Abstract
Horizontal transfer is the passage of genetic material between genomes by means other than parent-to-offspring inheritance. Although the transfer of genes is thought to be crucial in prokaryotic evolution, few instances of horizontal gene transfer have been reported in multicellular eukaryotes; instead, most cases involve transposable elements. With over 200 cases now documented, it is possible to assess the importance of horizontal transfer for the evolution of transposable elements and their host genomes. We review criteria for detecting horizontal transfers and examine recent examples of the phenomenon, shedding light on its mechanistic underpinnings, including the role of host-parasite interactions. We argue that the introduction of transposable elements by horizontal transfer in eukaryotic genomes has been a major force propelling genomic variation and biological innovation.
Keywords: horizontal transfer, transposable element, eukaryotic genome evolution
The importance of horizontal transfer of DNA in genome evolution
Horizontal transfer has long been recognized as a crucial mechanism driving bacterial evolution [1]. In contrast, the evolutionary significance of horizontal transfer between the nuclear genomes of multicellular eukaryotes has remained more obscure [2]. We believe this gap in perceived importance is attributable to the disproportionate attention given to the transfer of genes as opposed to non-genic DNA. A fundamental difference in the genomic composition of multicellular eukaryotes compared to prokaryotes is that genes represent a minor and relatively static component of most eukaryotic genomes. Instead, most eukaryotic genomes are littered with non-coding DNA and transposable elements (TEs), discrete segments of DNA capable of moving from one locus to another and often duplicating themselves in the process. Not only are TEs the single most abundant entity of large eukaryotic genomes (e.g. about half of the human genome and 85% of the maize genome; [3,4]), they are also one of their most dynamic components. The movement and accumulation of TEs introduces a prolific source of raw genomic and epigenomic variation among lineages that has both an immediate and lasting influence on the evolutionary trajectory of the host species (for recent reviews, see [5-8]). Given the known importance and abundance of TEs in eukaryotic genomes, an examination of their propensity for HT is overdue.
The role of horizontal transfer in the persistence of transposable elements
The question of how TEs and other forms of “selfish DNA” [9] persist in genomes while having no direct selective benefit to the host has long intrigued evolutionary biologists (e.g., [10,11]). Several models have been developed to assess the relative effects of transposition and excision rates, negative selection, and population genetic parameters (such as effective population size) on the long-term survival of TEs in the population (e.g., [12,13]). Most of these models aimed to identify conditions under which TEs would be at a transposition-selection balance (for review, see [14]). The unexpected discovery in the early 90's that some TEs, notably the P element of Drosophila, were able to colonize new genomes by means of horizontal transfer [15] unveiled an additional way TEs could persist over time. Horizontal escape of an active transposon into a new genomic background would allow the element to evade a seemingly inevitable vertical extinction in its original host lineage resulting from elimination (by drift or selection) or inactivation due to mutational decay ([16,17]; Box 1).
Although the inherent ability of TEs to mobilize and integrate into the genome suggested a proclivity for horizontal transfer [18], until recently it was unclear whether the process affected a broad range of TEs and organisms. Here we review the methods and criteria for detecting horizontal transposon transfer (HTT), examine trends and patterns revealed by the growing number of documented cases, and highlight the importance of HTT in the lifecycle of various types of TEs. We conclude that virtually all types of TEs may be subject to HTT, and that viruses and parasites may facilitate the spread of TEs across widely diverged species. Lastly, we argue that the importance of HTT in the evolution of multicellular eukaryotes has been largely overlooked and discuss the impact of such a process on the evolution of host genomes.
Detecting HTT in the genomic era
Traditionally, three criteria have been used to infer HTT: (i) patchy distribution of the TE within a group of taxa, (ii) high sequence similarity of the TE from different host species which exceeds levels that would be expected given the divergence time of the hosts, and (iii) incongruence of TE and host phylogeny (reviewed by [19,20]). It is, however, important to keep in mind that each of these patterns may also result from other evolutionary processes, such as stochastic loss and/or differential fixation of ancestral polymorphism [21], purifying selection acting to preserve TE sequences for cellular function [22,23], or variable rates and modes of evolution of the TEs [20,24]. Inferences about HTT, therefore, need to be based on more than one line of evidence, with the strongest cases being those for which alternative hypotheses can be confidently refuted by sampling additional taxa and loci, as well as performing tests to examine the role of selection in preserving TE sequences after their insertion in the genome.
The difficulty in establishing HTT is well illustrated by the alleged horizontal transfer of the Bov-B non-LTR retrotransposon between squamate reptiles and ruminants. Here the claim for HTT was originally made based on the high level of interspecific sequence similarity in a 550-bp region at the 3' end of the element and its patchy distribution among tetrapods [25]. This claim was later refuted based on sequence analyses of a longer segment of the reverse transcriptase region from a broader range of related non-LTR retrotransposons [26]. The case for HTT remained unsettled until additional taxa were sampled and further sequence analyses were conducted, which now strongly argue in favor of HTT [27,28]. The argument for HTT was further bolstered by the discovery of a Bov-B-derived short interspersed element from squamate reptiles integrated in the genome of a poxvirus known to infect mammals [29].
One method that may be used to infer HTT between host species is the comparison of rates of synonymous mutations (Ks) observed in TEs with those in orthologous genes. If the presence of a TE in two hosts is due to HTT, then it will be younger and will have accumulated fewer synonymous mutations than the host genes [30,31]. With many complete genome sequences now available, this approach can be implemented in a robust statistical framework taking into account the Ks distribution of hundreds of host genes in order to formally define the Ks threshold under which the presence of a TE is considered to be the result of HTT. An advantage of this approach is that it can be applied to the detection of HTTs between closely-related species, such as Drosophila melanogaster and D. simulans, which diverged less than 5 Myrs ago [31].
Access to whole genome sequences provides the opportunity to gather most TE copies from a given genome which can also be used to supply robust evidence for HTT. Phylogenetic analysis of individual TE copies extracted from multiple genomes involved in HTT, in conjunction with biogeographical and ecological data, can help decipher the direction of transfer (e.g., [32,33]). In addition, whole genomes provide a way to estimate the timing of amplification of TE families, which, in turn, can help strengthen the case for HTT. Both empirical data [34] and simulations [35] suggest that TE amplification occurs immediately after the initial introduction of an active founder copy. Thus, by dating TE amplification one can approximate the date at which HTT might have occurred. Dating can be performed by calculating the pairwise divergence between all individual TE copies and an ancestral founder copy, which can be approximated by a consensus sequence reconstructed using multiple copies belonging to the same TE family from a given species. Because TE sequences typically evolve neutrally after insertion in the genome [16,17], the date of amplification can be obtained by converting the average sequence divergence into absolute time using the neutral substitution rate of the host species [36,37]. A difference in the dates of TE amplification between different hosts may indicate that the TE was transferred in some species earlier than in others, which may also provide insight into the direction of HTT.
Known cases of HTT: the tip of the iceberg?
Data accumulated over the last two decades have shown that both RNA- and DNA-mediated elements have crossed species boundaries on many occasions (Table 1; see also Supplemental Table 1 and references therein). Our survey of the literature reveals 214 convincing cases of HTT, with 103, 97, and 14 cases affecting DNA transposons, LTR retrotransposons, and non-LTR retrotransposons, respectively (Table 1). The apparent difference in the success rate of HTT among TE types may stem from differences in the replication strategies of the elements (Box 1), mechanisms of transposition (Box 2), and/or may reflect historical and sampling biases. Because DNA transposons were the first type of elements reported to transfer horizontally, they may have subsequently been subject to closer investigation (see Supplemental Table 1). Similarly, the majority of HTT cases involve drosophilid flies (137 out of 214), but this is likely because this is a group in which some of the most famous, early cases of HTT were discovered and for which extensive genomic resources are available. Currently, the taxonomic sampling bias of whole genome sequencing projects among eukaryotes leans strongly towards animals and fungi, making it difficult to infer patterns among underrepresented groups, for example unicellular eukaryotes and plants. Thus, it is important to be cautious when making generalizations about patterns of HTT based on fewer than 1000 sequenced genomes until data for a more diverse assemblage of the >1.5 million extant eukaryotes become available.
Table 1.
