion and the UAS–GFP reporter. Similar patterns of glomerular targeting were seen in 0- to 1-day-old flies (not shown). Abbreviations: P, proximal; Do, dorsal; Di, distal; M, medial; L, lateral; V, ventral.
Figure 9.—
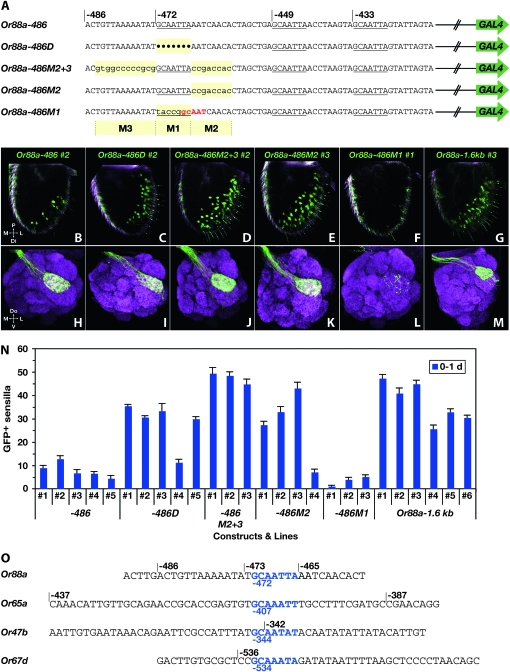
Mutation of sequences near −472 of Or88a. (A) Constructs. Mutated bases are in lowercase type and shaded. Dots indicate deleted base pairs. Red letters denote a novel GCAAT sequence in Or88a−486M1 created by mutating GCAATTA at –472. The mutated subregions of Or88a−486 are indicated below the sequence (M1, M2, M3). (B–G) Z-projected interior confocal sections of 0- to 1-day male antennae heterozygous for both the UAS–GFP reporter and the indicated Or88a–GAL4 construct. (H–M) Z-projections of 0- to 1-day male antennal lobes heterozygous for both the indicated Or88a–GAL4 construct and the UAS–GFP reporter. Antennal lobes are immunostained with anti-GFP (green) and nc82 (magenta). (N) Mean numbers of GFP-labeled sensilla (±SEM; 5 ≤ n ≤ 11). (O) The putative repressor element GCAATTA upstream of Or88a, and similar sequence elements upstream of other trichoid Or genes. In each case the element lies within or very near a region with negative regulatory function. Occurrences of these elements are aligned and shown in blue type. For A and O, positions are relative to the predicted translation start sites of the indicated genes. Abbreviations: d, days post-eclosion; P, proximal; Do, dorsal; M, medial; L, lateral; Di, distal; V, ventral.
Sensillar labeling patterns fall into two classes. The Or47b, Or65a, and Or88a drivers show expression in trichoid sensilla along the distolateral region of the antenna; by contrast, the Or67d driver shows expression in a band of trichoid sensilla that extends across the central region of the antenna. The differences in these two patterns can be seen most clearly in optical sections at the cuticle level (Figure 2A). These results are consistent with previous results from in situ hybridization and reporter gene studies showing that Or47b, Or65a, and Or88a are each expressed separately in one of the three ORNs that innervate at4 trichoid sensilla in the distolateral region of the antenna and that Or67d is expressed in singly innervated at1 sensilla in the central region of the antenna (Vosshall et al. 1999; Couto et al. 2005; Fishilevich and Vosshall 2005). We note that the Or47b, Or65a, and Or88a constructs label distolateral trichoid sensilla exclusively; the Or67d driver shows additional expression in a few nontrichoid sensilla in the vicinity of at1 sensilla, as well as in some nonneuronal cells, as considered below. Projections to the antennal lobe demonstrate that each of the four initial GAL4 constructs labels the appropriate antennal lobe glomerulus; the Or65a and Or67d constructs also label a small number of additional glomeruli (Figure S1 and Table S2), consistent with prior reports of Or65a and Or67d reporter construct expression (Couto et al. 2005; Fishilevich and Vosshall 2005; Ejima et al. 2007). The simplest interpretation of our results is that the upstream regions used in our constructs are sufficient to drive expression in the appropriate ORNs, with no misexpression or limited misexpression.
Or47b truncations reveal repressive functions:
Or47b encodes a receptor sensitive to extracts of both males and virgin females, suggesting that it may report the proximity of another fly (van der Goes van Naters and Carlson 2007). We generated a series of seven constructs containing successively smaller regions of DNA upstream of the Or47b coding region (Figure 3A). Each Or47b construct was fused to the GAL4 gene, and the expression of each GAL4 driver was examined by using it to drive a UAS–GFP reporter construct.
The three longest constructs, Or47b−596, Or47b−519, and Or47b−419, which extend from position −1 to positions −596, −519, and −419 bp, respectively, upstream of the translation start site, drove expression in patterns consistent with the distribution of at4 sensilla (Couto et al. 2005; Fishilevich and Vosshall 2005) and similar to the expression pattern of the Or47b driver containing 7.9 kb of upstream DNA (Figure 3B). We note that although the transcription start site has not been identified for Or47b, rapid amplification of 5′ cDNA ends (5′ RACE) for four other Or genes revealed that in every case the predicted transcriptional start sites, as defined by the longest RACE products, lie within 50 bp of the predicted translational start sites (Ray et al. 2007).
Constructs carrying less Or47b upstream DNA, however, show striking differences in expression. First, labeling of the antenna is not restricted to trichoid sensilla or to the distolateral region. Second, expression is observed in another olfactory organ, the maxillary palp. Third, expression patterns are more variable than are those of the longer constructs, in the sense that different lines, containing insertions at different positions, exhibit somewhat different expression patterns. Each of these effects is described below.
We examined 3−5 lines each of Or47b−342, Or47b−269, and Or47b−219, yielding a total of 11 lines (Table 1). In all of these lines, antennal expression was observed in basiconic sensilla as well as trichoid sensilla (Figure 3C). In 9 of these 11 cases, expression was observed in the large basiconic sensilla of the proximomedial region and in trichoid sensilla, but was largely excluded from a swath across the central region of the antenna, e.g., Figure 3C1. In the other two cases, sensilla were labeled in all or nearly all regions of the antenna, e.g., Figure 3C6. It is interesting to note that in some lines the amount of label is greater in the proximomedial region than in the distolateral region; thus, the amount of ectopic label is greater than the amount of label in the normal region.
TABLE 1.
Or47b reporter construct expression
| Antennal ORN expressiona |
||||||
|---|---|---|---|---|---|---|
| Construct name | Upstream DNA (bp) | Lines examined | Distolateral only | Distolateral + proximomedial | Entire antenna | Maxillary palp expressionb |
| Or47b−7.9kb | 7934 | 3 | 3/3 | 0/3 | 0/3 | 0/3 |
| Or47b−596 | 596 | 2 | 2/2 | 0/2 | 0/2 | 0/2 |
| Or47b−519 | 519 | 2 | 2/2 | 0/2 | 0/2 | 0/2 |
| Or47b−419 | 419 | 3 | 3/3 | 0/3 | 0/3 | 0/3 |
| Or47b−342 | 342 | 3 | 0/3 | 2/3 | 1/3 | 3/3 |
| Or47b−269 | 269 | 3 | 0/3 | 3/3 | 0/3 | 3/3 |
| Or47b−219 | 219 | 5 | 0/5 | 4/5 | 1/5 | 4/5 |
| Or47b−119 | 119 | 2 | 0/2 | 0/2 | 0/2 | 0/2 |
Fraction of lines exhibiting each pattern.
Fraction of lines that show any GFP-labeled olfactory sensilla on the maxillary palp.
