Abstract
Connective tissue growth factor (CTGF), a member of the cysteine-rich 61 (Cyr 61), CTGF, nephroblastoma overexpressed (NOV) (CCN) family of proteins, is synthesized by osteoblasts, and its overexpression inhibits osteoblastogenesis and causes osteopenia. The global inactivation of Ctgf leads to defective endochondral bone formation and perinatal lethality; therefore, the consequences of Ctgf inactivation on the postnatal skeleton are not known. To study the function of CTGF, we generated Ctgf+/LacZ heterozygous null mice and tissue-specific null Ctgf mice by mating Ctgf conditional mice, where Ctgf is flanked by lox sequences with mice expressing the Cre recombinase under the control of the paired-related homeobox gene 1 (Prx1) enhancer (Prx1-Cre) or the osteocalcin promoter (Oc-Cre). Ctgf+/LacZ heterozygous mice exhibited transient osteopenia at 1 month of age secondary to decreased trabecular number. A similar osteopenic phenotype was observed in 1-month-old Ctgf conditional null male mice generated with Prx1-Cre, suggesting that the decreased trabecular number was secondary to impaired endochondral bone formation. In contrast, when the conditional deletion of Ctgf was achieved by Oc-Cre, an osteopenic phenotype was observed only in 6-month-old male mice. Osteoblast and osteoclast number, bone formation, and eroded surface were not affected in Ctgf heterozygous or conditional null mice. In conclusion, CTGF is necessary for normal skeletal development but to a lesser extent for postnatal skeletal homeostasis.
Inactivation of Ctgf causes a developmental and adult skeletal phenotype in male mice characterized by osteopenia and indicative that CTGF is necessary to maintain skeletal homeostasis.
Cysteine-rich 61 (Cyr 61), connective tissue growth factor (CTGF), nephroblastoma overexpressed (NOV) (CCN) and Wnt-inducible secreted proteins (WISP) 1, 2, and 3 are a family of cysteine-rich secreted proteins (1,2). CCN proteins are structurally related and share four distinct modules: 1) an IGF-binding domain, 2) a von Willebrand type C domain, 3) a thrombospondin-1 domain, and 4) a C-terminal domain, the latter absent in WISP-2 and important for protein interactions (1,2). CCN proteins bear a structural relationship with certain bone morphogenetic protein (BMP) antagonists, such as twisted gastrulation and chordin, and can have important interactions with regulators of osteoblast cell growth and differentiation (3).
CTGF is expressed in bone and cartilage; and in osteoblasts, CTGF expression is induced by BMP, TGF-β, and Wnt (4,5). In addition, CTGF has important interactions with these signaling molecules. CTGF binds to BMP and Wnt coreceptors and can decrease BMP and Wnt signaling (6,7). CTGF enhances TGF-β activity and mediates effects of TGF-β on mesenchymal cell condensation (6,8). In addition to its interactions with members of the TGF-β superfamily and Wnt, CTGF inhibits Notch signaling in osteoblastic cells, and Notch receptors play a critical role in cell fate decisions (9,10,11).
CTGF regulates different cellular events, including adhesion, proliferation, migration, and differentiation. Targeted disruption of Ctgf in mice leads to perinatal lethality and severe skeletal developmental abnormalities as a result of impaired cartilage/bone development and defective growth plate angiogenesis (12). The function of CTGF in cells of the osteoblastic lineage is not well understood, and the study of the effects of CTGF in these cells has yielded controversial results (6,11,13,14). Down-regulation of CTGF using RNA interference revealed that CTGF may be required for osteoblastogenesis, but overexpression of CTGF or addition of CTGF protein to cells of the osteoblastic lineage were reported to both favor and oppose osteoblastogenesis (4,6,11,13,14). These observations suggest that different in vitro experimental conditions can lead to different interactions between CTGF and osteogenic signals and, as a consequence, to different biological events. Recently, we examined the effect of CTGF overexpression on skeletal cells in vivo. Transgenic mice overexpressing CTGF under the control of the osteocalcin promoter exhibited decreased bone formation causing osteopenia (15). Osteoblastic cells from CTGF transgenics exhibited decreased osteoblastogenesis and impaired BMP/Smad, Wnt, and IGF-I signaling. These observations demonstrate that CTGF in excess has the potential to act as a BMP, Wnt, and IGF-I antagonist. However, the consequences of Ctgf inactivation on adult skeletal homeostasis have not been defined, because the skeletal developmental phenotype of Ctgf-null mice leads to perinatal death (12).
The intent of the present study was to define the function of CTGF in skeletal tissue in vivo. For this purpose, we created Ctgf global and conditional null mice. In the conditional null model, Ctgf was inactivated by Cre recombination directed by either the paired-related homeobox 1 (Prx1) enhancer expressed in limb buds at d 10.5 of embryonic life (E10.5) or the osteocalcin promoter expressed at E18.5 in osteoblasts. This approach would allow the inactivation of Ctgf in the pre- and perinatal skeleton. The skeletal phenotype of Ctgf global and conditional null mice was determined by histomorphometric and structural analyses.
Materials and Methods
Generation of Ctgf-null mice
To generate a conditional null allele of Ctgf, we applied a conditional-by-inversion (COIN) approach using Velocigene (16). Briefly, a bacterial artificial chromosome (BAC) containing mouse genomic DNA encompassing Ctgf was selected from a BAC library of 129/SvJ mouse genomic DNA (id 460d11) containing approximately 170 kb of mouse genomic DNA. The COIN intron was introduced into exon 2 of Ctgf to generate the BAC-based targeting vector for the Ctgfe2COIN allele (Fig. 1). In this process, exon 2 (223 bp) of Ctgf is split into two exons so that the 5′ end of exon 2 to the COIN intron is 120 bp, and the 3′ end of exon 2 to the COIN intron is 103 bp (Fig. 1). This modification does not disrupt expression of Ctgf as evidenced by the fact that Ctgfe2COIN/e2COIN mice express Ctgf mRNA. The COIN intron is a modified intron derived from intron 2 of the rabbit β-globin gene. The COIN element contains a lox66_SA-egfp)-polyA_lox71 sequence placed in the antisense strand, where SA is the 5′ spliced region from rabbit β-globin intron 2 and polyA is from the 3′ untranslated region of the rabbit β-globin gene. The SA-EGFP-polyA cassette was optimized to block transcription when brought into the sense strand after Cre recombination. The COIN element also contained the selection cassette hygromycin phosphotransferase-Δ thymidine kinase mini gene (HygΔTK), which was used for the initial selection of embryonic stem (ES) cells and was flanked by flippase (FLP) recognition targets for its removal (17,18). Conversion of a COIN allele from silent to a null allele is brought about by the Cre recombinase that recognizes left/right mutant lox sites lox71 and lox66 (19,20). Because lox66 is in the reverse complement orientation with respect to lox71, upon exposure to the enzyme, the lox66 site recombines with the lox71 site, inverting the COIN sequence flanked by these sites (21). Within the COIN element, the lox66 and lox71 sites were engineered in a configuration enabling the permanent inversion of the loxP flanked sequences by Cre recombinase (22,23). After inversion, the transcriptional machinery does not access exons 2–5 of Ctgf, resulting in a message comprised of exons 1, a fraction of exon 2, and eGFP, containing minimal Ctgf coding sequences. This should result in the inactivation of Ctgf because CTGF is not active in conventional Ctgf-null mice containing an intact exon 2 (24).
Figure 1.
Engineering of the Ctgfe2COIN allele and strategy for the conditional inactivation of Ctgf. The upper panel reveals the exon and intron structure of Ctgf (adapted from Ensembl.org). Dark gray boxes indicate coding sequences, whereas white boxes indicate untranslated regions (UTR). The introns are shown as dotted lines. In the Ctgfe2COIN allele, exon 2 (223 bp) is split by inserting a COIN intron into two new exons of 120 and 103 bp upstream and downstream of the COIN intron, respectively. The COIN intron contains a COIN element that is comprised of lox66_SA-Egpf-polyA_lox71, placed in the antisense orientation with respect to the transcription of the Ctgf. A single FLP recognition target (FRT) site indicates the placement of the HygΔTK drug selection cassette, which was removed by the action of FLP. A normal CTGF mRNA is expressed by the Ctgfe2COIN allele. The middle panel shows that exposure to Cre recombinase results in the virtually irreversible inversion of the COIN element and conversion of the lox66-lox71 pair to lox71-loxP. A new message is expressed, comprised of exons 1, the new exon 2, and COIN element exon, which encodes for a transmembrane domain-eGFP fusion protein (TMeGFP). In the lower panel, a representative PCR analysis, using primers 1, 2, and 3 (P1, P2, and P3) depicted in the upper and middle panels and described in Supplemental Table 1, is shown. Calvarial DNA from Ctgf conditional null and control mice before and after recombination by Cre expressed under the control of the Prx1 enhancer (left panel) or of the osteocalcin promoter (right panel) is shown. A 710-bp band is detected in the CtgfINV allele, and a 650-bp band is detected in the noninverted allele.