Summary of known cases of HTT Cases listed by class and tallied by superfamily with the number of cases observed in drosophilids and involving cross-phyla transfer noted (see Supplemental Table for complete list and references).
| Class | Superfamily | Number of HTTs | Number of HTTs in Drosophila | Number of cross-phyla HTTs |
|---|---|---|---|---|
| Non-LTR retrotransposons | jockey | 3 | 3 | - |
| RTE | 6 | - | - | |
| CR1 | 1 | - | - | |
| Rex1 | 1 | - | - | |
| Tad | 2 | - | - | |
| Smal (SINE) | 1 | - | - | |
| LTR retrotransposons | Ty3/gypsy | 70 | 63 | - |
| Ty1/copia | 16 | 4 | - | |
| Penelope | Penelope | 11 | 11 | - |
| DNA transposons | P | 28 | 28 | - |
| Tc1/mariner | 34 | 17 | 3 | |
| PIF | 3 | 3 | - | |
| Mutator | 1 | - | - | |
| hAT | 38 | 10 | 9 | |
| IS5 | 1 | - | - | |
| PiggyBac | 2 | - | - | |
| TOTAL | 218 | 139 | 12 |
Despite these sampling limitations, several initial inferences can be made based on the first large-scale systematic studies of HTT (Table 2). First, there is evidence that HTT has occurred numerous times, at least in some taxa. For example, a genome-wide study across Drosophila estimates that approximately one HTT event per TE family occurs every 20 Myrs in this group [31]. Furthermore, there has been a steady accumulation of clear cases in opistokonts (fungi and animals) and plants, the two eukaryotic supergroups in which TEs are especially abundant and for which most whole genome sequence data are presently available. Remarkably, in several instances, nearly identical elements have been able to infiltrate species separated by more than 500 Myrs of evolution, including at least 12 instances of movement across animal phyla (Table 1 and Box 3). So far, all these ‘long jumps’ involve DNA transposons (Tc1/mariner and hAT superfamilies), suggesting that these elements are well adapted to invade a wide range of species. This is not surprising given that several eukaryotic DNA transposons (in particular, the Tc1/mariner, hAT, and piggyBac superfamilies) can transpose readily when introduced experimentally into the genome of heterologous species, even when the latter belong to a different phylum (e.g., [38-40]), a different kingdom (e.g., [41-43]), or even a different domain of life, i.e. bacteria or archea (e.g., [44,45]). HTT is commonplace among prokaryotes (e.g., [46]) and often serves as a vehicle to transfer genes between bacterial species [1], however only one putative example of transfer of a prokaryotic TE into a eukaryote has been reported. It involves the recent introduction of an IS5-like insertion sequence from an unknown bacterial species into a bdelloid rotifer [47]. Although the element is seemingly intact, it is only present as a single copy and appears to be transcriptionally inactive, suggesting that it failed to adapt to a eukaryotic host. Nonetheless, this example suggests that prokaryotic TEs may be delivered to eukaryotic hosts, which could explain the patchy distribution of some eukaryotic DNA transposons that are phylogenetically related to bacterial insertion sequences (e. g., the Merlin superfamily; [48]).
In search of the smoking gun: mechanisms underlying HTT
While the inherent mobility and replication abilities of TEs undoubtedly facilitate excision and integration, the precise mechanisms by which TEs can be transported between organisms, including potential vectors, remain largely mysterious. Successful transfer requires delivery of DNA from donor to host cell (and to the germline for multicellular organisms), followed by integration into the recipient host genome. It has long been established that “naked” DNA and RNA can circulate in animal bodily fluids such as blood, plasma, lymph, saliva, and milk (e.g., [49]); however, the half-life of such extrachromosomal nucleic acids has not been quantified in most species. Other proposed routes for HTT are through feeding (although this has never been demonstrated) or via some kind of vector (e.g., [50]). In the simplest case, TEs themselves may facilitate the transfer of other TEs, for example, non-LTR retrotransposons nesting in more HTT-prone DNA transposons might be able to shuttle between host species by hitchhiking. In addition, LTR retrotransposons can make their own virus-like particles, and several can encode envelope-like proteins (e.g., [51]), which could increase their stability in the environment and confer infectious properties facilitating HTT (see Box 2).
Other potential vehicles for HTT are the multitude of pathogens and parasites that infect eukaryotes. The two types of vectors discussed most frequently in the literature are bacteria and viruses because of their known propensity to transduce and recombine host DNA fragments and because they are often able to enter and exit eukaryotic cells. Although the involvement of microorganisms and viruses is easy to imagine, it remains difficult to prove. The main obstacle lies in the low likelihood of fixation and rapid removal of nonessential DNA in the genomes of viruses and bacteria (e.g., [52]), which would rapidly erase any traces of transient eukaryotic TEs in their genomes. However, there are several reported instances where a TE was essentially ‘caught in the act’ of a horizontal escape from eukaryotic host to viral genome. These include the discoveries of several active insect DNA transposons (e.g., piggyBac) and of one LTR retrotransposon (TED) after escaping the nuclear genome of lepidopteran cells into a baculovirus in the laboratory [53-55]. Host-to-virus transposition can also take place in nature, as revealed by the identification of a short interspersed element (a non-LTR retrotransposon) from the genome of a snake integrated into a poxvirus [29].
Another possible vector for HTT is Wolbachia, an intracellular parasitic bacterium known to transfer horizontally among individuals and species of insects (e.g., [56]) and capable of donating genetic material to its host [57,58]. Other common endoparasites, such as schistosomes (blood flukes) or trypanosomes (intracellular parasites), may also be capable of delivering or receiving fragments of DNA to and from their host (e.g., [59-60]), therefore opening the door for HTT. The most famous case of HTT among insects (the transfer of P elements among drosophilids) is thought to have been mediated by the ectoparasitic mite, Proctolaelaps regalis [50], although no P element sequence could be detected in the genome of the mite, making it difficult to unequivocally show that the parasite was involved in the transfer. While chromosomal integration of a TE in a parasite is not necessary for it to act as a vector for HTT, such an event would offer not only compelling evidence for the involvement of the parasite, but also a plausible mechanism for the recurrent delivery of the same element to multiple host species. This situation was recently encountered in the genome of a blood-sucking triatomine bug, Rhodnius prolixus, which harbors at least four DNA transposon families occurring in a diverse array of vertebrates, including some of its preferred hosts in nature ([61] and see Box 3).
Another convincing example of HTT between host and parasite is a report of nearly identical mariner-like elements in a parasitoid braconid wasp and its lepidopteran host [62]. Remarkably, this case might have also implicated a viral intermediate: the polydnaviruses (PDVs), a group of dsDNA viruses that have established a symbiotic relationship with many braconid parasitoids to suppress the immune response of their lepidopteran hosts [63]. PDVs reside in the wasp genome as integrated proviral sequences and produce viral particles in the ovary that are then injected into the host along with the parasitoid eggs. Thus, a TE landing into the wasp proviral PDV sequences could be co-packaged and delivered to the lepidopteran cells, essentially creating a delivery system for HTT [64]. Interestingly, several TE-related genes and TE fragments have been found nested in proviral PDV sequences, bringing support to this scenario (e.g., [65]). The intimate association between hosts and parasites (including viruses) makes it easy to envision multiple opportunities for HTT between widely-diverged organisms by a variety of mechanisms.
Consequences of HTT for eukaryotic evolution
Regardless of the precise mechanism(s) underlying HTT, the accumulation of cases in the literature clearly points to a recurrent phenomenon that may be viewed as an integral facet of the lifecycle of TEs. But does it matter for eukaryotic evolution? It has been argued that biological innovation in multicellular organisms is largely driven by changes in copy number or function of pre-existing genetic material, rather than by the sudden appearance of genes and pathways de novo [66]. Because TEs play a major role in the duplication and rearrangement of genes and regulatory DNA [5,6,8,67] and because HTT provides the gateway for many TE invasions, HTT can be seen as the trigger in a series of events that actively shapes genomic architecture and eventually catalyzes biological innovation (Box 4).
The most direct consequence of HTT is the addition of TE copies themselves; indeed, many of the episodes of HTT reviewed herein have given rise to massive waves of TE amplification resulting in substantial increases in genome size. In the case of the little brown bat, Myotis lucifugus, DNA transposons from at least four different TE families have horizontally entered the genome and amplified over the past 30 Myrs and these together account for the accumulation of ~21 Mb of DNA in the vespertilionid bat lineage [23,61]. The evolutionary consequences of the structural genomic variation resulting from these multiple waves of TE amplification have not been investigated, but it is intriguing that they coincide with one of the most dramatic episodes of speciation documented in mammals [6,68].
Beyond the mere addition and rearrangement of raw genomic material, HTT can also result in the birth of new cellular genes. This can occur via several processes (Box 4), including the ‘domestication’ of genes originally encoded by TEs that become co-opted for host functions [22,67]. For example, the transposase gene from a copy of SPACE INVADERS, a horizontally-transferred family of DNA transposon, was captured to form a new fusion gene specific to murine rodents [23]. In addition to domestication, HTT might lead to the evolution of novel genes or regulatory regions via the transduction of captured functional sequence from the host. In prokaryotes, HTT (involving a variety of mobile elements, including phages, integrons, conjugative, composite, and rolling-circle transposons [69]) commonly acts as a vehicle for gene transfer among species. Although TEs have not yet been shown to transfer host genes between different species in eukaryotes, they are capable of capturing and transducing sequences at high frequency within a species (e.g., [70,71]). Thus, it would not be surprising to discover that HTT was responsible for the direct lateral movement of functional DNA among eukaryotes.
In addition to the impact on individuals and species, HTT may have had a profound influence on eukaryotic evolution by contributing to the origins of the primordial eukaryotic cell and features of multicellularity. Martin and Koonin [72] speculated that selective pressure exerted by mobile introns favored the evolution of the nuclear membrane as a protective barrier to separate transcription and translation after mobile DNA invaded the genome. Following similar reasoning, Johnson [73] hypothesized that the selfish spread of TEs drove the evolution of cellular partitioning, leading to the division of germline and soma. This model is based on the assumption that mobilization of TEs in somatic tissue has a greater fitness cost to the host than TE activity in germ cells. Thus, mutations causing the sequestration of reproductive cells may be favored if, by restricting activity to the germline, they minimize deleterious phenotypic effects of TEs on the host. This hypothesis depends on the observation that TE activity can be restricted to the germline in some species (e.g., in maize, Drosophila, and mouse; [74-76]), however this has not been assessed systematically and, in some instances, TE activity is observed only or predominantly in the soma [77-79]. Further circumstantial evidence for the potential importance of germline sequestration as a line of defense against HTT comes from the observation that HTT appears to be rampant in planaria (Schmidtea mediterranea; [61, 80]), a species lacking a sequestered germline. Further investigation into the frequency of HTT, including the comparison of HTT between animals and plants (which also lack germline sequestration), will be required to untangle the importance of TE replication versus host genome vulnerability in explaining the patterns of HTT observed among species.
