Abstract
Examples of small self-cleaving RNAs embedded in noncoding regions already have been found to be involved in the control of gene expression, although their origin remains uncertain. In this work, we show the widespread occurrence of the hammerhead ribozyme (HHR) motif among genomes from the Bacteria, Chromalveolata, Plantae, and Metazoa kingdoms. Intergenic HHRs were detected in three different bacterial genomes, whereas metagenomic data from Galapagos Islands showed the occurrence of similar ribozymes that could be regarded as direct relics from the RNA world. Among eukaryotes, HHRs were detected in the genomes of three water molds as well as 20 plant species, ranging from unicellular algae to vascular plants. These HHRs were very similar to those previously described in small RNA plant pathogens and, in some cases, appeared as close tandem repetitions. A parallel situation of tandemly repeated HHR motifs was also detected in the genomes of lower metazoans from cnidarians to invertebrates, with special emphasis among hematophagous and parasitic organisms. Altogether, these findings unveil the HHR as a widespread motif in DNA genomes, which would be involved in new forms of retrotransposable elements.
Keywords: RNA world, satellite DNA, viroid, three-helical junction
INTRODUCTION
RNAs display a large variety of roles in biology, including the capability of chemical catalysis in the absence of proteins (Kruger et al. 1982), a feature that provided support for the hypothesis of a prebiotic RNA world (Gilbert 1986). Among these autocatalytic RNAs, the hammerhead ribozyme (HHR) has been extensively studied as a paradigm for small ribozymes after its discovery in viroids and other small RNA plant pathogens (Prody et al. 1986). HHRs catalyze a transesterification reaction of self-cleavage, a step required for the replication of these infectious circular RNAs (Flores et al. 2004). For the last 20 years, the HHR has been considered an oddity whose presence has been also exceptionally reported in the genomes of a few unrelated eukaryotes: the satellite DNA of newts (Epstein and Gall 1987), schistosomes (Ferbeyre et al. 1998) and crickets (Rojas et al. 2000); carnation (Daròs and Flores 1995), and Arabidopsis thaliana (Przybilski et al. 2005) genomes; some mammalian 3′ UTRs (Martick et al. 2008); and, more recently, widespread in the genomes of Xenopus tropicalis, lampreys, and as intronic motifs ultraconserved in Amniota species, including Homo sapiens (de la Peña and García-Robles 2010).
Although a vast amount of biochemical and structural data have been published about the HHR, an evolutionary framework to interpret its origin and exceptional presence in eukaryotic genomes has hitherto been lacking. In this work, and following a simple but powerful bioinformatic analysis, we have detected the ubiquitous presence of this ribozyme among most life kingdoms. Altogether, our results unveil a more general landscape for the role of HHRs within the retrotransposable elements in eukaryotes, whose detailed molecular characterization should be deciphered in the future.
RESULTS
Bioinformatic search of HHRs
Previously, extensive bioinformatic approaches have been devised to search HHR motifs among genomes (for a review, see Hammann and Westhof 2007). Basically, these methods considered the 15 catalytic conserved nucleotides as the only source of the phylogenetic signal, and when combined with the secondary structure algorithms allowed to detect the HHR fold (Ferbeyre et al. 1998, 2000; Rojas et al. 2000; Przybilski et al. 2005; Martick et al. 2008). In this work, an extra phylogenetical signal out of the conserved core was considered. More precisely, tertiary interactions between HHR helixes (de la Peña et al. 2003; Khvorova et al. 2003) show significant conservation in stem size and loop composition (Supplemental Fig. S1; Chi et al. 2008; Dufour et al. 2009; de la Peña et al. 2009). Thus, natural HHR motifs were split into two major helical motifs (seeds), each one composed of either the SC site/Helix I/U box (I-type seeds) or the U box/Helix II/P box (II-type seeds). Seeds were used for BLAST searches (Fig. 1), and when II-type seeds were employed, the retuned hits often fulfilled the criteria for a HHR fold: (1) sequences folded as canonical Helixes II; (2) catalytic boxes were preserved; and (3) 5′ and 3′ surrounding regions could adopt canonical Helixes I, III, and SC motifs. Despite the intrinsic low probability of a chance occurrence for the HHR motif (1 per 1013 nucleotide) (Ferbeyre et al. 1998), extra points of validation for our in silico searches were obtained from (4) their recurrent appearance within the telomeric and tandem repeats, like previously described in other organisms (Epstein and Gall 1987; Ferbeyre et al. 1998; Rojas et al. 2000); (5) examples of covariations and compensatory mutations reinforcing the RNA helixes; (6) the presence of compatible loop1–loop2 interactions; or (7) in the case of higher vertebrates, an evolutionary ultraconservation within intronic regions (de la Peña and García-Robles 2010).
