Abstract
Aging is the primary risk factor of neurodegenerative disorders such as Alzheimer's disease (AD). However, the molecular events occurring during brain aging are extremely complex and still largely unknown. For a better understanding of these age-associated modifications, animal models as close as possible to humans are needed. We thus analyzed the transcriptome of the temporal cortex of the primate Microcebus murinus using human oligonucleotide microarrays (Affymetrix). Gene expression profiles were assessed in the temporal cortex of 6 young adults, 10 healthy old animals and 2 old, “AD-like” animals that presented ß-amyloid plaques and cortical atrophy, which are pathognomonic signs of AD in humans. Gene expression data of the 14,911 genes that were detected in at least 3 samples were analyzed. By SAM (significance analysis of microarrays), we identified 47 genes that discriminated young from healthy old and “AD-like” animals. These findings were confirmed by principal component analysis (PCA). ANOVA of the expression data from the three groups identified 695 genes (including the 47 genes previously identified by SAM and PCA) with significant changes of expression in old and “AD-like” in comparison to young animals. About one third of these genes showed similar changes of expression in healthy aging and in “AD-like” animals, whereas more than two thirds showed opposite changes in these two groups in comparison to young animals. Hierarchical clustering analysis of the 695 markers indicated that each group had distinct expression profiles which characterized each group, especially the “AD-like” group. Functional categorization showed that most of the genes that were up-regulated in healthy old animals and down-regulated in “AD-like” animals belonged to metabolic pathways, particularly protein synthesis. These data suggest the existence of compensatory mechanisms during physiological brain aging that disappear in “AD-like” animals. These results open the way to new exploration of physiological and “AD-like” aging in primates.
Introduction
In EU, life expectancy is about 83 years for women and 76 for men, and it is expected to further increase. This trend will have major economic and social impacts due to the prevalence of age-related diseases. “Normal” age-related cognitive changes have been traditionally ascribed to cell loss in the brain. However, experimental evidence is now accumulating suggesting that synaptic rearrangement, rather than cell loss, causes critical age-related brain changes [1]–[3]. Most elderly individuals experience no major, or very limited functional impairment thus highlighting the human brain ability to compensate for potential cognitive decline. In other words, although aging may be a cause of cognitive alteration and is the main risk factor for Alzheimer's disease (AD) [4], they are clearly two separated processes. AD is not accelerated aging and therefore it is important to be able to distinguish physiological from pathological aging. The discovery of genes, which are differentially expressed in normally aging and AD brains, could provide a tool to identify biomarkers [5] that can differentiate between physiological and pathological brain aging and which might help to identify new therapeutic targets.
Non-human primates constitute a valuable tool for studies on brain aging because of their lifespan and their closeness to humans. We propose to take advantage of the grey mouse lemur, Microcebus murinus, which is a pro-simian primate whose lifespan in captivity can reach 12–13 years. In grey mouse lemurs, the 50% survival point occurs at 60 months (i.e., at the age of 5 years) and allows defining the adult and elderly part of the population [6]. Animals older than 5 years of age show desynchronized biological rhythms, motor activity and sleep-wake cycles [6], [7]. Importantly, during aging, some lemurs develop, like humans, pathognomonic signs of AD such as presence of ß-amyloid plaques [8] with a prevalence of 5% (data unpublished), tau protein aggregation [9] and cerebral atrophy [10]. Animals with cerebral atrophy present the most severe neuropathology associated with a progressive and consistent pattern of neuronal-glial alterations [10]. Moreover, this animal model can be used to investigate cognitive deficits [11]–[14].
We thus analyzed the gene expression profiles in the temporal cortex of young adult, healthy elderly and old lemurs with ß-amyloid plaques (“AD-like” group) in order to identify genes involved in physiological and pathological aging. Since Microcebus murinus microarrays do not exist, we decided to use Affymetrix human genome chips since recent studies have illustrated the feasibility of detecting non-human primate brain transcripts using human genome chips [15]–[18]. We chose the temporal cortex because this region is connected to the hippocampus and to the frontal cortex, which are critical structures for learning and memory and are altered in AD. Analysis of the microarray data indicates that the temporal cortex has different gene expression profiles in young, old and “AD-like” animals and that several genes are differentially expressed in healthy aged and “AD-like” animals. Specifically, genes involved in metabolism, particularly in protein synthesis, were up-regulated during physiological brain aging, whereas in “AD-like” brains genes involved in protein synthesis and nuclear activity were often down-regulated.
Results
Histological characterization of young adult, elderly and “AD-like” brain in Microcebus murinus
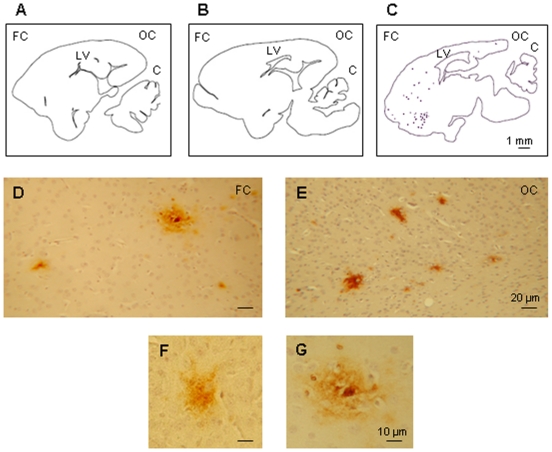
Before investigating the possible differences of gene expression in the temporal cortex of young adult, elderly and “AD-like” grey mouse lemurs, we compared the anatomical shape of the brains from the three groups (Fig. 1). We did not observe any difference in the thickness of the cerebral cortex and in the shape of the lateral ventricles between young (Fig. 1A) and healthy old animals (Fig. 1B). Conversely, in “AD-like” animals, we detected cortical atrophy, lateral ventricle dilatation (Fig. 1C) and ß-amyloid plaques (Fig. 1D,1E, 1F and 1G).
Figure 1. Brain sections and ß-amyloid plaques detection.
Brain sagittal sections, localized at L 2.80 and L 3.00 from the median line according to the stereotaxic atlas of Microcebus murinus [40]. The contour of the brain slice was drawn with the Mercator software (ExploraNova, La Rochelle, France). A. Young adult lemur (Nbr 986) B. Healthy old lemur (Nbr 43) C. AD-like old lemur (Nbr 896). Frontal cortex (FC), occipital cortex (OC), cerebellum (C), lateral ventricle (LV). Amyloid deposits representative of the two “AD-like” lemurs, in the frontal cortex (D) and the occipital cortex (E). Higher magnification showing labeling of ß-amyloid plaques (F and G).
Detection of transcripts with Affymetrix human genome chips
In order to assess the feasibility of using human genome arrays to study gene expression in the temporal cortex of Microcebus murinus, we first sequenced about 150 genes of the grey mouse lemur and compared them with their human homologues. They all presented a percentage of identity with their human counterparts that varied from 88% to 96% (data not shown). These results supported our choice of using the Affymetrix HG-U133 Plus 2.0 human microarray to investigate the pattern of gene expression in the temporal cortex of young adult, old and “AD-like” grey mouse lemurs (Table 1). The overall rate of genes with a “present” detection call (P) for each group was about 20%, with a slightly higher number of genes (9,456±1,378) detected in healthy aged animals, and a slightly lower number of genes (8,259±45) expressed in the “AD-like” group in comparison to the young adult group (8,914±1,482). Moreover, we noticed a lower SD in both “AD-like” animals, suggesting a better homogeneity of gene expression within this group. In human somatic tissues, which also included brain samples, the mean rate of genes with P call was 21278 (range: 7142–28389). Hence, although the percentage of genes detected by the HG-U133 Plus 2.0 microarray was lower in grey mouse lemur samples, it was still within the range of values observed in human samples.
Table 1. Genes detected in the temporal cortex of Microcebus murinus.
| Age (yr) | Males | Females | Number of detected genes | |
| Young | 1±0.1 | 2 | 4 | 8914.67±1482.68 |
| Old | 7.8±2.9 | 3 | 7 | 9456.20±1378.93 |
| AD-like | 10±3.3 | 0 | 2 | 8259.00±45.25 |
| All | 5 | 13 | 9142.67±1349.59 |
Transcripts detected in temporal cortex samples from young adult, old and “AD-like” animals by hybridization to the Affymetrix HG-U133 Plus 2.00 Array. The ages and the detected genes were expressed as mean ± SD.