Conclusions and Future Directions
Recognizing the prevalence of HTT and its importance for the long-term persistence of many TEs is a major step towards understanding the impact of this phenomenon on the evolution of eukaryotic genomes. Further progress will necessitate systematic, genome-wide scans to identify broad and unbiased patterns of HTT across different types of TEs and taxonomic groups. This effort should lead to a better understanding of the genetic, physiological and ecological factors influencing HTT. In addition, experiments are needed to i) delineate the mechanisms underlying HTT at the cellular and molecular levels, ii) assess the lines of genomic defense used by hosts to prevent infiltration by foreign DNA, and iii) uncover the biochemical loopholes that allow vectors of HTT to circumvent such defenses. Lastly, identifying cases of HTT with clear fitness costs or benefits will help clarify the phenotypic impacts of the process on eukaryotic evolution. The discovery of rampant horizontal gene transfer among bacteria has transformed our view of the prokaryotic tree of life into a tangled web or a ‘forest’ of life [81,82] where the genetic makeup of organisms reflect not only their ancestry but also their ecology. Similarly, we postulate that the widespread horizontal transfer of transposons in eukaryotes has the potential to shape genome content according to ecological interactions between species and to significantly distort the phylogenetic patterns expected from strict vertical inheritance.
Box 1. The role of horizontal transfer in the lifecycle of transposable elements.
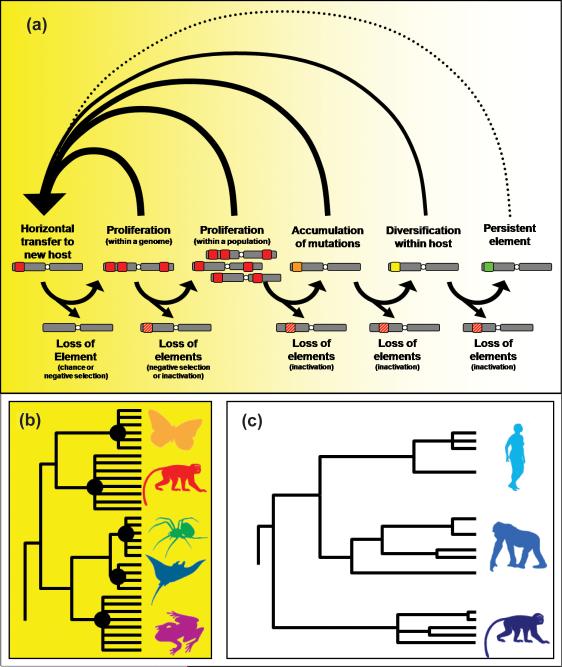
The lifecycle of a TE family is akin to a birth-and-death process: a new TE family is born when an active copy colonizes a novel host genome and it dies when all copies in a lineage are lost (by chance or negative selection) or inactivated, a process which may be driven by host-defense mechanisms and/or by the accumulation of disabling mutations in the TE sequence (see Figure I,a)). There are two major ways for TEs to escape extinction: the first is to horizontally transfer to a new host genome prior to inactivation and the second is to inflict minimal harmful effects (e.g., low replication rate), so as to evade the eye of selection in their current host. Like other parasites, it is possible that TEs will make use of different strategies over time (e.g., rely on high transmission rates initially [rapid replication and horizontal transfer], perhaps evolving towards a lower virulence strategy over time (‘the conventional wisdom’, according to [83]). The signature of each strategy (which do not represent a dichotomy as much as a continuum) is illustrated by looking at the relative congruence between TE phylogenies and that of their host (Figure I, b) and c)). In families of TEs where HTT is frequent, there should be dramatic incongruence between the phylogeny of the TE family and that of its various host species (Figure I, b)). In these cases, horizontal transfer might allow the TE to colonize a new genome in which host suppression mechanisms are inefficient [16,17], either because they have not had time to co-evolve or are copy number-dependent (e.g., [84,85]). In cases where TEs have persisted for long periods in a given host lineage, the reduced frequency of HTT can be inferred from the greater similarity between the TE and host phylogenies (Figure I, c)). For example, persistence could be achieved through self-regulatory mechanisms that limit copy number (proposed in [16]) or by evolving targeting preference for insertion into ‘safe havens’ in the genome (e.g., high copy number genes or heterochromatin [86,87]). The LINE-1 element of mammals provides an exceptional example of vertical endurance, having persisted and diversified over the past 100 million years with no evidence of HTT [36,88].
Figure I. (a) Simplified model of the lifecycle of TE families and the importance of horizontal transfer.
Grey bars represent chromosomes, colored squares represent TEs (with the original sequence in red, mutated in orange, diversified in yellow, persistent in green, and inactivated by orange and red lines). Arrows represent transitions between stages (not all possibilities are illustrated). (b and c) Expected phylogenetic patterns of TEs found among hosts when HTT is frequent (b) versus rare (c). Hypothetical TE phylogenies are depicted with black lines, black circles at nodes illustrate episodes of HTT, and host species are shown in colored silhouettes on the right of each tree with color similarity indicative of their phylogenetic relatedness.
Box 2. Transposition mechanisms and how they may influence HTT.
TEs are categorized into two major classes based on their mechanism of replication and are further clustered into superfamilies and families according to their sequence and structural similarities [89]. Retroelements (Class I) are referred to as “copy-and-paste” TEs because their method of mobilization typically involves replicative gain [90]. In contrast, DNA transposons (Class II) are typically characterized by “cut-and-paste” transposition, whereby the element is excised and reintegrated elsewhere [5,91]. The inherent ability of TEs to mobilize and integrate into chromosomes increases the possibility of their transfer compared to non-mobile sequences [18], however the propensity for HTT may differ based on the transposition mechanism of each group.
DNA transposons and LTR retroelements both have a double-stranded DNA intermediate that is thought to be more stable than the RNA intermediate of non-LTR retroelements and therefore more likely to be capable of HTT [26,92]. In addition, DNA transposons that have transferred across widely-diverged taxa may be more likely to function because only the transposase, but no specific host factors, are required for transposition to occur [38]. Structurally, most of these elements are extremely streamlined, often consisting of a single intronless gene encoding a transposase flanked by short terminal inverted repeats. In some cases, they lack a promoter and rely on read-through transcription from adjacent host promoters for expression (e.g., Tc1; [93]). It is tempting to interpret this minimal genetic organization as an adaptation for HTT. Other aspects of the transposition cycle are also likely to determine the probability of HTT, for example some LTR retrotransposons encode an envelope-like protein. As with retroviruses, such proteins could provide the TE with infection-like capabilities [51,94], although many cases of HTT involving envelope-less LTR elements have also been described (e.g., [95]).
Regardless of their class, TEs can be autonomous or non-autonomous, depending on whether they have the coding capacity for proteins required for their mobilization. Although both autonomous and non-autonomous elements can be activated if the necessary proteins are available and their cis-acting sequences are intact, non-autonomous elements may be less likely to transfer horizontally because they do not encode the proteins required for their own mobilization (but see [29] for an interesting case of host-to-virus transfer of a non-autonomous element).
Figure I. Schematics of the transposition mechanisms for the three major groups of TEs, with special reference to points in the process where the possibility of HTT is especially high.
White circles with solid borders are cells, gray circles with dashed borders are nuclei; bold lines indicate host DNA (black = donor site, grey = recipient site); TEs are represented by red boxes (DNA) or squiggly lines (mRNA); protein products are represented by shaded circles and ovals; thick black arrows indicate stage in transposition during which TEs are disassociated from host genomic DNA and thought to be most likely to horizontally transfer.
Box 3. Making the case: an example of widespread HTT.
Several lines of evidence can be used to make the case for HTT among organisms, including: a) A patchy taxonomic distribution is expected if TEs are moving horizontally rather than being vertically inherited. The timing of amplification can be used to help identify candidate vectors, given that species ranges (and their overlap) can shift significantly over evolutionary time periods. b) Identifying empty orthologous positions in other species helps verify that a given insertion has not been vertically inherited (in which case it would be found at the same genomic position in other taxa). c) The most frequently used criterion to uncover cases of HTT, however, is sequence similarity between TEs from species that exceeds the levels observed and expected based on the time elapsed since their divergence. d) Such sequence similarity can be analyzed in a phylogenetic framework and combined with information on the distribution and ecology of the species involved to make further inferences about the episodes of HTT. e) Lastly, the identification of a vector or mechanism of transfer represents a “holy grail” in terms of evidence for HTT. Despite mounting examples of HTT, the unequivocal confirmation of any specific mechanism acting to shuttle DNA among eukaryotes remains elusive.
Recently, Gilbert et al. [61] described a case of repeated, widespread HTT including data suggesting parasites may play a key role in facilitating HTT. The evidence showed HTT of four families of DNA transposons across four animal phyla and including exchanges among species on at least three different continents. In addition to the HTT observed in vertebrates, they identified two invertebrate species harboring the horizontally transferred TEs, both of which are associated with parasitic life cycles. The hemipteran, Rhodnius prolixus, which is an insect known to feed on the blood of mammals, birds, and reptiles, as well as the pond snail, Lymnaea stagnalis, which is an intermediate host for numerous trematodes parasitizing diverse vertebrates. In particular, two of the transposons identified in R. prolixus cluster phylogenetically with those found in the opossum and squirrel monkey, which rank high among this bug's preferred mammalian hosts in South America. Transposon DNA could have been directly ingested or delivered by the bug through the frequent exchanges of blood and saliva that occur between host and parasite during feeding. In addition, it is possible that trypanosomes, intracellular protozoan parasites transmitted to vertebrate hosts by triatomine bugs during feeding, acted as an intermediate vector for HTT.