Maxillary palp expression was observed in 10 of the 11 lines (Table 1). Labeling was observed in ORNs, as judged by the presence of GFP in dendrites and axons. Interestingly, expression in the maxillary palp appeared to correlate with expression in the proximomedial ORNs: the number of maxillary palp cells that were labeled appeared to correlate with the intensity of labeling in the proximomedial region of the antenna. Ectopic expression in the maxillary palp was not observed in lines carrying more than 342 bp of upstream sequences (Table 1 and Figure 3B).
To understand better the variation between different lines of the same construct, we examined the projections of labeled ORNs to the antennal lobe in the brain for the two lines of Or47b−219 shown in Figure 3C. ORNs expressing a given Or gene project to a unique glomerulus in the AL (Couto et al. 2005; Fishilevich and Vosshall 2005; Endo et al. 2007); thus, determining which glomeruli are labeled by each construct allows us to infer the identities of the associated ORNs.
We found that the Or47b−7.9kb construct exclusively labels the appropriate glomerulus, VA1v (Figures 3E, Figure S1, and Table S2). This finding is consistent with the faithful expression directed by this construct. The two Or47b−219 lines, referred to as 5 and 6 (corresponding to Figures 3C5 and 3C6, respectively), represent opposite ends of the range of expression patterns observed among Or47b truncation constructs: particularly restricted in the case of line 5 and particularly broad in the case of line 6. The glomeruli labeled in line 5 were a subset of those labeled in line 6 (Figure 3, F and G, and Table S3). While line 5 was expressed in at most one ORN class per sensillum and was restricted to antennal basiconic and trichoid sensilla, line 6 frequently labeled both ORN classes in a sensillum. Line 6 also labeled maxillary palp sensilla as well as all three morphological types of antennal sensilla.
The simplest interpretation of our results is that negative regulatory elements between −342 and −419 repress expression in basiconic sensilla in the more proximomedial portions of the antenna, and in the maxillary palp. No expression was observed from an Or47b−119 construct (Figure 3D), suggesting that sequences required for ORN expression lie upstream of −119.
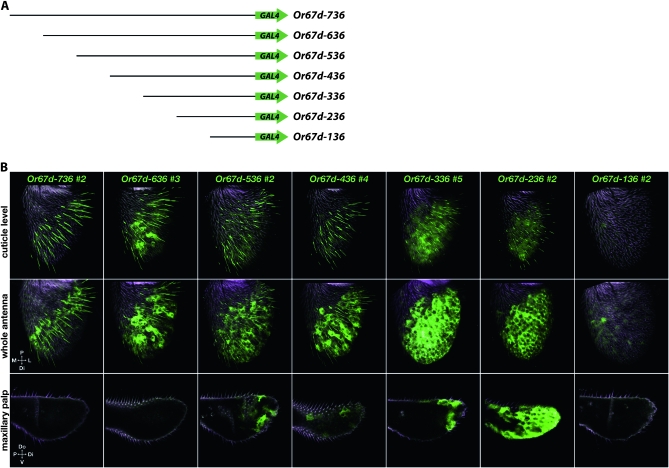
Or67d constructs respect spatial boundaries of antennal expression and reveal a function that restricts expression to ORNs:
Or67d encodes a receptor that is sensitive to the pheromone cVA, which can function as an anti-aphrodisiac to deter males from courting a recently mated female (Ha and Smith 2006; Ejima et al. 2007; Kurtovic et al. 2007; van der Goes van Naters and Carlson 2007). We generated eight constructs that include varying extents of DNA upstream of Or67d. The longest construct, containing 6.138 kb of upstream DNA, shows expression in a band of trichoid ORNs that extends across the central region of the antenna, a distribution similar to that of at1 (Figure 2). Examination of 10 independent insertion lines of Or67d−6.1kb revealed expression in a small number of basiconic sensilla and coeloconic sensilla in the vicinity of these trichoid sensilla, and in some nonneuronal cells. Some limited labeling of maxillary palp neurons is also observed (Table 2). Antibody staining confirms that glomerulus DA1, targeted by Or67d-expressing ORNs, is labeled (Figure S1 and Table S2); it also confirms the observation of some expression in additional ORN classes.
TABLE 2.
Or67d reporter construct expression
| Construct name | Upstream DNA (bp) | Lines examined | Antennal ORN expressiona | Maxillary palp expressionb |
|---|---|---|---|---|
| Or67d−6.1kb | 6138 | 10 | +++ | +c |
| Or67d−736 | 736 | 3 | +++ | — |
| Or67d−636 | 636 | 2 | +++ | — |
| Or67d−536 | 536 | 3 | +++ | +++ |
| Or67d−436 | 436 | 3 | +++ | +++ |
| Or67d−336 | 336 | 4 | +++ | +++ |
| Or67d−236 | 236 | 4 | +++ | +++ |
| Or67d−136 | 136 | 2 | — | — |
Qualitative assessment of expression level. We observe GFP-labeled ORNs in both trichoid and nontrichoid sensilla; +++ indicates the highest number of GFP-labeled olfactory sensilla and — indicates no GFP-labeled sensilla. All constructs except 67d−136 show expression in nonneuronal antennal cells.
+++ indicates consistent GFP expression and — indicates GFP expression in no more than one maxillary palp among all lines examined.
We analyzed maxillary palp expression in one line, which was homozygous for both Or67d−6.1kb and the UAS-GFP reporter, and observed modest GFP expression.
Six of the constructs containing less DNA also drive expression in ORNs of trichoid sensilla (Figure 4). The labeled trichoid sensilla in the truncation constructs are concentrated in the central region, as in Or67d−6.1kb. These six constructs also drive some limited expression in other sensillum types, including both basiconic and coeloconic sensilla.
Figure 4.—
Or67d–GAL4 truncation constructs. (A) Scheme of Or67d reporter constructs. Black lines indicate regulatory DNA and green arrows indicate the GAL4 coding region and vector sequences. (B) Expression of Or67d–GAL4 truncation constructs. Antennae and maxillary palps were dissected from male flies of various ages for Or67d−136, and at 7–8 days for all other constructs. Flies were heterozygous for both the indicated Or67d–GAL4 construct and the UAS–GFP reporter. Images are Z-projections of cuticle-level confocal sections or of entire antennae (top and middle rows, respectively) and confocal Z-projections of maxillary palps (bottom row). GFP-labeled ORNs lie primarily in a band across the midregion of the antenna (most evident in the top row). All Or67d–GAL4 truncation constructs with at least 236 bp of upstream DNA exhibited nonneuronal GFP expression in the antenna (most evident in middle row). Numbers after the construct names identify the specific transgenic line shown. Abbreviations: P, proximal; Do, dorsal; M, medial; L, lateral; Di, distal; V, ventral.
All six constructs also show nonneuronal expression (Figure 4). Although it is difficult to quantitate rigorously the level of nonneuronal expression, the shorter constructs appear to show particularly high levels of nonneuronal labeling. The cells have prominent unlabeled nuclei and broad cell bodies, and they do not extend processes into sensilla. Our observations are consistent with prior descriptions of antennal epidermal cells (Shanbhag et al. 2000). These cells lie in a range extending from the proximomedial limit of trichoid sensilla to the distolateral tip of the antenna, i.e., the entire range of trichoid sensilla. Interestingly, there is little if any labeling of these nonneuronal cells in the proximomedial region of the antenna. Thus for both neuronal and nonneuronal expression, certain spatial boundaries of antennal expression are still respected despite the removal of large amounts of upstream regulatory DNA. Or67d differs fundamentally from Or47b, in that constructs of Or47b show strong labeling of sensilla in the proximomedial region.
Although there is little or no maxillary palp expression in Or67d−636 or longer constructs, the shorter constructs showed high levels of maxillary palp labeling in some lines. Labeling is observed in both neuronal cells, as judged by the presence of axons and dendrites, and in nonneuronal cells.