Using restriction mapping, it was determined that the modified BAC had homology arms of approximately 120 and 40 kb flanking the COIN intron, and it was used as a vector to target Ctgf in a C57BL/6-129SvJ hybrid ES line, F1H4 286A-B8, that already harbors a null allele of Ctgf (24). ES cell clones were genotyped using a loss-of-allele assay, and 12 of 192 clones screened were targeted, indicating a targeting frequency of 6.25%. Targeted ES cell lines were used to generate chimeric male mice at the transgenic facility of the University of Connecticut Health Center (Farmington, CT). Chimeras that were complete transmitters of ES-derived sperm were bred to 129/SvJ mice expressing the FLP recombinase under the control of the Gt(ROSA)26Sor promoter (The Jackson Laboratory, Bar Harbor, ME) for the removal of the HygΔTK selection cassette (17,18). The excision of the selection cassette was confirmed by PCR, and the resulting FLP recombinase transgene was segregated by mating the mice with C57BL/6 wild-type mice. Heterozygous mice were intermated to create homozygous Ctgfe2COIN/e2COIN mice in a 129SvJ/C57BL/6 genetic background.
To study the consequences of the Ctgf inactivation during early limb development, Ctgfe2COIN/e2COIN mice were bred to homozygous Prx1-Cre mice in a C57BL/6 genetic background (The Jackson Laboratory) to create heterozygous Prx1-Cre/+;Ctgfe2COIN/+ mice (25). These were mated with Ctgfe2COIN/e2COIN mice to create Prx1-Cre/+;Ctgfe2COIN/e2COIN to be mated with Ctgfe2COIN/e2COIN to generate an experimental cohort, in which the COIN element is inverted by Cre (CtgfINV/INV) and a control group without Cre-mediated inversion (Ctgfe2COIN/e2COIN) and studied at 1 month of age. Ctgfe2COIN/e2COIN mice also were compared with wild-type littermate controls to ensure that before recombination they did not exhibit a skeletal phenotype. To study the inactivation of Ctgf in mature osteoblasts, transgenic mice expressing the Cre recombinase under the control of a 3.9-kb human osteocalcin promoter (Oc-Cre), created in a Friend virus B type (FVB) genetic background, were obtained from T. Clemens (Baltimore, MD) (26). Ctgfe2COIN/e2COIN mice were studied in a Ctgf heterozygous null background. For this purpose, Oc-Cre mice were mated to Ctgf heterozygous (Ctgf+/LacZ) null mice, backcrossed eight times into a C57BL/6 background after the excision of a neomycin selection cassette by breeding with mice expressing the Cre recombinase under the control of the cytomegalovirus (CMV) promoter (24). Oc-Cre and Ctgf heterozygous mice were intermated for the creation of Oc-Cre/Oc-Cre homozygous mice in a heterozygous Ctgf+/LacZ null background. These were mated with homozygous Ctgfe2COIN/e2COIN mice, generating an experimental cohort, where Cre inverts the COIN element from the Ctgfe2COIN allele and where a Ctgf-null allele is retained (CtgfINV/LacZ), and a control littermate cohort is carrying a Cre-inverted Ctgfe2COIN allele and a wild-type allele (CtgfINV/+). To ensure that the latter were appropriate controls, the skeletal phenotype of Ctgf+/LacZ mice was compared with that of wild-type littermate C57BL/6 mice. Conditional null mice were compared with littermate controls of identical genetic composition at 1, 4, and 6 months of age.
Genotyping of Oc-Cre, Prx1-Cre, Ctgfe2COIN, and CtgfLacZ alleles was carried out by PCR in tail DNA extracts (Supplemental Table 1 published on The Endocrine Society’s Journals Online web site at http://endo.endojournals.org). Deletion of the neo cassette in CtgfLacZ mice by Cre recombination and deletion of the HygΔTK cassette in Ctgfe2COIN/e2COIN mice by FLP recombination was determined by PCR in tail DNA and inversion of lox71-lox66 flanked sequences in Ctgfe2COIN/e2COIN mice by Cre recombination was documented by PCR in DNA extracted from calvariae (Fig. 1). The Ctgf-null state was confirmed by documenting suppressed Ctgf mRNA in calvarial extracts by real-time RT-PCR (27,28). All animal experiments were approved by the Animal Care and Use Committee of Saint Francis Hospital and Medical Center.
X-ray analysis, bone mineral density (BMD), and femoral length
X-rays were performed on eviscerated mice at an intensity of 30 kW for 20 sec on a Faxitron x-ray system (model MX 20; Faxitron X-Ray Corp., Wheeling, IL). Total BMD (grams per square centimeter) was measured on anesthetized mice using the PIXImus small-animal dual-energy x-ray absorptiometry system (GE Medical System/LUNAR, Madison, WI) (29). Femoral images were used to determine femoral length in millimeters. Calibrations were performed with a phantom of defined value, and quality assurance measurements were performed before each use. The coefficient of variation for total BMD is less than 1% (n = 9).
Bone histomorphometric analysis
Static and dynamic histomorphometry was carried out on experimental and control mice after they were injected with calcein (20 mg/kg) and demeclocycline (50 mg/kg) at an interval of 2 d for 1-month-old animals and 7 d for 4- and 6-month-old animals. Mice were killed by CO2 inhalation 2 d after the demeclocycline injection. Longitudinal sections of femurs, 5 μm thick, were cut on a microtome (Microm; Richards-Allan Scientific, Kalamazoo, MI) and stained with 0.1% toluidine blue or von Kossa. Static parameters of bone formation and resorption were measured in a defined area between 360 and 2160 μm from the growth plate, using an OsteoMeasure morphometry system (Osteometrics, Atlanta, GA) (30). For dynamic histomorphometry, mineralizing surface per bone surface and mineral apposition rate were measured on unstained sections under UV light, using a triple diamidino-2-phenylindole fluorescein set long-pass filter, and bone formation rate was calculated. The terminology and units used are those recommended by the Histomorphometry Nomenclature Committee of the American Society for Bone and Mineral Research (31).
Microcomputed tomography (μCT)
Bone microarchitecture of femurs from experimental and control mice was analyzed by μCT (MicroCT40; Scanco Medical AG, Bassersdorf, Switzerland) (32). The metaphyseal region of the distal femur was scanned for microarchitecture, and cortical thickness was obtained at the midshaft. The femurs were scanned at a resolution of 12 μm, energy level of 45 keV, and intensity of 177 μA. The distal trabecular scan started about 0.6 mm proximal to the growth plate and extended proximally 1.5 mm. One hundred fifty cross-sectional slices were obtained at 12-μm intervals at the distal end beginning at the edge of the growth plate and extending in a proximal direction, and 100 contiguous slices were selected for analysis. Trabecular regions were assessed for bone volume fraction (bone volume/total volume), trabecular thickness, trabecular number, trabecular separation, connectivity density, and structure model index. The midshaft cortical thickness values were obtained by averaging 18 slices at the midpoint of the femur.
Serum C-terminal cross-linked telopeptide of type I collagen (CTX)
The serum bone remodeling marker CTX was measured by ELISA using RatLaps ELISA kits (Nordic Bioscience Diagnostics, Herlev, Denmark), according to manufacturer’s instructions.
