Abstract
Limited availability of in vitro and in vivo model systems has hampered efforts to understand tumor biology and test novel therapies for ependymoma, the third most common malignant brain tumor that occurs in children. To develop clinically relevant animal models of ependymoma, we directly injected a fresh surgical specimen from a 9-year-old patient into the right cerebrum of RAG2/severe complex immune deficiency (SCID) mice. All five mice receiving the initial transplantation of the patient tumor developed intracerebral xenografts, which have since been serially subtransplanted in vivo in mouse brains for 4 generations and can be cryopreserved for long-term maintenance of tumorigenicity. The xenograft tumors shared nearly identical histopathological features with the original tumors, harbored 8 structural chromosomal abnormalities as detected with spectral karyotyping, maintained gene expression profiles resembling that of the original patient tumor with the preservation of multiple key genetic abnormalities commonly found in human ependymomas, and contained a small population (<2.2%) of CD133+ stem cells that can form neurospheres and display multipotent capabilities in vitro. The permanent cell line (BXD-1425EPN), which was derived from a passage II xenograft tumor and has been passaged in vitro more than 70 times, expressed similar differentiation markers of the xenograft tumors, maintained identical chromosomal abnormalities, and formed tumors in the brains of SCID mice. In conclusion, direct injection of primary ependymoma tumor cells played an important role in the generation of a clinically relevant mouse model IC-1425EPN and a novel cell line, BXD-1425EPN. This cell line and model will facilitate the biological studies and preclinical drug screenings for pediatric ependymomas.
Keywords: cancer stem cell, ependymoma, gene expression profiling, orthotopic xenograft model
Ependymoma is the third most common malignant brain tumor of childhood, accounting for 6%–12% of all intracranial tumors in children and up to 30% of brain tumors in those younger than 3 years.1–3 These tumors originate either from the wall of the cerebral ventricles or from the spinal canal.2 Histological examination often shows many perivascular pseudorosettes, the pathological hallmark of ependymomas. Despite gross total resection and radiation, 40%–60% of ependymomas recur at the primary tumor site or with metastatic spread,4–6 especially tumors in young children that show anaplastic features and/or display MIB-1 proliferation index >20%.7–9
To develop more effective therapies for the treatment of ependymomas, improved understanding of its biology is urgently needed. Recent identification of cancer stem cells in various human malignancies has provided new clues to tumor initiation, propagation, and therapy resistance.10–16 In ependymoma, restricted populations of radial glial cells have been identified as candidate stem cells of different subgroups of ependymomas.17,18 However, obstacles to preclinical studies of ependymoma include a limited availability of tumor tissues, and more importantly, a lack of in vitro and in vivo model systems. Efforts to establish permanent ependymoma cell lines have largely been unsuccessful, as many of the cultured ependymoma cells can only survive for a short period of times.19 Although Brisson et al.20 were able to maintain 2 of their 11 cultures long-term in vitro, these cells proliferated unusually slowly, requiring 2 years to reach passage 18. No genetically engineered mouse model of ependymoma has been established yet. Currently, only 5 subcutaneous xenograft mouse models exist, including D528EP, D612EP, and HxBr5,21–23 which have been established and are currently used in multiple preclinical drug screens. However, subcutaneous xenograft models often fail to replicate the complex interactions between tumors and their native microenvironment;24–28 attempts have therefore been made to subtransplant some of the subcutaneous ependymoma xenografts into the brains of immunodeficient animals to create orthotopic xenograft mouse models. Although direct injection of human primary tumors into anatomically matched locations in an immunodeficient animal has been shown to better replicate the biology of various human cancers and may have better predictability of future clinical success,29–31 no such models have been established for human ependymomas at this time.
In this study, we describe the establishment of an orthotopic ependymoma xenograft mouse model from a fresh surgical specimen of a recurrent anaplastic ependymoma. We determined the tumorigenicity and serial subtransplantability of xenograft tumors, and evaluated its clinical relevance by examining the histological, immunohistochemical (IHC), and invasive growth characteristics of the xenograft tumors when compared with the original patient tumor. To further examine the molecular fidelity of this xenograft model, we compared whole-genome gene expression profiles between xenografts (during serial passaging) and the patient tumor. We also examined whether cancer stem cells were preserved in the established xenografts, and whether the isolated CD133+ cells maintained the self-renewal and multilineage differentiation capabilities in vitro. Since there are still no ependymoma cell lines available, we further tested whether our xenograft tumor cells can be maintained in vitro long term to establish a cell line.
Materials and Methods
Tumor Tissue
The patient was a 9-year-old male undergoing craniotomy for the resection of a recurrent tumor of the right cerebrum at Texas Children's Hospital. This tumor recurred locally, despite complete tumor resection and radiation therapy when the boy was 5 years old. Both the original and recurrent tumors were consistent with an anaplastic ependymoma. Following patient consent to an Institutional Review Board-approved tissue bank protocol, tumor tissues were obtained from the cryostat laboratory and quickly transferred, in DMEM growth medium supplemented with 10% FBS on ice, to the tissue culture room, where they were washed and mechanically dispersed as described previously.32 After the cell suspension was passed through a 35 µm cell strainer, the live tumor cells as single cells and small clumps (∼5–10 cells) were counted with trypan blue staining and resuspended in growth medium (1 × 108/mL), followed by immediate transferring to animal facility on ice.
Orthotopic Transplantation into Rag2/Severe Complex Immune Deficiency Mice
Surgical transplantation of tumor cells into mouse cerebrum was performed as we described previously,29,32 following the Institutional Animal Care and Use Committee (IACUC) approved protocol. The Rag2/severe complex immune deficiency (SCID) mice were bred and maintained in a specific pathogen-free animal facility of Texas Children's Hospital. Mice, aged 6–8 weeks of both sex, were anesthetized with sodium pentobarbital (50 mg/kg). Tumor cells (1 × 105) were suspended in 2 µL of culture medium and injected into the cerebral hemisphere (1 mm to the right of the midline, 1.5 mm anterior to the lambdoid suture, and 3 mm deep) via a 10-µL 26-gauge Hamilton Gastight 1701 syringe needle. The animals were monitored daily until they developed signs of neurological deficit or became moribund, at which time they were euthanized and their brains removed for histopathological analysis.
IHC Staining
IHC staining was performed using a Vectastain Elite kit (Vector Laboratories) as described previously.29 Primary antibodies included the following: mouse antisynaptophysin (anti-SYP) (1:200), glial fibrillary acidic protein (GFAP) (1:200), vimentin (VIM) (1:200), epithelial membrane antigen (EMA) (1:100), α-smooth muscle actin (SMA) (1:100) (Dako), MAP-2 (1:200), Ki-67 (1:20) (Abcam, Inc.), neuron-specific class III B-tublin (TuJ-1) (1:100) (Fitzgerald Industries International, Inc.), human-specific mitochondria (MT) (1:50) (Chemicon International, Inc.); VEGF (c-1) (1:100) (Santa Cruz Biotechnology, Inc.); and rabbit anti-Nestin (NES) (1:500), anti-Von Willebrand Factor (vWF) (1:200) (Chemicon), and VEGFR-1 (1:150) (Epitomics, Inc.). After slides were incubated with primary antibodies, the appropriate biotinylated secondary antibodies were applied, and the final signal was developed using the 3,3′-diaminobenzidine substrate kit for peroxidase. A negative control was performed by replacing the primary antibody with 1% BSA in PBS. The IHC staining was assessed by combining the intensity, scored as negative (–), low (+), medium (++), and strongly positive (+++), and extent of immunopositivity expressed as the percentage of positive cells.
Spectral Karyotyping
Primary cultures from xenograft tumors were used to make chromosome preparations. To identify chromosome aberrations with spectral karyotyping (SKY), the cocktail of human chromosome paints obtained from Applied Spectral Imaging (ASI) was applied to the slides according to the manufacturer's protocol. Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Images were acquired with an SD200 Spectra cube (ASI) mounted on a Zeiss Axioplan II microscope using a custom designed optical filter (SKY-1) (Chroma Technology) and analyzed using SKY View 1.2 software (ASI). The breakpoints on the SKY-painted chromosomes were determined by comparison of the corresponding inverted-DAPI banding of the same chromosome.