Analysis of age-dependent gene expression changes in the temporal cortex
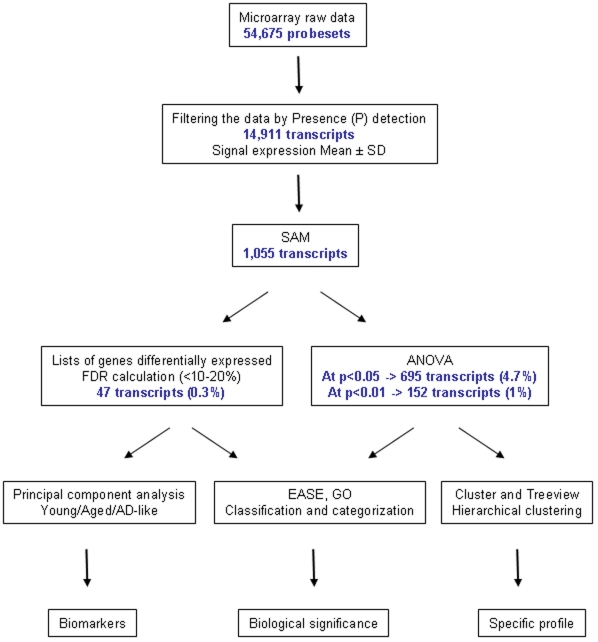
In order to characterize the expression profiles of adult, old and “AD-like” temporal cortex of Microcebus murinus, we first filtered the raw data (see Figure 2 for a schematic description for the analytical approach) and kept only the transcripts (n = 14,911) which had a “P” call in at least 3 samples. We then used the Significance Analysis of Microarrays (SAM) method to identify genes that were differentially expressed when comparing the three groups together or when assessing only healthy elderly and young or “AD-like” animals. The comparison of the three groups identified 1,055 transcripts with a false discovery rate (FDR) up to 45%. Since this risk of false positive was high, we assessed the significance of the changes in gene expression by ANOVA. The ANOVA analysis with a cut off at p<0.05 identified 695 genes with a significant gene expression change (i.e. 4.7% of expressed genes) and this number was reduced to 152 when the cut off was set at p<0.01 (i.e. 1% of the expressed genes) (Fig. 2 and Table S1).
Figure 2. Schematic representation of the different analyses.
Microarray data were filtered to detect the present transcripts (P). Then the filtered data were processed by SAM. ANOVA was performed with the (P) data and with the data sorted by SAM and the 47 P selected genes were analyzed by PCA. Finally, the 47 and the 695 genes were investigated by clustering and were classified by functional categories.
On the other hand, comparison of the gene expression data from the young and the old group by SAM sorted 44 transcripts (FDR about 10%, Table 2), which were significantly up-regulated (fold change between 1.37 and 3.82) in healthy aging animals (Table 3). These genes are mainly involved in protein synthesis, mitochondrial metabolism, and nuclear regulation. Conversely, comparison of the gene expression data from “AD-like” and healthy old animals indicated that 40 genes were strongly down-regulated (fold change between −1.6 and −3.3) in the “AD-like” group (Tables 4 and 5), with the exception of GK001 that was up-regulated. Among these genes, only a fraction plays a role in protein synthesis and nuclear regulation.
Table 2. Number of genes identified by SAM as differentially regulated in the temporal cortex of old animals.
| Delta | Significant genes | FDR% |
| 0.76 | 6 | 0.00 |
| 0.74 | 14 | 6.84 |
| 0.64 | 30 | 9.58 |
| 0.64 | 44 | 10.88 |
| 0.56 | 53 | 12.65 |
| 0.56 | 70 | 13.68 |
| 0.49 | 83 | 17.31 |
| 0.43 | 105 | 20.06 |
| 0.43 | 116 | 20.64 |
| 0.42 | 148 | 21.35 |
| 0.41 | 177 | 22.18 |
| 0.40 | 213 | 23.83 |
| 0.39 | 233 | 24.25 |
| 0.36 | 334 | 27.24 |
| 0.33 | 399 | 31.08 |
| 0.30 | 532 | 34.74 |
| 0.28 | 605 | 37.67 |
| 0.25 | 710 | 41.41 |
Delta table: number of genes with significant expression change in brain of aged lemurs detected by SAM analysis of expression data from temporal cortex samples of 10 healthy old lemurs and 6 young adults. The delta parameter is the threshold used to select genes that are significantly differentially expressed. The false discovery rate (FDR, expressed in %) is defined by the number of false positive genes divided by the number of genes identified as differentially expressed. Higher delta values lower the FDR but also reduce the number of selected genes. Delta table.
Table 3. Genes identified by SAM as differentially regulated in the temporal cortex of old animals.
| Probesets | Name | FC | p-value | Function |
| 205061_s_at | EXOSC9 | 1.77 | **** | Prot. Synthesis |
| 0200936_at | RPL8 | 1.58 | *** | Prot. Synthesis |
| 200817_x_at | RPS10 | 1.56 | *** | Prot. Synthesis |
| 226131_s_at | RPS16 | 1.59 | *** | Prot. Synthesis |
| 211487_x_at | RPS17 | 1.70 | *** | Prot. Synthesis |
| 212270_x_at | RPL17 | 1.54 | *** | Prot. Synthesis |
| 214003_x_at | RPS20 | 1.65 | **** | Prot. Synthesis |
| 200834_s_at | RPS21 | 1.65 | *** | Prot. Synthesis |
| 218830_at | RPL26L1 | 1.59 | ** | Prot. Synthesis |
| 219762_s_at | RPL36 | 1.85 | **** | Prot. Synthesis |
| 212863_x_at | CTBP1 | 1.47 | ** | Prot. Synthesis |
| 200757_s_at | CALU | 1.37 | **** | Prot. Maturation |
| 222578_s_at | UBE1DC1 | 1.58 | *** | Proteolysis |
| 201322_at | ATP5B | 1.38 | ** | Mitoch. Metabolism |
| 207507_s_at | ATP5G3 | 1.56 | * | Mitoch. Metabolism |
| 200818_at | ATP5O | 1.49 | ** | Mitoch. Metabolism |
| 201226_at | NDUFB8 | 1.63 | ** | Mitoch. Metabolism |
| 201227_s_at | NDUFB8 | 1.65 | *** | Mitoch. Metabolism |
| 35201_at | HNRPL | 1.34 | * | RNA related |
| 218117_at | RBX1 | 1.59 | *** | Nuclear factor |
| 226465_s_at | SON | 1.72 | *** | Nuclear factor |
| 204009_s_at | KRAS | 1.38 | *** | Epigenetic |
| 207721_x_at | HINT1 | 1.58 | ** | Epigenetic |
| 1555961_a_at | HINT1 | 1.68 | *** | Epigenetic |
| 224415_s_at | HINT2 | 1.47 | *** | Epigenetic |
| 222540_s_at | HBXAP | 1.38 | *** | Epigenetic |
| 204745_x_at | MT1G | 2.04 | *** | Cell cycle regulation |
| 206461_x_at | MT1H | 2.02 | *** | Cell cycle regulation |
| 218206_x_at | SCAND1 | 1.45 | ***** | Cell proliferation |
| 208864_s_at | TXN | 1.56 | *** | Cell proliferation |
| 205352_at | SERPINI1 | 1.50 | *** | Neurogenesis |
| 232520_s_at | NSFL1C | 1.48 | *** | Synapse |
| 202670_at | MAP2K1 | 1.59 | **** | Kinase |
| 226888_at | CSNK1G1 | 1.35 | ***** | Kinase |
| 217848_s_at | PP | 1.69 | ***** | Phosphatase |
| 205809_s_at | WASL | 1.80 | ** | Transduction |
| 219392_x_at | FLJ11029 | 1.36 | *** | Unknown |
| 229302_at | MGC33926 | 1.46 | ***** | Unknown |
| 230076_at | FLJ10156 | 1.47 | **** | Unknown |
| 225956_at | LOC153222 | 1.54 | * | Unknown |
| 218859_s_at | C20orf6 | 1.88 | ** | Unknown |
| 241993_x_at | — | 1.90 | ***** | Unknown |
| 218011_at | — | 1.57 | ***** | Unknown |
List of 44 transcripts detected with a delta value set at 0.64 and a FDR of about 10%. Genes were classified according to their function: metabolism, nuclear activity, neurotransmission–synaptic plasticity, signaling pathways and unknown functions. P-value of the Student's t-test: 0.05 (*); 0.01 (**); 0.05 (***); 0.001 (****); 0.0005 (*****); FC: fold change. The 6 genes detected with 0% FDR were noted in bold.