The shortest construct, Or67d−136, shows no GFP expression in either neurons or other cell types, indicating that essential cis-regulatory elements that drive expression of Or67d are located between −236 bp and −136 bp.
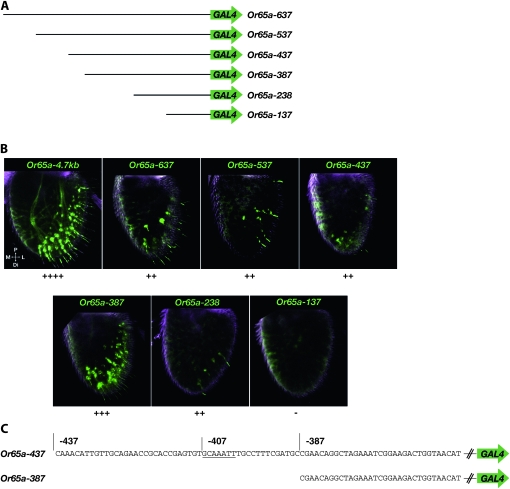
Dissection of Or65a regulatory sequences:
We next examined the Or65a gene, which encodes a receptor sensitive to extracts from males, male genital material, and genital material from mated females, but not from virgin females. Or65a may signal to a male the presence of an inappropriate mating partner (Ejima et al. 2007; van der Goes van Naters and Carlson 2007). We generated a series of seven constructs containing variable extents of DNA upstream of the Or65a coding sequences (Figure 5A). All but the shortest construct drive expression in antennal ORNs of trichoid sensilla (Figure 5B and Table 3). All show labeling in distolateral sensilla on the antennal surface, in a pattern similar to that of at4 sensilla, to which Or65a has been mapped (Couto et al. 2005; Fishilevich and Vosshall 2005). Examination of AL projections in one line of Or65a−4.7kb confirmed that this construct labels the Or65a-associated glomerulus, DL3 (Figure S1 and Table S2). Glomeruli targeted by the other two ORNs in at4 sensilla, as well as two of three glomeruli targeted by at3 ORNs, were also labeled, although at qualitatively lower levels than the Or65a glomerulus.
Figure 5.—
Expression of Or65a–GAL4 truncation constructs. (A) Constructs. (B) Z-projections of interior confocal antennal sections from flies heterozygous for both the indicated Or65a–GAL4 truncation construct and the UAS–GFP reporter. Expression levels are indicated beneath the images. (C) DNA sequence upstream of Or65a. A GCAAATT element is underlined. Positions relative to the predicted Or65a translation start site are indicated. Abbreviations: P, proximal; M, medial; L, lateral; Di, distal.
TABLE 3.
Or65a reporter construct expression
| Construct name | Upstream DNA (bp)a | Lines examined | Antennal ORN expressionb | Maxillary palp expression |
|---|---|---|---|---|
| Or65a−4.7kb | 4665 | 3 | ++++ | — |
| Or65a−637 | 636 | 2 | ++ | — |
| Or65a−537 | 536 | 2 | ++ | — |
| Or65a−437 | 436 | 5 | ++ | — |
| Or65a−387 | 386 | 7 | +++ | — |
| Or65a−238 | 237 | 5 | ++ | — |
| Or65a−137 | 136 | 2 | — | — |
The Or65a anchor primer (Ejima et al. 2007) extends to −2 relative to the Or65a translation start site; thus, the amount of upstream DNA included in each truncation is 1 bp shorter than suggested by the construct name.
Qualitative assessment of expression level. We observe GFP-labeled ORNs in both trichoid and nontrichoid sensilla; ++++ indicates the highest number of GFP-labeled olfactory sensilla and — indicates no GFP-labeled sensilla.
We observed differences in the number of trichoid sensilla labeled by various Or65a truncation constructs. The three longest truncation constructs, Or65a−637, Or65a−537, and Or65a−437, appear to label fewer sensilla than the longer Or65a−4.7kb construct. This suggests the presence of an enhancer element upstream of −637. Interestingly, the Or65a−387 construct appears to label more sensilla than other truncations, although not more than Or65a−4.7kb. One interpretation of this result is that there may be a repressor element between –387 and −437. We note the presence of a GCAAATT sequence in this interval (Figure 5C). The corresponding sequence is identical in D. simulans, D. yakuba, D. erecta, D. ananassae, and D. pseudoobscura and is discussed further below.
The shortest Or65a truncation construct that drove antennal expression, Or65a−238, did so only weakly (Figure 5B and Table 3). Fewer than five sensilla per antenna were labeled in two of the three transgenic lines, and no expression was detected in a third line.
We did not observe labeling of the maxillary palps in any Or65a construct, in contrast to our results when dissecting the regulatory regions of Or47b and Or67d.
Regulatory architecture of the Or88a gene:
The Or88a gene encodes a receptor that is sensitive to odor of both males and females and that may serve to report the proximity of another fly (van der Goes van Naters and Carlson 2007). We generated a series of constructs, each containing between 1621 and 160 bp upstream of the translation start site (Table 4 and Figure 6A).
TABLE 4.
Or88a reporter construct expression
| Construct name | Upstream DNA (bp) | Lines examined | Trichoid ORN expressiona | Maxillary palp expression |
|---|---|---|---|---|
| Or88a−1.6kb | 1621 | 6 | ++++ | — |
| Or88a−486 | 486 | 6 | ++ | — |
| Or88a−486M1 | 486 | 3 | +++ | — |
| Or88a−486M2 | 486 | 4 | +++ | — |
| Or88a−486M2+3 | 486 | 6 | +++ | — |
| Or88a−486M4 | 486 | 5 | — | — |
| Or88a−486D | 479 | 5 | +++ | — |
| Or88a−473 | 472b | 3 | ++ | — |
| Or88a−465 | 465 | 5 | ++ | — |
| Or88a−465M1 | 465 | 7 | — | — |
| Or88a−359 | 360 | 1 | — | — |
| Or88a−159 | 160 | 1 | — | — |
Qualitative assessment of expression level; ++++ indicates highest number of GFP-labeled trichoid sensilla, — indicates no GFP-labeled sensilla.
This construct extends to position −473 of the Or88a upstream region but carries a −458A deletion, making the total construct length 472 bp.
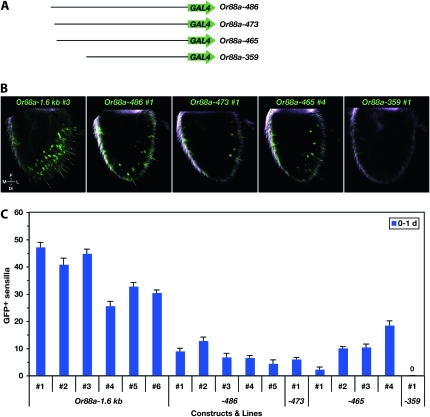
Figure 6.—
Expression of Or88a–GAL4 constructs. (A) Constructs. (B) Expression patterns of the indicated Or88a–GAL4 truncation constructs are shown in Z-projections of internal confocal sections. The three constructs with 486–465 bp of Or88a upstream DNA are expressed in the distolateral region of the antenna, but in fewer ORNs than Or88a−1.6kb. Constructs carrying 359 or 159 bp (not shown) do not drive detectable GFP expression in antennal ORNs. Antennae from flies heterozygous for both an Or88a–GAL4 driver and the UAS–GFP reporter were dissected 0–1 day post-eclosion. (C) Mean numbers of GFPlabeled sensilla per antenna (±SEM; 5 ≤ n ≤ 10 antennae) of 0- to 1-day males. Each bar represents data from an independent insertion of the indicated Or88a–GAL4 construct. 0 indicates that no GFP-labeled sensilla were detected. Abbreviations: P, proximal; M, medial; L, lateral; Di, distal.