Whole-Genome Gene Expression Profiling
The genome-wide expression analysis was performed using Illumina's Human-6 v2 BeadChips, which contains more than 48K transcript probes, following the manufacturer's instructions. Briefly, total RNA was extracted from original patients tumor tissues and intracerebral (IC) xenograft tumors with TRIZOL reagent (Invitrogen) as described previously.33,34 Normal human cerebral cortex RNA, which is pooled from 10 male and female donors, was procured from Clontech Laboratories, Inc. RNA concentrations were measured using a Nanodrop 1000 spectrophotometer. RNA integrity was checked by electrophoresis using a 2% agarose gel with 6% formaldehyde. Half a microgram of total RNA was used to synthesize biotinylated cRNA using the Totalprep RNA amplification kit (Ambion, Inc.), and 1.5 µg of biotinylated cRNA was applied to the HumanWG-6 v2 Beadchips and processed according to the vendor's instruction (Illumina). Each sample was loaded in triplicates on the chips. The Beadchips were scanned using Beadstation 500 GX scanner.
The raw image files from the scanner were imported into the Bead Studio software Gene Expression module version 3.2.7 (Illumina, Inc.) and were processed using the quantile normalization algorithm. This method assumed that the distribution of the expression values did not change dramatically between arrays. All arrays were adjusted so that they showed an almost identical intensity distribution from all the genes. All 42 620 elements shown in the gene profile tab in Bead Studio were used in the normalization. The log intensity values were exported and analyzed in Bioconductor.35
Quantitative Real-Time RT–PCR
This was performed with SYBR green master mix in an ABI 7000 DNA detection system (ABI), as we described previously.33,34 Complementary DNA was synthesized with MuLV reverse transcriptase and random hexamers (Perkin Elmer) in a total volume of 20 µL from 1 µg of total RNA extracted with TRIZOL reagent. Primers for PCR amplification were designed to flank more than one exon (Supplementary Material, Table S1). Expression levels of selected genes were normalized to the internal standard GAPDH using the standard ΔΔCt method. All reactions were performed in duplicate and repeated twice, and the specificities of PCR products were confirmed by analyzing dissociation curves from individual reactions and visualizations on 2% agarose gel.
Fluorescence-Activated Cell Sorting and In Vitro Characterization of CD133+ Cancer Stem Cells
Cancer stem cells were labeled with phycoerythin (PE)-conjugated monoclonal antibodies against human CD133 (CD133/2-PE, Milteny Bio, Inc.) at 4°C for 10 minutes per the manufacturer's instruction.32 Cells were then resuspended in stem cell growth medium, which consisted of Neurobasal media (Invitrogen), N2 and B27 supplements (0.5× each; Invitrogen), human recombinant basic fibroblast growth factor (bFGF) (50 ng/mL; R&D Systems), epidermal growth factor (EGF) (40 ng/mL; R&D Systems),36 penicillin G, and streptomycin sulfate (1:100; Invitrogen), and flow-sorted with Cytomation MoFlo (Dako). To maximize our chances of isolating real CD133+ cells, a stringent gate setting was applied using non-stained cells as a reference. Dead cells were excluded by propidium iodide staining.
The isolated CD133+ cells were incubated in the aforementioned stem cell growth medium for the formation of neurospheres. To determine the multilineage differentiation capabilities of the CD133+ cells, the neurospheres were transferred into poly-d-lysine–coated chamber slides and incubated with DMEM growth medium supplemented with 10% FBS. Immunofluorescent staining was performed as described previously32 by probing the slides with mouse anti-GFAP, MAP-2, SYP, VIM, MT (as listed above), and mouse against A2B5 (1:200) (Chemicon), rabbit antioligodendrocyte marker O4 (O4) (R&D systems), and RC2 (the University of Iowa) antibodies. The secondary antibodies were FITC- or Texas Red-conjugated antimouse or antirabbit IgG (Vector Laboratories). Cells were counterstained with DAPI (Vector Laboratories), examined with a Nikon microscope, and their images captured with a color CCD camera at specific magnifications.
Long-Term Growth of Xenograft Tumor Cells In Vitro
After the xenograft tumor from passage II was mechanically dissociated into single cells as described above, tumor cells were plated into T75 flasks and incubated in DMEM supplemented with 10% FBS in a CO2 incubator at 37°C. The growth culture medium was changed every 3–4 days. When cells reached more than 80% confluence, they were passaged at 1:2 through trypsin digestion. Mycoplasma contamination was monitored with MycoAlert Mycoplasma Detection kit (Cambrex Bio Science).
Statistical Analysis
Animal survival times were examined through log-rank analysis followed by pairwise multiple comparison with the Holm–Sidak method with SigmaStat 3.5 (Systat Software, Inc.) and graphed with SigmaPlot 10 (Systat Software, Inc.).
Microarray Data Analysis
Clustering
The sample clustering was performed only for 8863 genes that showed a detection P value <.001, using the hierarchical clustering algorithm provided in the “stats” R library. The clustering method was complete linkage and the metric was 1 − r2.
Correlations
All 42 620 elements were used to calculate the correlation coefficient (r2 values) of 4 groups of samples hybridized in triplicates (the normal sample, the patient sample, passage I with 2 samples, and passage III with 2 samples). Intragroup correlation illustrated the reproducibility of the array experiments. Intergroup correlation values demonstrated the similarities of gene expression profiles between groups of samples. The overall correlations matrix were generated using R software with the package ‘lattice’ in default parameters.37
Differential Analysis using the Normal Sample as a Reference
All samples that contained genes with a detection P value <.001 (8863 total) were subjected to a differential analysis using the Illumina build-in differential gene expression analysis function in Beadstudio software with false discovery correction.38 Four groups of samples went through 3 comparison tests: (i) normal vs patient, (ii) normal vs passage I, and (iii) normal vs passage III. The total number of differential expressed genes in all 3 comparisons was 7668 (differential P value <.001). For visualization purposes, we used the Top Score Pair (TSP) gene selection method39 to reduce the number of genes to 670. The TSP method ranked with high score pairs of genes whose expression levels inverted from one condition to another. The standardized score of the log expression intensities was used for clustering purposes. Gene clustering was performed using single linkage and Canberra metric, whereas sample clustering was performed using complete linkage and the Euclidean distance.
Gene Ontology Analysis for Differential Expression Data Generated using the Normal Triplicates as References
The significant genes found by the TSP gene selection method were subjected to gene enrichment analysis using Metacore program version 5.3 from GeneGo, Inc. Enrichment analysis consisted of matching the list of significant genes with gene IDs from the GeneGO Pathway Maps functional ontology in Metacore. The canonical pathway maps used in the estimation of the P value represented a set of about 650 signaling and metabolic maps (GeneGO, Inc.). The probability of random matching was calculated in a P value using hypergeometric distribution.
Differential Analysis Using the Patient Tissue Sample as a Reference
Given the large difference between the patient and the normal samples, an additional differential expression analysis was conducted using all samples except the normal replicates. There were 3 groups of samples (the patient in triplicates, passage I with 2 samples each in triplicates, and passage II with 2 samples each in triplicates) and 2 comparison tests were performed: (i) patient vs. passage I and (ii) patient vs. passage III. To find differentially expressed genes that were due solely to changes from patient to passages I and III, an additional differential analysis using the Illumina build-in differential gene expression analysis function with false discovery correction38 was performed using the patient as a reference.
The differential expression test was performed using the significant analysis of microarray (SAM) which employs a standardized mean difference applied on repeated permutations of the data to rank the genes according to their statistical significance.40 Given the 5 groups in the experiment (the patient in triplicates, passage I(#1) in triplicates, passage I(#2) in triplicates, passage III(#1) in triplicates, and passage III(#2) in triplicates), a multiclass SAM analysis with a modified F-statistic was used.41 A total of 100 genes showed false discovery rate (FDR) values <0.00012%. A clustogram was generated using the “gplots” R library.42 Whereas gene clustering was performed using single linkage and Canberra metric, sample clustering with all genes was performed using complete linkage and the Euclidean distance.