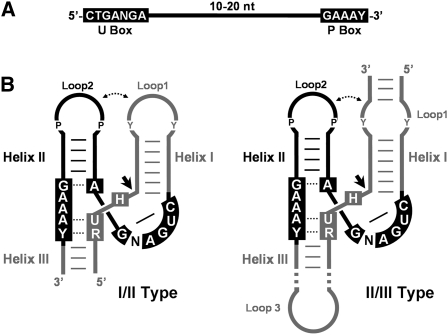
FIGURE 1.
Small sequence strings of 22–32 nt were used for the HHR motif searches. (A) Schematic representation of seeds used for the bioinformatic searches. The conserved sequence motifs among all seeds (black boxes) corresponded to the region containing the U-turn (U Box) and a purine-rich motif (P Box). (B) Schematic representation of the I/II-type (left) and II/III-type (right) HHRs considered in this study, sharing Helix II, U, and P boxes (in black color). Consensus self-cleavage site (RUH box), Helix I, and Helix III domains not included in the searches, but required to form a catalytically active RNA, are depicted in gray color. Purine and pyrimidine nucleotides involved in most naturally occurring loop1–loop2 interactions are indicated as P and Y, respectively.
Presence of HHRs in bacteria and metagenomic data
Up to four HHR motifs were detected in three different bacterial genomes. A first I/II-type HHR was found in the genome of the plant endosymbiont Azorhizobium caulinodans (Lee et al. 2008). The ribozyme mapped within a small intergenic region of 99 nucleotides (nt) and in the same strand as both genes surrounding the motif (Fig. 2A, left). A second ribozyme was found in the genomic plasmid pGOX1 of the plant-associated bacteria Gluconobacter oxydans (Prust et al. 2005). This II/III-type HHR mapped in an intergenic region of 560 nt preceded by a coding region in the same strand (Fig. 2A, right). Two identical II/III-type HHRs were also detected in the genome of the halophilic anaerobic bacterium Desulfotomaculum reducens (Fig. 2B, left). Both motifs were located at similar intergenic regions of 1 and 2 kb, respectively. The genes preceding both ribozymes were also coded in the same strand as the HHR.
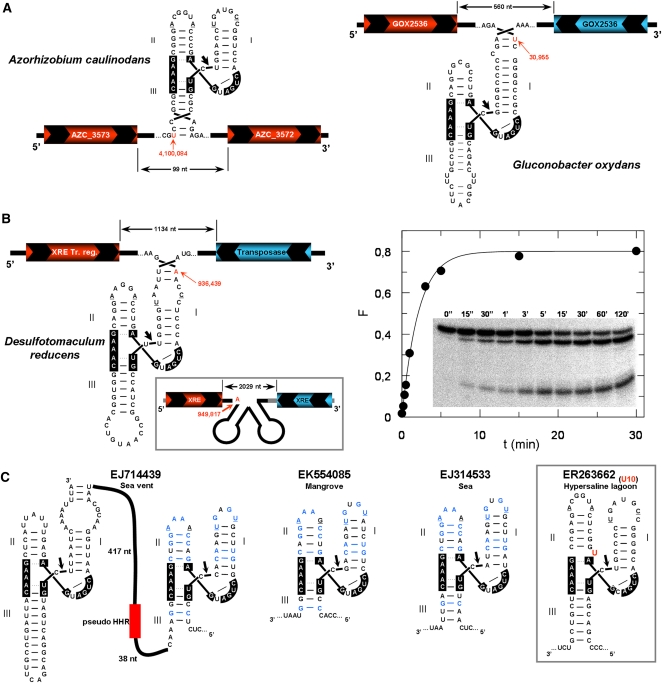
FIGURE 2.
Bacterial genomic data showing the presence of HHRs. (A) The HHR motifs obtained in the plant-associated bacteria Azorhizobium caulinodans, (left) and Gluconobacter oxydans (right). Positions of the ribozymes within each genome are shown in red with an arrow. The hypothetical ORFs coded in the sense and antisense strands are shown in red and blue, respectively. Nucleotides involved in the conserved loop–loop interaction are underlined. (B) The HHR motifs detected within the genome of the Desulfotomaculum reducens bacteria. Kinetic analysis of the in vitro self-cleavage capabilities of this ribozyme is shown at the right. (C) Some examples of HHR motifs detected from the Global Ocean metagenomics project (Rusch et al. 2007). A case showing a U insertion at the HHR catalytic core is shown in the right inset.