The longest Or88a–GAL4 construct expresses in sensilla along the distolateral edge of the antenna (Figures 6B and 2B); this expression pattern matches that of the at4 sensilla, consistent with previous reports of Or88a expression based on reporter gene expression and in situ hybridization (Vosshall 2001; Couto et al. 2005; Fishilevich and Vosshall 2005). We quantitatively compared the expression of three independent Or88a−1.6kb lines at two different ages: 0–1 day and 7–9 days. We found no significant age-dependent difference in the number of labeled sensilla in lines 1 and 2 (P > 0.05, t-test) and a modest difference in line 3 (P < 0.05) (Figure 7A). For line 3, we then compared flies heterozygous for one autosomal copy of the GAL4 driver and one autosomal copy of UAS–GFP to flies homozygous for the GAL4 driver and for the UAS–GFP. We found no effect of dosage on the number of labeled sensilla either at 0–1 day or at 7–9 days (P > 0.05, t-test) (Figure 7B; compare bars of the same color).
Next we compared the expression of Or88a−1.6kb to that of truncated constructs (Figure 6). We found that Or88a−486, Or88a−473, and Or88a−465 drive reporter gene expression in patterns similar to that of Or88a−1. 6kb: sensilla along the distolateral edge of the antenna are labeled, with one ORN labeled per sensillum (Figure 6B). However, relative to Or88a−1.6kb, the mean numbers of labeled cells at 0–1 day were smaller in Or88a−486 (P < 0.001, t-test) and Or88a−465 (P < 0.001) (Figure 6, B and C, and Table 4). A subset of Or88a−486 and Or88a−465 lines were examined at 7–9 days and, in contrast to the Or88a−1.6kb results, all showed significant age-dependent increases in the mean numbers of labeled sensilla (P < 0.05) (Figure 7, A and C, and Figure S2). Constructs containing still less DNA, Or88a−359 and Or88a−159, showed no expression in the antenna (Figure 6, B and C, Table 4, and data not shown), although the number of examined lines was limited. These results provide evidence that sequences necessary for driving the general pattern of Or88a expression are located within 486 bp of the translation start site.
A sequence element that acts in regulation of Or88a:
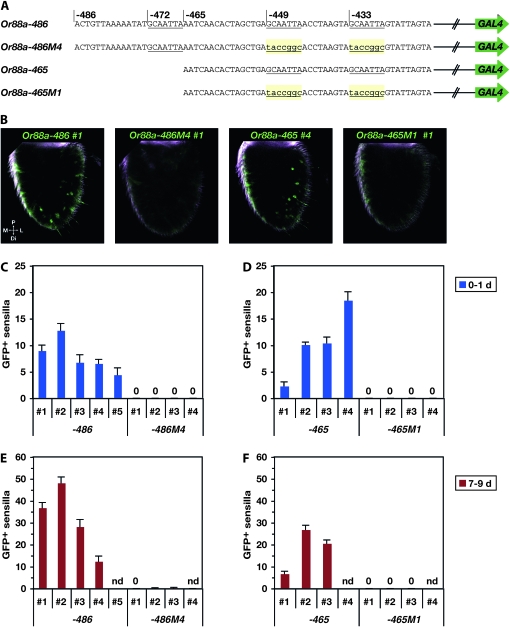
The expression patterns we observed with the series of Or88a–GAL4 truncation constructs, and in particular the differences in expression level between Or88a−465 and Or88a−359, suggested the presence of a positive regulatory element between −465 and −359. We noted with interest a sequence, GCAATTA, which occurs at three positions between −465 and −359: at −449, −433, and −376; moreover, it also occurs at –472 (three of these iterations are underlined in Figure 8A). The three most-upstream occurrences of this sequence are identical to the corresponding sequences in D. simulans, D. yakuba, and D. erecta. The occurrence at −376 differs by a single base in D. yakuba. We wondered whether any of these iterations of the sequence played a role in the regulation of Or88a.
Figure 8.—
Mutation of two GCAATTA elements upstream of Or88a. (A) Constructs. Each Or88a promoter fragment extends to, but does not include, the ATG of Or88a. Hatched lines represent downstream Or88a promoter sequence; green arrows represent vector and GAL4 coding sequence. Mutated base pairs are in lowercase type and shaded, while the positions of three iterations of GCAATTA are underlined. (B) Confocal Z-projections of 0- to 1-day male antennae heterozygous for both the indicated Or88a–GAL4 construct and the UAS–GFP reporter. (C–F) Numbers of labeled sensilla in flies heterozygous for one of the Or88a–GAL4 constructs in A and the UAS–GFP reporter. 5 ≤ n ≤ 13 antennae for all lines except that for −486M4, line 1 and −465M1, line 1, n = 3. The scales in C and D are different from those in E and F. 0 indicates that no GFP-labeled sensilla were detected. Abbreviations: nd, not determined; d, days post-eclosion; P, proximal; M, medial; L, lateral; Di, distal.
We examined the role of the GCAATTA repeats at −449 and −433 by simultaneously mutating them (Figure 8A). We quantitated the effects of these mutations in two different contexts: in the context of the Or88a−486 construct (Or88a−486M4) and in the context of the Or88a−465 construct (Or88a−465M1). The effects were evaluated at two different ages: 0–1 day and 7–9 days.
We found that mutation of these sequences ablated expression of both constructs, at both ages (Figure 8, B–F). The simplest interpretation of these results is that this sequence acts as a positive regulatory element, in one or both positions, and that the identical element at −472 cannot substitute for them.
We next examined the role of the GCAATTA motif at −472. When this element was deleted in the context of Or88a−486, to generate Or88a−486D (D represents deletion), there was a striking increase in the number of labeled trichoid sensilla (Figure 9, B, C, and N). We quantitated the number of labeled sensilla in five independent insertion lines of Or88a−486D and compared the numbers to those from the five independent insertion lines of Or88a−486. The mean counts for four of the five Or88a−486D lines exceeded those of all five of the Or88a−486 lines at 0–1 day (Figure 9N) and the two sets of means differ significantly (t-test, P < 0.01). The simplest interpretation of these results is that in the context of Or88a−486, the GCAATTA at −472 acts in negative regulation.
Since ablation of the other GCAATTA motifs had indicated a role in positive regulation, we had not expected to find a role for the GCAATTA motif at −472 in negative regulation. To explore the possibility that other sequences in the vicinity of −472 contribute to the negative regulation, we mutated the 12 bp immediately upstream (M3) and the 8 bp immediately downstream from the GCAATTA (M2), to generate Or88a−486M2+3. In each of three independent insertions examined, there was a major increase in the number of labeled trichoid sensilla compared to the Or88a−486 control at 0–1 day, and the means of these two sets differed significantly (P < 0.0001, t-test) (Figure 9, B, D, and N, and Figure S3). We note that the M2 sequence, like the GCAATTA sequence at −472, is identical in D. simulans, D. yakuba, and D. erecta; the M3 sequence is less evident.
We next asked whether an increase in expression would be observed if we mutated only the 8 bp downstream. We generated Or88a−486M2 to address this question and found that levels were elevated in three of four independent insertion lines (Figure 9, B, E, and N, and Figure S3).
Finally, we generated a construct, Or88a−486M1, that mutated the GCAATTA motif at −472, but that created a partial motif, GCAAT, slightly displaced from its position in wild type (Figure 9A). This construct, like Or88a−486, drove very low levels of expression at 0–1 day. These levels are statistically indistinguishable from the levels of Or88a−486 expression (P = 0.05) (Figures 9, B, F, and N, and Figure S3).
We found that ORNs labeled by Or88a−486 and by all mutation constructs derived from it targeted only the Or88a glomerulus (Figure 9, H–M), as do ORNs labeled by Or88a−1.6kb (Table S2), confirming that all these constructs label a single class of trichoid sensilla and a single ORN in those sensilla.