Table 4. Number of genes identified by SAM as differentially regulated in “AD-like” temporal cortex samples.
| Delta | Significant genes | FDR% |
| 0.46 | 2 | 0.00 |
| 0.38 | 27 | 17.42 |
| 0. 36 | 37 | 19.06 |
| 0.35 | 40 | 21.16 |
| 0.28 | 52 | 23.51 |
| 0.24 | 84 | 30.79 |
| 0.24 | 94 | 29.51 |
| 0.23 | 110 | 30.78 |
| 0.21 | 172 | 35.27 |
| 0.20 | 208 | 35.72 |
| 0.20 | 244 | 36.61 |
| 0.19 | 341 | 37.78 |
| 0.19 | 460 | 40.07 |
| 0.17 | 569 | 42.15 |
| 0.17 | 765 | 43.89 |
| 0.16 | 836 | 45.28 |
| 0.15 | 968 | 48.92 |
| 0.14 | 1073 | 51.27 |
Delta table: number of genes with significant expression changes in aged animals detected by SAM analysis of expression data from temporal cortex samples from 2 “AD-like” lemurs and 10 healthy old animals.
Table 5. Genes identified by SAM as differentially regulated in “AD-like” temporal cortex samples.
| Probesets | Name | FC | p-value | Function |
| 211487_x_at | RPS17 | −2.07 | *** | Prot. synthesis |
| 200834_s_at | RPS21 | −1.60 | **** | Prot. synthesis |
| 225672_at | GOLGA2 | −1.88 | * | Prot. maturation |
| 38710_at | OTUB1 | −1.51 | * | Proteolysis |
| 35201_at | HNRPL | −1.61 | ****** | RNA related |
| 203752_s_at | JUND | −1.86 | * | Transcription |
| 227772_at | LATS1 | −1.98 | * | Transcription |
| 200055_at | TAF10 | −1.90 | *** | RNA related |
| 209431_s_at | ZNF278 | −2.53 | ****** | Nuclear factor |
| 218977_s_at | SECP43 | −2.71 | ****** | Nuclear factor |
| 48580_at | CXXC1 | −2.84 | * | Nuclear factor |
| 223132_s_at | TRIM8 | −1.74 | * | Nuclear factor |
| 208676_s_at | PA2G4 | −1.90 | * | Cell cycle regulation |
| 218034_at | TTC11 | −1.65 | ** | Apoptosis |
| 219888_at | SPAG4 | −3.34 | ** | Cytoskeleton |
| 203069_at | SV2A | −1.64 | ** | Synapses |
| 225058_at | GPR108 | −2.23 | ** | Synapses |
| 209982_s_at | NRXN2 | −2.59 | *** | Synapses |
| 229309_at | ADRB1 | −1.79 | * | Synapses |
| 206397_x_at | GDF1 | −2.01 | * | Growth factor |
| 210185_at | CACNB1 | −1.70 | * | Ion channel |
| 222432_s_at | GK001 | 2.65 | ****** | Signal. Transd. |
| 213108_at | CAMK2A | −1.65 | ****** | Kinase |
| 215903_s_at | MAST2 | −2.22 | ***** | Kinase |
| 228302_x_at | CaMKIINalpha | −3.09 | *** | Kinase |
| 209945_s_at | GSK3B | −1.97 | * | Kinase |
| 200822_x_at | TPI1 | −1.87 | *** | Phosphate metabol. |
| 33197_3_at | GAPD | −1.33 | * | Phosphate metabol. |
| 225519_at | PPP4R2 | −2.08 | ****** | Phosphatase |
| 227325_at | LOC255783 | −5.11 | ****** | Unknown |
| 228935_at | LOC283400 | −5.20 | ****** | Unknown |
| 210408_s_at | CPNE6 | −2.06 | ****** | Unknown |
| 217033_x_at | — | −1.68 | ***** | Unknown |
| 1554429_a_at | DMWD | −1.87 | **** | Unknown |
| 219392_x_at | FLJ11029 | −1.79 | **** | Unknown |
| 48106_at | — | −2.01 | *** | Unknown |
| 219910_at | HYPE | −1.57 | * | Unknown |
| 207435_s_at | SRRM2 | −1.97 | * | Unknown |
| 218089_at | C20orf4 | −1.92 | * | Unknown |
| 218429_s_at | FLJ11286 | −1.72 | * | Unknown |
List of the 40 transcripts sorted using a delta set at 0.35 and a FDR of 21%. All were down-regulated in AD, but for GK001. Genes were classified according to their function: metabolism, nuclear activity, neurotransmission–synaptic plasticity, signaling pathways and unknown functions. FC: fold change; P value: 0.05 (*); 0.01 (**); 0.05 (***); 0.001 (****); 0.0005 (*****). The 2 genes detected with 0% FDR were noted in bold.
When for the comparison of the three groups a more stringent delta cutoff was chosen (from 0.058 to 0.03) the number of genes identified as differentially expressed in the three groups was reduced to 47 and among them were included the 28 genes that had been already sorted by the two SAM analyses using the data from young and healthy old animals or healthy old and both “AD-like” animals (Tables 6 and 7).
Table 6. SAM analysis of the expression data from the three groups.
| Probesets | Name | FCA/Y | p-value | FCAD/Y | p-value | FCAD/A | p- value | Functions | |
| A | 226131_s_at | RPS16 | 1.59 | *** | 1.19 | NS | −1.34 | * | prot synthesis |
| 214003_x_at | RPS20 | 1.65 | **** | 1.03 | NS | −1.60 | *** | prot synthesis | |
| 218830_at | RPL26L1 | 1.59 | *** | 1.26 | * | −1.26 | * | prot synthesis | |
| 219762_s_at | RPL36 | 1.85 | **** | −1.02 | NS | −1.88 | * | prot synthesis | |
| 200757_s_at | CALU | 1.37 | ***** | −1.07 | * | −1.47 | ***** | prot maturation | |
| 201227_s_at | NDUFB8 | 1.65 | **** | 1.20 | ** | −1.37 | **** | mitoch metabol. | |
| 218206_x_at | SCAND1 | 1.45 | ***** | −1.04 | NS | −1.51 | *** | cell prolif. | |
| 207721_x_at | HINT1 | 1.58 | *** | 1.13 | NS | −1.40 | NS | Epigenetic | |
| 224415_s_at | HINT2 | 1.47 | *** | −1.04 | NS | −1.53 | ***** | Epigenetic | |
| 222540_s_at | HBXAP | 1.38 | *** | 1.09 | NS | −1.27 | ** | Epigenetic | |
| 204009_s_at | KRAS | 1.38 | *** | 1.04 | NS | −1.32 | * | Epigenetic | |
| 202670_at | MAP2K1 | 1.59 | ***** | 1.21 | NS | −1.32 | NS | Kinase | |
| 217848_s_at | PP | 1.69 | ***** | 1.34 | *** | −1.26 | * | Phosphatase | |
| 230076_at | FLJ10156 | 1.47 | ***** | 1.10 | NS | −1.34 | *** | Unknown | |
| 241993_x_at | — | 1.90 | **** | 1.35 | NS | −1.41 | NS | Unknown | |
| 218011_at | — | 1.57 | ***** | 1.09 | NS | −1.45 | NS | Unknown | |
| B | 223132_s_at | TRIM8 | −1.06 | NS | −1.85 | * | −1.74 | * | nuclear fact. |
| 222432_s_at | GK001 | −1.19 | NS | 2.23 | ****** | 2.65 | ***** | signal transd. | |
| 213108_at | CAMK2A | 1.42 | * | −1.16 | **** | −1.65 | ***** | Kinase | |
| 209945_s_at | GSK3B | 1.43 | * | −1.38 | NS | −1.97 | * | Kinase | |
| C | 211487_x_at | RPS17 | 1.70 | **** | −1.22 | NS | −2.07 | *** | prot synthesis |
| 200834_s_at | RPS21 | 1.65 | **** | −1.03 | NS | −1.60 | **** | prot synthesis | |
| 35201_at | HNRPL | 1.34 | * | −1.20 | * | −1.61 | ***** | RNA related | |
| 219392_x_at | FLJ11029 | 1.36 | *** | −1.32 | NS | −1.79 | **** | Unknown |
Genes with significant expression changes identified when data from the cortex samples of young (Y), healthy aged (A) and “AD-like” (AD) were compared. FC: Fold Change. These genes have been already sorted as regulated by age (see Table 3); B: genes already identified as regulated by AD (Table 5); C: genes regulated by age and by AD (Table 3 and 5).