Results
Formation of Orthotopic Xenograft Tumors in the Brains of SCID Mice
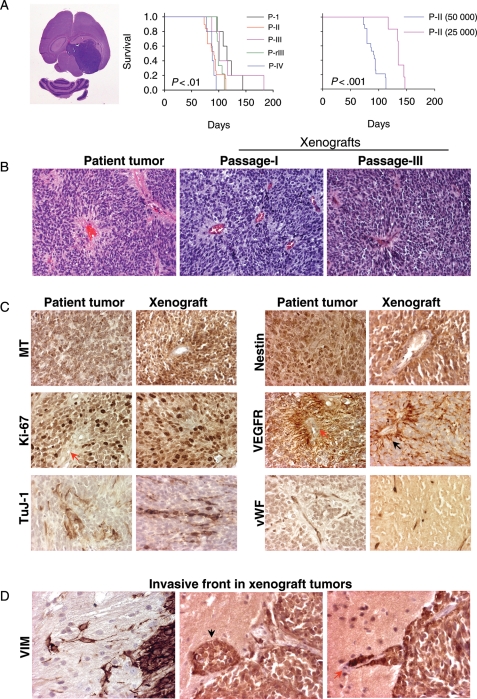
To minimize the potential selective, adaptive, and progressive changes that are associated with long-term in vitro culture of human cancer cells, we utilized mechanical dispersion techniques to prepare the cell suspension from a freshly resected ependymoma specimen and injected these tumor cells into mouse brains within 60 minutes of tumor resection. All 5 Rag2/SCID mice injected with 1 × 105 primary tumor cells developed signs of neurological deficit or became moribund within 97–144 (117 ± 17.6) days postinjection. In all of these mice, the growth of xenograft tumor was confirmed (Fig. 1A). Grossly, the mouse brains were invariably enlarged, which, when sectioned, often reveal a huge IC xenograft tumor (Fig. 1A). This model was therefore designated as IC-1425EPN. The xenograft tumors exhibited medium firmness, formed relatively sharp margins from the surrounding normal mouse brains, and were easily dissected.
Fig. 1.
Ependymoma xenograft in the mouse brain. (A) Gross appearance of H&E-stained mouse brain with huge xenograft tumors (left panel), and changes of animal survival times during serial subtransplantation from passage I (P-I) to passage III (P-III) and from cryopreserved xenograft tumor cells at passage III (P-rIII) (middle panel) as well as reduced cell numbers (right panel). P < .01 between passages I and IV, and P < .001 between mice injected with 50 000 and 25 000 cells. (B) H&E staining showing the replication of pseudorosettes in xenograft tumors (×10). (C) Representative images of IHC showing the positive staining of human-specific mitochondrial (MT) antigen, cell proliferation marker (Ki-67), neuronal marker (TuJ), neural progenitor marker (Nestin), VEGFR, as well as microvessel density (vWF) (×40). (D) Detection of invasive tumor cells using human-specific antibodies against VIM (×40).
In Vivo Subtransplantation and Long-Term Cryopreservation of Xenograft Tumors
Since all the tumor-bearing mice would ultimately die of the disease, it is important to develop strategies that will maintain and expand the animal model populations. To determine whether we could achieve these goals through subtransplantation, we harvested the xenograft tumors from donor mice and retransplanted tumor cells into the brains of recipient mice. All animals injected with 105 tumor cells at the passages II (n = 32), III (n = 5), and IV (n = 20) developed IC tumors. A single xenograft tumor normally yielded 10–15 × 106 live tumor cells, which was sufficient for injecting 100–150 mice. To determine whether repeated subtransplantations would cause major changes of tumor growth velocity, we first examined the animal survival times using log-rank analysis and pairwise comparison. Compared with the median survival times of 115 days in mice receiving primary patient tumors (passage I), mice injected with xenograft tumor cells at passage II, III, and IV survived for 94, 101, and 88 days, respectively (Fig. 1A). The only significant difference was found between passages IV and I (P < .01), suggesting that repeated subtransplantations are prone to induce changes of tumor growth in vivo.
To examine the tumorigenic capabilities of decreased cell numbers, we injected 2.5 × 104 and 5 × 104xenograft cells from a passage I mouse into the brains of 6 and 5 recipient mice, respectively. Tumor formation was confirmed in all the animals (Fig. 1A), indicating that once established, these xenograft tumors would remain tumorigenic in SCID mice even with remarkably reduced cell numbers.
To provide sustainable supply of tumor-bearing animal models and to minimize the genetic drift associated with prolonged and repeated in vivo subtransplantation, we tested whether freshly isolated xenograft tumor cells could be cryopreserved for long-term storage while preserving their tumorigenicity. All 15 mice receiving the injection of xenograft tumor cells that have been prestored in liquid nitrogen for 5 months developed tumors, and their survival times were similar to those injected with fresh xenograft tumors (Fig. 1A), suggesting that long-term cryopreservation did not introduce significant changes in tumor growth in this model.
Replication of the Histopathological Features of the Original Tumor
To determine whether the xenograft tumors replicated the histopathological phenotypes of the parent tumor, particularly during repeated subtransplantation, paraffin sections of xenograft tumors were reviewed. From H&E-stained sections, the xenograft tumors from passages I to IV displayed characteristic histological features of the original anaplastic ependymoma, including high cellularity, poor differentiation, microvascular proliferation, brisk mitotic activity, and perivascular pseudorosettes (Fig. 1B).
Immunohistochemically, the xenograft tumors also displayed a profile similar to that of the patient tumor (Fig. 1C and Table 1), including a high proliferative (Ki-67) index (60%–70%) homogeneous high-level expression of neuronal precursor marker nestin, and patches of cells expressing early neuronal differentiation marker TuJ-1(Fig. 1C). Expression of mature neuronal marker MAP2 was seen in isolated xenograft tumor cells. The glial marker GFAP was only expressed in isolated xenograft cells as well, although strong expression was readily detected in the reactive astrocytes trapped in and surrounded the xenograft tumors (Supplementary Material, Fig. S1). The overall expression of VEGF was low in the tumor cells, but strong VEGFR1 positivity was detected in tumor cells forming the perivascular pseudorosettes. Nearly, all the xenograft tumor cells stained strongly positive for intermediate neurofilament VIM, including tumor cells infiltrating into normal parenchyma (Fig. 1D). Similar to the patient tumor, staining for markers of mesenchymal (EMA) and smooth muscle (SMA) differentiation was absent in the xenograft tumors.
Table 1.
Summary of immunohistochemical features of the xenograft tumors during in vivo subtransplantations
| Target phenotypes | Marker | Patient tumor | Xenograft tumors |
|
|---|---|---|---|---|
| P-I | P-III | |||
| Human identity | MT | +100% | +100% | +100% |
| Proliferation | Ki67 | +60% | ++ 70% | ++ 70% |
| Neuron precursor | Nestin | +80% | ++ 100% | ++ 100% |
| Neuron-early | TuJ-1 | ++ 50%–80% | ++ 50%–70% | ++ 50% |
| Neuron mature | MAP2 | +isolated | ++ 50% | ++ 50% |
| Neuron mature | SYP | (−) | +30% | +5%–10% |
| Astrocyte | GFAP | +isolated | +40% | ++ 10%, +60% |
| Epithelial membrane antigen | EMA | (−) | (−) | (−) |
| Angiogenesis | VWF | 5–10/HP | 5–10/HP | >10/HP |
| Angiogenesis | VEGF | +70% | ++ 20%, + 60% | +80% |
| Angiogenesis | VEGFR1 | ++ 30%–50% | ++ 60% | ++ 50% |
| Intermediate filament | VIM | +100% | ++ + 100% | ++ + 100% |
Detection of Invasive Xenograft Tumor Cells in Mouse Brains
Human ependymoma often form a clear-cut edge toward the adjacent nervous tissue, although anaplastic ependymomas can occasionally be frankly invasive. To positively identify infiltrating ependymoma cells, we performed IHC staining using human-specific antibodies against mitochondria. Since all the xenograft cells expressed robust VIM, we also applied human-specific VIM antibodies to detect invasive tumor cells. As shown in Fig. 1D, although the xenograft tumors were relatively well demarcated, occasional invasion into surrounding normal brain was observed. In addition to invading single cells, formation of microsatellite tumors and migration along blood vessels were also seen (Fig. 1D). Development of metastatic foci from cerebrospinal fluid spread was rare.