Primary osteoblast cell cultures and adenoviral infection
Osteoblastic cells were isolated from parietal bones of 3- to 5-d-old Ctgfe2COIN/e2COIN mice. Cells were obtained by five sequential digestions of the parietal bones using bacterial collagenase (CLS II; Worthington Biochemical, Freehold, NJ) (33). Cell populations harvested from the third to the fifth digestions were cultured as a pool and were previously shown to have osteoblast characteristics. Osteoblastic cells were cultured in DMEM (Life Technologies, Inc., Grand Island, NY) supplemented with nonessential amino acids, 20 mm HEPES, 100 μg/ml ascorbic acid, and 10% fetal bovine serum (FBS) (Atlanta Biologicals, Lawrenceville, GA) at 37 C in a humidified 5% CO2 incubator. Subconfluent Ctgfe2COIN/e2COIN cells were trypsinized and plated at a density of 25,000 cells/cm2 and cultured to subconfluence (∼35,000 cells/cm2). Cells were transferred to DMEM containing 2% FBS and transduced with 100 multiplicity of infection of replication-defective recombinant adenovirus. An adenoviral vector expressing Cre recombinase under the control of the CMV promoter (Ad-CMV-Cre; Vector Biolabs, Philadelphia, PA) was used to induce recombination of lox sequences in vitro, and an adenoviral vector expressing green fluorescent protein (GFP) under the control of the CMV promoter (Ad-CMV-GFP) was used as a control (34). After 24 h, cells were washed with versene (Invitrogen, Carlsbad, CA), trypsinized, plated, and cultured in DMEM containing 10% FBS. Ctgf and alkaline phosphatase mRNA were measured by real-time RT-PCR. Alkaline phosphatase activity was determined in 0.5% Triton X-100 cell extracts by the hydrolysis of p-nitrophenol phosphate to p-nitrophenol and measured by spectroscopy at 405 nm after 10 min of incubation at room temperature according to manufacturer’s instructions (Sigma-Aldrich, St. Louis, MO). Data are expressed as nanomoles of p-nitrophenol released per minute per microgram of protein. Total protein content was determined in cell extracts by the DC protein assay, in accordance with manufacturer’s instructions (Bio-Rad, Hercules, CA).
Real-time RT-PCR
Total RNA was extracted from calvariae or from osteoblast cultures and mRNA levels determined by real-time RT-PCR (27,28). For this purpose, RNA was reverse transcribed using SuperScript III Platinum Two-Step qRT-PCR kit (Invitrogen), according to manufacturer’s instructions. Product amplification was conducted in the presence of 5′-CACTCCGGGAAATGCTGCAAGGAG[FAM]G-3′ and 5′-GTTGGGTCTGGGCCAAATGT-3′ primers for CTGF (GenBank accession number NM_010217), which binds at base 772 and 840 of the CTGF reverse-transcribed DNA; 5′-CGGTTAGGGCGTCTCCACAGTAAC[FAM]G-3′ and 5′-CTTGGAGAGGGCCACAAAGG-3′ primers for alkaline phosphatase (GenBank accession no. NM_007431), which binds at base 439 and 514 of the alkaline phosphatase reverse-transcribed DNA; and 5′-CGAACCGGATAATGTGAAGTTCAAGGTT[FAM]G-3′ and 5′-CTGCTTCAGCTTCTCTGCCTTT-3′ primers for ribosomal protein L38 (RPL38) (GenBank accession no. NM_001048057), which binds at base 223 and 268 of the RPL38 reverse-transcribed DNA. Primers were mixed with Platinum Quantitative PCR SuperMix-UDG (Invitrogen) and amplification conducted at 60 C for 45 cycles (35). Transcript copy number was estimated by comparison with a standard curve constructed using CTGF (R. P. Rysek, Princeton, NJ), alkaline phosphatase, or RPL38 (both from American Type Culture Collection, Manassas, VA) cDNA (36). Reactions were conducted in a 96-well spectrofluorometric thermal iCycler (Bio-Rad), and fluorescence was monitored during every PCR cycle at the annealing step. Data are expressed as copy number corrected for Rpl38.
Statistical analysis
Data are expressed as means ± sem. Statistical differences were determined by unpaired Student’s t test or ANOVA.
Results
Ctgf heterozygous null mice
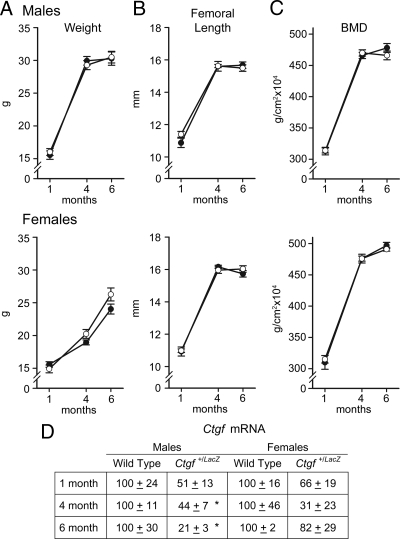
To study Ctgf heterozygous null mice, Ctgf+/LacZ mice were mated with wild-type mice to obtain Ctgf+/LacZ mice and wild-type littermate controls. Ctgf mRNA levels were 20–80% lower in calvariae from 1-, 4-, and 6-month-old Ctgf+/LacZ than in calvarial extracts from wild-type controls (Fig. 2). Ctgf+/LacZ heterozygous null mice appeared normal and not different from their wild-type littermates. Contact radiography at 1, 4, and 6 months of age revealed no apparent skeletal abnormalities in Ctgf+/LacZ mice (not shown). Ctgf+/LacZ heterozygous null mice had normal body weight, femoral length, and BMD at 1, 4, and 6 months of age (Fig. 2). Static and dynamic histomorphometric analysis revealed a 30–45% decrease in bone volume/tissue volume at 1 month of age in male and female Ctgf+/LacZ mice (Table 1 and Fig. 3). In accordance with the endochondral skeletal developmental phenotype of homozygous Ctgf-null mice, the osteopenia appeared to be secondary to a decreased number of trabeculae by 27% in male and 36% in female mice (P < 0.05). Trabecular thickness was not decreased significantly (6–14%), and the osteopenia was transient and not observed in 4- or 6-month-old Ctgf+/LacZ heterozygous mice. Osteoblast and osteoclast number per perimeter and parameters of bone formation or bone resorption were not different between Ctgf+/LacZ and wild-type controls at 1–6 months of age.
Figure 2.
Weight, femoral length, BMD, and Ctgf mRNA expression in male (upper panels) and female (lower panels) Ctgf+/LacZ heterozygous null mice (•) and wild-type littermate controls (○). The weight in grams (A), femoral length in millimeters (B), total BMD in grams per square centimeter (C), and Ctgf mRNA levels in total calvarial extracts (D), expressed as Ctgf copy number corrected for Rpl38 and normalized to 100 at 1, 4, and 6 months of age, are shown. Values are means ± sem (n = 5–17) except for mRNA levels, which are expressed as percentage of control for each independent age (n = 3–4). * Significantly different from control mice, P < 0.05.
Table 1.