G. oxydans and D. reducens II/III-type ribozymes showed an atypically extended Helix III. However, this helix is very short in most naturally occurring II/III-type HHRs (Supplemental Fig. S1) that largely prevents self-cleavage. It has been proposed that the adoption of double HHRs placed in tandem allows extending Helix III and reaching regular levels of cleavage (Forster et al. 1988). For the case of D. reducens HHR, in vitro self-cleavage resulted in a moderate activity at 25°C in 1 mM Mg2+ (kobs = 0.23 ± 0.07 min−1) (Fig. 2B, right), indicating that this ribozyme could efficiently self-cleave as a single motif.
A last and puzzling observation was obtained from the metagenomics project of the Sorcerer II Global Ocean Sampling expedition (Rusch et al. 2007). Ten different HHRs were detected in the DNA sequences exclusively obtained from microbial samples at the volcanic Galapagos Islands, from either marine or hypersaline lagoon origin (Fig. 2C; Supplemental Fig. S2). Ribozymes were I/II-type HHRs, with the exception of a single DNA entry showing both I/II- and II/III-type HHRs. Noticeably, we also detected a HHR showing an unpaired uracil at the base of Helix II (U10) in a similar way as previously described for some viroidal ribozymes (Fig. 2C, inset; de la Peña and Flores 2001).
Widespread presence of HHRs in oomycete and plant genomes
II/III-type HHR motifs were detected in expressed sequence tags (ESTs) and genomic sequences from three eukaryotic plant pathogens (oomycetes) of the Chromalveolata kingdom: Phytophtora infestans, Phytophtora sojae, and Hyaloperonospora parasitica. The whole ribozyme motifs from these water molds were all very similar, with most of the sequence variability reinforcing the three-helical structure (Fig. 3A). Like most natural II/III-HHRs (see above), Helixes III were short and capped by palindromic loops, an indication of a double-HHR self-cleavage mechanism (Forster et al. 1988). This possibility is already suggested for some of the P. infestans motifs, which appeared as tandem repeats of 710 nt (Fig. 3A; Supplemental Fig. S3). As a common feature for the Chromalveolata HHRs, the stems of Helices I and II were extended and reduced by 2–3 base pairs, respectively, compared with most natural HHRs, indicating that HHR tertiary interactions, if any, could be different.
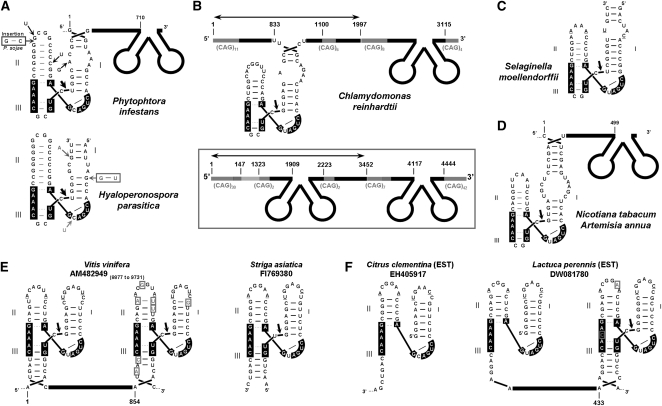
FIGURE 3.
HHRs in Chromalveolata and Plantae genomic data. (A) The HHR motifs detected in sequences of water molds from the Chromalveolata kingdom (Phytophtora infestans, Phytophtora sojae, and Hyaloperonospora parasitica). Some of the intraspecies nucleotide variability is highlighted with arrows. (B) Examples of genomic HHRs found in the algae Chlamydomonas reindhartii. The number of CAG repetitions is shown by subscripts. Monomer units are indicated by arrows. (C) HHR found in the club moss Selaginella moellendorfii associated with telomeric DNA sequences. (D) II/III-type HHR motifs found in ESTs from Nicotiana tabacum and Artemisia annua plants. (E) Examples of I/II-type HHR motifs found in sequences from grape vine Vitis vinifera (left). Sequence heterogeneity within HHRs is highlighted with boxed nucleotides) or the parasitic witchweed Striga asiatica (right). (F) Evidences of in vivo activity for HHR motifs mapping at the 5′ end of several plant ESTs.