Identification of Chromosomal Abnormalities with SKY
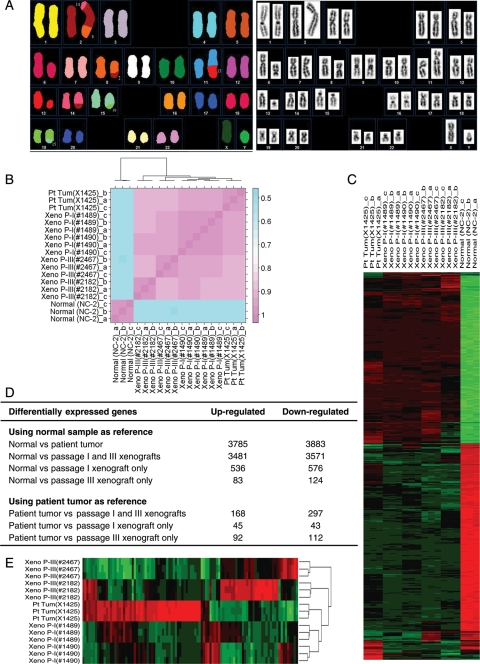
Clinical cytogenetic analysis of the patient tumor with G-banding revealed a normal male karyotype (46, XY), but this result may be due to the contaminating fibroblasts in the cultures. However, the tumors derived from xenograft displayed 8 structural aberrations in their genomic compliment (Fig. 2A). All the analyzed metaphase spreads showed a karyotype of 46, XY, t(2;14)(p21;q24), t(2;8)(q32;q21), del(6)(q13), t(11;13)(q25;q14), t(15;19)(q21;q13)[15]. These 8 chromosomal abnormalities were maintained during serial in vivo passaging from I to III, and no new abnromalities were observed. Since the resolution of SKY is limited, it is possible that small regions of homozygous deletion or amplfication have been missed.
Fig. 2.
Cellular and molecular genetic features of IC-1425EPN xenograft tumors. (A) Identification of structural chromosomal abnormalities with SKY (left panel) and inverted-DAPI banding (right panel). (B) The heat map of the correlation coefficients among samples. The scale bar at the right illustrated the r2 value. Clustering of the samples was illustrated at the dendrogram at the top. (C) The hierarchical clustering of samples including the normal samples. Green rectangles represented down-expression, whereas red rectangles signified up-regulation. (D) List of gene numbers that are differentially expressed using normal brain tissue and patient tumor as references. (E) The hierarchical clustering of samples excluding the normal samples. The dendrogram at the right identified the grouping of samples. Green rectangles represented down-expression, whereas red rectangles signified up-regulation.
Comparison of Gene Expression Profiles Between Original Ependymoma and Xenograft Tumors During In Vivo Subtransplantations
To determine whether the xenograft tumors maintained global gene expression characteristics of the original tumor and to identify signaling pathways that were deregulated in this model, we compared the gene expression profiles of xenograft tumors from passages I and III with those of the original patient tumor using Illumina's gene expression microarray. Since the cell of origin of ependymoma has yet to be unequivocally established,43 we utilized RNAs from a pooled normal cerebral tissue that was analyzed simultaneously as a normal control. The normalized and raw data have been deposited in GEO (www.ncbi.nlm.nih.gov/geo/) (accession no. GSE17516). Using all 42 620 elements on the microarray, we calculated the correlation coefficients and found a strong positive correlation (r2 = 0.99, P < .0001) among the replicates (Fig. 2B). The correlation coefficient of normal pooled sample and the patient tumor was 0.56 (P < .0001). The correlations between the profiles of the patient tumor and xenografts were high (r2 = 0.96 at passage I and 0.93 at passage III, P < .0001) (Fig. 2B). These results suggest that repeated in vivo subtransplantations maintained the overall genomic similarity of the xenografts.
To identify genes that are significantly deregulated in this model, the expression profiles of the patient tumor and the xenograft tumors were compared with the pooled RNA from normal human cerebral tissues (Fig. 2C). In the original patient ependymoma, 3875 and 3883 genes were found to be up-regulated and down-regulated, respectively. Orthotopic transplantation of this tumor into the mouse brain has led to the preservation of 3481 up-regulated genes and 3571 down-regulated genes in both passages I and III of the xenograft tumors (Fig. 2D). There were 536 up-regulated genes and 576 down-regulated genes that were uniquely induced during passage I. There were 83 and 124 down-regulated genes that were uniquely induced during passage III. There were 304 up-regulated and 312 down-regulated genes lost in passages I and III.
To examine whether growth of IC-1425EPN in mouse brains introduced new gene expression changes, differential expression analysis was run for xenograft tumors from passages I and III using the patient tumor as a reference. The number of differentially expression genes among the patient primary tumor tissue and the passages was much reduced compared with those using the normal sample as a reference. A total of only 168 up-regulated genes and 297 down-regulated genes were identified (Fig. 2D). Compared with the 45 up-regulated and 43 down-regulated genes found exclusively in passage I xenografts, there were 92 up-regulated and 112 down-regulated new genes in passage III (Fig. 2D), suggesting that long-term growth and repeated subtransplantations in vivo were prone to induce more genetic changes. However, the new genetic changes occurring in passage III might be due to tumor's genetic instability and tumor progression. Further experiments are needed to establish the causes of these new genetic changes associated with serial subtransplantations. These results were also supported by the significant analysis of microarray (SAM) applied to all samples except the normal replicates. As shown by the clustogram (Fig. 2E) generated from the 100 genes found by SAM, passage I groups cluster together and close to the patient samples, whereas both passage III groups cluster separately further apart. The SAM analysis and the differential gene analysis in Beadstudio were in high concordance. Among the 100 significant genes found by SAM, 98 of them were also found to be differentially expressed in the Beadstudio differential gene expression analysis.
Gene ontology analysis using the normal sample as a reference showed that a list of 10 pathways were found to have a P value <.001 (−log10(P value) > 3) and FDR < 0.05 (Supplementary Material, Fig. S2). The genes were involved in chromosome condensation, DNA damage ATM/ATR regulation of G1/S checkpoint, cell-cycle control via APC pathway, CDK5 pathway in presynaptic signaling, calcium signaling, platelet-derived growth factor signaling via STATs and NF-κB, and G-protein signaling (Supplementary Material, Fig. S3). Gene ontology analysis using the patient primary tissue as a reference showed that a list of 10 pathways were found to be significantly enriched and showed a P value <.005 (−log10(P value) > 2.3) and FDR < 0.05 (Supplementary Material, Fig. S2). The major pathways being enriched are sphingosine-1 phosphate-dependant G-protein–coupled receptor, interleukin 6 signaling, Oncostatin M signaling via MAPK, JAK-Stat, transcription control of VDR, trobopoetin signaling, cytoskeleton remodeling, and G-protein signaling via Ras family cascade.