Femoral histomorphometry of 1-, 4-, and 6-month-old male and female Ctgf+/LacZ heterozygous mice and wild-type controls
| 1 month
|
4 months
|
6 months
|
||||
|---|---|---|---|---|---|---|
| Wild type | Ctgf+/LacZ | Wild type | Ctgf+/LacZ | Wild type | Ctgf+/LacZ | |
| Males | ||||||
| Bone volume/tissue volume (%) | 7.7 ± 0.7 | 5.2 ± 0.6a | 9.2 ± 0.7 | 7.7 ± 0.8 | 7.1 ± 1.1 | 6.7 ± 1.1 |
| Trabecular separation (μm) | 292 ± 25 | 482 ± 70a | 252 ± 19 | 266 ± 13 | 409 ± 51 | 372 ± 33 |
| Trabecular number (mm−1) | 3.4 ± 0.2 | 2.5 ± 0.2a | 3.7 ± 0.2 | 3.5 ± 0.2 | 2.5 ± 0.3 | 2.6 ± 0.2 |
| Trabecular thickness (μm) | 22.2 ± 1.0 | 20.8 ± 0.6 | 24.7 ± 0.6 | 23.2 ± 0.6 | 27.4 ± 1.7 | 25.4 ± 2.6 |
| Osteoblast surface/bone surface (%) | 24.5 ± 1.1 | 25.9 ± 1.5 | 16.4 ± 1.5 | 18.4 ± 1.4 | 15.5 ± 1.2 | 14.2 ± 0.6 |
| Number of osteoblasts/bone perimeter (mm−1) | 27 ± 1 | 30 ± 2 | 13.3 ± 1.4 | 15.0 ± 1.1 | 11.9 ± 1 | 11.1 ± 1 |
| Number of osteoblasts/tissue area (mm−2) | 142 ± 9 | 112 ± 11a | 77 ± 7 | 82 ± 5 | 47 ± 8 | 45 ± 4 |
| Osteoclast surface/bone surface (%) | 16.1 ± 0.6 | 17.0 ± 0.6 | 4.4 ± 0.4 | 4.8 ± 0.5 | 5.2 ± 0.4 | 5.6 ± 0.5 |
| Number of osteoclasts/bone perimeter (mm−1) | 7.3 ± 0.3 | 8.2 ± 0.2 | 2.1 ± 0.2 | 2.3 ± 0.2 | 2.2 ± 0.1 | 2.5 ± 0.2 |
| Number of osteoclasts/tissue area (mm−2) | 43 ± 3 | 32 ± 3a | 12 ± 1 | 13 ± 2 | 9 ± 1 | 10 ± 1 |
| Eroded surface/bone surface (%) | 28 ± 1 | 29 ± 1 | 10 ± 1 | 11 ± 1 | 10 ± 1 | 11 ± 1 |
| Mineral apposition rate (μm/d) | 2.20 ± 0.11 | 2.38 ± 0.09 | 0.77 ± 0.04 | 0.85 ± 0.04 | 0.64 ± 0.03 | 0.70 ± 0.04 |
| Mineralizing surface/bone surface (%) | 1.86 ± 0.35 | 2.56 ± 0.28 | 3.69 ± 0.55 | 3.73 ± 0.49 | 3.43 ± 0.63 | 3.15 ± 0.91 |
| Bone formation rate (μm3/μm2/d) | 0.043 ± 0.009 | 0.061 ± 0.007 | 0.029 ± 0.005 | 0.032 ± 0.005 | 0.023 ± 0.004 | 0.023 ± 0.007 |
| Females | ||||||
| Bone volume/tissue volume (%) | 7.1 ± 0.6 | 4.0 ± 0.8a | 3.1 ± 3.3 | 3.3 ± 0.3 | 3.0 ± 0.4 | 3.2 ± 0.4 |
| Trabecular separation (μm) | 273 ± 31 | 468 ± 69a | 695 ± 62 | 631 ± 54 | 752 ± 97 | 704 ± 81 |
| Trabecular number (mm−1) | 3.6 ± 0.3 | 2.3 ± 0.4a | 1.5 ± 0.2 | 1.6 ± 0.1 | 1.4 ± 0.2 | 1.4 ± 0.1 |
| Trabecular thickness (μm) | 19.9 ± 0.8 | 17.2 ± 1.2 | 19.8 ± 0.4 | 20.1 ± 0.9 | 21.5 ± 0.7 | 21.7 ± 1.1 |
| Osteoblast surface/bone surface (%) | 27.7 ± 1.7 | 27.3 ± 2.1 | 23.3 ± 2.1 | 17.8 ± 2.2 | 26.6 ± 3.2 | 21.0 ± 1.6 |
| Number of osteoblasts/bone perimeter (mm−1) | 31 ± 1 | 32 ± 3 | 20 ± 2 | 14 ± 2 | 19.7 ± 2.5 | 16.0 ± 1.1 |
| Number of osteoblasts/tissue area (mm−2) | 171 ± 14 | 110 ± 10a | 47 ± 5 | 34 ± 3a | 41 ± 5 | 36 ± 5 |
| Osteoclast surface/bone surface (%) | 16.0 ± 0.9 | 15.6 ± 0.7 | 8.4 ± 0.6 | 9.7 ± 0.5 | 6.8 ± 0.7 | 7.4 ± 1.4 |
| Number of osteoclasts/bone perimeter (mm−1) | 8.0 ± 0.4 | 7.9 ± 0.4 | 4.3 ± 0.3 | 4.9 ± 0.3 | 2.8 ± 0.3 | 3.2 ± 0.7 |
| Number of osteoclasts/tissue area (mm−2) | 44 ± 3 | 28 ± 5a | 10 ± 1 | 12 ± 1 | 6 ± 1 | 7 ± 1 |
| Eroded surface/bone surface (%) | 29 ± 2 | 29 ± 1 | 18 ± 1 | 22 ± 0 | 13 ± 1 | 15 ± 3 |
| Mineral apposition rate (μm/d) | 2.12 ± 0.14 | 2.13 ± 0.36 | 1.01 ± 0.11 | 1.14 ± 0.07 | 1.11 ± 0.04 | 1.14 ± 0.07 |
| Mineralizing surface/bone surface (%) | 3.9 ± 0.6 | 3.8 ± 0.5 | 1.84 ± 0.40 | 1.57 ± 0.29 | 6.68 ± 1.61 | 10.55 ± 1.03 |
| Bone formation rate (μm3/μm2/d) | 0.084 ± 0.014 | 0.080 ± 0.009 | 0.019 ± 0.004 | 0.018 ± 0.003 | 0.076 ± 0.020 | 0.118 ± 0.009 |
Bone histomorphometry was performed on femurs from 1-, 4-, and 6-month-old male and female Ctgf+/LacZ heterozygous mice and wild-type littermate controls. Values are means ± sem (n = 5–13).
Significantly different from controls, P < 0.05 by unpaired t test.
Figure 3.
Representative histological sections and calcein/demeclocycline labeling of bone femoral sections from 1-month-old Ctgf+/LacZ heterozygous mice (A), 1-month-old Prx1-Cre/+;CtgfINV/INV conditional null mice and littermate Ctgfe2COIN/e2COIN controls (B), and 6-month-old Oc-Cre/+;CtgfINV/LacZ conditional null mice and Oc-Cre/+;CtgfINV/+ littermate controls (C). Sections from male mice were stained with von Kossa without counterstain (final magnification, ×40) or unstained and examined under fluorescence microscopy (final magnification, ×400). WT, Wild type.
Conditional Ctgf-null mice
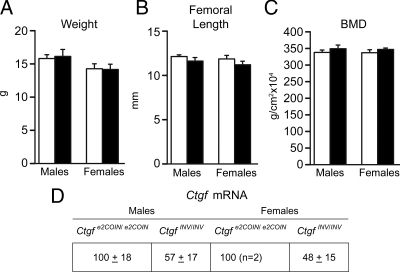
To induce the conditional inactivation of Ctgf in the limb bud at E10.5, Prx1-Cre/+;Ctgfe2COIN/e2COIN mice were mated with Ctgfe2COIN/e2COIN mice to create limb bud-specific Prx1-Cre/+;CtgfINV/INV conditional null and Ctgfe2COIN/e2COIN to serve as littermate controls (25). In preliminary experiments, we documented that Ctgfe2COIN/e2COIN mice were not different from wild-type controls by bone histomorphometric analysis (not shown). Ctgf mRNA levels in calvarial extracts from conditional Prx1-Cre/+;CtgfINV/INV-null mice were about 50% lower than in control mice (Fig. 4). The conditional inactivation of Ctgf in the developing limb bud caused a similar phenotype as that described for heterozygous Ctgf+/LacZ mice at 1 month of age. Conditional Prx1-Cre/+;CtgfINV/INV mice appeared normal; their weight, femoral length, and BMD were not different from controls (Fig. 4), and contact radiography did not reveal skeletal abnormalities (not shown). Bone histomorphometric analysis revealed osteopenia secondary to decreased trabecular number in male, but not in female, mice, suggesting impaired formation of bone trabeculae during development (Table 2 and Fig. 3). The number of osteoblasts and osteoclasts in conditional Prx1-Cre/+;CtgfINV/INV-null mice were not different from control mice, and eroded surface and bone formation were not affected. Serum levels of the marker of bone remodeling CTX were not different between Ctgf conditional null mice and controls (not shown). μCT revealed a 25% decrease in trabecular bone volume in male Ctgf conditional null mice, but that decrease was not statistically significant, and other parameters of bone structure were not affected (Supplemental Table 2).
Figure 4.
Weight, femoral length, BMD, and Ctgf expression in male and female Prx1-Cre/+;CtgfINV/INV conditional null mice (black bars) and littermate Ctgfe2COIN/e2COIN controls (white bars). The weight in grams (A), femoral length in millimeters (B), total BMD in grams per square centimeter (C), and Ctgf mRNA levels in total calvarial extracts (D), expressed as Ctgf copy number corrected for Rpl38 and normalized to 100, are shown. Values are means ± sem (n = 3–6), except for RNA levels, which are expressed as percentage of control (n = 2–6).
Table 2.