Table 7. SAM analysis of the expression data from the three groups.
| Probesets | Name | FCA/Y | p-value | FCAD/Y | p-value | FCAD/A | p- value | Functions |
| 200029_at | RPL19 | 1.69 | *** | 1.07 | NS | −1.58 | NS | prot synthesis |
| 222991_s_at | UBQLN1 | −1.27 | NS | 1.61 | ** | 2.04 | *** | Proteolysis |
| 204528_s_at | NAP1L1 | 1.09 | NS | 2.15 | ****** | 2.34 | ***** | RNA related |
| 213470_s_at | HNRPH1 | −1.52 | *** | −1.45 | * | 1.05 | NS | RNA related |
| 204847_at | ZBTB11 | 1.34 | ** | 2.17 | ****** | 1.62 | ***** | nuclear fact. |
| 203138_at | HAT1 | 1.17 | NS | 2.14 | ** | 1.83 | ** | Epigenetic |
| 203845_at | PCAF | 1.13 | NS | 1.93 | NS | 1.71 | NS | Epigenetic |
| 202260_s_at | STXBP1 | 1.08 | NS | 1.50 | ****** | 1.40 | ***** | Synapse |
| 214441_at | STX6 | −1.67 | ***** | −1.50 | ** | 1.12 | NS | Synapse |
| 232520_s_at | NSFL1C | 1.48 | **** | 1.20 | ** | −1.23 | * | Synapse |
| 226683_at | SNAG1 | 1.01 | NS | 1.51 | ****** | 1.50 | ***** | Synapse |
| 205352_at | SERPINI1 | 1.50 | *** | 1.64 | * | 1.09 | NS | Neurogenes. |
| 222960_at | CACNA1H | −2.04 | ****** | −1.81 | * | 1.13 | NS | Ion channel |
| 223208_at | KCTD10 | −1.86 | *** | 1.52 | ****** | 2.83 | ***** | Ion channel |
| 211302_s_at | PDE4B | −2.29 | *** | −4.68 | *** | −2.05 | NS | Phosphatase |
| 1563674_at | SPAP1 | 1.68 | NS | 4.78 | *** | 2.84 | ** | Phosphatase |
| 218021_at | DHRS4 | −1.49 | ***** | 1.10 | NS | 1.64 | ** | Dehydrogen |
| 229302_at | MGC33926 | 1.46 | ****** | 1.28 | *** | −1.14 | * | Unknown |
| 65635_at | FLJ21865 | −1.30 | *** | −1.43 | *** | −1.10 | NS | Unknown |
| 215672_s_at | KIAA0828 | −1.72 | ****** | −1.66 | NS | 1.04 | NS | Unknown |
| 48106_at | — | −1.18 | NS | −2.37 | **** | −2.01 | *** | Unknown |
| 240835_at | — | 1.16 | NS | 2.26 | ****** | 1.95 | ***** | Unknown |
| 1553705_a_at | — | −1.50 | *** | −1.07 | NS | 1.41 | *** | Unknown |
Genes with significant expression changes identified when data from the cortex samples of young (Y), healthy aged (A) and “AD-like” (AD) were compared. FC: Fold Change. These genes have been identified by SAM using the data from the three groups.
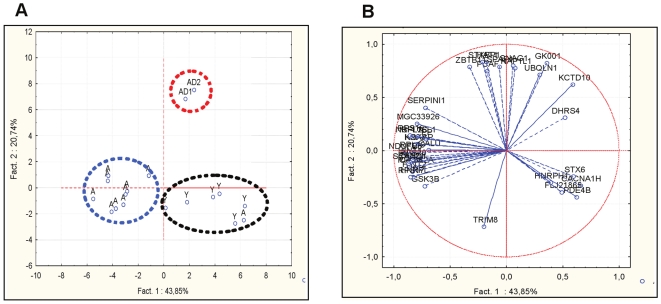
Principal component analysis of the 47 genes, which were identified as differentially expressed in the three groups by SAM, confirmed that they could clearly separate young, healthy old and “AD-like” animals (Fig. 3). The first component represented the gene expression signature with a score of 64.59% of the variance in the three groups. Healthy aged animals could be differentiated from the young ones with a score of 43.85% (Fig. 3A, axis 1), and the “AD-like” group could be distinguished from the healthy old group with a score of 20.74% (Fig. 3A, axis 2). Genes with a high correlation coefficient were considered to be important for discriminating the three groups (Fig. 3B). Genes located to the left of axis 1 characterized the group of healthy aged animals. Most of them are involved in protein synthesis (RPS16, 17, 19, 20, 21, RPL26L1 and 36), mitochondrial metabolism (NADH dehydrogenase complex, NDUFB8) or in the regulation of transcription (SCAND1, HNRPL, HINT1-2, KRAS). Genes located at the top of axis 2 characterized the “AD-like” group. Some of these genes play a role in neurotransmission (STXBP1), intracellular trafficking (SNAG1), regulation of transcription (ZBTB11) or in epigenetic processes (HAT1). However, also genes, which are not yet reported as having brain-related functions, such as SPAP1, which is expressed in mature lymphocyte B cells, contributed to delineate the “AD-like” group. Calumenin (CALU), a protein which is involved in the regulation of vitamin K-dependent carboxylation of multiple amino-terminal glutamate residues and normally is present at very low levels in the brain, also was up-regulated in the 2 “AD-like” animals. Others encode proteins that are detected in neurofibrillary tangles, such as Ubiquilin-1 (UBQLN1), which promotes the accumulation of uncleaved PSEN1 and PSEN2 that are involved in AD pathology.
Figure 3. Principal component analysis of the 47 transcripts that are differentially regulated in aging or “AD-like” brain detected by SAM.
A. Projection of the individuals shows that healthy aged animals can be discriminated from the young ones (axis 1), and “AD-like” can be differentiated from healthy old animals (axis 2). B. Projection of the genes. Genes located to the left of axis 1 characterize healthy aged animals, whereas genes located at the top of axis 2 characterize the “AD-like” group. Young (Y), aged (A) and “AD-like” (AD) animals.
The 47 genes sorted by SAM were included in the 695 genes identified by ANOVA as differentially expressed in the three groups. Among these 695 genes, the expression of 350 was modified by age, the expression of 427 by the “AD-like” pathology (Table 8) and some by both. Among the 350 genes modified by age, 209 were up-regulated and 141 down-regulated, and among the 427 genes modified by the pathology, 250 were up-regulated and 177 down-regulated. Moreover, 28% of the genes that were differentially regulated in “AD-like” samples showed a similar change in expression also in healthy old animals, whereas 72% showed an opposite change of expression in comparison to the healthy old group. We also analyzed the expression data by taking into account only the females. In this case the number of transcripts with significant changes was higher: 1,079 instead of 695 because of less variance due to a better homogeneity of the populations.
