Abstract
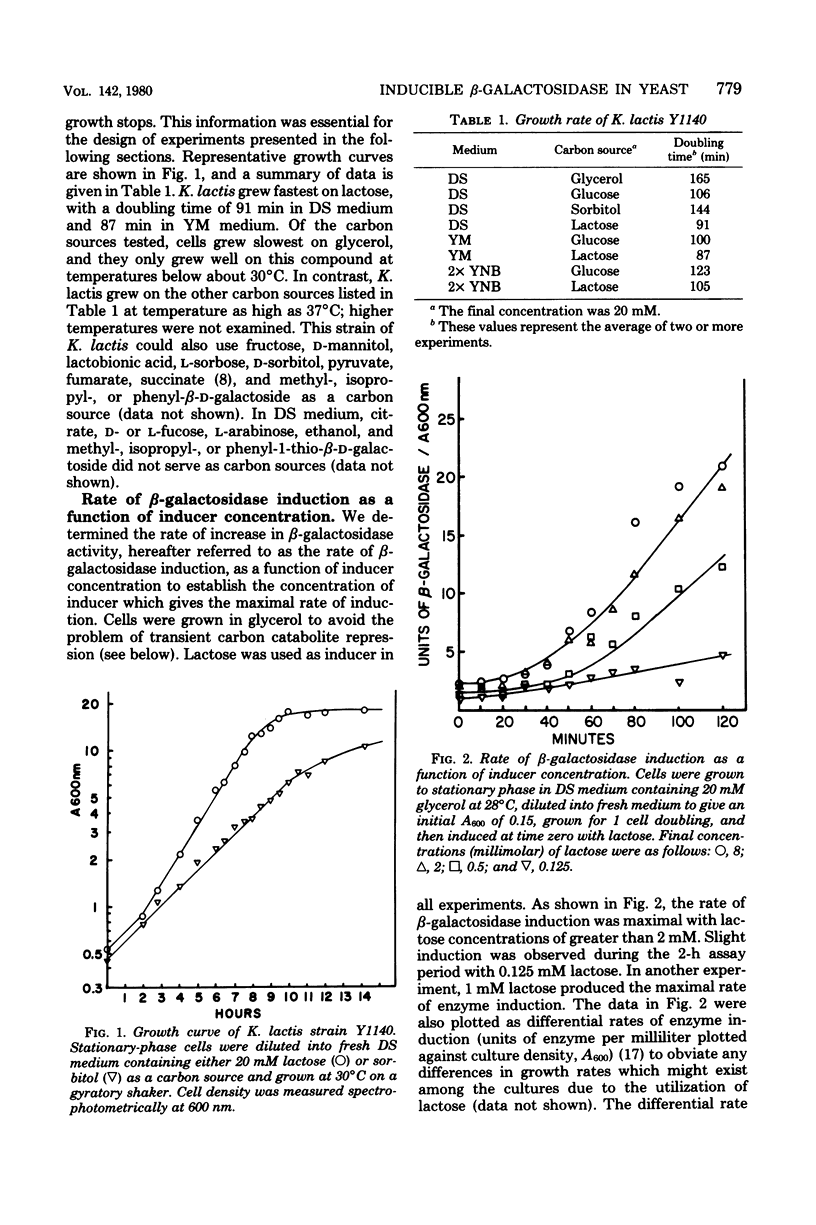
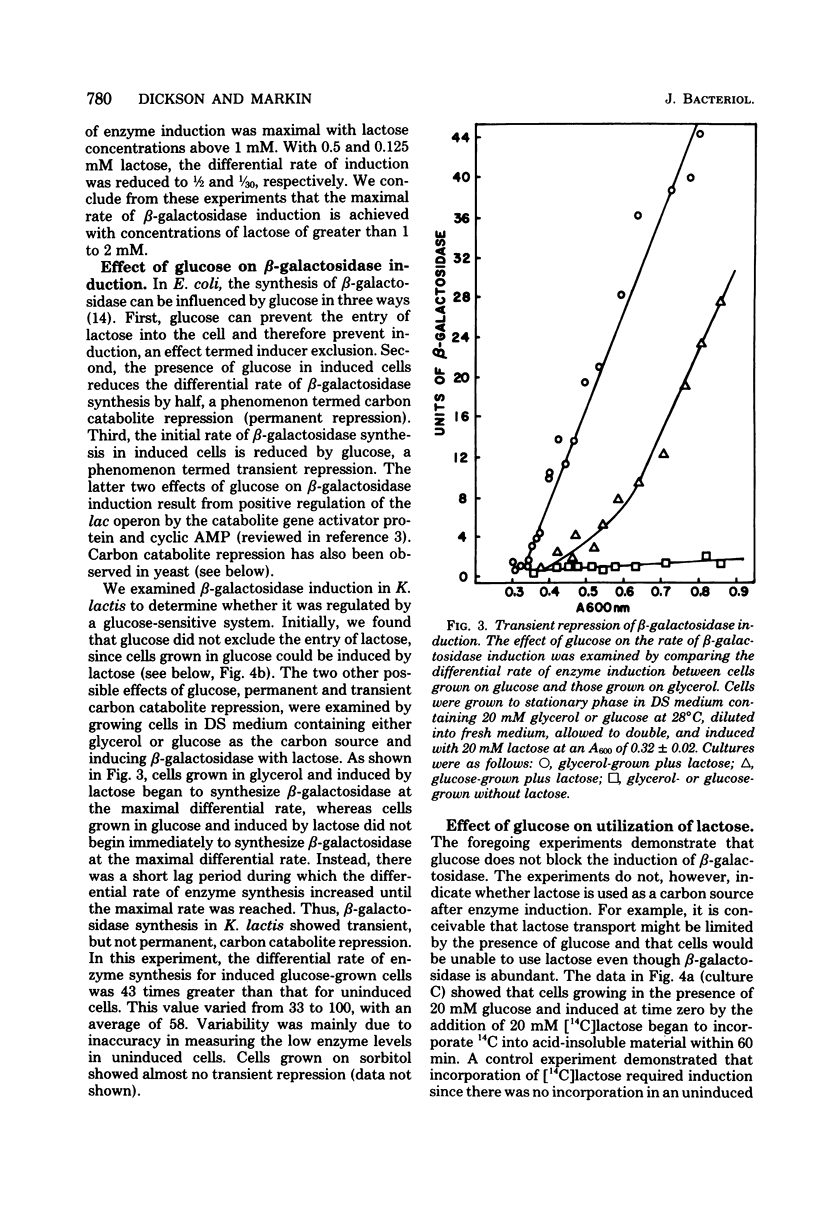
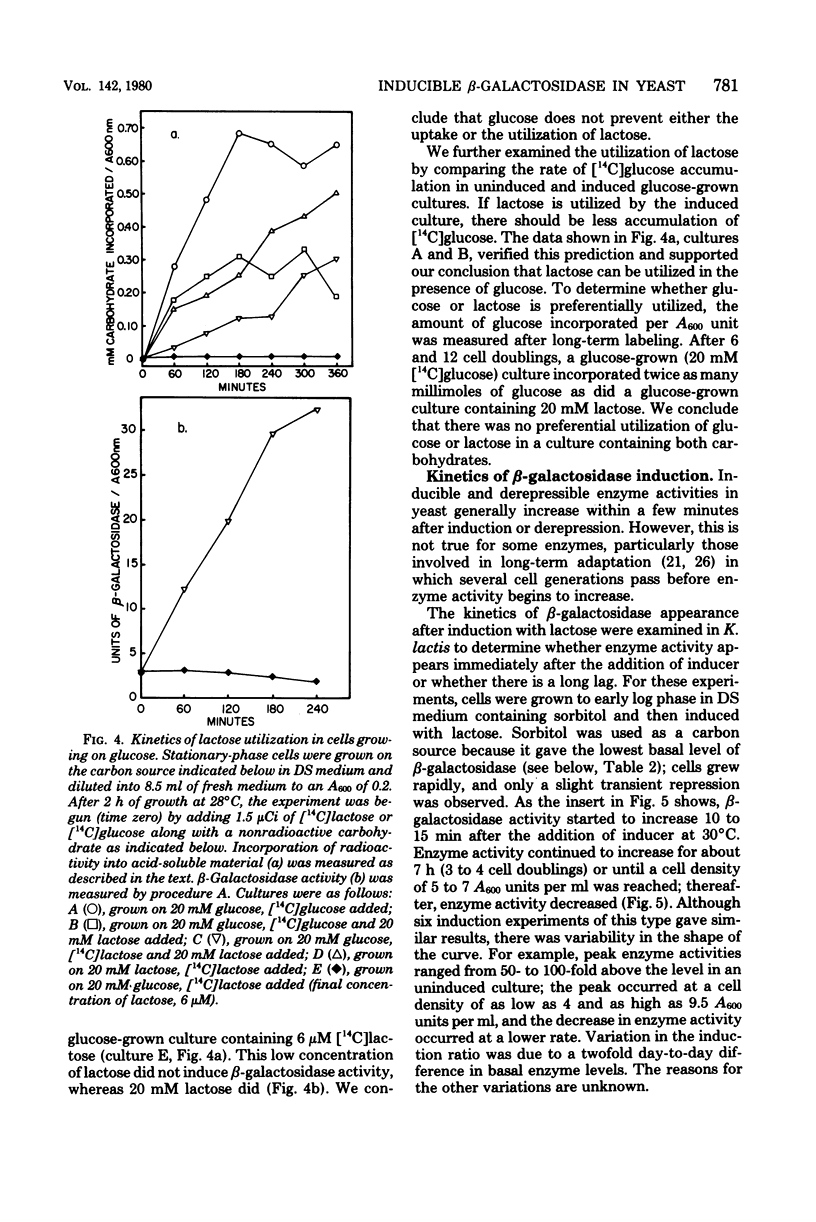
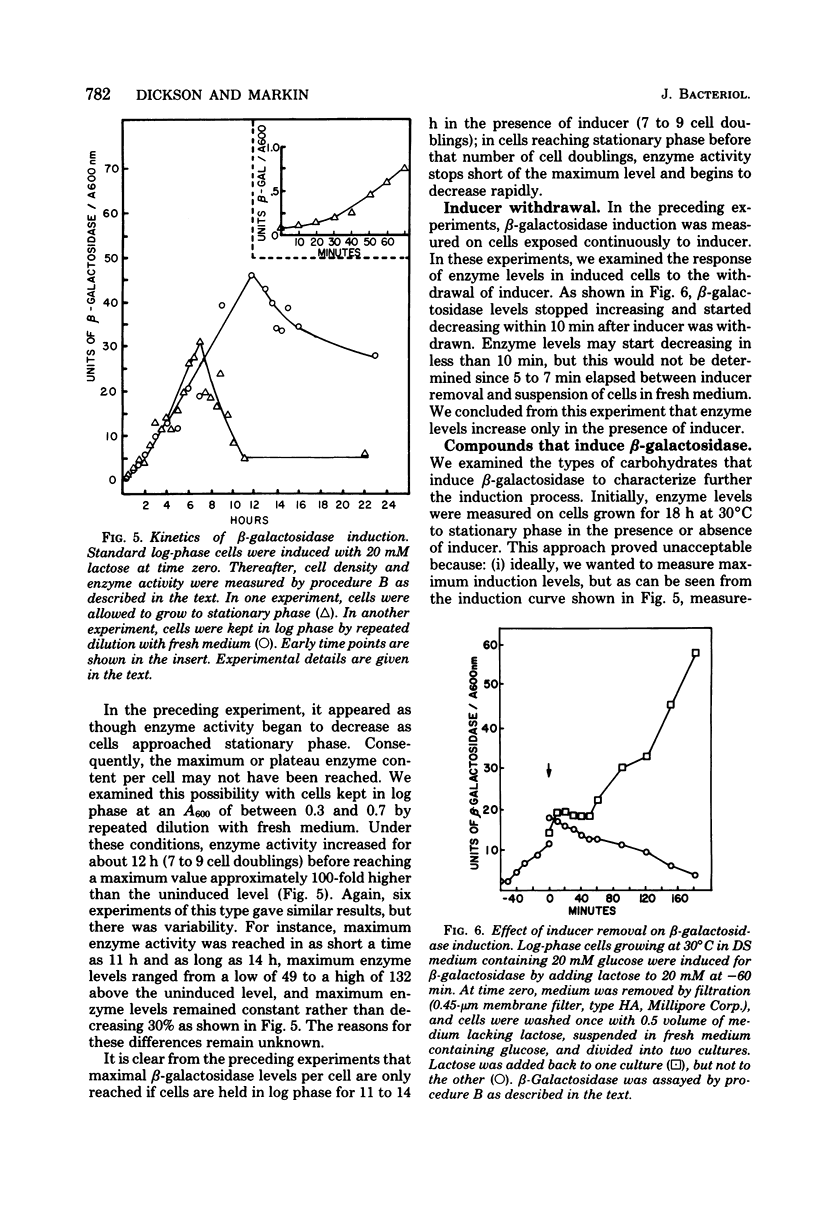
We examined the kinetics of β-galactosidase (EC 3.2.1.23) induction in the yeast Kluyveromyces lactis. Enzyme activity began to increase 10 to 15 min, about 1/10 of a cell generation, after the addition of inducer and continued to increase linearly for from 7 to 9 cell generations before reaching a maximum, some 125- to 150-fold above the basal level of uninduced cells. Thereafter, as long as logarithmic growth was maintained, enzyme levels remained high, but enzyme levels dropped to a value only 5- to 10-fold above the basal level if cells entered stationary phase. Enzyme induction required the constant presence of inducer, since removal of inducer caused a reduction in enzyme level. Three nongratuitous inducers of β-galactosidase activity, lactose, galactose, and lactobionic acid, were identified. Several inducers of the lac operon of Escherichia coli, including methyl-, isopropyl- and phenyl-1-thio-β-d-galactoside, and thioallolactose did not induce β-galactosidase in K. lactis even though they entered the cell. The maximum rate of enzyme induction was only achieved with lactose concentrations of greater than 1 to 2 mM. The initial differential rate of β-galactosidase appearance after induction was reduced in medium