Figure I. An example of the lines of evidence used to infer the horizontal transfer of a transposable element family, OposCharlie1 (OC1), across 3 phyla and 3 continents.
(a) Phylogenetic evidence based on a tree showing the patchy distribution and timing of amplification of OC1 across phyla. Presence indicated with an orange lightning bolt, timing of amplification (mya) estimated for vertebrates based on sequence divergence of copies from the consensus shown below tree. (b) Lack of orthologous insertions among taxa illustrated by an alignment showing empty sites at orthologous positions across taxa sharing recently transferred copies of OC1 (target site duplications of insert shown in yellow). (c) Sequence identity shown by a plot of the percent identity at the nucleotide level across all aligned regions of the OC1 consensus sequence across taxa, including the transposase open-reading frame (indicated by the blue rectangle), using 10 bp window and 3 bp steps. (d) Biogeographical evidence represented by a phylogeny of OC1 elements superimposed on a map to illustrate the presumed distribution of the host species at the inferred time of transfer which shows higher identity among geographically overlapping species. (e) Candidate vectors include i. naked DNA or RNA, ii. TEs, iii. viruses, iv. bacteria (e.g., Wolbachia), v. cellular parasites (e.g., trypanosomes), vi. internal parasites (e.g., schistosomes), vii. obligate endoparasitoids (e.g., parasitoid wasps), viii. ectoparasites (e.g., R. prolixus, the blood sucking triatomine bug, which has OC1 copies 95% similar to those found in its preferred host, the opossum [based on Gilbert et al. 2010]).
Box 4. Impact of horizontal transfer and amplification of TEs on genome evolution.
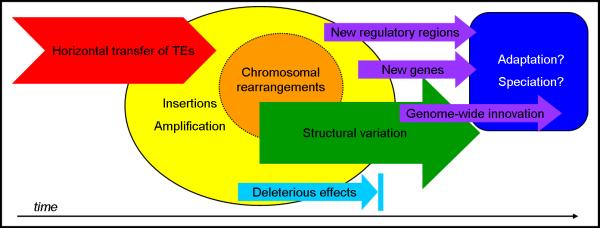
When TEs invade the genome by HTT or other means, their ability to amplify (replicate and mobilize) can lead to a broad spectrum of effects on the host. Most fundamentally, TE amplification can lead to the accumulation of DNA and an increase in genome size. In addition, TE insertions increase genetic variation. If TEs insert into genic regions, they can cause mutations or changes in genes or gene expression patterns. These are typically thought to be deleterious, but in some cases have been shown to be adaptive [96]. If a TE confers a selective benefit to the host, it can be retained and is said to be domesticated [22,67]. Different forms of domestication include the formation of novel gene chimeras between host and TE genes (e.g., [97]) or the co-option of an intact TE's function for the host's benefit (e.g., [98]). In addition, TE sequences contain a myriad of cis-regulatory elements, including transcription factor binding sites, which can be co-opted for the wiring of gene regulatory networks (for review, [67]). TEs can occasionally pick up fragments of the host genome during transposition and amplification, thereby leading to massive gene duplication or shuffling of exons (e.g., [70,71]; see [5] for review). Another way by which TE activity may lead to the formation of new genes is through the accidental recognition of cellular transcripts by the transposition machinery encoded by retrotransposons. During mammalian evolution, for example, the process has generated thousands of retroposed gene duplicates, many of which have evolved to take on new cellular functions (reviewed by [99]). The amplification and dispersion of TEs throughout the genome also generate an abundant substrate for subsequent rearrangements, for example through illegitimate recombination between TE copies leading to chromosomal duplications, deletions or inversions. Like other mutational events, these rearrangements may provide the raw material for adaptive genomic innovations. For example, TE-mediated rearrangements have been implicated in the formation of segmental duplications, which are an important source of genetic novelty and phenotypic variation in primate genomes [100].
Figure I. Schematic of the impact of HTT on the genome.
Red chevron represents HTT, yellow and orange circles represent proximate physical effects of HTT, colored arrows represent consequences (purple = beneficial, green = neutral, and light blue = deleterious), and the rectangle represents potential downstream outcomes of effects for which there is a selective benefit.
Supplementary Material
Supplementary Table 1. Compilation of all known cases of HTT involving eukaryotes classified by TE class, TE superfamily, TE family, and organism. Iincludes all reported cases of eukaryotic HTTs supported by sequence evidence. HT of viruses are not considered in this study. The column “Evidence” indicates which line(s) of evidence were used to infer HTT. The abbreviations follow Loreto et al. (2008): ss = sequence similarity; Ks = comparison between the number of synonymous mutations observed at orthologous genes and the number of synonymous mutations observed in TEs; dN/dS = test for purifying selection; phyl = phylogeny of the TE incongruent with the phylogeny of the host; pd = patchy taxonomic distribution of the TE; * = cross-phyla HTT.
References
1. Alberola, T.M., and de Frutos, R. (1996) Molecular structure of a gypsy element of Drosophila subobscura (gypsyDs) constituting a degenerate form of insect retroviruses. Nucleic Acids Res. 24, 914-923
2. Arca, B., and Savakis, C. (2000) Distribution of the transposable element Minos in the genus Drosophila. Genetica 108, 263-267
3. Bartolome, C., et al. (2009) Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes. Genome Biol. 10
4. Biedler, J.K., et al. (2007) Evolution and horizontal transfer of a DD37E DNA transposon in mosquitoes. Genetics 177, 2553-2558
5. Brunet, F., et al. (1999) Phylogenetic analysis of Mos1-like transposable elements in the Drosophilidae. J. Mol. Evol. 50, 760-768
6. Casola, C., et al. (2007) PIF-like Transposons are common in Drosophila and have been repeatedly domesticated to generate new host genes. Mol. Biol. Evol.24, 1872-1888
7. Clark, J.B., and Kidwell, M.G. (1997) A phylogenetic perspective on P transposable element evolution in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 94, 11428-11433
8. Clark, J.B., et al. (1994) Phylogenetic analysis supports horizontal transfer of P transposable elements. Mol. Biol. Evol.11, 40-50
9. Daboussi, M.J., et al. (2002) Evolution of the Fot1 transposons in the genus Fusarium: Discontinuous distribution and epigenetic inactivation. Mol. Biol. Evol.19, 510-520
10. Daniels, S.B., et al. (1990) Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics 124, 339-355
11. Daniels, S.B., et al. (1984) Sequences homologous to P elements occur in Drosophila paulistorum. Proc. Natl. Acad. Sci. U.S.A.81, 6794-6797
12. de Almeida, L.M., and Carareto, C.M.A. (2005) Multiple events of horizontal transfer of the Minos transposable element between Drosophila species. Mol. Phylogenet. Evol. 35, 583-594
13. de Almeida, L.M., and Carareto, C.M.A. (2006) Sequence heterogeneity and phylogenetic relationships between the copia retrotransposon in Drosophila species of the repleta and melanogaster groups. Genet. Sel. Evol. 38, 535–550
14. de Castro, J.P., and Carareto, C.M.A. (2004) Canonical P elements are transcriptionally active in the saltans group of Drosophila. J. Mol. Evol. 59, 31-40
15. de Setta, N., et al. (2009) Multiple invasions of Gypsy and Micropia retroelements in genus Zaprionus and melanogaster subgroup of the genus Drosophila. BMC Evol. Biol. 9, 18
16. Deprá, M., et al. (2010) hosimary: a new hAT transposon group involved in horizontal transfer. Mol. Genet. Genomics 283, 451-459
17. Diao, X.M., et al. (2006) Horizontal transfer of a plant transposon. PLoS Biol. 4, 119-128