Preservation of Ependymoma Gene Expression Signature in Xenograft Tumors
Several recent studies have demonstrated that ependymomas arising within different regions of the CNS are molecularly distinct diseases.17,44,45 In agreement with previous reports, a subset of the differentially expressed genes that were shown to be a characteristic of supratentorial ependymomas have been identified in our xenografts (both passages I and III) as well as the original ependymoma tumor. As listed in Table 2, the NOTCH signaling pathway was activated as evidenced by high-level (>2-fold) up-regulation of NOTCH1, NOTCH2NL, LFNG, HES1, HES4, JAG1, JAG2, and PSENEN and down-regulation of NOTCH inhibitor NUMB. Overexpression of SMO, MSI1, MSI2, and GLI3 strongly indicated that the sonic hedgehog (SHH) signaling pathway was activated as well. Additional genes associated with ependymoma, such as overexpression of MSX1 (>10-fold), IGF2 (>30-fold), GGH (>4-fold), MERTK (>2-fold), were also detected in our xenograft tumors. Moreover, several genes involved in WNT/FREZZLED signaling pathway (AXIN2, DKK1, DVL2, FRZB, FZD1, FZD2, FZD6, FZD10, as well as WNT2b) were also overexpressed, suggesting that the pathway was activated in our models. We also found that genes that are known to be associated with cancer stem cells, that is, prominin (CD133),14,17,46,47 nestin (NES),17,48 integrin beta-1 (ITGB1),49 and aldehyde dehydrogenase (ALDH4A1, ALDH3A1, ALDH16A1, and ALDH1A3)50 were also up-regulated (Table 2). There were, however, some previously reported gene expression abnormalities in ependymoma that were not detected in the xenografts or the patient tumor, including EGFR and PAX3,17,44,51 although up-regulation of EFNB1 (>3-fold), EFNB3 (5-fold), and PAX6 (>2-fold) were observed.
Table 2.
Partial list of dysregulated genes that were originally present in patient tumor and maintained in the xenografts during serial subtransplantations
| Accession | Target ID | Definition | Log 2 ratio (tumor/normal) |
||
|---|---|---|---|---|---|
| Patient tumor | Xeno (P-I) | Xeno (P-III) | |||
| NOTCH pathway | |||||
| NM_017617.3 | NOTCH1 | Notch homolog 1, translocation-associated | 2.89 | 3.01 | 2.35 |
| NM_203458.3 | NOTCH2NL | Notch homolog 2 (Drosophila) N-terminal like | 1.35 | 1.38 | 1.35 |
| NM_005524.2 | HES1 | Hairy and enhancer of split 1 (Drosophila) | 1.85 (1.13)a | 1.90 (1.5) | 2.08 (1.25) |
| NM_001024598 | HES3 | Hairy and enhancer of split 3 (Drosophila) | 1.61 | 2.21 | 2.75 |
| NM_021170.2 | HES4 | Hairy and enhancer of split 4 (Drosophila) | 5.41 (1.26) | 5.65 (1.13) | 5.59 (1.49) |
| NM_000214.1 | JAG1 | Jagged 1 (Alagille syndrome) | 2.03 | 1.31 | 0.79 |
| NM_002226.3 | JAG2 | Jagged 2 (JAG2), transcript variant 1 | 1.64 (1.47) | 2.57 (1.96) | 2.83 (1.64) |
| NM_001040167 | LFNG | Lunatic fringe homolog (Drosophila) | 5.51 | 5.32 | 5.22 |
| NM_172341.1 | PSENEN | Presenilin enhancer 2 homolog (Caenorhabditis elegans) | 1.02 (1.77) | 0.96 (2.26) | 2.41 (1.62) |
| NM_001005744 | NUMB | Numb homolog (Drosophila) | −1.14 (−0.97) | −1.11 (−0.66) | −1.01 (−1.24) |
| PTCH-SHH pathway | |||||
| NM_005270.3 | GLI2 | GLI-Kruppel family member GLI2 | 1.56 | 2.08 | 1.94 |
| NM_000168.3 | GLI3 | GLI-Kruppel family member GLI3 | 2.60 (20.5) | 2.94 (2.38) | 2.86 (0.93) |
| NM_152629.3 | GLIS3 | GLIS family zinc finger 3 | 2.02 | 1.34 | 1.29 |
| NM_005631.3 | SMO | Smoothened homology (Drosophila) | 1.91 | 2.32 | 2.32 |
| NM_002442.2 | MSI1 | Musashi homolog 1 (Drosophila) | 2.33 (1.74) | 2.49 (2.54) | 2.98 (1.27) |
| NM_170721.1 | MSI2 | Musashi homolog 2 (Drosophila) | 1.24 | 1.30 | 0.98 |
| WNT pathway | |||||
| NM_004655.2 | AXIN2 | Axin 2 (conductin, axil) | 2.36 | 1.89 | 2.27 |
| NM_024494.1 | WNT2B | Wingless-type MMTV integration site family, member 2B | 3.50 | 3.12 | 2.45 |
| NM_012242.2 | DKK1 | Dickkopf homolog 1 (Xenopus laevis) | 2.29 | 0.72 | 1.31 |
| NM_014421.2 | DKK2 | Dickkopf homolog 2 (Xenopus laevis) | 2.65 | 2.58 | 2.90 |
| NM_004422.2 | DVL2 | Dishevelled, dsh homolog 2 (Drosophila) | 1.07 | 0.92 | 0.97 |
| NM_001463.2 | FRZB | Frizzled-related protein | 4.82 (3.64) | 4.77 (2.45) | 4.65 (3.05) |
| NM_003505.1 | FZD1 | Frizzled homolog 1 (Drosophila) | 2.19 (2.14) | 2.44 (2.35) | 2.35 (2.54) |
| NM_007197.2 | FZD10 | Frizzled homolog 10 (Drosophila) | 1.70 | 2.11 | 2.16 |
| NM_001466.2 | FZD2 | Frizzled homolog 2 (Drosophila) | 3.26 | 3.48 | 3.02 |
| NM_003506.2 | FZD6 | Frizzled homolog 6 (Drosophila) | 1.84 | 1.99 | 1.94 |
| NM_003507.1 | FZD7 | Frizzled homolog 7 (Drosophila) | 1.61 | 1.49 | 1.28 |
| Stem cell marker | |||||
| NM_006017.1 | PROM1 | Prominin 1 | 3.44 (3.83) | 2.81 (6.05) | 3.39 (2.40) |
| NM_006617.1 | NES | Nestin | 3.11 (1.99) | 3.39 (3.15) | 3.15 (0.55) |
| NM_000693.2 | ALDH1A3 | Aldehyde dehydrogenase 1 family, member A3 | 1.97 | 2.17 | 1.37 |
| NM_000691.3 | ALDH3A1 | Aldehyde dehydrogenase 3 family, member A1 | 1.78 | 1.81 | 2.12 |
| NM_170726.1 | ALDH4A1 | Aldehyde dehydrogenase 4 family, member A1 | 2.02 | 2.25 | 1.57 |
| NM_002211.2 | ITGB1 | Integrin, beta 1, transcript variant 1A | 2.43 (0.72) | 2.57 (1.73) | 2.41 (0.51) |
| NM_033668.1 | ITGB1 | Integrin, beta 1, transcript variant 1D | 2.20 | 2.16 | 2.22 |
| Oncogenes | |||||
| NM_005157.3 | ABL1 | v-abl Abelson murine leukemia viral oncogene | 2.24 | 2.37 | 1.82 |
| NM_031934.3 | RAB34 | RAB34, member RAS oncogene family | 3.41 | 3.37 | 3.63 |
| NM_012250.3 | RRAS2 | Related RAS viral (r-ras) oncogene homolog 2 | 1.48 | 1.32 | 1.43 |
| Drugable targets | |||||
| NM_004964.2 | HDAC1 | Histone deacetylase 1 | 2.23 | 1.91 | 1.53 |
| NM_006037.3 | HDAC4 | Histone deacetylase 4 | 2.28 | 2.23 | 1.95 |
| NM_001067.2 | TOP2A | Topoisomerase (DNA) II alpha 170 kDa | 5.56 | 5.67 | 6.03 |
| NM_006437.3 | PARP4 | Poly(ADP-ribose) polymerase family, member 4 | 2.40 | 2.27 | 2.50 |
| NM_031458.1 | PARP9 | Poly(ADP-ribose) polymerase family, member 9 | 2.10 | 1.40 | 1.68 |
| NM_022750.2 | PARP12 | Poly(ADP-ribose) polymerase family, member 12 | 2.05 | 1.44 | 1.61 |
| NM_017554.1 | PARP14 | Poly(ADP-ribose) polymerase family, member 14 | 2.16 | 2.29 | 2.49 |
| Differentiation marker | |||||
| NM_003380.2 | VIM | Vimentin | 5.54 | 5.31 | 5.12 |
| NM_003179.2 | SYP | Synaptophysin | −3.17 | −2.97 | −2.59 |
| NM_031845.2 | MAP2 | Microtubule-associated protein 2 | −1.83 | −1.99 | −2.44 |
| NM_002055.2 | GFAP | Glial fibrillary acidic protein | −1.41 | −4.95 | −4.97 |
aRatio of quantitative reverse transcriptase–polymerase chain reaction.