Table 8. Genes that are differentially regulated during physiological or pathological aging sorted by ANOVA.
| Comparison | Young vs Old | AD-like vs Old |
| Up-regulation | 209 | 250 |
| Down-regulation | 141 | 177 |
| All | 350 | 427 |
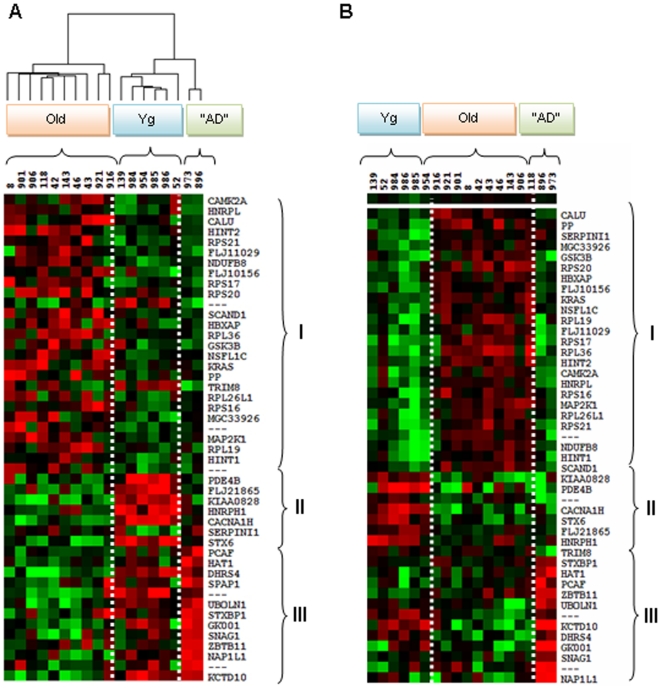
Finally, hierarchical clustering indicated that each group had a distinct profile whatever the statistical test (SAM (Fig. 4) or ANOVA (Fig. 5)) used to sort and select the genes and that each profile was homogenous within the specific group, particularly in the “AD-like” animals. Hierarchical clustering identified clusters of genes that were down-regulated in old and up-regulated in “AD-like” animals and genes, which were up-regulated in young adults and down-regulated in old and “AD-like” animals, in good agreement with the PCA data and also with the ANOVA analysis that showed opposite changes (Fig. 4II; 5II) and similar changes (Fig. 4 I; 5III) in gene expression.
Figure 4. Transcriptional profiles in the temporal cortex of Microcebus murinus.
Hierarchical clustering obtained with the 47 genes sorted by SAM. The transcriptional profiles of the temporal cortex of the 18 Microcebus murinus (i.e., 6 young adults (Yg), 10 old lemurs (Old) and 2 “AD-like” (AD)) showed three distinct profiles for the 3 groups. Each profile could be separated in three distinct regions which were conserved in the three groups. (A) Dendogram and clustering showing that the animals were clustered by age or by pathology. (B) Hierarchical clustering of lemurs according to age and pathology. The clusters show that the expression of some genes in “AD-like” lemurs is similar to that in young animals. The profiles of healthy elderly animals are drastically different from those of the two “AD-like” lemurs. In the three different parts: I- genes that are down-regulated in aging and in “AD-like” animals; II- genes that are up-regulated with young adults but are down-regulated in “AD-like” animals; III- genes that are down-regulated in old and up-regulated in “AD-like” animals. Red: over-expressed genes; green: down-regulated genes; black: genes without expression changes.
Figure 5. Transcriptional profiles in the temporal cortex of Microcebus murinus.
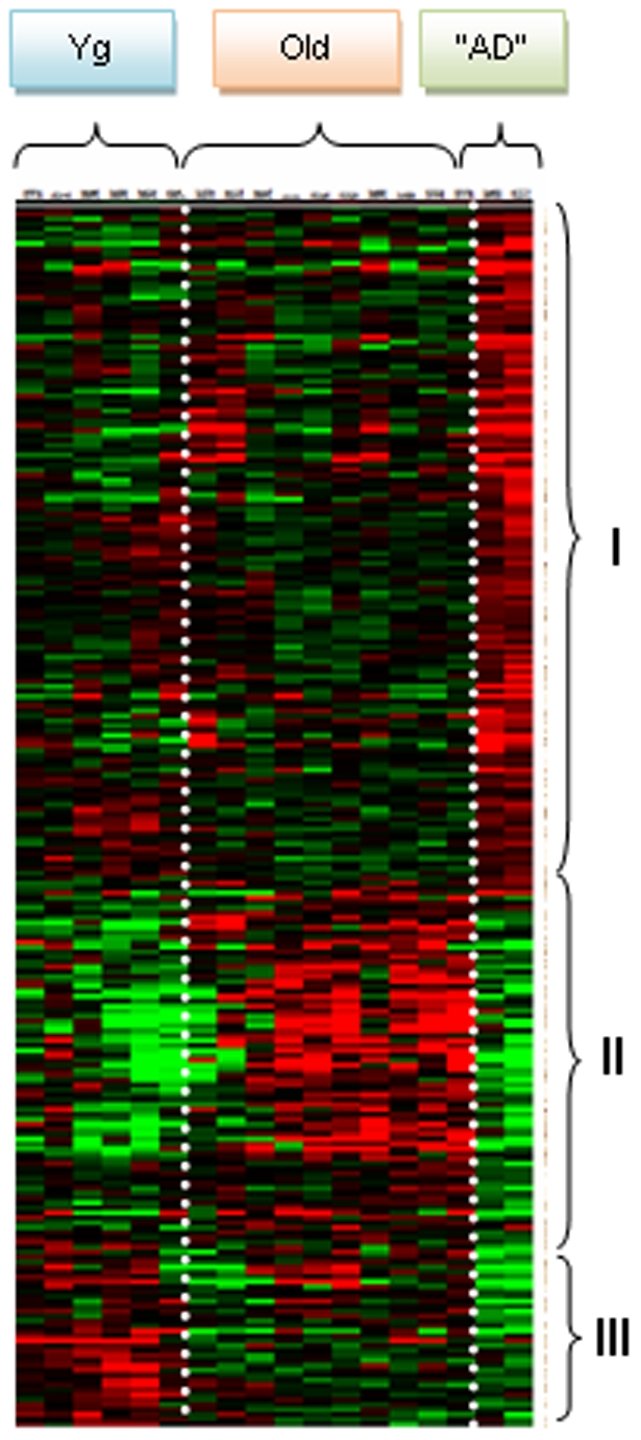
Hierarchical clustering obtained with the 695 genes sorted by ANOVA. The transcriptional profiles of the 18 Microcebus murinus (i.e., 6 young adults (Yg), 10 old lemurs (Old) and 2 “AD-like” (AD)) showed 3 distinct regions: I- genes that are up-regulated in “AD-like” temporal cortex; II- genes that are down-regulated in “AD-like” temporal cortex and preferentially up-regulated in aged cortex; III- genes that are down-regulated in “AD-like” and preferentially up-regulated in young adult cortex. In red are shown genes that are over-expressed, in green genes that are under-expressed and in black genes without expression changes.
Functional categorization and sexually dimorphic changes in aged Microcebus murinus males and females
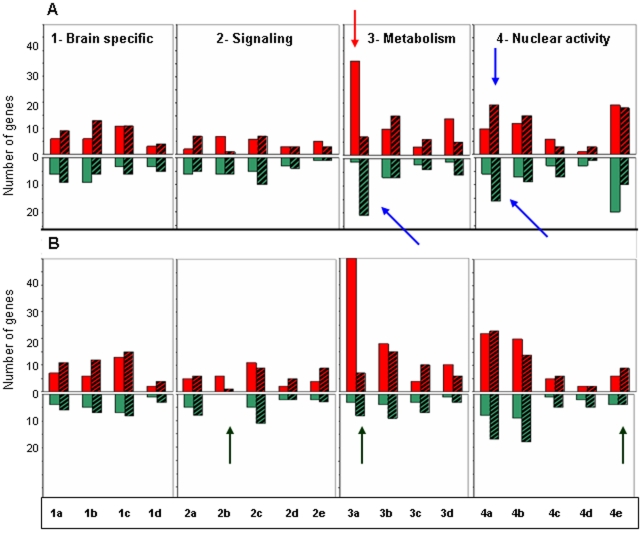
By assessing the relative changes in expression within gene ontology categories, we could classify the differentially expressed genes according to their cellular functions and distribute them in 4 main modules: 1- Brain plasticity; 2- Signaling pathways; 3- Metabolism and protein synthesis; 4- Nuclear activity (Fig. 6 and Table S2_gene functional categorization). Most of the genes that were up-regulated with age and down-regulated in “AD-like” animals (especially those involved in protein synthesis) belonged to the “metabolism and protein synthesis” module (Fig. 6A, module 3a). Nuclear factors (Fig. 6A, module 4a) were differentially expressed particularly in the “AD-like” group. Analysis of the data from the 2 females only showed that the general profiles were maintained when compared with the data from all animals (Fig. 6B). Nevertheless, the number of genes that were up-regulated with age was higher, whereas the number of down-regulated genes involved in signaling transduction (Fig. 6B, module 2b), metabolism (Fig. 6B, module 3a) and in epigenetic regulations (Fig. 6B, module 4e) was lower.