18. Evgen'ev, M., et al. (2000) Invasion of Drosophila virilis by the Penelope transposable element. Chromosoma 109, 350-357
19. Fablet, M., et al. (2007) Evolutionary pathways of the tirant LTR retrotransposon in the Drosophila melanogaster subgroup of species. J. Mol. Evol. 64, 438-447
20. Garcia-Planells, J., et al. (1998) Molecular evolution of P transposable elements in the genus Drosophila. II. The obscura species group. J. Mol. Evol. 47, 282-291
21. Gilbert C., et al. (2010) A role for host-parasite relationships in horizontal transfer of DNA transposons across animal phyla. Nature 464, 1347-1350
22. Gladyshev, E.A., and Arkhipova I.R., (2009) A single copy IS5-like transposon in the genome of a bdelloid rotifer. Mol. Biol. Evol. 26, 1921-1929
23. Gogolevsky, K.P., et al. (2008) Bov-B-mobilized SINEs in vertebrate genomes. Gene 407, 75-85
24. Gomulski, L.M., et al. (2001) A new basal subfamily of mariner elements in Ceratitis rosa and other tephritid flies. J. Mol. Evol. 53, 597-606
25. Gonzalez, P., and Lessios H.A., Evolution of Sea Urchin Retroviral-Like (SURL) Elements: Evidence from 40 Echinoid Species. Mol. Biol. Evol. 16, 938–952
26. Hagemann, S., et al. (1998) Horizontal transmission versus vertical inheritance of P elements in Drosophila and Scaptomyza: Has the M-type subfamily spread from East Asia? J. Zool. Syst. Evol. Research 36, 75-83
27. Hagemann, S., et al. (1996) Repeated horizontal transfer of P transposons between Scaptomyza pallida and Drosophila bifasciata. Genetica 98, 43-51
28. Hagemann, S., et al. (1992) Identification of a complete P element in the genome of Drosophila bifasciata. Nucleic Acids Res.20, 409-413
29. Hamada, M., et al. (1997) A newly isolated family of short interspersed repetitive elements (SINEs) in coregonid fishes (whitefish) with sequences that are almost identical to those of the SmaI family of repeats: Possible evidence for the horizontal transfer of SINEs. Genetics 146, 355-367
30. Haring, E., et al. (2000) Ancient and recent horizontal invasions of drosophilids by P elements. J. Mol. Evol. 51, 577-586
31. Heredia, F., et al. (2004) Complex evolution of gypsy in drosophilid species. Mol. Biol. Evol.21, 1831-1842
32. Ivics, Z., et al. (1996) Identification of functional domains and evolution of Tc1-like transposable elements. Proc. Natl. Acad. Sci. U.S.A. 93, 5008-5013
33. Jordan, I.K., et al. (1999) Evidence for the recent horizontal transfer of long terminal repeat retrotransposon. Proc. Natl. Acad. Sci. U.S.A. 96, 12621-12625
34. Jordan, I.K., and McDonald, J.F. (1998) Evolution of the copia retrotransposon in the Drosophila melanogaster species subgroup. Mol. Biol. Evol. 15, 1160-1171
35. Koga, A., et al. (2000) Evidence for recent invasion of the medaka fish genome by the Tol2 transposable element. Genetics 155, 273-281
36. Konieczny, A., et al. (1991) A superfamily of Arabidopsis thaliana retrotransposons. Genetics 127, 801-809
37. Leaver, M.J. (2001) A family of Tc1-like transposons from the genomes of fishes and frogs: evidence for horizontal transmission. Gene 271, 203-214
38. Lohe, A.R., et al. (1995) Horizontal transmission, vertical inactivation, and stochastic loss of mariner-like transposable elements. Mol. Biol. Evol. 12, 62-72
39. Loreto, E.L.D., et al. (2001) Drosophila mediopunctata P elements: A new example of horizontal transfer. J. Hered. 92, 375-381
40. Ludwig, A., and Loreto, E.L.S. (2007) Evolutionary pattern of the gtwin retrotransposon in the Drosophila melanogaster subgroup. Genetica 130, 161-168
41. Lyozin, G.T., et al. (2001) The structure and evolution of Penelope in the virilis species group of Drosophila: an ancient lineage of retroelements. J. Mol. Evol. 52, 445-456
42. Maruyama, K., and Hartl, D.L. (1991) Evidence for interspecific transfer of the transposable element mariner between Drosophila and Zaprionus. J. Mol. Evol. 33, 514-524
43. Maside, X., et al. (2003) Inferences on the evolutionary history of the S element family of Drosophila melanogaster. Mol. Biol. Evol. 20, 1183-1187
44. Morales-Hojas, R., et al. (2006) The evolutionary history of the transposable element Penelope in the Drosophila virilis group of species. J. Mol. Evol. 63, 262-273
45. Mota, N.R., et al. (2009) harrow: new Drosophila hAT transposons involved in horizontal transfer. Insect Mol. Biol. (In press).
46. Novick, P., et al. Independent and parallel lateral transfer of DNA transposons in tetrapod genomes. Gene 450, 85-94
47. Novikova, O., et al. (2009) Non-LTR retrotransposons in fungi. Funct. Integr. Genomics 9, 27-42
48. Novikova, O., et al. (2007) CRI clade of non-LTR retrotransposons from Maculinea butterflies (Lepidoptera : Lycaenidae): evidence for recent horizontal transmission. BMC Evol. Biol. 7, 19
49. Pace, J.K., et al. (2008) Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc. Natl. Acad. Sci. U.S.A. 105, 17023-17028
50. Pagan, H.J.T., et al. (2010) PiggyBac-ing on a primate genome: novel elements, recent activity and horizontal transfer. Genome Biol. Evol. doi:10.1093/gbe/evq021
51. Robertson, H.M. (1997) Multiple mariner Transposons in flatworms and hydras are related to those of insects. J. Hered. 88, 195-201
52. Robertson, H.M., and Lampe, D.J. (1995) Recent horizontal transfer of a mariner transposable element among and between Diptera and Neuroptera. Mol. Biol. Evol. 12, 850-862
53. Roulin, A., et al. (2009) Whole genome surveys of rice, maize and sorghum reveal multiple horizontal transfers of the LTR-retrotransposon Route66 in Poaceae. BMC Evol. Biol. 9, 10
54. Roulin, A., et al. (2008) Evidence of multiple horizontal transfers of the long terminal repeat retrotransposon RIRE1 within the genus Oryza. Plant J. 53, 950-959
55. Sanchez-Gracia, A.., et al. (2005) High rate of horizontal transfer of transposable elements in Drosophila. Trends Genet. 21, 200-203
56. Silva, J.C., and Kidwell, M.G. (2000) Horizontal transfer and selection in the evolution of P elements. Mol. Biol. Evol. 17, 1542-1557
57. Simmons, G.M. (1992) Horizontal transfer of hobo transposable elements within the Drosophila melanogaster species complex: Evidence from DNA sequencing. Mol. Biol. Evol. 9, 1050-1060
58. Smit, A.F.A., and Riggs, A.D. (1996) Tiggers and other DNA transposon fossils in the human genome. Proc. Natl. Acad. Sci. U.S.A. 93, 1443-1448
59. Terzian, C., et al. (2000) Evolution of the gypsy endogenous retrovirus in the Drosophila melanogaster subgroup. Mol. Biol. Evol. 17, 908-914
60. Torti, C., et al. (2005) Cchobo, a hobo-related sequence in Ceratitis capitata. Genetica 123, 313-325
61. Vazquez-Manrique, R.P., et al. (2000) Evolution of gypsy endogenous retrovirus in the Drosophila obscura species group. Mol. Biol. Evol. 17, 1185-1193
62. Volff, J.N., et al. (2000) Multiple lineages of the non-LTR retrotransposon Rex1 with varying success in invading fish genomes. Mol. Biol. Evol. 17, 1673-1684
63. Waterston, R.H., et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520-562
64. Yoshiyama, M., et al. (2001) Possible horizontal transfer of a transposable element from host to parasitoid. Mol. Biol. Evol. 18, 1952-1958
65. Zupunski, V., et al. (2001) Evolutionary dynamics and evolutionary history in the RTE clade of non-LTR retrotransposons. Mol. Biol. Evol. 18, 1850-1863
Acknowledgments
We would like to acknowledge the research conducted on this topic by many colleagues that could not be cited or discussed due to space constraints. We also wish to thank the following for helpful discussions and comments: E.J. Pritham, J. Meik, B. Koskella, and three anonymous reviewers. This work was supported by NSF award 0805546 to SS and NIH grant R01GM77582 to CF.