The simplest interpretation of these Or88a−486 mutations is that the GCAATTA motif at −472 acts in negative regulation in conjunction with sequences that flank it. Sufficiently severe disruption of these sequences increases the number of labeled sensilla to levels comparable to those driven by the Or88a−1.6kb construct, which contains all the upstream sequences up to the next annotated gene (Figure 9, B–G, and N, and Figure S3).
We note that there is a motif related to GCAATTA, in all cases of the form GCAAWWW, upstream of all four Or genes considered in this study (Figure 9O). Each of these instances lies within or extremely close to regions that possess negative cis-regulatory activity, as judged from experiments using truncation and mutation constructs. Moreover, in each case, corresponding sequences in D. simulans, D. yakuba, and D. erecta are identical. These observations are consistent with a functional role for GCAATTA and its flanking sequences in trichoid Or gene regulation.
DISCUSSION
In this study we have investigated the logic of Or gene regulation. We have focused on four receptors recently shown to respond to fly odors in an in vivo expression system. These four receptors form the basis of a model for the olfactory recognition of mates by males (van der Goes van Naters and Carlson 2007). The proper expression of these receptors is thus likely to underlie a function critical to the reproduction of the species. We have attempted to gain insight into the logic of this regulation by subjecting the upstream regions of the four genes to a mutational analysis, using deletion and base substitution mutations. This approach has revealed several features of the regulation of these Or genes.
Compactness of regulatory regions:
For each of the four Or genes, relatively short regions of upstream DNA were sufficient to confer substantial expression in patterns resembling those of the endogenous genes. Regions of 419 bp (Or47b), 736 bp (Or67d), 637 bp (Or65a), and 465 bp (Or88a) all drove reporter gene expression in patterns similar to those of the sensilla in which they are expressed, although in some cases the expression levels were reduced. This finding is consistent with the observation that short regions are sufficient for faithful expression of two maxillary palp Or genes, Or85e (450 bp) and Or46a (400 bp) (Ray et al. 2007).
The shortest truncation constructs derived from the four genes showed no expression. In the case of Or88a, 360 bp of upstream DNA was not sufficient to drive expression in trichoid sensilla or elsewhere in the antenna, nor was 160 bp sufficient. Few if any sensilla of any kind were labeled in constructs containing 137 bp of Or65a DNA, 136 bp of Or67d DNA, or 119 bp of Or47b DNA. The labeling observed in somewhat larger constructs provides evidence that ORN expression requires DNA in the following intervals: −465 to −360 of Or88a, −387 to −238 of Or65a, −236 to −136 of Or67d, and −219 to −119 of Or47b.
The small size of these regions may reflect a relative simplicity of temporal regulation: there is no evidence for expression of these genes other than at the adult stage (Fishilevich et al. 2005; Kreher et al. 2008). There is also no evidence for expression of these four genes in organs other than the antenna (Couto et al. 2005; Fishilevich and Vosshall 2005; Goldman et al. 2005). Expression in the correct subset of antennal cells, however, requires the action of multiple regulatory elements that act in concert to dictate proper Or gene expression, as discussed below.
Small regulatory regions may be advantageous in gene families that undergo expansion through duplication and divergence. Since the distance between start and stop codons in these four genes is also short, in no case exceeding 1.7 kb, even small duplications may create fully functional receptor genes that can then undergo changes in odor sensitivity or expression pattern (Ray et al. 2007).
Positive regulation of expression level:
Truncations of the Or88a promoter region reduced the numbers of labeled sensilla (Figure 6). This reduction is likely due to a reduction in the expression of the GFP reporter driven by the truncated promoters. For both Or88a−486 and Or88a−465, as well as all constructs derived from Or88a−486 (except Or88a−486M4, which was not expressed), more GFP-labeled sensilla were counted in 7- to 9-day flies than in 0- to 1-day flies for all lines examined, presumably reflecting the accumulation of GFP with time, the perdurance of GFP driven by the GAL4-UAS system (Osterwalder et al. 2001), and the sensitivity threshold of the analysis (Figures S2 and Figure S3). By contrast, the number of sensilla labeled by the longest Or88a construct, Or88a−1.6kb, exhibited an age-dependent increase in only one of three lines examined—and the increase in that case was very small. Moreover, the number of expressing sensilla was not greater in flies doubly homozygous for the driver and reporter, suggesting that this long construct drives expression above the detection threshold virtually everywhere it is expressed, even when present in a single copy (Figure 7). A simple explanation of these results is that Or88a constructs that include 486 bp or less of upstream sequences lack regulatory elements that positively regulate the level of gene expression. Interestingly, a recent study detected more Or88a transcript in 6-week-old flies than in ∼1-week-old flies (Zhou et al. 2009). This observation raises the possibility that Or88a-intrinsic age-dependent dynamics may also contribute to the increased number of sensilla labeled by Or88a truncation constructs.
Negative regulation restricts the pattern of Or gene expression:
Each antennal Or gene requires information dictating its expression in the antenna, in the proper subtype of sensilla of the antenna, and in a single ORN of each sensillum. In principle, such expression could be mediated solely by positive regulatory mechanisms. However, we found evidence that negative regulation restricts the pattern of Or gene expression to the correct trichoid ORNs. Deletion of upstream regions produced ectopic expression in another organ, in another region of the antenna, or in another cell type, providing evidence for negative regulatory elements whose action limits expression of Or genes to the correct cells. Below we consider each of these restrictions in turn.
Restriction to the antenna:
Deletion of Or47b–GAL4 sequences in the interval between −419 and −342 resulted in ectopic expression in the maxillary palp, indicating the presence of an element that represses maxillary palp expression. Previously, two cis-regulatory elements, Oligo-1 and Dyad-1, were defined in a study of maxillary palp Or gene regulation and shown to promote maxillary palp expression (Ray et al. 2007). Moreover, Oligo-1 also represses antennal expression of maxillary palp genes, a function reciprocal to the repression of maxillary palp expression that we have defined here for antennal genes.
A repressive function limiting expression to the antenna was also identified in the case of Or67d. Although there was some limited labeling of maxillary palp neurons in the Or67d−6.1kb construct, the deletion of sequences in the interval between −636 and −536 markedly elevated ectopic expression in the maxillary palp, particularly in nonneuronal cells.
These results raise interesting questions about the relationship between antennal and maxillary palp Or gene expression. The expression of antennal Or drivers in the maxillary palp and vice versa suggests the sharing of regulatory mechanisms between these two organs and between the Or genes that they express. The sharing of regulatory mechanisms among olfactory organs may facilitate evolutionary switching of expression from one organ to another. In this regard, we note two pairs of Or genes (Or85b, Or85d) and (Or59b, Or59c), which are close to each other in the genome and closely related phylogenetically (Robertson et al. 2003). In each pair, one gene is expressed in the antenna and one in the maxillary palp (Couto et al. 2005; Fishilevich and Vosshall 2005; Goldman et al. 2005). Perhaps each pair of genes arose through duplication and then diverged in expression pattern. Our results suggest that a small deletion of upstream DNA may be sufficient to alter the organ specificity of expression. We note that misexpression of an antennal receptor in the maxillary palp was found to alter the chemosensory behavior of a fly (Shiraiwa 2008), illustrating the possibility that a change in organ specificity of Or expression could confer a selective advantage to the animal. Moreover, CO2 reception apparently moved from one organ to the other during the evolution of the Dipterans; it also moved following experimental mutation of a microRNA in Drosophila (Cayirlioglu et al. 2008).