Our bioinformatic search also unveiled more than 50 different HHRs from 20 species of the Plantae kingdom. Organisms ranged from a unicellular algae (Chlamydomonas reinhardtii) to a primitive club moss (Selaginella moellendorffii) and vascular plants (herbaceous and woody species), suggesting a generalized occurrence of HHRs in plants (Supplemental Table S1). Up to three different II/III-HHR motifs were detected in 11 Chlamydomonas genomic scaffolds (Fig. 3B; Supplemental Table S1). Some of the ribozymes were embedded within dimeric repeats of 1–3 kb, with each monomer flanked by CAG-triplet repeats. Genomic sequences from the Selaginella moss showed a couple of II/III-type HHRs (Fig. 3C), both associated with tandem repeats of the sequence 5′-CCCTAAA-3′, a typical motif in telomeric and subtelomeric regions of plants (Richards and Ausubel 1988). Similar II/III-type HHRs were detected in sequence entries of the vascular plants Nicotiana tabacum and Artemisia annua (Fig. 3D), which again appeared embedded in tandem repeats of 499 nt (Supplemental Fig. S4).
But most of the detected HHRs in the plants corresponded to the characteristic I/II-type from plant virus satellites and viroids (Fig. 3E; Supplemental Table S1). Interestingly, several ESTs from Citrus clementina or Lactuca perennis showed a perfect match between the 5′ end of these cloned RNAs and the expected 5′ end of the self-cleaved ribozyme, strongly suggesting that these HHRs self-cleave in vivo (Fig. 3F).
Presence of HHRs in metazoan genomes: From Cnidaria to Arthropoda
HHRs of the II/III-type were previously described in the Smα satellite DNA of Schistosoma mansoni and related platyhelminth parasites like Schistosoma haematobium or Schistosoma douthitti (Ferbeyre et al. 1998). Thanks to the accomplishment of the S. mansoni and Schistosoma japonicum genomes, the presence of thousands of HHR entries in these organisms has been recently reported (de la Peña and García-Robles 2010). For many entries, HHR-inactivating mutations were detected within the catalytic boxes, indicating possible cases of fossilized ribozymes.
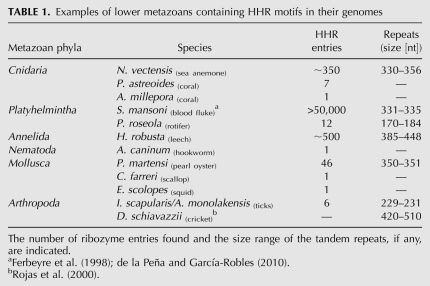
Our searches revealed the widespread presence of similar HHRs in the metazoan genomes (Table 1). Examples of HHRs were already found in very simple animals like the cnidarians Nematostella vectensis (sea anemone) or Porites astreoides and Acropora millepora corals (Fig. 4A; Supplemental Table S1). BLAST searches with the whole HHR motif of N. vectensis against its genome (Putnam et al. 2007) resulted in hundreds of highly similar entries. In many cases, double or even triple ribozymes were found within 350-nt repeats (Fig. 4A).
TABLE 1.
Examples of lower metazoans containing HHR motifs in their genomes
FIGURE 4.
Examples of II/III-type HHR motifs associated to repetitive and telomeric DNA in metazoans. Examples of metazoan II/III-type HHR motifs detected in (A) Cnidaria, (B) Rotifera, (C) Annelida, and (D) Arthropoda species. Natural HHR heterogeneity for each entry is highlighted with boxed nucleotides.
The II/III-type HHRs were also detected in the genomes of more evolved metazoans. HHR motifs were found within tandem repetitions of 174 nt at the telomeric repeats and subterminal DNA junctions of the rotifer Philodina roseola (Fig. 4B). Zooparasites like the dog hookworm Ancylostoma caninum or the leech Helobdella robusta also revealed the presence of II/III-type HHRs. For this latter case, genomic searches with the whole HHR showed hundreds of entries, which in some cases appeared again as tandem HHR motifs separated by 385–450 nt (Fig. 4C). The presence of HHRs was also revealed in sequences from molluscs like the pearl oyster Pinctada martensis, the scallop Chlamys farreri, and the squid Euprymna scolopes (Table 1; Supplemental Table S1). Finally, and in a similar way as previously described for the satellite DNA of some Dolichopoda species (Rojas et al. 2000), HHRs were also detected in the genomes from different orders among the arthropods: Diptera (three mosquito species), Coleoptera (one beetle species), Hemiptera (one psyllid species), Himenoptera (one termite species), and Ixodida and other Aracnida species. For this latter case, up to three different tick species were detected, with noticeable examples of triple tandem HHRs obtained from analysis of the salivary transcriptome (Fig. 4D; Supplemental Table S1).