Figure 6. Gene changes in relation to their cellular function identified by gene ontology.
The 695 genes sorted by ANOVA were classified into four main modules: 1- genes involved in brain plasticity [1a-neurotransmission, 1b-neurogenesis, 1c-adhesion and extracellular matrix, 1d-cytoskeleton]; 2- genes involved in transduction and signaling [2a-ion channels, 2b-kinases, 2c-phosphatases, 2d-transferases and 2e-growth factors]; 3- genes involved in metabolism and catabolism [3a-protein synthesis and maturation, 3b-proteolysis, 3c-glucidic and lipid metabolism and 3d-mitochondrial metabolism]; 4- genes involved in nuclear activity [4a-nuclear factors, 4b-transcription regulation, 4c-cell cycle regulation, 4d-apoptosis and 4e- epigenetic control]. Red bars represent genes that are up-regulated and the green bars genes that are down-regulated in the temporal cortex of aging animals; hatched red bars represent genes that are up-regulated and hatched green bars genes that are down-regulated in the temporal cortex of “AD-like” lemurs. A. Classification with males and females together. B. Classification with females only. In aging animals genes involved in protein synthesis were more frequently up-regulated (red arrow), and the most important differences between aging and “AD-like” profiles concerned genes that have a role in protein synthesis and nuclear activity (blue arrows).The major differences between A and B are indicated by black arrows.
Discussion
Microcebus murinus is a useful primate model of cerebral aging and AD. We already showed that, not only they display pathognomonic lesions for AD and as observed in humans, both lemurs with AD-like pathology presented cortical atrophy. Recent studies have shown that brain measurement by MRI could be an important biomarker of degeneration like in the case of AD [19], [20]. They also represent a tool for anti-amyloid vaccination studies [21], MRI studies [10] and for developing cognitive tests [12], [14]. Therefore, to further develop the potentialities of this model, we wanted to identify in the temporal cortex of Microcebus murinus modifications in gene expression that were associated with physiological or pathological aging. Using human genome microarrays the overall rate of gene detection was about 20%. This value could appear to be rather low, but it has to be compared with other heterologous (Rhesus monkey with human microarrays) or homologous (human with human microarray) studies where only 30–35% and 30–40% of genes were detected ([18]; JDV, personal communication). Our results also confirm earlier works [15]–[18] that human chips can be used in non-human primates.
Among more than 14,000 genes, we have identified 695 genes (around 4.9%) with significant expression changes in young, aged and “AD-like” animals. Analysis of the cortex transcriptome from human brain samples at different ages indicates that the expression of about 400 genes among 11,000 (around 3.6%) is significantly changed [22]. Our results are in the same order of magnitude.
We then compared the list of genes that changed in the human frontal cortex sorted by Lu et coll. (2004) with the genes changing in the lemurian temporal cortex. We found 13 common genes (out of 150) and also 58 (out of 150) that belong to the same family and have similar biological function (e.g. VAMP1/VAMP2 or Synapsin II and synapsin IIb). Most of the genes that are differentially expressed in both works appeared to be involved in the synaptic transmission (8/15), Ca2+ homeostasis (5/10), cAMP signalling (2/2)) and in transcription (6/8). In the study by Lu and coll. the expression of these genes was modified by age, whereas in our study, they are modified by age and/or by pathology.
The same applies to the fold changes that varied between 1.5 and 3. Moreover, analysis of the microarray data indicates that each group of animals (young adult, healthy old, and “AD-like”) has a distinctive gene expression profile in the temporal cortex, a structure involved in integrative functions such as memory. Previous studies have already concluded that the expression of many genes is modified during aging (reviewed in [2], [23], [24]. Specific profiles in relation with age were observed in human brain [22], [25] and in Rhesus monkey brain, particularly in the corpus callosum [18] and in the frontal cortex [25]. Interestingly, each brain region of adult or healthy elderly subjects had a distinct gene expression profile [26]–[28]. Altered gene expression was also observed between healthy elderly individuals and AD patients, for review [29], particularly in the hippocampus [30], [31] and in neurons of several brain structures [27].
Our results support the hypothesis that the events occurring during physiological brain aging and AD development are distinct. However, an important issue is to understand the transition from physiological to pathological brain aging which leads to neurodegenerative disorders. We observed that many of the genes up-regulated in healthy aged animals are involved in protein synthesis such as the genes coding for ribosomal proteins. These results support the idea that physiological aging, at least in the brain, is associated with permanent compensatory effects via increased protein synthesis. Conversely, these genes were down-regulated in the 2 “AD-like” animals, indicating a failure to compensate a decline in the metabolic activity in the brain of animals with amyloid deposits. Moreover, the expression of UBQLN1, an ubiquitin-like protein that plays a role in the regulation of the degradation of proteins by proteasome [32], was significantly decreased in both “AD-like” animals. This observation is important as accumulation of misfolded proteins, which are not degraded by the proteasome, may be involved in the development of neurodegenerative diseases. The expression of other genes that are related to the regulation of neural plasticity, an important process involved in brain aging [3], [33], also was changed in “AD-like” animals in comparison to healthy old ones. Particularly, GRIN1, the NR1 subunit of the NMDA glutamate receptor, and GRIA1, a subunit of the AMPA glutamate receptor, which activate pathways involved in the induction of long-term potentiation (LTP) were markedly down-regulated. AD-related pathologic processes lead to synaptic loss and dysfunction, particularly of glutamatergic transmission [34], [35] that can cause inhibition of LTP [36] as it has been shown also in rodents following acute treatment with Aβ oligomers [37]. Moreover, recent data have also demonstrated that changes in the expression of nuclear proteins, which induce epigenetic modifications of chromatin, play an important role in regulating synaptic plasticity and memory processes [38]. In our study, we observed that several genes implicated in epigenetic processes and transcription, such as histone acetyltransferases (HAT1, PCAF), histidine triad nucleotide binding proteins (HINT1, HINT2) and the cAMP responsive element binding protein (CREB1), were down-regulated during physiological and pathological brain aging. Conversely, KRAS and HBXAP, which mediates nucleosome assembly and chromatin remodeling, were up-regulated in healthy aged Microcebus murinus. Finally, the expression of CLU (APOJ, Clusterin), which is considered as a risk factor for AD [39], [40] was significantly increased in both “AD-like” animals.
Our data also indicate that changes in gene profiles during aging in Microcebus murinus temporal cortex might show gender-specificity, suggesting that brain continues to present sexual differences during the entire life. This observation needs, however, to be confirmed since both “AD-like” animals were females. Nevertheless, sexual dimorphism in gene expression in aging human brain has been reported by Berchtold et al. [28] and in primate brain by Reinius et al. [41], whereas Lu and colleagues [22] did not evidence significant gender differences in the transcriptomic profiles of the human frontal cortex.
Overall, our expression data support the relevance of Microcebus murinus as a valuable non-human primate model for studying molecular brain changes in relation with age and neurodegenerative diseases such as AD.
Materials and Methods
Animals
The Microcebus murinus animals were all born in captivity in our breeding colony (CECEMA, University of Montpellier 2). They were divided into three groups: the first (young animals) included six 1 year/old adults (4 females and 2 males), the second (healthy old animals) included ten 8 year/old animals (7 females and 3 males), and the third (“AD-like” animals) two old females with cerebral ß-amyloid plaques. About 5% of old lemurs in our colony develop ß-amyloid plaques in the brain (unpublished data). Animals were euthanized with 150 mg/Kg ketamine. Animal care was in accordance with institutional guidelines and the animal protocol was approved by the Ethic Committee, Comité Régional d'Ethique pour l'Expérimentation Animale - Languedoc Roussillon (authorization #CE-LR-0816, 2008) in accordance with the European Community Council directives of November 24, 1986.