containing glucose, indicating transient carbon catabolite repression. However, glucose did not exclude lactose from K. lactis, it did not cause permanent carbon catabolite repression of β-galactosidase synthesis, and it did not prevent lactose utilization. These three results are in direct contrast to those observed for lactose utilization in E. coli. Furthermore, these results, along with our observation that K. lactis grew slightly faster on lactose than on glucose, indicate that this organism has evolved an efficient system for utilizing lactose.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams B. G. Induction of galactokinase in Saccharomyces cerevisiae: kinetics of induction and glucose effects. J Bacteriol. 1972 Aug;111(2):308–315. doi: 10.1128/jb.111.2.308-315.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews K. J., Lin E. C. Thiogalactoside transacetylase of the lactose operon as an enzyme for detoxification. J Bacteriol. 1976 Oct;128(1):510–513. doi: 10.1128/jb.128.1.510-513.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriacy M. Cis-dominant regulatory mutations affecting the formation of glucose-repressible alcohol dehydrogenase (ADHII) in Saccharomyces cerevisiae. Mol Gen Genet. 1976 Jun 15;145(3):327–333. doi: 10.1007/BF00325831. [DOI] [PubMed] [Google Scholar]
- Dickson R. C., Dickson L. R., Markin J. S. Purification and properties of an inducible beta-galactosidase isolated from the yeast Kluyveromyces lactis. J Bacteriol. 1979 Jan;137(1):51–61. doi: 10.1128/jb.137.1.51-61.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. C., Markin J. S. Molecular cloning and expression in E. coli of a yeast gene coding for beta-galactosidase. Cell. 1978 Sep;15(1):123–130. doi: 10.1016/0092-8674(78)90088-0. [DOI] [PubMed] [Google Scholar]
- HERMAN A., HALVORSON H. GENETIC CONTROL OF BETA-GLUCOSIDASE SYNTHESIS IN SACCHAROMYCES LACTIS. J Bacteriol. 1963 Apr;85:901–910. doi: 10.1128/jb.85.4.901-910.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HERMAN A., HALVORSON H. IDENTIFICATION OF THE STRUCTURAL GENE FOR BETA-GLUCOSIDASE IN SACCHAROMYCES LACTIS. J Bacteriol. 1963 Apr;85:895–900. doi: 10.1128/jb.85.4.895-900.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber R. E., Kurz G., Wallenfels K. A quantitation of the factors which affect the hydrolase and transgalactosylase activities of beta-galactosidase (E. coli) on lactose. Biochemistry. 1976 May 4;15(9):1994–2001. doi: 10.1021/bi00654a029. [DOI] [PubMed] [Google Scholar]
- Jobe A., Bourgeois S. lac Repressor-operator interaction. VI. The natural inducer of the lac operon. J Mol Biol. 1972 Aug 28;69(3):397–408. doi: 10.1016/0022-2836(72)90253-7. [DOI] [PubMed] [Google Scholar]
- Langridge J. Mutations conferring quantitative and qualitative increases in beta-galactosidase activity in Escherichia coli. Mol Gen Genet. 1969;105(1):74–83. doi: 10.1007/BF00750315. [DOI] [PubMed] [Google Scholar]