Glossary
- Autonomous elements
Transposable elements that encode the proteins necessary to perform a complete transposition reaction on their own, i.e., to move from one genomic locus to another
- DNA transposons (Class 2)
TEs that transpose via a DNA intermediate, also often referred to as “cut-and-paste” elements because they excise and integrate elsewhere, unlike retroelements which do not excise
- LTR elements
one of two major subclasses of retroelements comprised of several superfamilies (e.g., Ty1/Copia), some of which produce virus-like particles; characterized by long terminal repeats (LTRs) which are generated upon chromosomal integration
- Non-autonomous
Transposable element that do not encode the transposition machinery and are therefore not able to transpose on their own. In order to move, these elements must utilize the proteins encoded by autonomous elements
- Non-LTR elements
the second of two major subclasses of retroelements (characterized by the lack of terminal repeats), also comprised of numerous superfamilies (e.g., L1, RTE and Alu)
- Retroelements (or retrotransposons; Class 1)
TEs that replicate based on the reverse transcription of an RNA intermediate, also referred to as “copy-and-paste” elements
- Transposable elements (TEs)
pieces of DNA characterized by their ability to move from one locus to another in the genome
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Frost LS, et al. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Micro. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 2.Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 3.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 4.Schnable, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;20:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 5.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliver KR, Greene WK. Transposable elements: powerful facilitators of evolution. Bioessays. 2009;31:703–714. doi: 10.1002/bies.200800219. [DOI] [PubMed] [Google Scholar]
- 7.Zeh DW, et al. Transposable elements and an epigenetic basis for punctuated equilibria. Bioessays. 2009;31:715–726. doi: 10.1002/bies.200900026. [DOI] [PubMed] [Google Scholar]
- 8.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orgel LE, Crick FHC. Selfish DNA- the ultimate parasite. Nature. 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 10.Hickey DA. Selfish DNA: a sexually-transmitted nuclear parasite. Genetics. 1982;101:519–531. doi: 10.1093/genetics/101.3-4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pritham EJ. Transposable elements and facotros influencing their success in eukaryotes. J. Heredity. 2009;100:648–655. doi: 10.1093/jhered/esp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brookfield JFY, Badge RM. Population genetics models of transposable elements. Genetica. 1997;100:281–294. [PubMed] [Google Scholar]
- 13.Le Rouzic A, et al. Long-term evolution of transposable elements. Proc. Nat. Acad. Sci. 2007;104:19375–19380. doi: 10.1073/pnas.0705238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charlesworth B, et al. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- 15.Daniels SB, et al. Evidence for the horizontal transmission of the P transposable element between Drosophila species. Genetics. 1990;124:339–355. doi: 10.1093/genetics/124.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartl DL, et al. Modern thoughts on an ancyent marinere: function, evolution, regulation. Annu. Rev. Genet. 1997;31:337–358. doi: 10.1146/annurev.genet.31.1.337. [DOI] [PubMed] [Google Scholar]
- 17.Roberston HM. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. ASM Press; Washington: 2002. pp. 1093–1110. [Google Scholar]
- 18.Kidwell MG. Horizontal transfer of P-elements and other short inverted repeat transposons. Genetica. 1992;86:275–286. doi: 10.1007/BF00133726. [DOI] [PubMed] [Google Scholar]
- 19.Silva JC, et al. Factors that affect the horizontal transfer of transposable elements. Curr. Issues Mol. Biol. 2004;6:57–71. [PubMed] [Google Scholar]
- 20.Loreto ELS, et al. Revisiting horizontal transfer of transposable elements in Drosophila. Heredity. 2008;100:545–554. doi: 10.1038/sj.hdy.6801094. [DOI] [PubMed] [Google Scholar]
- 21.Shedlock AM, et al. SINEs of speciation: tracking lineages with retroposons. Trends Ecol. Evol. 2004;19:545–553. doi: 10.1016/j.tree.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Volff JN. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays. 2006;28:913–922. doi: 10.1002/bies.20452. [DOI] [PubMed] [Google Scholar]
- 23.Pace JK, et al. Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc. Natl. Acad. Sci. U.S.A. 2008;105:17023–17028. doi: 10.1073/pnas.0806548105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capy P, et al. The strange phylogenies of transposable elements - are horizontal transfers the only explanation? Trends Genet. 1994;10:7–12. doi: 10.1016/0168-9525(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 25.Kordis D, Gubensek F. Unusual horizontal transfer of a long interspersed nuclear element between distant vertebrate classes. Proc. Nat. Acad. Sci. 1998;95:10704–10709. doi: 10.1073/pnas.95.18.10704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malik HS, et al. The age and evolution of non-LTR retrotransposable elements. Mol. Biol. Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 27.Zupunski V, et al. Evolutionary dynamics and evolutionary history in the RTE clade of non-LTR retrotransposons. Mol. Biol. Evol. 2001;18:1849–1863. doi: 10.1093/oxfordjournals.molbev.a003727. [DOI] [PubMed] [Google Scholar]
- 28.Gogolevsky KP, et al. Bov-B-mobilized SINEs in vertebrate genomes. Gene. 2008;407:75–85. doi: 10.1016/j.gene.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 29.Piskurek O, Okada N. Poxviruses as possible vectors for horizontal transfer of retroposons from reptiles to mammals. Proc. Natl. Acad. Sci. U.S.A. 2007;104:12046–12051. doi: 10.1073/pnas.0700531104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez-Gracia A, et al. High rate of horizontal transfer of transposable elements in Drosophila. Trends Genet. 2005;21:200–203. doi: 10.1016/j.tig.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Bartolome C, et al. Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes. Genome Biol. 2009;10 doi: 10.1186/gb-2009-10-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roulin A, et al. Evidence of multiple horizontal transfers of the long terminal repeat retrotransposon RIRE1 within the genus Oryza. Plant J. 2008;53:950–959. doi: 10.1111/j.1365-313X.2007.03388.x. [DOI] [PubMed] [Google Scholar]
- 33.Roulin A, et al. Whole genome surveys of rice, maize and sorghum reveal multiple horizontal transfers of the LTR-retrotransposon Route66 in Poaceae. BMC Evol. Biol. 2009;9:10. doi: 10.1186/1471-2148-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anxolabehere D, et al. Molecular characteristics of diverse populations are consistent with the hypothesis of a recent invasion of Drosophila melanogaster by mobile P elements. Mol. Biol. Evol. 1988;5:252–269. doi: 10.1093/oxfordjournals.molbev.a040491. [DOI] [PubMed] [Google Scholar]
- 35.Le Rouzic A, Capy P. The first steps of transposable elements invasion: Parasitic strategy vs. genetic drift. Genetics. 2005;169:1033–1043. doi: 10.1534/genetics.104.031211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan H, et al. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pace JK, Feschotte C. The evolutionary history of human DNA transposons: Evidence for intense activity in the primate lineage. Genome Res. 2007;17:422–432. doi: 10.1101/gr.5826307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plasterk RHA, et al. Resident aliens - the Tc1/mariner superfamily of transposable elements. Trends Genet. 1999;15:326–332. doi: 10.1016/s0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- 39.Ding S, et al. Efficient transposition of the piggyBac resource (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 40.Kodama K, et al. The Tol1 element of the medaka fish, a member of the hAT transposable element family, jumps in Caenorhabditis elegans. Heredity. 2008;101:222–227. doi: 10.1038/hdy.2008.47. [DOI] [PubMed] [Google Scholar]
- 41.Weil CF, Kunze R. Transposition of maize Ac/Ds transposable elements in the yeast Saccharomyces cerevisiae. Nat. Genet. 2000;26:187–190. doi: 10.1038/82827. [DOI] [PubMed] [Google Scholar]
- 42.Emelyanov A, et al. Trans-kingdom transposition of the maize Dissociation element. Genetics. 2006;174:1095–1104. doi: 10.1534/genetics.106.061184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evertts AG, et al. The Hermes transposon of Musca domestica is an efficient tool for the mutagenesis of Schizosaccharomyces pombe. Genetics. 2007;177:2519–2523. doi: 10.1534/genetics.107.081075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rubin EJ, et al. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl. Acad. Sci. U.S.A. 1999;96:1645–1650. doi: 10.1073/pnas.96.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang JK, et al. In vivo transposon mutagenesis of the methanogenic archaeon Methanosarcina acetivorans C2A using a modified version of the insect mariner-family transposable element Himar1. Proc. Natl. Acad. Sci. U.S.A. 2000;97:9665–9670. doi: 10.1073/pnas.160272597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Touchon M, Rocha EPC. Causes of insertion sequences abundance in prokaryotic genomes. Mol. Biol. Evol. 2007;24:969–981. doi: 10.1093/molbev/msm014. [DOI] [PubMed] [Google Scholar]
- 47.Gladyshev EA, Arkhipova IR. A single-copy IS5-like transposon in the genome of a bdelloid rotifer. Mol Biol Evol. 2009;8:1921–1999. doi: 10.1093/molbev/msp104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feschotte C. Merlin, a new superfamily of DNA transposons identified in diverse animal genomes and related to bacterial IS1016 insertion sequences. Mol. Biol. Evol. 2004;21:1769–1780. doi: 10.1093/molbev/msh188. [DOI] [PubMed] [Google Scholar]
- 49.Stroun M, et al. Alu repeat sequences are present in increased proportions compared to a unique gene in plasma/serum DNA: Evidence for a preferential release from viable cells? Ann. N. Y. Acad. Sci. 2001;945:258–264. doi: 10.1111/j.1749-6632.2001.tb03894.x. [DOI] [PubMed] [Google Scholar]