The expression levels of a selected group of genes were validated with quantitative real-time RT–PCR and IHC. As shown in Table 2, increased expressions were validated in 14 of the 16 (87.5%) genes selected, and decreased expression of 1 gene was also validated.
Detection and Isolation of CD133+ Tumor Stem Cells in the Orthotopic Xenograft Tumors
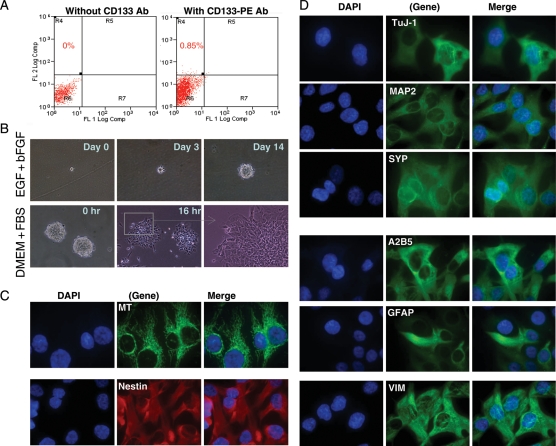
To determine whether cancer stem cells were maintained in the IC-1425EPN xenografts, we utilized antibodies against CD133, an established cell surface marker of both normal neuronal and cancer stem cells,14,46,47 including ependymomas,17 to detect and isolate CD133+ cells by fluorescence-activated cell sorting (FACS) in xenografts from passages I (n = 2), II (n = 1), and III (n = 2) as described previously.32 As shown in Fig. 3A, rare populations of CD133+ cells were present in passages I (0.25% and 0.46%), II (0.85%), and III (1.5% and 2.2%) xenografts. When these cells were incubated in stem cell growth medium, containing neurobasal medium, bFGF (50 ng/mL), EGF (40 ng/mL), N2, and B27 (Invitrogen), they formed neurospheres. Upon withdrawal of growth factors, the neurospheres attached to the poly-d-lysine–coated chamber glasses (Fig. 3B). From the cells that spread out, protein expression of neuronal markers of early (TuJ) and mature (MAP2 and SYP) stages, common glial precursor marker A2B5, as well as glial marker GFAP was detected. The majority of the cells expressed neuronal markers. The GFAP-positive cells, however, were mostly located in the inner circles of the attached “spheroid”, scattered in between cells with neuronal features. Strong expression of NES, a precursor cell marker expressed in ependymoma stem cells,17 was detected in nearly all the attached cells even after longer (3–5 days) incubations (Fig. 3C and D). The expression of RC2, the radial glial cell marker expressed in ependymoma stem cells,17 was also detected, albeit at low-to-medium levels.
Fig. 3.
CD133+ cells in IC-1425EPN xenograft tumors. (A) Representative graphs showing the isolation of CD133+ cells from an IC-1425EPN xenograft tumor (passage II) with FACS using PE-conjugated monoclonal antibodies against human CD133. (B) In vitro formation of neurosphere from isolated CD133+ cells in the presence of EGF and bFGF (upper panel), and induced differentiation of preformed neurosphere with FBS (lower panel). (C and D) Representative IMF images showing the expression of human-specific mitochondrial (MT) antigen, markers of neuroprogenitor (Nestin), early (TuJ-1), and mature (MAP2 and SYP) neurons, glial precursor (A2B5), astrocyte (GFAP), as well as intermediate filament VIM (×60).
Establishment of Permanent Cell Line BXD-1425EPN
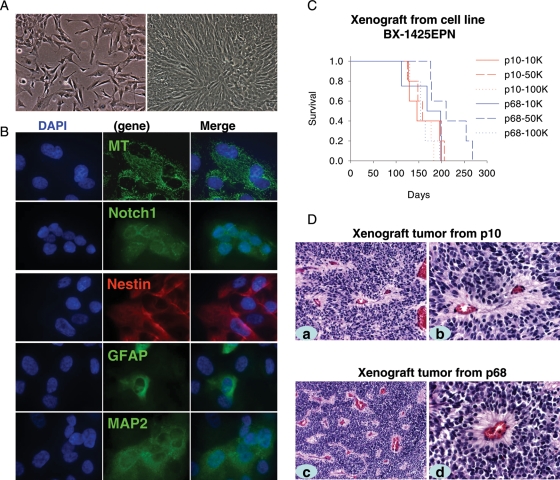
Since there are still no ependymoma cell lines established, we hypothesized that the xenograft tumor cells, having gone through a rigorous selection in vivo in mouse brains, may have better chances of surviving in vitro. We therefore incubated mechanically dissociated xenograft tumor cells from IC-1425EPN (passage II) with DMEM supplemented with 10% FBS. The xenograft cells attached overnight forming monolayer cells that displayed triangle or satellite-like in shape, which tend to pile up displaying a spindle-like morphology if allowed to grow into confluence (Fig. 4A). These cells have since been serially passaged every 3–4 days in vitro for more than 80 times, during which time they remained mycoplasma-free. Immunocytochemical staining at passages 40 and 75 showed that these cells expressed MT (100% cells), NES (100%), GFAP (20%), MAP2 (100%), as well as Notch1 (100%) (Fig. 4B), resembling the parent xenograft tumor. Chromosomal analysis with SKY identified the same structural abnormalities as well. This cell line is designated as BXD (Baylor xenograft derived)-1425EPN.
Fig. 4.
Characteristics of the ependymoma cell line BXD-1425EPN. (A) Morphology of cells with 50% (left panel) and 100% (right panel) confluence. (B) Representative images of IMF staining. Cells were processed for immunofluorescent labeling to detect the presence of MT, Notch1, Nestin, GFAP, and MAP2. Nuclei were counterstained with DAPI. (C) Log-rank analysis of animal survival times of the mice injected with different amount of cells, ranging from 1 × 104 (10K) to 1 × 105(100K), from passage 10 (p10) and passage 68 (p68) into right cereberum. (D) H&E staining of orthotopic xenograft tumors generated from passage 10 (p10) and passage 68 cells (a and c, ×20; b and d, ×40).
To determine whether the cultured cells can form tumors in mouse brains, we injected cultured cells (1 × 105, 5 × 105, and 1 × 106) from passages 10 and 68 into the right cerebrum of RAG2/SCID mice (n = 5 per group), respectively. Tumor formation was confirmed in all the mice. The survival times in mice receiving different numbers of tumor cells, however, were not significant (P = .75) as determined with log-rank analysis (Fig. 4C). Histopathological examination of these cell line-based xenografts on H&E-stained sections showed that they shared similar histological features as the original tumor and primary tumor-based orthotopic xenografts, including the presence of perivascular pseudorosettes (Fig. 4D).
Discussion
A major impediment to preclinical studies of ependymoma has been the lack of in vitro and in vivo models. We report here the creation and characterization of a clinically relevant orthotopic xenograft mouse model of human ependymoma (IC-1425EPN) and the establishment of a permanent cell line (BXD-1425EPN) from this model. Our xenograft model replicated the histopathological phenotypes and genomic profiles of the original tumor, and it also preserved the cancer stem cell pool during repeated in vivo subtransplantations. Critical to creating such a model is the direct orthotopic injection of tumor cells from a fresh surgical specimen, and strict subtransplantation of xenograft tumors in vivo in mouse brains, thereby maximizing the preservation of biological phenotypes of the original patient tumor.