Femoral histomorphometry of 1-month-old male and female Prx1-Cre/+;CtgfINV/INV conditional null mice and controls
| Ctgfe2COIN/e2COIN | CtgfINV/INV | |
|---|---|---|
| Males | ||
| Bone volume/tissue volume (%) | 13.2 ± 1.2 | 9.2 ± 0.8a |
| Trabecular separation (μm) | 189 ± 15 | 250 ± 18a |
| Trabecular number (mm−1) | 4.7 ± 0.3 | 3.7 ± 0.2a |
| Trabecular thickness (μm) | 27.7 ± 1.7 | 24.6 ± 1.2 |
| Osteoblast surface/bone surface (%) | 36.2 ± 2.5 | 35.9 ± 1.8 |
| Number of osteoblasts/bone perimeter (mm−1) | 36.5 ± 2.3 | 37.4 ± 2.5 |
| Number of osteoblasts/tissue area (mm−2) | 272 ± 26 | 221 ± 24 |
| Osteoclast surface/bone surface (%) | 10.8 ± 0.7 | 10.1 ± 0.8 |
| Number of osteoclasts/bone perimeter (mm−1) | 5.2 ± 0.3 | 4.8 ± 0.3 |
| Number of osteoclasts/tissue area (mm−2) | 39 ± 3 | 28 ± 2a |
| Eroded surface/bone surface (%) | 23 ± 2 | 21 ± 1 |
| Mineral apposition rate (μm/d) | 3.33 ± 0.18 | 3.35 ± 0.11 |
| Mineralizing surface/bone surface (%) | 1.67 ± 0.28 | 3.52 ± 1.17 |
| Bone formation rate (μm3/μm2/d) | 0.055 ± 0.009 | 0.123 ± 0.042 |
| Females | ||
| Bone volume/tissue volume (%) | 10.2 ± 0.6 | 9.7 ± 1.1 |
| Trabecular separation (μm) | 293 ± 12 | 309 ± 26 |
| Trabecular number (mm−1) | 3.1 ± 0.1 | 3.0 ± 0.2 |
| Trabecular thickness (μm) | 33.2 ± 0.8 | 31.6 ± 1.1 |
| Osteoblast surface/bone surface (%) | 36.1 ± 2.1 | 32.8 ± 1.4 |
| Number of osteoblasts/bone perimeter (mm−1) | 34.2 ± 0.9 | 33.4 ± 1.2 |
| Number of osteoblasts/tissue area (mm−2) | 210 ± 2 | 202 ± 17 |
| Osteoclast surface/bone surface (%) | 7.1 ± 0.1 | 8.0 ± 0.6 |
| Number of osteoclasts/bone perimeter (mm−1) | 4.4 ± 0.3 | 5.2 ± 0.4 |
| Number of osteoclasts/tissue area (mm−2) | 27 ± 2 | 31 ± 3 |
| Eroded surface/bone surface (%) | 19 ± 2 | 22 ± 1 |
| Mineral apposition rate (μm/d) | 4.01 ± 0.13 | 3.95 ± 0.21 |
| Mineralizing surface/bone surface (%) | 1.33 ± 0.42 | 1.84 ± 0.45 |
| Bone formation rate (μm3/μm2/d) | 0.052 ± 0.015 | 0.071 ± 0.015 |
Bone histomorphometry was performed on femurs from 1-month-old male and female Prx1-Cre/+;CtgfINV/INV conditional null mice and Ctgfe2COIN/e2COIN littermate controls. Values are means ± sem (n = 6–7).
Significantly different from controls, P < 0.05 by unpaired t test.
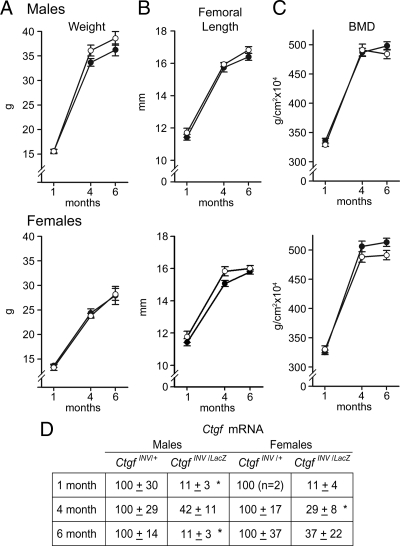
For the conditional deletion of Ctgf in mature osteoblasts, Oc-Cre/Oc-Cre;Ctgf+/LacZ were mated with homozygous Ctgfe2COIN/e2COIN mice to create Oc-Cre/+;CtgfINV/LacZ as an experimental group and Oc-Cre/+;CtgfINV/+ as littermate controls. CTGF mRNA levels in calvarial extracts from Oc-Cre/+;CtgfINV/LacZ conditional null mice were suppressed by 60–90% in relation to those measured in littermate controls. Although heterozygous Ctgf+/LacZ mice had an osteopenic phenotype at 1 month of age (Table 1), it was transient and not observed at 4 and 6 months of age. Consequently, heterozygous Oc-Cre/+;CtgfINV/+ mice were considered to be comparable to wild-type mice at 4 and 6 months of age. When compared with heterozygous Oc-Cre/+;CtgfINV/+ littermates, Oc-Cre/+; CtgfINV/LacZ conditional null mice appeared visually normal and had normal weight, femoral length, and BMD (Fig. 5), and contact radiography did not reveal obvious skeletal abnormalities (not shown). Although 1-month-old heterozygous Ctgf+/LacZ mice were osteopenic compared with wild-type mice, removal of a second allele in conditional Oc-Cre/+;CtgfINV/LacZ mice did not accentuate the phenotype (Table 3). Female 1-month-old conditional Ctgf-null mice exhibited decreased trabecular separation, but bone volume was not significantly affected. Bone histomorphometric analysis of femurs from 4-month-old male and female Oc-Cre/+;CtgfINV/LacZ conditional null mice revealed no skeletal phenotype when compared with Oc-Cre/+;CtgfINV/+ mice (Table 3 and Fig. 3). A decrease in trabecular bone volume was observed at 6 months of age in male Oc-Cre/+;CtgfINV/LacZ-null mice. The decrease in bone volume was secondary to a decrease in trabecular number. Osteoblast number/perimeter and osteoblast surface were not different from controls. Fluorescence microscopy of Oc-Cre/+;CtgfINV/LacZ conditional null male and female mice did not reveal changes in bone formation rate. Changes in trabecular bone volume in 6-month-old male mice were not associated with changes in bone resorption, because osteoclast number and eroded surface were normal, and serum CTX levels were not different from control mice (not shown). μCT of Oc-Cre/+;CtgfINV/LacZ revealed microarchitectural changes in male mice consistent with the histomorphometric data (Table 4). Bone volume and trabecular number were decreased, and connectivity was reduced by 70%. The changes affected the trabecular compartment only, and cortical thickness was not different between Oc-Cre/+;CtgfINV/LacZ and controls.
Figure 5.
Weight, femoral length, BMD, and Ctgf expression in male (upper panels) and female (lower panels) Oc-Cre/+;CtgfINV/LacZ conditional null mice (•) and Oc-Cre/+;CtgfINV/+ littermate controls (○). The weight in grams (A), femoral length in millimeters (B), total BMD in grams per square centimeter (C), and Ctgf mRNA levels in total calvarial extracts (D), expressed as Ctgf copy number corrected for Rpl38 and normalized to 100 at 1, 4, and 6 months of age, are shown. Values are means ± sem (n = 4–13) except for mRNA levels, which are expressed as percentage of control for each independent age (n = 2–6). *, Significantly different from controls by unpaired t test, P < 0.05.
Table 3.