- 50.Houck MA, et al. Possible horizontal transfer of Drosophila genes by the mite Proctolaelaps regalis. Science. 1991;253:1125–1129. doi: 10.1126/science.1653453. [DOI] [PubMed] [Google Scholar]
- 51.Malik HS, et al. Poised for contagion: Evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 2000;10:1307–1318. doi: 10.1101/gr.145000. [DOI] [PubMed] [Google Scholar]
- 52.Mira A, et al. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- 53.Fraser MJ, et al. Acquisition of host-cell DNA-sequences by baculoviruses: Relationship between host DNA insertions and FP mutants of Autographa californica and Galleria mellonella nuclear polyhedrosis viruses. J. Virol. 1983;47:287–300. doi: 10.1128/jvi.47.2.287-300.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Friesen PD, Nissen MS. Gene organization and transcription of ted, a lepidopteran retrotransposon integrated within the baculovirus genome. Mol. Cell. Biol. 1990;10:3067–3077. doi: 10.1128/mcb.10.6.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jehle JA, et al. Horizontal escape of the novel Tc1-like lepidopteran transposon TCp3.2 into Cydia pomonella granulovirus. J. Mol. Evol. 1998;46:215–224. doi: 10.1007/pl00006296. [DOI] [PubMed] [Google Scholar]
- 56.Raychoudhury R, et al. Modes of acquisition of Wolbachia: horizontal transfer, hybrid introgression, and codivergence in the Nasonia species complex. Evolution. 2009;63:165–183. doi: 10.1111/j.1558-5646.2008.00533.x. [DOI] [PubMed] [Google Scholar]
- 57.Hotopp JCD, et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 2007;317:1753–1756. doi: 10.1126/science.1142490. [DOI] [PubMed] [Google Scholar]
- 58.Klasson L, et al. Horizontal gene transfer between Wolbachia and the mosquito Aedes aegypti. BMC Genomics. 2009;10:9. doi: 10.1186/1471-2164-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Steglich C, Schaeffer SW. The ornithine decarboxylase gene of Trypanosoma brucei: Evidence for horizontal gene transfer from a vertebrate source. Infect. Genet. Evol. 2006;6:205–219. doi: 10.1016/j.meegid.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 60.Hecht MM, et al. Inheritance of DNA transferred from American trypanosomes to human hosts. PLoS ONE. 2010;5:e9181. doi: 10.1371/journal.pone.0009181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gilbert C, et al. A role for host-parasite interactions in the horizontal transfer of DNA transposons across animal phyla. Nature. 2010;464:1347–1350. doi: 10.1038/nature08939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoshiyama M, et al. Possible horizontal transfer of a transposable element from host to parasitoid. Mol. Biol. Evol. 2001;18:1952–1958. doi: 10.1093/oxfordjournals.molbev.a003735. [DOI] [PubMed] [Google Scholar]
- 63.Turnbull M, Webb B. Perspectives on polydnavirus origins and evolution. Adv. Virus Res. 2002;58:203–254. doi: 10.1016/s0065-3527(02)58006-4. [DOI] [PubMed] [Google Scholar]
- 64.Doucet D, et al. In vitro integration of an ichnovirus genome segment into the genomic DNA of lepidopteran cells. J. Gen. Virol. 2007;88:105–113. doi: 10.1099/vir.0.82314-0. [DOI] [PubMed] [Google Scholar]
- 65.Desjardins CA, et al. Comparative genomics of mutualistic viruses of Glyptapanteles parasitic wasps. Genome Biol. 2008;9:17. doi: 10.1186/gb-2008-9-12-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shubin N, et al. Deep homology and the origins of evolutionary novelty. Nature. 2009;457:818–823. doi: 10.1038/nature07891. [DOI] [PubMed] [Google Scholar]
- 67.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ray DA, et al. Multiple waves of recent DNA transposon activity in the bat, Myotis lucifugus. Genome Res. 2008;18:717–728. doi: 10.1101/gr.071886.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Toleman MA, et al. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev. 2006;70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang N, et al. Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004;431:569–73. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- 71.Morgante M, et al. Gene duplication and exon shuffling by Helitron-like transposons generate intraspecies diversity in maize. Nat. Genet. 2005;37:997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- 72.Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 2006;440:41–45. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- 73.Johnson LJ. Selfish genetic elements favor the evolution of a distinction between soma and germline. Evolution. 2008;62:2122–2124. doi: 10.1111/j.1558-5646.2008.00433.x. [DOI] [PubMed] [Google Scholar]
- 74.Donlin MJ, et al. Tissue-specific accumulation of MURB, a protein encoded by MuDR, the autonomous regulator of the Mutator transposable element family. Plant Cell. 1995;7:1989–2000. doi: 10.1105/tpc.7.12.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Siebel CW, et al. Soma-specific expression and cloning of PSI, a negative regulator of P element pre-messenger-RNA splicing. Genes Dev. 1995;9:269–283. doi: 10.1101/gad.9.3.269. [DOI] [PubMed] [Google Scholar]
- 76.Trelogan SA, Martin SL. Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 1995;92:1520–1524. doi: 10.1073/pnas.92.5.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Emmons SW, Yesner L. High-frequency excision of transposable element Tc1 in the nematode Caenorhabditis elegans is limited to somatic-cells. Cell. 1984;36:599–605. doi: 10.1016/0092-8674(84)90339-8. [DOI] [PubMed] [Google Scholar]
- 78.Coufal NG, et al. L1 retrotranspoition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kano H, et al. L1 transposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Novick P, et al. Independent and parallel lateral transfer of DNA transposons in tetrapod genomes. Gene. 2009;449:85–94. doi: 10.1016/j.gene.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 81.Doolittle WF, Bapteste E. Pattern pluralism and the Tree of Life hypothesis. Proc. Nat. Acad. Sci. 2007;104:2043–2049. doi: 10.1073/pnas.0610699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Koonin EV, Wolf YI. The fundamental units, processes and patterns of evolution, and Tree of Life conundrum. Biol. Direct. 2009;4:33. doi: 10.1186/1745-6150-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.May RM, Anderson RM. Epidemiology and genetics in the coevolution of hosts and parasites. Proc Roy Soc B. 1983;219:281–313. doi: 10.1098/rspb.1983.0075. [DOI] [PubMed] [Google Scholar]
- 84.Jensen S, et al. Taming of transposable elements by homology-dependent gene silencing. Nat. Genet. 1999;21:209–212. doi: 10.1038/5997. [DOI] [PubMed] [Google Scholar]
- 85.Galagan JE, Selker EU. RIP: the evolutionary cost of genome defense. Trends Genet. 2004;20:417–423. doi: 10.1016/j.tig.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 86.Bushman FD. Targeting survival: integration site selection by retroviruses and LTR-retrotransposons. Cell. 2003;115:135–8. doi: 10.1016/s0092-8674(03)00760-8. [DOI] [PubMed] [Google Scholar]
- 87.Zhou J, Eickbush TH. The pattern of R2 retrotransposon activity in natural populations of Drosophila simulans reflects the dynamic nature of the rDNA locus. PLoS Genet. 2009;5:e1000386. doi: 10.1371/journal.pgen.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Furano AV, et al. L1 (LINE-1) retrotransposon diversity differs dramatically between mammals and fish. Trends Genet. 2004;20:9–14. doi: 10.1016/j.tig.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 89.Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 90.Eickbush TH, Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Research. 2008;134:221–234. doi: 10.1016/j.virusres.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Curcio MJ, Derbyshire KM. The outs and ins of transposition: From mu to kangaroo. Nat. Rev. Mol. Cell Biol. 2003;4:865–877. doi: 10.1038/nrm1241. [DOI] [PubMed] [Google Scholar]
- 92.Silva JC, Kidwell MG. Horizontal transfer and selection in the evolution of P elements. Mol Biol Evol. 2000;10:1542–1557. doi: 10.1093/oxfordjournals.molbev.a026253. [DOI] [PubMed] [Google Scholar]
- 93.Sijen T, Plasterk RHA. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature. 2003;426:310–314. doi: 10.1038/nature02107. [DOI] [PubMed] [Google Scholar]
- 94.Song SU, et al. An env-like protein encoded by a Drosophila retroelement: Evidence that Gypsy is an infectious retrovirus. Genes Dev. 1994;8:2046–2057. doi: 10.1101/gad.8.17.2046. [DOI] [PubMed] [Google Scholar]
- 95.de Setta N, et al. Multiple invasions of Gypsy and Micropia retroelements in genus Zaprionus and melanogaster subgroup of the genus Drosophila. BMC Evol. Biol. 2009;9:18. doi: 10.1186/1471-2148-9-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gonzalez J, Petrov DA. The adaptive role of transposable elements in the Drosophila genome. Gene. 2009;15:124–133. doi: 10.1016/j.gene.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cordaux R, et al. Birth of a chimeric primate gene by apture of the transposase gene from a mobile element. Proc. Nat. Acad. Sci. 2006;103:8101–8106. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pardue M-L, DeBaryshe PG. Retrotransposons provide an evolutionarily robust non-telomerase mechanism to maintain telomeres. Ann. Rev. Genetics. 2003;37:485–511. doi: 10.1146/annurev.genet.38.072902.093115. (2003) [DOI] [PubMed] [Google Scholar]
- 99.Kaessmann H, et al. RNA-based gene duplication: mechanistic and evolutionary insights. Nat. Rev. Genet. 2009;10:19–31. doi: 10.1038/nrg2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marques-Bonet T, et al. The origins and impacto f primate segmental duplications. Trends Genet. 2009;25:443–454. doi: 10.1016/j.tig.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Compilation of all known cases of HTT involving eukaryotes classified by TE class, TE superfamily, TE family, and organism. Iincludes all reported cases of eukaryotic HTTs supported by sequence evidence. HT of viruses are not considered in this study. The column “Evidence” indicates which line(s) of evidence were used to infer HTT. The abbreviations follow Loreto et al. (2008): ss = sequence similarity; Ks = comparison between the number of synonymous mutations observed at orthologous genes and the number of synonymous mutations observed in TEs; dN/dS = test for purifying selection; phyl = phylogeny of the TE incongruent with the phylogeny of the host; pd = patchy taxonomic distribution of the TE; * = cross-phyla HTT.