Clinical relevance is important for animal models of human cancers.23 Using human-specific antibodies and chromosomal analysis, we showed that the xenograft tumor cells were all of human origin. Subsequent analysis confirmed the preservation of histopathological features of the original patient tumor, including pseudorosettes, a histological hallmark of ependymoma. In agreement with clinical observations, we showed that although the anaplastic ependymoma xenograft appeared to be well demarcated, infiltrating tumor cells were observed in neighboring brain parenchyma. We further documented that in our xenograft model, tumor cells either infiltrated as single cells into normal brain parenchyma or migrated along blood vessels. Since tumor cells infiltrating normal brain parenchyma may have a better chance of escaping surgical resection and become a source of tumor recurrence, further identification of cellular and molecular mechanisms governing the migration of these invading cells in our xenograft model should facilitate development of more effective therapies against ependymomas. Worthy of note is that this model was developed from a recurrent anaplastic ependymoma; treatment selection in favor of aggressive cells may have contributed to the increased tumor-forming capacity of the original tumor and introduced new biological changes as well.
Like in many other human cancers, cancer stem cells have also been identified in ependymomas.17 Since this small population of tumor cells are hypothesized to be essential for the initiation of most, if not all, human cancers and play important roles in drug resistance and tumor recurrence,52 it is important to determine whether cancer stem cells are preserved in animal models. In this study, we showed that CD133+ cells, although a small fraction (0.25%–2.2%), were maintained in IC-1425EPN xenografts during serial passaging. These cells were functionally active as well, forming neurospheres and displaying multilineage differentiation capabilities. We also found persistent expression of progenitor markers of neuronal (Nestin), radial glial (RC2), and common glial (A2B5) lineage, suggesting that the induced differentiation is incomplete, which is in agreement with the malignant nature of cancer stem cells.
A detailed analysis of gene expression profiles of xenograft tumors should provide critical information about the molecular fidelity of the xenograft tumors. No such studies, however, have been described in primary tumor-based orthotopic xenograft mouse models of human cancers. In this report, we showed the strong positive correlations of gene expression profiles between patient tumor and xenografts (r2 = 0.96 at passage I and 0.93 at passage III). This result combined with preservation of more than 80% of up- and down-regulated genes of patient tumor in the xenograft tumors provided molecular evidences to support that direct orthotopic engraftment of the human ependymoma tumor specimen is an effective technique to create clinically accurate animal models. However, additional studies of xenograft tumors and a spectrum of original patient brain tumors are needed to confirm the consistency of genomic profile preservation.
Gene expression profiling also allows identification of dysregulated genetic pathways, which is particularly important for the development of targeted therapies. Using normal human cerebral tissues as a control, we identified several key genetic abnormalities in both the patient tumor and xenograft tumors. Many of these deregulated genes have been previously described in human supratentorial ependymomas.18,43,44,53,54 These include the activation of NOTCH signaling, SHH pathway, as well as the WNT/Frizzles pathway. Therefore, our model provides a platform for preclinical testing of novel inhibitors of these pathways, acting alone or in combination, and functional studies of these complex pathways.
Our successful establishment of the ependymoma cell line BXD-1425 from the xenograft tumor IC-1425EPN provided an additional asset for ependymoma study. Since it grows very rapidly in vitro, expresses similar patterns of differentiation markers, maintains the identical chromosomal abnormalities as the parent xenografts, and is tumorigenic, this cell line should serve as a very useful tool for the initial screening of new compounds or preliminary functional study of candidate genes. Whether cell lines can be routinely established from primary tumor-based orthotopic xenograft mouse models, however, remains to be determined.
In summary, our study showed that direct orthotopic implantation of a fresh surgical specimen led to the development of a clinically relevant animal model of pediatric ependymoma, a tumor for which no permanent cell lines or genetically engineered animal models are available. We demonstrated that this IC-1425EPN model maintained key histopathological and growth characteristics of the original patient tumor, and it preserved the cancer stem cells during serial in vivo subtransplantations. More importantly, we confirmed that the xenograft tumors replicated key genetic abnormalities in the originating tumor and preserved its gene expression profile. Our technique provides a potential approach for creating a clinically accurate animal model for other brain tumors lacking established cell line or a relevant animal model.
Supplementary Material
Supplementary material is available at Neuro-Oncology Journal online.
Conflict of interest statement. None declared.
Funding
Cancer Fighters of Houston (X.-N.L), Clayton Foundation (X.-N.L).
Supplementary Material
References
- 1.Duncan JA, III, Hoffman HJ. Intracranial ependymomas. In: Kaye AH, Lows ERJ, editors. Brain Tumors. Edinburgh: Churchill Livingstone; 1995. pp. 493–504. [Google Scholar]
- 2.Kleihues P, Louis DN, Scheithauer BW, et al. The WHO classification of tumors of the nervous system. J Neuropathol Exp Neurol. 2002;61:215–225. doi: 10.1093/jnen/61.3.215. [DOI] [PubMed] [Google Scholar]
- 3.Ridley L, Rahman R, Brundler MA, et al. Multifactorial analysis of predictors of outcome in pediatric intracranial ependymoma. Neuro-Oncology. 2008;10:675–689. doi: 10.1215/15228517-2008-036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merchant TE, Mulhern RK, Krasin MJ, et al. Preliminary results from a phase II trial of conformal radiation therapy and evaluation of radiation-related CNS effects for pediatric patients with localized ependymoma. J Clin Oncol. 2004;22:3156–3162. doi: 10.1200/JCO.2004.11.142. [DOI] [PubMed] [Google Scholar]
- 5.Foreman NK, Love S, Thorne R. Intracranial ependymomas: analysis of prognostic factors in a population-based series. Pediatr Neurosurg. 1996;24:119–125. doi: 10.1159/000121027. [DOI] [PubMed] [Google Scholar]
- 6.Pollack IF, Gerszten PC, Martinez AJ, et al. Intracranial ependymomas of childhood: long-term outcome and prognostic factors. Neurosurgery. 1995;37:655–666. doi: 10.1227/00006123-199510000-00008. [DOI] [PubMed] [Google Scholar]
- 7.Ritter AM, Hess KR, McLendon RE, Langford LA. Ependymomas: MIB-1 proliferation index and survival. J Neurooncol. 1998;40:51–57. doi: 10.1023/a:1006082622699. [DOI] [PubMed] [Google Scholar]
- 8.Gatta G, Capocaccia R, Coleman MP, Ries LA, Berrino F. Childhood cancer survival in Europe and the United States. Cancer. 2002;95:1767–1772. doi: 10.1002/cncr.10833. [DOI] [PubMed] [Google Scholar]
- 9.Zacharoulis S, Levy A, Chi SN, et al. Outcome for young children newly diagnosed with ependymoma, treated with intensive induction chemotherapy followed by myeloablative chemotherapy and autologous stem cell rescue. Pediatr Blood Cancer. 2007;49:34–40. doi: 10.1002/pbc.20935. [DOI] [PubMed] [Google Scholar]
- 10.Bao S, Wu Q, Sathornsumetee S, et al. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 2006;66:7843–7848. doi: 10.1158/0008-5472.CAN-06-1010. [DOI] [PubMed] [Google Scholar]