- Lawther R. P., Cooper T. G. Kinetics of induced and repressed enzyme synthesis in Saccharomyces cerevisiae. J Bacteriol. 1975 Mar;121(3):1064–1073. doi: 10.1128/jb.121.3.1064-1073.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MONOD J., PAPPENHEIMER A. M., Jr, COHEN-BAZIRE G. La cinétique de la biosynthèse de la beta-galactosidase chez E. coli considérée comme fonction de la croissance. Biochim Biophys Acta. 1952 Dec;9(6):648–660. doi: 10.1016/0006-3002(52)90227-8. [DOI] [PubMed] [Google Scholar]
- Matern H., Holzer H. Catabolite inactivation of the galactose uptake system in yeast. J Biol Chem. 1977 Sep 25;252(18):6399–6402. [PubMed] [Google Scholar]
- Polakis E. S., Bartley W. Changes in the enzyme activities of Saccharomyces cerevisiae during aerobic growth on different carbon sources. Biochem J. 1965 Oct;97(1):284–297. doi: 10.1042/bj0970284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytka J., Sledziewski A., Lukaszkiewicz J., Biliński T. Haemoprotein formation in yeast. III. The role of carbon catabolite repression in the regulation of catalase A and T formation. Mol Gen Genet. 1978 Mar 20;160(1):51–57. [PubMed] [Google Scholar]