References
1. Alberola, T.M., and de Frutos, R. (1996) Molecular structure of a gypsy element of Drosophila subobscura (gypsyDs) constituting a degenerate form of insect retroviruses. Nucleic Acids Res. 24, 914-923
2. Arca, B., and Savakis, C. (2000) Distribution of the transposable element Minos in the genus Drosophila. Genetica 108, 263-267
3. Bartolome, C., et al. (2009) Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes. Genome Biol. 10
4. Biedler, J.K., et al. (2007) Evolution and horizontal transfer of a DD37E DNA transposon in mosquitoes. Genetics 177, 2553-2558
5. Brunet, F., et al. (1999) Phylogenetic analysis of Mos1-like transposable elements in the Drosophilidae. J. Mol. Evol. 50, 760-768
6. Casola, C., et al. (2007) PIF-like Transposons are common in Drosophila and have been repeatedly domesticated to generate new host genes. Mol. Biol. Evol.24, 1872-1888
7. Clark, J.B., and Kidwell, M.G. (1997) A phylogenetic perspective on P transposable element evolution in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 94, 11428-11433
8. Clark, J.B., et al. (1994) Phylogenetic analysis supports horizontal transfer of P transposable elements. Mol. Biol. Evol.11, 40-50
9. Daboussi, M.J., et al. (2002) Evolution of the Fot1 transposons in the genus Fusarium: Discontinuous distribution and epigenetic inactivation. Mol. Biol. Evol.19, 510-520
10. Daniels, S.B., et al. (1990) Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics 124, 339-355
11. Daniels, S.B., et al. (1984) Sequences homologous to P elements occur in Drosophila paulistorum. Proc. Natl. Acad. Sci. U.S.A.81, 6794-6797
12. de Almeida, L.M., and Carareto, C.M.A. (2005) Multiple events of horizontal transfer of the Minos transposable element between Drosophila species. Mol. Phylogenet. Evol. 35, 583-594
13. de Almeida, L.M., and Carareto, C.M.A. (2006) Sequence heterogeneity and phylogenetic relationships between the copia retrotransposon in Drosophila species of the repleta and melanogaster groups. Genet. Sel. Evol. 38, 535–550
14. de Castro, J.P., and Carareto, C.M.A. (2004) Canonical P elements are transcriptionally active in the saltans group of Drosophila. J. Mol. Evol. 59, 31-40
15. de Setta, N., et al. (2009) Multiple invasions of Gypsy and Micropia retroelements in genus Zaprionus and melanogaster subgroup of the genus Drosophila. BMC Evol. Biol. 9, 18
16. Deprá, M., et al. (2010) hosimary: a new hAT transposon group involved in horizontal transfer. Mol. Genet. Genomics 283, 451-459
17. Diao, X.M., et al. (2006) Horizontal transfer of a plant transposon. PLoS Biol. 4, 119-128
18. Evgen'ev, M., et al. (2000) Invasion of Drosophila virilis by the Penelope transposable element. Chromosoma 109, 350-357
19. Fablet, M., et al. (2007) Evolutionary pathways of the tirant LTR retrotransposon in the Drosophila melanogaster subgroup of species. J. Mol. Evol. 64, 438-447
20. Garcia-Planells, J., et al. (1998) Molecular evolution of P transposable elements in the genus Drosophila. II. The obscura species group. J. Mol. Evol. 47, 282-291
21. Gilbert C., et al. (2010) A role for host-parasite relationships in horizontal transfer of DNA transposons across animal phyla. Nature 464, 1347-1350
22. Gladyshev, E.A., and Arkhipova I.R., (2009) A single copy IS5-like transposon in the genome of a bdelloid rotifer. Mol. Biol. Evol. 26, 1921-1929
23. Gogolevsky, K.P., et al. (2008) Bov-B-mobilized SINEs in vertebrate genomes. Gene 407, 75-85
24. Gomulski, L.M., et al. (2001) A new basal subfamily of mariner elements in Ceratitis rosa and other tephritid flies. J. Mol. Evol. 53, 597-606
25. Gonzalez, P., and Lessios H.A., Evolution of Sea Urchin Retroviral-Like (SURL) Elements: Evidence from 40 Echinoid Species. Mol. Biol. Evol. 16, 938–952
26. Hagemann, S., et al. (1998) Horizontal transmission versus vertical inheritance of P elements in Drosophila and Scaptomyza: Has the M-type subfamily spread from East Asia? J. Zool. Syst. Evol. Research 36, 75-83
27. Hagemann, S., et al. (1996) Repeated horizontal transfer of P transposons between Scaptomyza pallida and Drosophila bifasciata. Genetica 98, 43-51
28. Hagemann, S., et al. (1992) Identification of a complete P element in the genome of Drosophila bifasciata. Nucleic Acids Res.20, 409-413
29. Hamada, M., et al. (1997) A newly isolated family of short interspersed repetitive elements (SINEs) in coregonid fishes (whitefish) with sequences that are almost identical to those of the SmaI family of repeats: Possible evidence for the horizontal transfer of SINEs. Genetics 146, 355-367
30. Haring, E., et al. (2000) Ancient and recent horizontal invasions of drosophilids by P elements. J. Mol. Evol. 51, 577-586
31. Heredia, F., et al. (2004) Complex evolution of gypsy in drosophilid species. Mol. Biol. Evol.21, 1831-1842
32. Ivics, Z., et al. (1996) Identification of functional domains and evolution of Tc1-like transposable elements. Proc. Natl. Acad. Sci. U.S.A. 93, 5008-5013
33. Jordan, I.K., et al. (1999) Evidence for the recent horizontal transfer of long terminal repeat retrotransposon. Proc. Natl. Acad. Sci. U.S.A. 96, 12621-12625
34. Jordan, I.K., and McDonald, J.F. (1998) Evolution of the copia retrotransposon in the Drosophila melanogaster species subgroup. Mol. Biol. Evol. 15, 1160-1171
35. Koga, A., et al. (2000) Evidence for recent invasion of the medaka fish genome by the Tol2 transposable element. Genetics 155, 273-281
36. Konieczny, A., et al. (1991) A superfamily of Arabidopsis thaliana retrotransposons. Genetics 127, 801-809
37. Leaver, M.J. (2001) A family of Tc1-like transposons from the genomes of fishes and frogs: evidence for horizontal transmission. Gene 271, 203-214
38. Lohe, A.R., et al. (1995) Horizontal transmission, vertical inactivation, and stochastic loss of mariner-like transposable elements. Mol. Biol. Evol. 12, 62-72
39. Loreto, E.L.D., et al. (2001) Drosophila mediopunctata P elements: A new example of horizontal transfer. J. Hered. 92, 375-381
40. Ludwig, A., and Loreto, E.L.S. (2007) Evolutionary pattern of the gtwin retrotransposon in the Drosophila melanogaster subgroup. Genetica 130, 161-168
41. Lyozin, G.T., et al. (2001) The structure and evolution of Penelope in the virilis species group of Drosophila: an ancient lineage of retroelements. J. Mol. Evol. 52, 445-456
42. Maruyama, K., and Hartl, D.L. (1991) Evidence for interspecific transfer of the transposable element mariner between Drosophila and Zaprionus. J. Mol. Evol. 33, 514-524
43. Maside, X., et al. (2003) Inferences on the evolutionary history of the S element family of Drosophila melanogaster. Mol. Biol. Evol. 20, 1183-1187
44. Morales-Hojas, R., et al. (2006) The evolutionary history of the transposable element Penelope in the Drosophila virilis group of species. J. Mol. Evol. 63, 262-273
45. Mota, N.R., et al. (2009) harrow: new Drosophila hAT transposons involved in horizontal transfer. Insect Mol. Biol. (In press).
46. Novick, P., et al. Independent and parallel lateral transfer of DNA transposons in tetrapod genomes. Gene 450, 85-94
47. Novikova, O., et al. (2009) Non-LTR retrotransposons in fungi. Funct. Integr. Genomics 9, 27-42
48. Novikova, O., et al. (2007) CRI clade of non-LTR retrotransposons from Maculinea butterflies (Lepidoptera : Lycaenidae): evidence for recent horizontal transmission. BMC Evol. Biol. 7, 19
49. Pace, J.K., et al. (2008) Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc. Natl. Acad. Sci. U.S.A. 105, 17023-17028
50. Pagan, H.J.T., et al. (2010) PiggyBac-ing on a primate genome: novel elements, recent activity and horizontal transfer. Genome Biol. Evol. doi:10.1093/gbe/evq021
51. Robertson, H.M. (1997) Multiple mariner Transposons in flatworms and hydras are related to those of insects. J. Hered. 88, 195-201
52. Robertson, H.M., and Lampe, D.J. (1995) Recent horizontal transfer of a mariner transposable element among and between Diptera and Neuroptera. Mol. Biol. Evol. 12, 850-862
53. Roulin, A., et al. (2009) Whole genome surveys of rice, maize and sorghum reveal multiple horizontal transfers of the LTR-retrotransposon Route66 in Poaceae. BMC Evol. Biol. 9, 10
54. Roulin, A., et al. (2008) Evidence of multiple horizontal transfers of the long terminal repeat retrotransposon RIRE1 within the genus Oryza. Plant J. 53, 950-959
55. Sanchez-Gracia, A.., et al. (2005) High rate of horizontal transfer of transposable elements in Drosophila. Trends Genet. 21, 200-203
56. Silva, J.C., and Kidwell, M.G. (2000) Horizontal transfer and selection in the evolution of P elements. Mol. Biol. Evol. 17, 1542-1557
57. Simmons, G.M. (1992) Horizontal transfer of hobo transposable elements within the Drosophila melanogaster species complex: Evidence from DNA sequencing. Mol. Biol. Evol. 9, 1050-1060
58. Smit, A.F.A., and Riggs, A.D. (1996) Tiggers and other DNA transposon fossils in the human genome. Proc. Natl. Acad. Sci. U.S.A. 93, 1443-1448
59. Terzian, C., et al. (2000) Evolution of the gypsy endogenous retrovirus in the Drosophila melanogaster subgroup. Mol. Biol. Evol. 17, 908-914
60. Torti, C., et al. (2005) Cchobo, a hobo-related sequence in Ceratitis capitata. Genetica 123, 313-325
61. Vazquez-Manrique, R.P., et al. (2000) Evolution of gypsy endogenous retrovirus in the Drosophila obscura species group. Mol. Biol. Evol. 17, 1185-1193
62. Volff, J.N., et al. (2000) Multiple lineages of the non-LTR retrotransposon Rex1 with varying success in invading fish genomes. Mol. Biol. Evol. 17, 1673-1684
63. Waterston, R.H., et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520-562
64. Yoshiyama, M., et al. (2001) Possible horizontal transfer of a transposable element from host to parasitoid. Mol. Biol. Evol. 18, 1952-1958
65. Zupunski, V., et al. (2001) Evolutionary dynamics and evolutionary history in the RTE clade of non-LTR retrotransposons. Mol. Biol. Evol. 18, 1850-1863