- 11.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 12.Zeppernick F, Ahmadi R, Campos B, et al. Stem cell marker CD133 affects clinical outcome in glioma patients. Clin Cancer Res. 2008;14:123–129. doi: 10.1158/1078-0432.CCR-07-0932. [DOI] [PubMed] [Google Scholar]
- 13.Liu G, Yuan X, Zeng Z, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemmati HD, Nakano I, Lazareff JA, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 16.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 17.Taylor MD, Poppleton H, Fuller C, et al. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell. 2005;8:323–335. doi: 10.1016/j.ccr.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Poppleton H, Gilbertson RJ. Stem cells of ependymoma. Br J Cancer. 2007;96:6–10. doi: 10.1038/sj.bjc.6603519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jennings MT, Kaariainen IT, Gold L, Maciunas RJ, Commers PA. TGF beta 1 and TGF beta 2 are potential growth regulators for medulloblastomas, primitive neuroectodermal tumors, and ependymomas: evidence in support of an autocrine hypothesis. Hum Pathol. 1994;25:464–475. doi: 10.1016/0046-8177(94)90118-x. [DOI] [PubMed] [Google Scholar]
- 20.Brisson C, Lelong-Rebel I, Mottolese C, et al. Establishment of human tumoral ependymal cell lines and coculture with tubular-like human endothelial cells. Int J Oncol. 2002;21:775–785. [PubMed] [Google Scholar]
- 21.McLendon RE, Fung KM, Bentley RC, et al. Production and characterization of two ependymoma xenografts. J Neuropathol Exp Neurol. 1996;55:540–548. doi: 10.1097/00005072-199605000-00007. [DOI] [PubMed] [Google Scholar]
- 22.Horowitz ME, Parham DM, Douglass EC, Kun LE, Houghton JA, Houghton PJ. Development and characterization of human ependymoma xenograft HxBr5. Cancer Res. 1987;47:499–504. [PubMed] [Google Scholar]
- 23.Houghton PJ, Adamson PC, Blaney S, et al. Testing of new agents in childhood cancer preclinical models: meeting summary. Clin Cancer Res. 2002;8:3646–3657. [PubMed] [Google Scholar]
- 24.Gilbertson RJ, Gutmann DH. Tumorigenesis in the brain: location, location, location. Cancer Res. 2007;67:5579–5582. doi: 10.1158/0008-5472.CAN-07-0760. [DOI] [PubMed] [Google Scholar]
- 25.Gutmann DH, Maher EA, Van Dyke T. Mouse models of human cancers consortium workshop on nervous system tumors. Cancer Res. 2006;66:10–13. doi: 10.1158/0008-5472.CAN-05-3180. [DOI] [PubMed] [Google Scholar]
- 26.Gutmann DH, Hunter-Schaedle K, Shannon KM. Harnessing preclinical mouse models to inform human clinical cancer trials. J Clin Invest. 2006;116:847–852. doi: 10.1172/JCI28271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morton CL, Houghton PJ. Establishment of human tumor xenografts in immunodeficient mice. Nat Protoc. 2007;2:247–250. doi: 10.1038/nprot.2007.25. [DOI] [PubMed] [Google Scholar]
- 28.Suggitt M, Bibby MC. 50 years of preclinical anticancer drug screening: empirical to target-driven approaches. Clin Cancer Res. 2005;11:971–981. [PubMed] [Google Scholar]
- 29.Shu Q, Antalffy B, Su JM, et al. Valproic acid prolongs survival time of severe combined immunodeficient mice bearing intracerebellar orthotopic medulloblastoma xenografts. Clin Cancer Res. 2006;12:4687–4694. doi: 10.1158/1078-0432.CCR-05-2849. [DOI] [PubMed] [Google Scholar]
- 30.Hoffman RM. Orthotopic metastatic mouse models for anticancer drug discovery and evaluation: a bridge to the clinic. Invest New Drugs. 1999;17:343–359. doi: 10.1023/a:1006326203858. [DOI] [PubMed] [Google Scholar]
- 31.Hoffman RM. Orthotopic metastatic (MetaMouse) models for discovery and development of novel chemotherapy. Methods Mol Med. 2005;111:297–322. doi: 10.1385/1-59259-889-7:297. [DOI] [PubMed] [Google Scholar]
- 32.Shu Q, Wong KK, Su JM, et al. Direct orthotopic transplantation of fresh surgical specimen preserves cancer stem cell pool in clinically relevant mouse models of medulloblastoma and glioma. Stem Cells. 2008;26:1414–1424. doi: 10.1634/stemcells.2007-1009. [DOI] [PubMed] [Google Scholar]
- 33.Li XN, Shu Q, Su JM, Perlaky L, Blaney SM, Lau CC. Valproic acid induces growth arrest, apoptosis, and senescence in medulloblastomas by increasing histone hyperacetylation and regulating expression of p21Cip1, CDK4, and CMYC. Mol Cancer Ther. 2005;4:1912–1922. doi: 10.1158/1535-7163.MCT-05-0184. [DOI] [PubMed] [Google Scholar]
- 34.Li XN, Parikh S, Shu Q, et al. Phenylbutyrate and phenylacetate induce differentiation and inhibit proliferation of human medulloblastoma cells. Clin Cancer Res. 2004;10:1150–1159. doi: 10.1158/1078-0432.ccr-0747-3. [DOI] [PubMed] [Google Scholar]
- 35.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5 doi: 10.1186/gb-2004-5-10-r80. R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 37.Lattice: Lattice Graphics [computer program] 2008 [Google Scholar]
- 38.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 39.Geman D, d'Avignon C, Naiman DQ, Winslow RL. Classifying gene expression profiles from pairwise mRNA comparisons. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1071. Article19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siggenes: Multiple Testing using SAM and Efron's Empirical Bayes Approaches [computer program] 2008 [Google Scholar]
- 42.Gplots: Various R Programming Tools for Plotting Data [computer program] 2008 [Google Scholar]
- 43.Zangen I, Kneitz S, Monoranu CM, et al. Ependymoma gene expression profiles associated with histological subtype, proliferation, and patient survival. Acta Neuropathol (Berl). 2007;113:325–337. doi: 10.1007/s00401-006-0190-5. [DOI] [PubMed] [Google Scholar]
- 44.Modena P, Lualdi E, Facchinetti F, et al. Identification of tumor-specific molecular signatures in intracranial ependymoma and association with clinical characteristics. J Clin Oncol. 2006;24:5223–5233. doi: 10.1200/JCO.2006.06.3701. [DOI] [PubMed] [Google Scholar]
- 45.Korshunov A, Neben K, Wrobel G, et al. Gene expression patterns in ependymomas correlate with tumor location, grade, and patient age. Am J Pathol. 2003;163:1721–1727. doi: 10.1016/S0002-9440(10)63530-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 47.Singh SK, Clarke ID, Hide T, Dirks PB. Cancer stem cells in nervous system tumors. Oncogene. 2004;23:7267–7273. doi: 10.1038/sj.onc.1207946. [DOI] [PubMed] [Google Scholar]
- 48.Mellodew K, Suhr R, Uwanogho DA, et al. Nestin expression is lost in a neural stem cell line through a mechanism involving the proteasome and Notch signalling. Brain Res Dev Brain Res. 2004;151:13–23. doi: 10.1016/j.devbrainres.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 49.Hall PE, Lathia JD, Miller NG, Caldwell MA, ffrench-Constant C. Integrins are markers of human neural stem cells. Stem Cells. 2006;24:2078–2084. doi: 10.1634/stemcells.2005-0595. [DOI] [PubMed] [Google Scholar]
- 50.Corti S, Locatelli F, Papadimitriou D, Donadoni C, Del BR, Crimi M, et al. Transplanted ALDHhiSSClo neural stem cells generate motor neurons and delay disease progression of nmd mice, an animal model of SMARD1. Hum Mol Genet. 2006;15:167–187. doi: 10.1093/hmg/ddi446. [DOI] [PubMed] [Google Scholar]
- 51.Gilbertson RJ, Bentley L, Hernan R, et al. ERBB receptor signaling promotes ependymoma cell proliferation and represents a potential novel therapeutic target for this disease. Clin Cancer Res. 2002;8:3054–3064. [PubMed] [Google Scholar]
- 52.Clarke MF, Dick JE, Dirks PB, et al. Cancer stem cells—perspectives on current status and future directions: AACR workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 53.Nicolis SK. Cancer stem cells and “stemness” genes in neuro-oncology. Neurobiol Dis. 2007;25:217–229. doi: 10.1016/j.nbd.2006.08.022. [DOI] [PubMed] [Google Scholar]
- 54.Suarez-Merino B, Hubank M, Revesz T, et al. Microarray analysis of pediatric ependymoma identifies a cluster of 112 candidate genes including four transcripts at 22q12.1-q13.3. Neuro Oncol. 2005;7:20–31. doi: 10.1215/S1152851704000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.