- SPIEGELMAN S., SUSSMAN R. R., PINSKA E. On the cytoplasmic nature of "long-term adaptation" in yeast. Proc Natl Acad Sci U S A. 1950 Nov;36(11):591–606. doi: 10.1073/pnas.36.11.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian J., Carter B. L., Halvorson H. O. Induction capacity of enzyme synthesis during the cell cycle of Saccharomyces cerevisiae. Eur J Biochem. 1973 Sep 3;37(3):516–522. doi: 10.1111/j.1432-1033.1973.tb03013.x. [DOI] [PubMed] [Google Scholar]
- Stewart P. R. Analytical methods for yeasts. Methods Cell Biol. 1975;12:111–147. doi: 10.1016/s0091-679x(08)60955-3. [DOI] [PubMed] [Google Scholar]
- Tingle M., Halvorson H. O. Mutants in Saccharomyces lactis controlling both -glucosidase and -galactosidase activities. Genet Res. 1972 Feb;19(1):27–32. doi: 10.1017/s0016672300014233. [DOI] [PubMed] [Google Scholar]
- Yashphe J., Halvorson H. O. beta-D-galactosidase activity in single yeast cells during cell cycle of Saccharomyces lactis. Science. 1976 Mar 26;191(4233):1283–1284. doi: 10.1126/science.1257751. [DOI] [PubMed] [Google Scholar]
- Zimmermann F. K., Kaufmann I., Rasenberger H., Haubetamann P. Genetics of carbon catabolite repression in Saccharomycess cerevisiae: genes involved in the derepression process. Mol Gen Genet. 1977 Feb 28;151(1):95–103. doi: 10.1007/BF00446918. [DOI] [PubMed] [Google Scholar]
- Zimmermann F. K., Scheel I. Mutants of Saccharomyces cerevisiae resistant to carbon catabolite repression. Mol Gen Genet. 1977 Jul 7;154(1):75–82. doi: 10.1007/BF00265579. [DOI] [PubMed] [Google Scholar]