Brain samples preparation
After euthanasia, each brain was cut in half along the midline. The right hemisphere was used for the microarray experiments and the left one for immunohistological experiments. For transcriptional analysis, the right hemisphere was dissected and cortex samples, weighing 25–35 mg, were flash-frozen and stored at −80°C until use. The left hemisphere was fixed in paraformaldehyde and embedded in paraffin as previously described [8], [14] and 6 µm serial sagittal sections were prepared.
Immunohistochemistry
Presence of amyloid-ß-42 (Aß42) peptide aggregates was assessed using the rabbit polyclonal FCA3542 antibody (Calbiochem, Merck-bio, Germany). Sections were deparaffinized and hydrated through ethanol gradient. After formic acid pretreatment, sections were pre-incubated with 10% goat serum in Tris-buffered saline for 30 min. Then, sections were incubated with anti-Aß42 antibodies (1∶1000) at 4°C overnight. Immunological complexes were detected with biotinylated immunoglobulins (1∶1000) and horseradish peroxidase-labelled avidin (Vector Laboratories, Burlingame, VT). Immunoreactivity was revealed with 0.005% diaminobenzidine tetrahydrochloride (DAB, 0.35 mg/mL, Sigma, St Louis, MO). The primary antibody was omitted in the negative control and sections from Microcebus murinus brain specimens known to contain ß-amyloid plaques were used as positive controls. The Microcebus murinus brain atlas was used to localize the brain area of each section [42].
RNA isolation and Affymetrix GeneChip processing
Total RNA was extracted from each temporal cortex sample (25–35 mg) using the RNeasy mini kit (Qiagen, Santa Clarina, CA) according to the manufacturer's protocol. The quality of each purified RNA sample was checked with a NanoDrop Agilent BioAnalyzer (Agilent Technologies, Santa Clara, CA). An average of 2 µg of total RNA from each cortex sample was prepared for hybridization with Affymetrix HG-U133 Plus 2.00 GeneChip (Santa Clara, CA) according to the manufacturer's protocol. This microarray contains 54,656 probes corresponding to 47,400 human transcripts. At the end of the experiment, the hybridized probes were scanned at a resolution of 3 µm in a confocal scanner (Affymetrix GeneChip Scanner 3000 7G). Affymetrix microarrays were processed in the Microarray Core Facility of the Institute of Research of Biotherapy, CHRU-INSERM-UM1 Montpellier (http://irb.chu-montpellier.fr/).
Affymetrix GeneChip data analysis and filtering
Data were normalized with the GeneChip Operating Software (GCOS) algorithms. Then, the converted digital intensity values were stored as image data files and converted into cell intensity files using the Microarray Affymetrix Software 5.0 (MAS 5). MAS5 labels each transcript as “present” (P), “marginal” (M) or “absent” (A) by taking into account the average difference value calculated between the perfect match (PM) and the mismatch (MM) as detailed in the Affymetrix Genechip procedure (www.Affymetrix.com). A p-value risk was associated with the signal intensity. For each gene, the mean signal expression and standard deviation (SD) were calculated for each group. The fold change was determined by the ratio between old and young, and between “AD-like” and healthy old in order to study gene expression during physiological and pathological aging, respectively. All primary expression data files (*.cel) and (*.xls) are available at the authors' web site (http://www.mmdn.univ-montp2.fr/) and at GEO/NCBI (provisional accession number GSE21779). All steps were conducted according to the MIAME (Minimum Information About a Microarray Experiment) checklist [43].
Detection of genes that were differently expressed in the three groups
To identify genes differently expressed in relation with brain aging or pathology, filtered data were analyzed with the SAM (Significance Analysis of Microarrays) method [44], which uses permutations of repeated measurements to protect against non-normal data distribution. For that purpose, each group was compared to the other groups using 1,000 permutations. Because each animal may show intrinsic individual variability, the threshold for determining the rate of change was set at 1.5. SAM calculates the false discovery rate (FDR) with a q-value according to the delta cutoffs. The statistical significance of the genes was also investigated by Student t-test or by ANOVA with the “R” software. Finally, to construct a predictive model of the changes observed, a projection by principal component analysis (PCA) was carried out.
Gene profiling
The gene expression profile of each group was established using the Cluster and Treeview software programs [45]. These analyses allowed us to build a hierarchical clustering of the genes that are significantly associated with brain aging.
Functional categorization of the identified genes
To categorize the genes that were differentially expressed, we used two approaches: the first approach consisted in searching the literature using PubMed (NCBI), GenBank (www.ncbi.nlm.nih.gov/Genbank/), or specific databases such as Gene Expression Omnibus (GEO) (www.ncbi.nlm.nih.gov/geo/), the KEEG pathway database (www.genome.jp/kegg/pathway.html), UniProt (www.uniprot.org), and the Allen Brain Atlas (www.brain-map.org; http://humancortex.alleninstitute.org/has/human/docs.html). The second approach consisted in assigning genes to biological and molecular functions using the hierarchical database of the Gene Ontology (GO) consortium and EASE (Expression Analysis Systematic Explorer software; www.geneontology.org/). Finally, PathwayArchitect (Stratagene, Genome Exploration, UK) and IPA-Ingenuity (www.ingenuity.com) allowed us to build interactive networks.
Supporting Information
Genes sorted by anova
(0.05 MB XLS)
Gene functional categorization
(0.19 MB XLS)
Acknowledgments
We would like to thank also A. Ménigoz, T. Imberdis, R. Penanguer-Alonso and E. Benattar students who participated to this study during their training period, and N. Devau for his help and his support in statistical analyses.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by Projet Exploratoire Pluridisciplinaires-Sciences et Tecnologies de l'Information et de Ingenierie (PEPS-ST2I) du Centre National de la Recherche Scientifique (CNRS), France; Fondation Jerome Lejeune, France; Fond de Cooperation Scientifique Alzheimer, France; Fond Unique Interministeriel et Region Languedoc-Roussillon, France. The funders had no role in the study design, collection and analyses of the data, decision to publish or preparation of the manuscript.