We did not observe ectopic maxillary palp expression for any construct derived from Or88a or Or65a. It is possible that these two genes contain a negative regulatory element that represses expression in the maxillary palp, but that lies close to sequences essential for expression, thereby precluding identification by our deletion analysis. It is also possible that these genes lack a positive cis-regulatory element essential for expression in maxillary palps.
Restriction to a region of the antenna:
The third segment of the antenna is patterned in zones along the proximal–distal axis, as revealed through anatomical (Shanbhag et al. 1999), physiological (de Bruyne et al. 2001; van der Goes van Naters and Carlson 2007), and molecular analysis (Shanbhag et al. 2001; Couto et al. 2005; Fishilevich and Vosshall 2005). The ectopic labeling patterns of the Or47b derivatives revealed different regions of antennal expression that are consistent with this organization: a distolateral region, a central region, and a proximomedial region. The simplest interpretation of our results is that in wild type the confinement of Or47b expression to the distolateral region depends on its repression in the other two regions of the antenna. The Or67d–GAL4 patterns can also be interpreted in terms of the proximal–distal axis. The longest constructs show expression primarily in a swath across the central region, but in shorter constructs, expression appears in the distolateral region as well.
Or67d–GAL4 showed some ectopic expression in nontrichoid sensilla even in the longest constructs we examined, as has been observed with other Or67d–GAL4 lines constructed by others (Couto et al. 2005; Fishilevich and Vosshall 2005). A recent report substituted the Or67d coding region with that of GAL4 by gene replacement technology and showed that reporter expression was limited to trichoid sensilla (Kurtovic et al. 2007). One interpretation of these results is that there is a cis-regulatory element downstream of Or67d that contributes to the repression of expression in the basiconic sensilla.
In no line did we observe expression in the sacculus, a sensory pit containing ∼45 coeloconic sensilla. Perhaps expression in sensilla of the sacculus requires cis-regulatory sequences not present upstream of these trichoid Or genes. We note also the lack of obvious sexual dimorphism in the antennal expression patterns of any construct (not shown), consistent with the lack of dimorphism in physiological responses of trichoid sensilla (van der Goes van Naters and Carlson 2007).
Restriction to ORNs:
A different kind of restriction, a restriction of expression to ORNs, is violated by deletions of the Or67d regulatory region. There are two particularly interesting aspects to this restriction. First, it does not appear to map exclusively to a single discrete interval in the case of Or67d. There is a very limited extent of nonneuronal labeling even in the Or67d−6.1kb line, but the truncated constructs show higher levels of nonneuronal labeling, with the highest levels observed in the shortest constructs. One possible explanation for these results is that restriction to neurons may depend on the action of multiple negative regulatory elements that are distributed within the upstream region. We expect that unambiguous identification of the nonneuronal cell type(s) labeled by Or67d constructs will be essential to understanding the significance of the misexpression.
Second, even among the nonneuronal cells, the restriction to the trichoid region of the antenna prevails. One interpretation of these results is that the establishment of regional specificity lies above the establishment of cell specificity in a hierarchy of Or67d gene control.
Restriction to a single ORN per sensillum:
In wild type, expression of individual Or genes is limited to a single ORN per sensillum. This limitation depends on Notch signaling (Endo et al. 2007; Ray et al. 2007). The initial constructs for both Or88a and Or47b exhibited this limitation (Figures 4E, 9M), as did mutated Or88a−486 constructs (Figure 9, H–L). However, the Or65a−4.7kb construct and one line of the Or47b−219 construct did not. In the case of Or65a−4.7kb, two of three at3 ORNs were labeled as were all three at4 ORNs (Figure S1 and Table S2). Such misexpression may result from regulatory mechanisms shared by ORNs of both at3 and at4. The case of the Or47b−219 construct is particularly interesting and is considered next.
Modulation of restriction:
Shorter Or47b truncations were striking in their variability of expression pattern: different lines carrying the same construct showed highly variable extents of ectopic expression (Figure 3C). Detailed examination of two independent lines of Or47b−219 revealed variation with respect (1) to the restriction of expression from the maxillary palp, (2) to the range of ORNs expressing this construct within the antenna, and (3) to the restriction of expression to one ORN per sensillum (Figure 3, E–G and Table S3). One possible interpretation of this variability is that, in addition to losing a negative regulatory element, the constructs have lost an insulator or boundary element (Gaszner and Felsenfeld 2006). The lack of the insulator element would render the constructs highly sensitive to genomic position effects. Our results suggest that such effects may be large enough to overwhelm many levels of negative regulation.
We note that there is a well-conserved region between 419 and 342 bp upstream of Or47b (Figure 3). Constructs carrying a mutation of a well-conserved 17-bp sequence within this region, from position −388 to −373, showed neither the ectopic expression nor the interline variability exhibited by constructs carrying 342–219 bp of upstream DNA (Figure S4), suggesting that this sequence is not required for such insulation.
To date, no other cases of insulator activity have been reported near other Drosophila Or genes, perhaps because cis-regulatory regions of only a few Or genes have been examined in detail (Ray et al. 2008, 2007). However, it is also possible that Or47b is particularly sensitive to the loss of an insulator and to position effects, perhaps because Or47b contains a regulatory element that is particularly susceptible to the influence of other regulatory elements in the genome.
Large families of odor receptor genes such as the Or family are believed to have evolved via duplication and dispersion (Robertson et al. 2003). A gene that requires an insulator may require either that the duplicating genetic unit contains an insulator or that it moves to a region that contains one. Alternatively, gene duplication events that retain gene-proximal cis-elements but omit upstream insulator elements could, in principle, serve as the first step of an evolutionary process resulting in a novel Or gene expression pattern.
Regulatory elements:
We were interested to find a GCAATTA sequence at four positions within 500 bp upstream of Or88a. Mutation of this sequence at −449 and −433 provided evidence that this sequence acts in the positive regulation of Or88a at one or both of these positions.
By contrast, mutation of GCAATTA at −472 provided evidence that the sequence may function in negative regulation: when these 7 bp were precisely deleted from the Or88a−486 construct to yield Or88a−486D, a striking increase in expression was observed (Figure 9). However, we acknowledge that the role of the GCAATTA in such negative regulation is not clear, for several reasons. First, a similar increase in expression was observed when the GCAATTA element was left intact but the 8 bp immediately downstream from it were mutated, in Or88a−486M2. Second, no such increase in expression was found when this GCAATTA element and sequences upstream of it were deleted, but the downstream sequences were left intact, in Or88a−465. One possible interpretation of these results is that the GCAATTA at −472 is capable of contributing to the negative regulation of Or88a in conjunction with sequences immediately downstream of it, but in a context-dependent manner. Another possibility is that the GCAATTA at −472 has a positive effect and that the flanking M2 and M3 regions have a negative effect that requires proper spacing between them. A detailed molecular explanation will require further experimentation.
In addition to the evidence for a negative regulatory function near −472 of Or88a, our deletion studies have also provided evidence for negative regulatory functions upstream of Or65a (between −437 and −387), Or47b (between −419 and −342), and Or67d (in multiple intervals). We note that in all of these small regions upstream of Or88a, Or65a, Or47b, and upstream of Or67d, there is a sequence similar to GCAATTA, of the form GCAAWWW (Figure 9O). In each case the sequence is identical in the corresponding regions of D. simulans, D. yakuba, and D. erecta genomes. The presence of GCAAWWW sequences in or very near regions with negative regulatory activity could reflect a common mechanism used to repress expression of pheromone receptor genes.
How could the GCAATTA function as a positive regulator at −449 and −433 of Or88a and a negative regulator at −472 of Or88a and perhaps other trichoid Or genes? There is ample precedent for sequence elements that activate some genes and repress others. For example, the Drosophila transcription factor Dorsal binds to a site that mediates activation at one promoter and repression at another promoter (Jiang et al. 1992; Pan and Courey 1992). Also, the regulatory effect of lozenge (lz), a transcription factor involved in eye and antennal development (Gupta et al. 1998; Voas and Rebay 2004), can be modulated by cofactors that bind sequences adjacent to the lz binding site (Canon and Banerjee 2003). Perhaps the GCAATTA elements at −449 and −433, which have been implicated in positive regulation, are flanked by sequences that bind a factor that modulates the function of a GCAATTA-binding protein, lending it a positive function.