Femoral histomorphometry of 1-, 4-, and 6-month-old male and female Oc-Cre/+;CtgfINV/LacZ conditional null mice and controls
| 1 Month
|
4 Months
|
6 Months
|
||||
|---|---|---|---|---|---|---|
| CtgfINV/+ | CtgfINV/LacZ | CtgfINV/+ | CtgfINV/LacZ | CtgfINV/+ | CtgfINV/LacZ | |
| Males | ||||||
| Bone volume/tissue volume (%) | 10.0 ± 1.4 | 9.2 ± 1.0 | 7.6 ± 1.2 | 6.4 ± 0.7 | 8.3 ± 0.8 | 5.4 ± 0.6a |
| Trabecular separation (μm) | 205 ± 27 | 234 ± 27 | 328 ± 51 | 347 ± 26 | 305 ± 21 | 444 ± 43a |
| Trabecular number (mm−1) | 4.6 ± 0.6 | 4.3 ± 0.4 | 3.0 ± 0.4 | 2.8 ± 0.2 | 3.1 ± 0.2 | 2.3 ± 0.2a |
| Trabecular thickness (μm) | 21.7 ± 1.4 | 21.6 ± 0.6 | 25.1 ± 1.7 | 22.8 ± 1.2 | 26.6 ± 1.6 | 23.4 ± 1.2 |
| Osteoblast surface/bone surface (%) | 22.4 ± 1.8 | 20.2 ± 1.3 | 12.6 ± 0.9 | 13.4 ± 1.2 | 15.0 ± 1.8 | 18.1 ± 1.0 |
| Number of osteoblasts/bone perimeter (mm−1) | 24.4 ± 2.5 | 22.0 ± 1.5 | 12.2 ± 0.8 | 13.5 ± 1.0 | 11.9 ± 1.4 | 15.0 ± 1.1 |
| Number of osteoblasts/tissue area (mm−2) | 183 ± 40 | 145 ± 15 | 58 ± 10 | 58 ± 5 | 57 ± 7 | 52 ± 4 |
| Osteoclast surface/bone surface (%) | 13.4 ± 1.6 | 14.4 ± 0.8 | 8.4 ± 0.7 | 9.9 ± 0.7 | 4.6 ± 0.5 | 5.2 ± 0.5 |
| Number of osteoclasts/bone perimeter (mm−1) | 5.9 ± 0.7 | 6.4 ± 0.4 | 4.8 ± 0.4 | 5.5 ± 0.3 | 2.1 ± 0.2 | 2.5 ± 0.2 |
| Number of osteoclasts/tissue area (mm−2) | 41 ± 3 | 42 ± 4 | 22 ± 2 | 24 ± 3 | 10 ± 1 | 9 ± 1 |
| Eroded surface/bone surface (%) | 21.8 ± 2.0 | 25.0 ± 1.6 | 23.2 ± 2.3 | 25.9 ± 1.1 | 10.5 ± 1.1 | 11.6 ± 1.1 |
| Mineral apposition rate (μm/d) | 1.93 ± 0.20 | 1.86 ± 0.09 | 0.62 ± 0.06 | 0.50 ± 0.05 | 0.60 ± 0.03 | 0.60 ± 0.03 |
| Mineralizing surface/bone surface (%) | 2.62 ± 0.60 | 3.68 ± 0.42 | 5.35 ± 2.14 | 3.47 ± 1.35 | 3.35 ± 0.71 | 2.08 ± 0.55 |
| Bone formation rate (μm3/μm2/d) | 0.053 ± 0.017 | 0.067 ± 0.006 | 0.03 ± 0.02 | 0.02 ± 0.01 | 0.020 ± 0.005 | 0.013 ± 0.004 |
| Females | ||||||
| Bone volume/tissue volume (%) | 3.8 ± 0.3 | 5.9 ± 0.7 | 3.4 ± 0.5 | 2.5 ± 0.7 | 1.8 ± 0.4 | 2.6 ± 0.5 |
| Trabecular separation (μm) | 554 ± 32 | 366 ± 52a | 662 ± 73 | 913 ± 167 | 1188 ± 204 | 982 ± 237 |
| Trabecular number (mm−1) | 1.8 ± 0.1 | 2.8 ± 0.4 | 1.5 ± 0.2 | 1.2 ± 0.2 | 0.9 ± 0.1 | 1.2 ± 0.2 |
| Trabecular thickness (μm) | 21.6 ± 1.1 | 21.5 ± 0.7 | 22.2 ± 1.8 | 19.8 ± 1.5 | 18.6 ± 1.5 | 20.9 ± 2.0 |
| Osteoblast surface/bone surface (%) | 24.3 ± 3.5 | 21.8 ± 1.1 | 16.1 ± 1.5 | 16.7 ± 4.7 | 19.1 ± 2.1 | 16.7 ± 1.7 |
| Number of osteoblasts/bone perimeter (mm−1) | 25.0 ± 3.8 | 23.1 ± 1.2 | 16.2 ± 1.4 | 15.8 ± 3.8 | 22.0 ± 2.3 | 18.9 ± 1.8 |
| Number of osteoblasts/tissue area (mm−2) | 68 ± 10 | 102 ± 18 | 40 ± 7 | 33 ± 12 | 33 ± 7 | 36 ± 6 |
| Osteoclast surface/bone surface (%) | 15.4 ± 2.1 | 15.8 ± 0.7 | 9.4 ± 0.2 | 9.8 ± 0.8 | 13.3 ± 1.7 | 13.1 ± 0.8 |
| Number of osteoclasts/bone perimeter (mm−1) | 7.1 ± 1.0 | 7.0 ± 0.4 | 6.0 ± 0.2 | 6.8 ± 0.4 | 6.8 ± 0.8 | 6.3 ± 0.4 |
| Number of osteoclasts/tissue area (mm−2) | 19 ± 3 | 31 ± 5 | 14 ± 2 | 13 ± 3 | 10 ± 2 | 12 ± 2 |
| Eroded surface/bone surface (%) | 27.9 ± 3.4 | 25.6 ± 0.9 | 24.6 ± 0.5 | 28.6 ± 2.9 | 21.9 ± 2.5 | 23.3 ± 1.4 |
| Mineral apposition rate (μm/d) | 1.84 ± 0.01 | 2.14 ± 0.17 | 0.87 ± 0.05 | 0.99 ± 0.20 | 0.66 ± 0.08 | 0.81 ± 0.06 |
| Mineralizing surface/bone surface (%) | 5.09 ± 1.00 | 2.99 ± 0.19a | 6.02 ± 0.91 | 4.43 ± 1.03 | 12.45 ± 2.96 | 11.25 ± 1.77 |
| Bone formation rate (μm3/μm2/d) | 0.094 ± 0.019 | 0.064 ± 0.007 | 0.053 ± 0.01 | 0.035 ± 0.01 | 0.093 ± 0.031 | 0.10 ± 0.02 |
Bone histomorphometry was performed on femurs from 1-, 4-, and 6-month-old male and female Oc-Cre/+;CtgfINV/LacZ conditional null mice and Oc-Cre/+;CtgfINV/+ littermate controls. Values are means ± sem (n = 3–10).
Significantly different from controls, P < 0.05 by unpaired t test.
Table 4.
Femoral bone microarchitecture assessed by μCT of 6-month-old male Oc-Cre/+;CtgfINV/LacZ conditional null mice and controls
| CtgfINV/+ | CtgfINV/LacZ | |
|---|---|---|
| Bone volume/tissue volume (%) | 9.7 ± 2.0 | 5.9 ± 0.9b |
| Trabecular separation (μm) | 221 ± 9 | 288 ± 13a |
| Trabecular number (mm−1) | 4.4 ± 0.2 | 3.5 ± 0.1a |
| Trabecular thickness (μm) | 48 ± 2 | 52 ± 3 |
| Connectivity density (1/mm3) | 60.7 ± 14.2 | 17.2 ± 4.7a |
| Structure model index | 3.0 ± 0.2 | 3.5 ± 0.3 |
| Cortical thickness (μm) | 224 ± 4 | 216 ± 4 |
Bone μCT was performed on femurs from Oc-Cre/+;CtgfINV/LacZ conditional null mice and Oc-Cre/+;CtgfINV/+ littermate controls.
Significantly different from controls, P < 0.05 by unpaired t test.
P < 0.057 by unpaired t test.
Inactivation of Ctgf in osteoblast cultures
To investigate the consequences of the Ctgf inactivation in vitro, calvarial osteoblasts from Ctgfe2COIN/e2COIN mice were cultured and transduced either with Ad-CMV-Cre to ablate Ctgf or with Ad-CMV-GFP as a control. Ad-CMV-Cre decreased the expression of Ctgf mRNA by 50–70% in cultured osteoblasts, and down-regulation of Ctgf resulted in decreased expression of alkaline phosphatase mRNA and alkaline phosphatase activity, suggesting that CTGF is required for normal osteoblastic function (Supplemental Table 3).