References
- 1.Hof PR, Morrison JH. The aging brain: morphomolecular senescence of cortical circuits. Trends Neurosci. 2004;27:607–613. doi: 10.1016/j.tins.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 2.Kelly KM, Nadon NL, Morrison JH, Thibault O, Barnes CA, et al. The neurobiology of aging. Epilepsy Res. 2006;68:S5–S20. doi: 10.1016/j.eplepsyres.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 3.Burke SN, Barnes CA. Senescent synapses and hippocampal circuit dynamics. Trends Neurosci. 2010;33:153–161. doi: 10.1016/j.tins.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mucke L. Alzheimer's disease. Nature. 2009;461:895–897. doi: 10.1038/461895a. [DOI] [PubMed] [Google Scholar]
- 5.Geschwind DH, Konopka G. Neuroscience in the era of functional genomics and systems biology. Nature. 2009;461:908–915. doi: 10.1038/nature08537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perret M. Change in photoperiodic cycle affects life span in a prosimian primate (Microcebus murinus). J Biol Rhythms. 1997;12:136–45. doi: 10.1177/074873049701200205. [DOI] [PubMed] [Google Scholar]
- 7.Cayetanot F, Nygård M, Perret M, Kristensson K, Aujard F. Plasma levels of interferon-gamma correlate with age-related disturbances of circadian rhythms and survival in a non-human primate. Chronobiol Int. 2009;26:1587–601. doi: 10.3109/07420520903398518. [DOI] [PubMed] [Google Scholar]
- 8.Mestre-Frances N, Keller E, Calenda A, Barelli H, Checler F, et al. Immunohistochemical analysis of cerebral cortical and vascular lesions in the primate Microcebus murinus reveal distinct amyloid beta 1–42 and beta1–40 immunoreactivity profiles. Neurobiol Dis. 2000;7:1–8. doi: 10.1006/nbdi.1999.0270. [DOI] [PubMed] [Google Scholar]
- 9.Delacourte A, Sautière PE, Wattez A, Mourton-Gilles C, Petter A, et al. Biochemical characterization of Tau proteins during cerebral aging of the lemurian primate Micorcebus murinus. CR Acad Sci III. 1995;318:85–9. [PubMed] [Google Scholar]
- 10.Kraska A, Dorieux O, Picq J-L, Petit F, Bourrin E, et al. Age-associated cerebral atrophy in mouse lemur primates. Neurobiol Aging. 2009 doi: 10.1016/j.neurobiolaging.2009.05.018. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joly M, Michel B, Deputte B, Verdier JM. Odor discrimination assessment with an automated olfactometric method in a prosimian primate, Microcebus murinus. Physiol Behav. 2004;82:325–329. doi: 10.1016/j.physbeh.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 12.Joly M, Deputte B, Verdier JM. Age effect on olfactory discrimination in a non-human primate, Microcebus murinus. Neurobiol Aging. 2006;27:1045–1049. doi: 10.1016/j.neurobiolaging.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Picq J-L. Aging affects executive functions and memory in mouse lemur primates. Experiment Gerontol. 2007;42:223–232. doi: 10.1016/j.exger.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Trouche SG, Maurice T, Rouland S, Verdier JM, Mestre-Frances N. The three panel runway maze adapted to Microcebus murinus reveals age-related differences in memory and perseverance performances. Neurobiology Learning Memory. 2010;94:100–106. doi: 10.1016/j.nlm.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marvanova M, Menager J, Bezard E, Bontrop RE, Pradier L, et al. Microarray analysis of nonhuman primates: validation of experimental models in neurological disorders. Faseb J. 2003;17:929–931. doi: 10.1096/fj.02-0681fje. [DOI] [PubMed] [Google Scholar]
- 16.Cáceres M, Lachuer J, Zapala MA, Redmond JC, Kudo L, et al. Elevated gene expression levels distinguish human from non-human primate brains. Proc Natl Acad Sci U S A. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fraser HB, Khaitovich P, Plotkin JB, Pääbo S, Eisen MB. Aging and gene expression in the primate brain. PLoS Biology. 2005;3:1653–1661. doi: 10.1371/journal.pbio.0030274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duce J, Podvin S, Hollander W, Kipling D, Rosene D, et al. Gene Profile Analysis Implicates Klotho as an Important Contributor to Aging Changes in Brain White Matter of the Rhesus Monkey. Glia. 2008;56:106–117. doi: 10.1002/glia.20593. [DOI] [PubMed] [Google Scholar]
- 19.Sullivan EV, Pfefferbaum A, Adalsteinsson E, Swan GE, Carmelli D. Differential rates of regional brain cahnage in callosal and ventricular size: a 4-year longitudinal MRI study of elderly men. Cerebral Cortex. 2002;12:438–445. doi: 10.1093/cercor/12.4.438. [DOI] [PubMed] [Google Scholar]
- 20.Salat DH, Lee SY, van der Kouwe AJ, Greve DN, Fischl B, et al. Age-associated alterations in cortical gray and white matter signal intensity and gray to white matter contrast. NeuroImage. 2009;48:21–28. doi: 10.1016/j.neuroimage.2009.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trouche SG, Asuni A, Rouland S, Wisniewski T, Frangione B, et al. Antibody response and plasma Abeta1–40 levels in young Microcebus murinus primates immunized with Abeta1–42 and its derivatives. Vaccine. 2009;27:957–64. doi: 10.1016/j.vaccine.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu T, Pan Y, Kao S-Y, Li C, Kohane I, et al. Gene regulation and DNA damage in the ageing brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 23.Partridge L, Gems D. Mechanisms of ageing: public or private? Nature Rev Genetics. 2002;3:165–175. doi: 10.1038/nrg753. [DOI] [PubMed] [Google Scholar]
- 24.Yankner BA, Lu T, Loerch P. The aging brain. Ann Rev Pathol-Mechanims of Disease. 2008;3:41–66. doi: 10.1146/annurev.pathmechdis.2.010506.092044. [DOI] [PubMed] [Google Scholar]
- 25.Loerch PM, Lu T, Dakin KA, Vann JM, Isaacs A, et al. Evolution of the aging brain transcriptome and synaptic regulation. PLoS One. 2008;3:e3329. doi: 10.1371/journal.pone.0003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, et al. Regional Patterns of gene expression in Human and Chimpanzee brains. Genome Res. 2004;14:1462–1473. doi: 10.1101/gr.2538704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang WS, Dunckley T, Beach TG, Grover A, Mastroeni D, et al. Altered neuronal gene expression in brain regions differentially affected by Alzheimer's disease: a reference data set. Physiol Genomics. 2008;33:240–56. doi: 10.1152/physiolgenomics.00242.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berchtold NC, Cribbs DH, Coleman PD, Rogers J, Head E, et al. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci. 2008;105:15605–15610. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papassotiropoulos A, Fountoulakis M, Dunckley T, Stephan DA, Reiman EM. Genetics, transcriptomics and proteomics of Alzheimer's disease. J Clin Psychiatry. 2006;67:652–670. doi: 10.4088/jcp.v67n0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, et al. Incipient Alzheimer's disease: Microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci USA. 2004;101:2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller JA, Oldham MC, Geschwind DH. A system level analysis of transcriptional changes in Alzheimer's disease and normal aging. J Neurosci. 2008;28:1410–1420. doi: 10.1523/JNEUROSCI.4098-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu A, Hiltunen M, Romano DM, Soininen H, Hyman BT, et al. Effects of ubiquilin 1 on the unfolded protein response. J Mol Neurosci. 2009;38:19–30. doi: 10.1007/s12031-008-9155-6. [DOI] [PubMed] [Google Scholar]
- 33.Burke SN, Barnes CA. Neural plasticity in the ageing brain. Nature Rev Neurosci. 2006;7:30–40. doi: 10.1038/nrn1809. [DOI] [PubMed] [Google Scholar]
- 34.Venkitaramani DV, Chin J, Netzer WJ, Gouras GK, Lesne S, et al. ß-Amyloid modulation of synaptic transmission and plasticity. J Neurosci. 2007;27:11832–11837. doi: 10.1523/JNEUROSCI.3478-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parameshwaran K, Dhanasekaran M, Suppiramaniam V. Amyloid beta peptides and glutamatergic synaptic dysregulation. Experimental Neurol. 2008;210:7–13. doi: 10.1016/j.expneurol.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 36.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, et al. Naturally secreted oligomers of amyloid ß protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 37.Ondrejcak T, Klyubin I, Hu NW, Barry AE, Cullen WK, et al. Alzheimer's Disease Amyloid beta-Protein and Synaptic Function. Neuromolecular Med. 2010;12:13–26. doi: 10.1007/s12017-009-8091-0. [DOI] [PubMed] [Google Scholar]
- 38.Roth TL, Sweatt JD. Regulation of chromatin structure in memory formation. Curr Opinion Neurobiol. 2009;19:1–7. doi: 10.1016/j.conb.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bertram L, Tanzi RE. Genome wide association studies in Alzheimer's disease. Human Mol Genetics. 2009;18:R137–R145. doi: 10.1093/hmg/ddp406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lambert JC, Heath S, Even G, Campion D, Sleegers K, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nature Genetics. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 41.Reinius B, Saetre P, Leonard JA, Blekhman R, Merino-Martinez R, et al. An evolutionarily conserved sexual signature in the primate brain. PLoS Genetics. 2008;4:e1000100. doi: 10.1371/journal.pgen.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bons N, Silhol S, Barbié V, Mestre-Francès N, Albe-Fessard D. A stereotaxic atlas of the grey lesser mouse lemur brain (Microcebus murinus). Brain Res Bull. 1998;46:1–173. doi: 10.1016/s0361-9230(97)00458-9. [DOI] [PubMed] [Google Scholar]
- 43.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nature Genetics. 2001;29(4):365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 44.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing data analysis radiation response. Proc Natl Acad Sci. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes sorted by anova
(0.05 MB XLS)
Gene functional categorization
(0.19 MB XLS)