Concluding remarks:
In this study we have used a genetic approach to investigate how, in a coordinated fashion, individual neurons of an olfactory organ select individual receptors to express. This process underlies the function of the entire olfactory system, which relies on the coordinated, parallel input of many individual sensory cells that each signal the presence of the odors detected by their receptors.
The problem of receptor gene choice can be considered from two perspectives: how an individual neuron selects which receptor to express, and how the sequences flanking an individual receptor gene dictate its expression in particular neurons. The first perspective has been illuminated by the identification of genes required by olfactory neurons to express particular receptors, in several species (Fuss and Ray 2009). In Drosophila these genes include the transcription factor genes acj6, pdm3, lozenge, and Scalloped (Clyne et al. 1999a; Ray et al. 2007, 2008; Tichy et al. 2008; Bai et al. 2009). As for the second perspective, previous work in Drosophila examined Or genes expressed in the maxillary palp and began with a computational analysis to identify candidate regulatory elements (Ray et al. 2007, 2008). This study has examined Or genes of the antenna and has taken a different point of departure: a genetic analysis.
The results of this genetic analysis highlight the dimensionality of the problem and reflect the prominence of negative regulation as a means of solving it. An odor receptor gene must be expressed in the correct olfactory organ, in the correct region of that organ, and in the correct cell type within that region. The simplest interpretation of our results is that flanking regulatory sequences restrict expression of an individual receptor gene to the antenna, to the proper region of the antenna, and to neurons.
Other systems, such as the visual and gustatory systems, also depend on the coordinated expression of distinct receptors in different sensory cells. Elegant work on regulation of opsin gene expression in the Drosophila eye has recently identified a number of genes that act positively or negatively in this process (Morante et al. 2007). Analysis of the Rhodopsin (Rh) gene regulatory regions has revealed a proximal region that acts as a core promoter and a distal region that confers cell-type specificity (Mismer and Rubin 1987; Fortini and Rubin 1990), a pattern of organization that is consistent with our results. Taste receptors of the Gustatory receptor (Gr) family (Clyne et al. 2000) show more extensive coexpression than the Or or Rh genes, but different subsets of receptors are expressed in distinct subsets of cells, and little is known about the mechanisms that govern their specificity of expression (Dunipace et al. 2001; Scott et al. 2001; Dahanukar et al. 2007). It seems clear that a combination of molecular, computational, and genetic approaches will be necessary to unravel the mechanisms of receptor gene regulation in these diverse systems and to thereby ascertain to what extent their regulation shares a common logic.
Acknowledgments
We are grateful to Takashi Shiraiwa for assistance in generating Or88a−1.6kb fly lines as well as for helpful discussions. We thank Chih-Ying Su, C. Andrea Yao, and Anand Ray for comments and advice, which improved this work, and Paul Graham for technical assistance. Akira Chiba's lab graciously donated flies carrying UAS-gapGFP. Wynand van der Goes van Naters back-crossed w1118 flies into the Canton-S background. This work was supported by a National Institutes of Health Training Grant in Genetics 5 T32 GM07499 (C.M.) and National Institutes of Health grants to J.C.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.117622/DC1.
References
- Bai, L., A. L. Goldman and J. R. Carlson, 2009. Positive and negative regulation of odor receptor gene choice in Drosophila by acj6. J. Neurosci. 29 12940–12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 401–415. [DOI] [PubMed] [Google Scholar]
- Canon, J., and U. Banerjee, 2003. In vivo analysis of a developmental circuit for direct transcriptional activation and repression in the same cell by a Runx protein. Genes Dev. 17 838–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, J. R., 1996. Olfaction in Drosophila: from odor to behavior. Trends Genet. 12 175–180. [DOI] [PubMed] [Google Scholar]
- Cayirlioglu, P., I. G. Kadow, X. Zhan, K. Okamura, G. S. Suh et al., 2008. Hybrid neurons in a microRNA mutant are putative evolutionary intermediates in insect CO2 sensory systems. Science 319 1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba, A., 1998. Personal communication to FlyBase. http://flybase.org/reports/FBti0010578.html.
- Clyne, P., A. Grant, R. O'Connell and J. R. Carlson, 1997. Odorant response of individual sensilla on the Drosophila antenna. Invert. Neurosci. 3 127–135. [DOI] [PubMed] [Google Scholar]
- Clyne, P. J., S. J. Certel, M. de Bruyne, L. Zaslavsky, W. A. Johnson et al., 1999. a The odor specificities of a subset of olfactory receptor neurons are governed by Acj6, a POU-domain transcription factor. Neuron 22 339–347. [DOI] [PubMed] [Google Scholar]
- Clyne, P. J., C. G. Warr, M. R. Freeman, D. Lessing, J. Kim et al., 1999. b A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 22 327–338. [DOI] [PubMed] [Google Scholar]
- Clyne, P. J., C. G. Warr and J. R. Carlson, 2000. Candidate taste receptors in Drosophila. Science 287 1830–1834. [DOI] [PubMed] [Google Scholar]
- Couto, A., M. Alenius and B. J. Dickson, 2005. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr. Biol. 15 1535–1547. [DOI] [PubMed] [Google Scholar]
- Dahanukar, A., Y. T. Lei, J. Y. Kwon and J. R. Carlson, 2007. Two Gr genes underlie sugar reception in Drosophila. Neuron 56 503–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, S. R., M. L. Vasconcelos, V. Ruta, S. Luo, A. Wong et al., 2008. The Drosophila pheromone cVA activates a sexually dimorphic neural circuit. Nature 452 473–477. [DOI] [PubMed] [Google Scholar]
- de Bruyne, M., K. Foster and J. R. Carlson, 2001. Odor coding in the Drosophila antenna. Neuron 30 537–552. [DOI] [PubMed] [Google Scholar]
- Dobritsa, A. A., W. van der Goes van Naters, C. G. Warr, R. A. Steinbrecht and J. R. Carlson, 2003. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron 37 827–841. [DOI] [PubMed] [Google Scholar]
- Dubchak, I., M. Brudno, G. G. Loots, L. Pachter, C. Mayor et al., 2000. Active conservation of noncoding sequences revealed by three-way species comparisons. Genome Res. 10 1304–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunipace, L., S. Meister, C. McNealy and H. Amrein, 2001. Spatially restricted expression of candidate taste receptors in the Drosophila gustatory system. Curr. Biol. 11 822–835. [DOI] [PubMed] [Google Scholar]
- Ejima, A., B. P. Smith, C. Lucas, W. van der Goes van Naters, C. J. Miller et al., 2007. Generalization of courtship learning in Drosophila is mediated by cis-vaccenyl acetate. Curr. Biol. 17 599–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, K., T. Aoki, Y. Yoda, K. Kimura and C. Hama, 2007. Notch signal organizes the Drosophila olfactory circuitry by diversifying the sensory neuronal lineages. Nat. Neurosci. 10 153–160. [DOI] [PubMed] [Google Scholar]
- Fischer, J. A., E. Giniger, T. Maniatis and M. Ptashne, 1988. GAL4 activates transcription in Drosophila. Nature 332 853–856. [DOI] [PubMed] [Google Scholar]