Discussion
Our findings demonstrate that heterozygous Ctgf-null mice survive postnatally and confirm that Ctgf is required for normal skeletal development because Ctgf haploinsufficiency caused reduced trabecular number (12). However, Ctgf haploinsufficiency did not appear to influence postnatal growth beyond 1 month of age, because the skeletal phenotype observed at 1 month resolves at 4 months of age. The skeletal phenotype of heterozygous Ctgf-null mice was reproduced by the conditional inactivation of Ctgf in male mice using the Prx1 enhancer to direct the Cre recombinase. This is in agreement with the expression of the Prx1 enhancer at E10.5 in the limb bud (25). The osteopenic phenotype of both the global and the conditional inactivation of Ctgf was characterized by a decreased number of bone trabeculae, confirming that CTGF is required for the formation of normal trabeculae and for endochondral bone formation (12). The conditional inactivation of Ctgf in the adult skeletal environment was achieved by expressing the Cre recombinase under the control of the osteocalcin promoter. Ctgf inactivation caused a decrease in trabecular bone volume secondary to a decrease in the number of trabeculae in older male mice. It is of interest that the conditional deletion of Ctgf caused a skeletal phenotype in male, but not in female, mice when directing Cre under either the osteocalcin promoter or the Prx1 enhancer. There is no immediate explanation for the sexual dimorphism observed in the skeletal phenotype of Ctgf conditional null mice, although probably it is due to inherent differences in the skeletal architecture of male and female mice. In this study, we confirm earlier observations demonstrating a more rapid age-dependent decline in trabecular bone volume in C57BL/6 female than in male mice, so that at the same age, the bone architecture differs between the two sexes (32). The lack of a skeletal phenotype at 6 months of age in Ctgf-null female mice may be because they have little trabecular bone structure, possibly precluding an additional decrease by the Ctgf inactivation. Another alternative is that the Cre recombination is more efficient in skeletal cells from male than from female mice. This does not seem probable because the decrease in skeletal Ctgf mRNA levels was not appreciably different between male and female Ctgf conditional null mice. The in vivo phenotype we describe indicates that CTGF is required not only for skeletal development but also to maintain adult skeletal homeostasis in male mice. Cyr 61, NOV, WISP-1, and WISP-2 are expressed by osteoblasts but did not appear to compensate for the absence of CTGF in the postnatal skeleton of male mice (5).
The osteopenia observed in adult Ctgf conditional null male mice is to an extent contradictory to previous work demonstrating that transgenic overexpression of CTGF under the control of the osteocalcin or the type XI collagen promoter causes osteopenia (15,37). This was secondary to decreased bone formation and interpreted to be secondary to the binding of BMP, Wnt, and IGF-I by CTGF, resulting in decreased activity of these osteogenic signals. A mechanism of action of CTGF entails the inhibition of BMP-2 activity by direct binding of CTGF to BMP-2 (6,7). Other CCN proteins, such as NOV, appear to act by similar mechanisms, because NOV binds BMPs (38). This is not surprising in view of the structural similarities among CCN proteins, and between CCN proteins and classic BMP antagonists, such as twisted gastrulation and chordin (1,2,3). Recently, we demonstrated that whereas NOV overexpression causes osteopenia, its global inactivation does not cause an obvious skeletal phenotype (39). These results bear similarities to those obtained in mice misexpressing CTGF and suggest that overexpression of CCN proteins prevent the actions of the osteogenic signals they bind; but until now, CCN proteins appear to be mostly dispensable for skeletal homeostasis. These observations do not exclude an important role of CCN proteins in skeletal homeostasis during conditions of induction. For CTGF, this occurs after BMP signaling and may serve to temper the activity of the osteogenic signal. It is of interest that neither the inactivation nor the overexpression of WISP-3 in an array of tissues caused a phenotype (40,41).
An expectation of the Ctgf inactivation would have been enhanced activity of BMP, Wnt, and IGF-I and, as a consequence, increased bone formation and bone volume. An alternate explanation of the findings is that the Ctgf inactivation caused a sensitization to BMP-2 and IGF-I activity and, as a consequence, increased bone resorption (42,43,44,45). However, if the phenotype observed in conditional Ctgf-null mice was caused by increased BMP and IGF-I activity and bone resorption, this would have been modest and transient because neither histomorphometric parameters nor biochemical markers of bone resorption revealed any differences between experimental and control mice under the conditions of this study.
The in vivo phenotype of the conditional inactivation of Ctgf also could be due to direct actions of CTGF on skeletal homeostasis. In accordance with this possibility, short-term cultures of Ctgfe2COIN/e2COIN osteoblasts where Ctgf was inactivated in vitro after the transduction of an Ad-CMV-Cre adenoviral vector exhibited decreased alkaline phosphatase expression, suggesting that CTGF is required for normal osteoblastic function. These results are in agreement with previous work in ST-2 stromal cells, where CTGF induces osteoblastogenesis (11). CTGF inhibits Notch signaling in ST-2 stromal cells, and CTGF may be necessary to temper the activity of Notch in vivo, a signal that inhibits osteoblastogenesis and causes osteopenia (34,46). Therefore, in the absence of CTGF, Notch activity may be enhanced and cause the osteopenia observed in Ctgf-null male mice. However, there were no changes in osteoblast number or significant changes in bone formation in Ctgf-null mice, suggesting that if enhanced Notch signaling was a factor, it played either a transient or a modest role.
In conclusion, our studies reveal that CTGF is necessary for normal skeletal development and postnatal skeletal homeostasis in male, but not female, mice.
Supplementary Material
Acknowledgments
We thank T. Clemens for Osteocalcin-Cre transgenics; R. P. Rysek for Ctgf cDNA; Melissa Burton, Harold Coombs, Deena Durant, Trung X. Le, and Kristen Parker for technical assistance; and Mary Yurczak for secretarial assistance.
Footnotes
This work was supported by Grants AR021707 (to E.C.) and AR043618 (to W.B.) from the National Institute of Arthritis and Musculoskeletal and Skin Diseases and Grants DK045227 (to E.C.) and DK042424 (to E.C.) from the National Institute of Diabetes and Digestive and Kidney Diseases.
Disclosure Summary: A.N.E. is employed by Regeneron Pharmaceuticals and owns Regeneron Pharmaceuticals stock. Other authors have nothing to disclose.
First Published Online June 9, 2010
Abbreviations: BAC, Bacterial artificial chromosome; BMD, bone mineral density; BMP, bone morphogenetic protein; CCN, Cyr61, CTGF, NOV; CMV, cytomegalovirus; COIN, conditional by inversion; μCT, microcomputed tomography; CTGF, connective tissue growth factor; CTX, C-terminal cross-linked telopeptide of type I collagen; Cyr 61, cysteine-rich 61; E10.5, d 10.5 of embryonic life; ES, embryonic stem; FBS, fetal bovine serum; FLP, flippase; GFP, green fluorescent protein; NOV, nephroblastoma overexpressed; RPL38, ribosomal protein L38; WISP, Wnt-inducible secreted protein.
References
- Brigstock DR 2003 The CCN family: a new stimulus package. J Endocrinol 178:169–175 [DOI] [PubMed] [Google Scholar]
- Brigstock DR, Goldschmeding R, Katsube KI, Lam SC, Lau LF, Lyons K, Naus C, Perbal B, Riser B, Takigawa M, Yeger H 2003 Proposal for a unified CCN nomenclature. Mol Pathol 56:127–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Abreu J, Coffinier C, Larraín J, Oelgeschläger M, De Robertis EM 2002 Chordin-like CR domains and the regulation of evolutionarily conserved extracellular signaling systems. Gene 287:39–47 [DOI] [PubMed] [Google Scholar]
- Luo Q, Kang Q, Si W, Jiang W, Park JK, Peng Y, Li X, Luu HH, Luo J, Montag AG, Haydon RC, He TC 2004 Connective tissue growth factor (CTGF) is regulated by Wnt and bone morphogenetic proteins signaling in osteoblast differentiation of mesenchymal stem cells. J Biol Chem 279:55958–55968 [DOI] [PubMed] [Google Scholar]
- Parisi MS, Gazzerro E, Rydziel S, Canalis E 2006 Expression and regulation of CCN genes in murine osteoblasts. Bone 38:671–677 [DOI] [PubMed] [Google Scholar]
- Abreu JG, Ketpura NI, Reversade B, De Robertis EM 2002 Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-β. Nat Cell Biol 4:599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercurio S, Latinkic B, Itasaki N, Krumlauf R, Smith JC 2004 Connective-tissue growth factor modulates WNT signalling and interacts with the WNT receptor complex. Development 131:2137–2147 [DOI] [PubMed] [Google Scholar]
- Song JJ, Aswad R, Kanaan RA, Rico MC, Owen TA, Barbe MF, Safadi FF, Popoff SN 2007 Connective tissue growth factor (CTGF) acts as a downstream mediator of TGF-β1 to induce mesenchymal cell condensation. J Cell Physiol 210:398–410 [DOI] [PubMed] [Google Scholar]
- Iso T, Kedes L, Hamamori Y 2003 HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol 194:237–255 [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX 2009 The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137:216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smerdel-Ramoya A, Zanotti S, Deregowski V, Canalis E 2008 Connective tissue growth factor enhances osteoblastogenesis in vitro. J Biol Chem 283:22690–22699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivkovic S, Yoon BS, Popoff SN, Safadi FF, Libuda DE, Stephenson RC, Daluiski A, Lyons KM 2003 Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development 130:2779–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida T, Nakanishi T, Asano M, Shimo T, Takigawa M 2000 Effects of CTGF/Hcs24, a hypertrophic chondrocyte-specific gene product, on the proliferation and differentiation of osteoblastic cells in vitro. J Cell Physiol 184:197–206 [DOI] [PubMed] [Google Scholar]
- Safadi FF, Xu J, Smock SL, Kanaan RA, Selim AH, Odgren PR, Marks Jr SC, Owen TA, Popoff SN 2003 Expression of connective tissue growth factor in bone: its role in osteoblast proliferation and differentiation in vitro and bone formation in vivo. J Cell Physiol 196:51–62 [DOI] [PubMed] [Google Scholar]
- Smerdel-Ramoya A, Zanotti S, Stadmeyer L, Durant D, Canalis E 2008 Skeletal overexpression of connective tissue growth factor (CTGF) impairs bone formation and causes osteopenia. Endocrinology 149:4374–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Economides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, Yasenchak J, Chernomorsky R, Boucher M, Elsasser AL, Esau L, Zheng J, Griffiths JA, Wang X, Su H, Xue Y, Dominguez MG, Noguera I, Torres R, Macdonald LE, Stewart AF, DeChiara TM, Yancopoulos GD 2003 High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol 21:652–659 [DOI] [PubMed] [Google Scholar]
- Buchholz F, Ringrose L, Angrand PO, Rossi F, Stewart AF 1996 Different thermostabilities of FLP and Cre recombinases: implications for applied site-specific recombination. Nucleic Acids Res 24:4256–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz F, Angrand PO, Stewart AF 1998 Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat Biotechnol 16:657–662 [DOI] [PubMed] [Google Scholar]
- Hoess RH, Abremski K 1990 The Cre-lox recombination system. In: Eckstein F, Lilley DMJ, eds. Nucleic acids and molecular biology. Berlin and Heidelberg: Springer-Verlag [Google Scholar]
- Sauer B, Henderson N 1988 Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA 85:5166–5170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert H, Dale EC, Lee E, Ow DW 1995 Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J 7:649–659 [DOI] [PubMed] [Google Scholar]
- Oberdoerffer P, Otipoby KL, Maruyama M, Rajewsky K 2003 Unidirectional Cre-mediated genetic inversion in mice using the mutant loxP pair lox66/lox71. Nucleic Acids Res 31:e140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lutz B 2002 Cre recombinase-mediated inversion using lox66 and lox71: method to introduce conditional point mutations into the CREB-binding protein. Nucleic Acids Res 30:e90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford LA, Guney MA, Oh YA, Deyoung RA, Valenzuela DM, Murphy AJ, Yancopoulos GD, Lyons KM, Brigstock DR, Economides A, Gannon M 2009 Connective tissue growth factor (CTGF) inactivation leads to defects in islet cell lineage allocation and β-cell proliferation during embryogenesis. Mol Endocrinol 23:324–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ 2002 Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 33:77–80 [DOI] [PubMed] [Google Scholar]
- Zhang M, Xuan S, Bouxsein ML, von Stechow D, Akeno N, Faugere MC, Malluche H, Zhao G, Rosen CJ, Efstratiadis A, Clemens TL 2002 Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J Biol Chem 277:44005–44012 [DOI] [PubMed] [Google Scholar]
- Nazarenko I, Lowe B, Darfler M, Ikonomi P, Schuster D, Rashtchian A 2002 Multiplex quantitative PCR using self-quenched primers labeled with a single fluorophore. Nucleic Acids Res 30:e37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarenko I, Pires R, Lowe B, Obaidy M, Rashtchian A 2002 Effect of primary and secondary structure of oligodeoxyribonucleotides on the fluorescent properties of conjugated dyes. Nucleic Acids Res 30:2089–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy TR, Prince CW, Li J 2001 Validation of peripheral dual-energy x-ray absorptiometry for the measurement of bone mineral in intact and excised long bones of rats. J Bone Miner Res 16:1682–1687 [DOI] [PubMed] [Google Scholar]
- Gazzerro E, Pereira RC, Jorgetti V, Olson S, Economides AN, Canalis E 2005 Skeletal overexpression of gremlin impairs bone formation and causes osteopenia. Endocrinology 146:655–665 [DOI] [PubMed] [Google Scholar]
- Parfitt AM, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, Ott SM, Recker RR 1987 Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res 2:595–610 [DOI] [PubMed] [Google Scholar]
- Glatt V, Canalis E, Stadmeyer L, Bouxsein ML 2007 Age-related changes in trabecular architecture differ in female and male C57BL/6J mice. J Bone Miner Res 22:1197–1207 [DOI] [PubMed] [Google Scholar]
- McCarthy TL, Centrella M, Canalis E 1988 Further biochemical and molecular characterization of primary rat parietal bone cell cultures. J Bone Miner Res 3:401–408 [DOI] [PubMed] [Google Scholar]
- Zanotti S, Smerdel-Ramoya A, Stadmeyer L, Durant D, Radtke F, Canalis E 2008 Notch inhibits osteoblast differentiation and causes osteopenia. Endocrinology 149:3890–3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouadjo KE, Nishida Y, Cadrin-Girard JF, Yoshioka M, St-Amand J 2007 Housekeeping and tissue-specific genes in mouse tissues. BMC Genomics 8:127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryseck RP, Macdonald-Bravo H, Mattéi MG, Bravo R 1991 Structure, mapping, and expression of fisp-12, a growth factor-inducible gene encoding a secreted cysteine-rich protein. Cell Growth Differ 2:225–233 [PubMed] [Google Scholar]
- Nakanishi T, Yamaai T, Asano M, Nawachi K, Suzuki M, Sugimoto T, Takigawa M 2001 Overexpression of connective tissue growth factor/hypertrophic chondrocyte-specific gene product 24 decreases bone density in adult mice and induces dwarfism. Biochem Biophys Res Commun 281:678–681 [Google Scholar]
- Rydziel S, Stadmeyer L, Zanotti S, Durant D, Smerdel-Ramoya A, Canalis E 2007 Nephroblastoma overexpressed (Nov) inhibits osteoblastogenesis and causes osteopenia. J Biol Chem 282:19762–19772 [DOI] [PubMed] [Google Scholar]
- Canalis E, Smerdel-Ramoya A, Durant D, Economides AN, Beamer WG, Zanotti S 2010 Nephroblastoma overexpressed (NOV) inactivation sensitizes osteoblasts to bone morphogenetic protein-2 but NOV is dispensable for skeletal homeostasis. Endocrinology 151:221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutz WE, Gong Y, Warman ML 2005 WISP3, the gene responsible for the human skeletal disease progressive pseudorheumatoid dysplasia, is not essential for skeletal function in mice. Mol Cell Biol 25:414–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Cui Y, Fernando C, Kutz WE, Warman ML 2009 Normal growth and development in mice over-expressing the CCN family member WISP3. J Cell Commun Signal 3:105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal CC, Ghosh Choudhury G, Ghosh-Choudhury N 2009 Phosphatidylinositol 3 kinase/Akt signal relay cooperates with smad in bone morphogenetic protein-2-induced colony stimulating factor-1 (CSF-1) expression and osteoclast differentiation. Endocrinology 150:4989–4998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki H, Hakeda Y, Wakatsuki N, Usui N, Akashi S, Sato T, Tanaka K, Kumegawa M 1992 Insulin-like growth factor-I supports formation and activation of osteoclasts. Endocrinology 131:1075–1080 [DOI] [PubMed] [Google Scholar]
- Okamoto M, Murai J, Yoshikawa H, Tsumaki N 2006 Bone morphogenetic proteins in bone stimulate osteoclasts and osteoblasts during bone development. J Bone Miner Res 21:1022–1033 [DOI] [PubMed] [Google Scholar]
- Sotillo Rodriguez JE, Mansky KC, Jensen ED, Carlson AE, Schwarz T, Pham L, MacKenzie B, Prasad H, Rohrer MD, Petryk A, Gopalakrishnan R 2009 Enhanced osteoclastogenesis causes osteopenia in twisted gastrulation-deficient mice through increased BMP signaling. J Bone Miner Res 24:1917–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti S, Canalis E 2010 Notch and the skeleton. Mol Cell Biol 30:886–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.