- Fishilevich, E., A. I. Domingos, K. Asahina, F. Naef, L. B. Vosshall et al., 2005. Chemotaxis behavior mediated by single larval olfactory neurons in Drosophila. Curr. Biol. 15 2086–2096. [DOI] [PubMed] [Google Scholar]
- Fishilevich, E., and L. B. Vosshall, 2005. Genetic and functional subdivision of the Drosophila antennal lobe. Curr. Biol. 15 1548–1553. [DOI] [PubMed] [Google Scholar]
- Fortini, M. E., and G. M. Rubin, 1990. Analysis of cis-acting requirements of the Rh3 and Rh4 genes reveals a bipartite organization to rhodopsin promoters in Drosophila melanogaster. Genes Dev. 4 444–463. [DOI] [PubMed] [Google Scholar]
- Frazer, K. A., L. Pachter, A. Poliakov, E. M. Rubin and I. Dubchak, 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32 W273–W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss, S. H., and A. Ray, 2009. Mechanisms of odorant receptor gene choice in Drosophila and vertebrates. Mol. Cell Neurosci. 41 101–112. [DOI] [PubMed] [Google Scholar]
- Gao, Q., and A. Chess, 1999. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics 60 31–39. [DOI] [PubMed] [Google Scholar]
- Gaszner, M., and G. Felsenfeld, 2006. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 7 703–713. [DOI] [PubMed] [Google Scholar]
- Goldman, A. L., W. Van der Goes van Naters, D. Lessing, C. G. Warr and J. R. Carlson, 2005. Coexpression of two functional odor receptors in one neuron. Neuron 45 661–666. [DOI] [PubMed] [Google Scholar]
- Gupta, B. P., G. V. Flores, U. Banerjee and V. Rodrigues, 1998. Patterning an epidermal field: Drosophila lozenge, a member of the AML-1/Runt family of transcription factors, specifies olfactory sense organ type in a dose-dependent manner. Dev. Biol. 203 400–411. [DOI] [PubMed] [Google Scholar]
- Ha, T. S., and D. P. Smith, 2006. A pheromone receptor mediates 11-cis-vaccenyl acetate-induced responses in Drosophila. J. Neurosci. 26 8727–8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem, E. A., and J. R. Carlson, 2006. Coding of odors by a receptor repertoire. Cell 125 143–160. [DOI] [PubMed] [Google Scholar]
- Hallem, E. A., M. G. Ho and J. R. Carlson, 2004. The molecular basis of odor coding in the Drosophila antenna. Cell 117 965–979. [DOI] [PubMed] [Google Scholar]
- Jiang, J., C. A. Rushlow, Q. Zhou, S. Small and M. Levine, 1992. Individual dorsal morphogen binding sites mediate activation and repression in the Drosophila embryo. EMBO J. 11 3147–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreher, S. A., D. Mathew, J. Kim and J. R. Carlson, 2008. Translation of sensory input into behavioral output via an olfactory system. Neuron 59 110–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtovic, A., A. Widmer and B. J. Dickson, 2007. A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature 446 542–546. [DOI] [PubMed] [Google Scholar]
- Laissue, P. P., C. Reiter, P. R. Hiesinger, S. Halter, K. F. Fischbach et al., 1999. Three-dimensional reconstruction of the antennal lobe in Drosophila melanogaster. J. Comp. Neurol. 405 543–552. [PubMed] [Google Scholar]
- Manoli, D. S., M. Foss, A. Villella, B. J. Taylor, J. C. Hall et al., 2005. Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature 436 395–400. [DOI] [PubMed] [Google Scholar]
- Mismer, D., and G. M. Rubin, 1987. Analysis of the promoter of the ninaE opsin gene in Drosophila melanogaster. Genetics 116 565–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts, P., 2004. Odorant receptor gene choice in olfactory sensory neurons: the one receptor-one neuron hypothesis revisited. Curr. Opin. Neurobiol. 14 31–36. [DOI] [PubMed] [Google Scholar]
- Morante, J., C. Desplan and A. Celik, 2007. Generating patterned arrays of photoreceptors. Curr. Opin. Genet. Dev. 17 314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder, T., K. S. Yoon, B. H. White and H. Keshishian, 2001. A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl. Acad. Sci. USA 98 12596–12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, D., and A. J. Courey, 1992. The same dorsal binding site mediates both activation and repression in a context-dependent manner. EMBO J. 11 1837–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Python, F., and R. F. Stocker, 2002. Adult-like complexity of the larval antennal lobe of D. melanogaster despite markedly low numbers of odorant receptor neurons. J. Comp. Neurol. 445 374–387. [DOI] [PubMed] [Google Scholar]
- Rasband, W. S., 1997. ImageJ. U. S. National Institutes of Health, Bethesda, MD.
- Ray, A., W. G. van Naters, T. Shiraiwa and J. R. Carlson, 2007. Mechanisms of odor receptor gene choice in Drosophila. Neuron 53 353–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, A., W. van der Goes van Naters and J. R. Carlson, 2008. A regulatory code for neuron-specific odor receptor expression. PLoS Biol. 6 e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzenthaler, S., E. Suzuki and A. Chiba, 2000. Postsynaptic filopodia in muscle cells interact with innervating motoneuron axons. Nat. Neurosci. 3 1012–1017. [DOI] [PubMed] [Google Scholar]
- Robertson, H. M., C. G. Warr and J. R. Carlson, 2003. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 100(Suppl. 2): 14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, K., R. Brady, Jr., A. Cravchik, P. Morozov, A. Rzhetsky et al., 2001. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell 104 661–673. [DOI] [PubMed] [Google Scholar]
- Shanbhag, S. R., B. Müller and R. A. Steinbrecht, 1999. Atlas of olfactory organs of Drosophila melanogaster 1. Types, external organization, innervation and distribution of olfactory sensilla. Int. J. Insect Morphol. Embryol. 28 377–397. [Google Scholar]
- Shanbhag, S. R., B. Muller and R. A. Steinbrecht, 2000. Atlas of olfactory organs of Drosophila melanogaster 2. Internal organization and cellular architecture of olfactory sensilla. Arthropod Struct. Dev. 29 211–229. [DOI] [PubMed] [Google Scholar]
- Shanbhag, S. R., D. Hekmat-Scafe, M. S. Kim, S. K. Park, J. R. Carlson et al., 2001. Expression mosaic of odorant-binding proteins in Drosophila olfactory organs. Microsc. Res. Tech. 55 297–306. [DOI] [PubMed] [Google Scholar]
- Shiraiwa, T., 2008. Multimodal chemosensory integration through the maxillary palp in Drosophila. PLoS One 3 e2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichy, A. L., A. Ray and J. R. Carlson, 2008. A new Drosophila POU gene, pdm3, acts in odor receptor expression and axon targeting of olfactory neurons. J. Neurosci. 28 7121–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Goes van Naters, W., and J. R. Carlson, 2007. Receptors and neurons for fly odors in Drosophila. Curr. Biol. 17 606–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voas, M. G., and I. Rebay, 2004. Signal integration during development: insights from the Drosophila eye. Dev. Dyn. 229 162–175. [DOI] [PubMed] [Google Scholar]
- Vosshall, L. B., 2001. The molecular logic of olfaction in Drosophila. Chem. Senses 26 207–213. [DOI] [PubMed] [Google Scholar]
- Vosshall, L. B., H. Amrein, P. S. Morozov, A. Rzhetsky and R. Axel, 1999. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 96 725–736. [DOI] [PubMed] [Google Scholar]
- Vosshall, L. B., A. M. Wong and R. Axel, 2000. An olfactory sensory map in the fly brain. Cell 102 147–159. [DOI] [PubMed] [Google Scholar]
- Zhou, S., E. A. Stone, T. F. Mackay and R. R. Anholt, 2009. Plasticity of the chemoreceptor repertoire in Drosophila melanogaster. PLoS Genet. 5 e1000681. [DOI] [PMC free article] [PubMed] [Google Scholar]