DISCUSSION
Although the main scope of this communication is to make known the widespread presence of HHR motifs among genomes, some implications and roles can be already advanced. Both the currently available genomic data and the iterative nature of our search method make regarding some of these generalizations with caution. In any case, our data would confirm that HHRs and small self-cleaving ribozymes in general (Webb et al. 2009; de la Peña and García-Robles 2010) are widespread in the genomes of most life beings, and together with their regular association within satellite DNAs, telomeric regions and RT ORFs point to a role for these ribozymes in mobile genetic elements.
Concerning the HHRs detected in the genomes of two bacterial species (A. caulinodans and G. oxydans) and three water molds, it has to be noticed that these organisms are either obligate plant parasites or symbionts. This observation, together with the large set of HHRs found in plant genomes, suggests that HHRs in these organisms could have come from a horizontal gene transfer from plants. However, this putative plant origin would not fit with the intergenic HHRs found in D. reducens, an anaerobic and sulfate-reducing bacterium initially described in marine sediments (Tebo and Obraztsova 1998) and hypersaline waters (Nevin et al. 2003). The case of D. reducens HHRs could be, in fact, connected with the collection of HHRs exclusively found in marine and hypersaline metagenomic data from the volcanic Galapagos Islands. Altogether, these data would suggest a more common presence for these self-cleaving RNAs in primitive bacterial or archaeal forms, which could be considered as the direct relics from an ancient RNA world. In any case, the possibility of multiple origins for the HHR during evolution cannot be ruled out (Salehi-Ashtiani and Szostak 2001)
Among the HHRs detected in plant and lower metazoan genomes, two major groups could be differentiated. Ribozymes appearing as isolated motifs within nonannotated genomic regions would require further characterization to conclude their origin and functions, if any. But for many cases, the HHR motifs recurrently appeared within the tandem repeats of satellite and telomeric DNA. A similar situation was described for HHRs in newts (Epstein and Gall 1987), schistosomes (Ferbeyre et al. 1998), and crickets (Rojas et al. 2000), indicating a role for this ribozyme in the biogenesis of such repetitive DNA. We could then assume that HHR-containing repeats would be involved in a form of retrotransposable elements of the SINE class (short interspersed repetitive elements). SINE retrotransposons are short DNA sequences of a few hundred nucleotides originating through the reverse transcription of small RNA molecules, exemplified by the Alu elements in primates. Interestingly, both the Alu and HHR RNA motifs share similar three-helical junction structures (Supplemental Fig. S5; Weichenrieder et al. 2000; de la Peña et al. 2009), a feature that could advance some hints of the molecular mechanisms involved in their successful genomic integration. Moreover, another striking trait detected in many of the tandem HHRs was the presence of inactivating mutations in those motifs located at the 3′ side, whereas the catalytically competent domains remain at the 5′ side.
The large occurrence of HHRs detected in plant genomes suggests that HHR-containing RNA plant pathogens (viroids and plant virus satellites) could have, in fact, arisen from the host plant transcriptome in a similar way as previously proposed for the HDV satellite in humans (Salehi-Ashtiani et al. 2006). Supporting this hypothesis, the noninfectious retroviroid described in carnation plants (Daròs and Flores 1995) would be an example of an intermediate step between genomic and free HHR-containing RNA. Also in this line, de novo emergence of a plant virus satellite has been recently involved with a transcriptomic origin (Hajimorad et al. 2009).
A last point about the presence of HHRs in metazoans concerns the intriguing absence of HHRs in well-known genomes from model invertebrate organisms. This feature, together with a puzzling preponderance among the parasitic and hematofagous species and their hosts (molluscs and vertebrates), suggests that these HHR-containing mobile elements could have intrinsic capabilities for breaking interspecies barriers through horizontal transfer. Future bioinformatic and molecular approaches together with new genomic data may allow us to refine these observations.
MATERIALS AND METHODS
HHR motif search methodology
Iterative bioinformatic searches for HHR motifs were performed through the NCBI-BLAST2 nucleotide tool at the European Bioinformatics Institute, as recently described (de la Peña and García-Robles 2010). Basically, sequence seeds for the queries corresponded to U-box/Helix II/P-box motifs obtained from naturally occurring HHR motifs described so far in the literature (Fig. 1; Supplemental Fig. S1). The obtained targets were manually inspected to strictly fulfill two initial criteria: The observed changes with respect to the introduced query should not affect any of the 11 totally conserved nucleotides of the HHR, and the changes detected in the targets either should be compensatory within the putative Helix II or should be located in loop 2. Selected hits were chosen for further analysis where three extra criteria were applied in order to define their capabilities of adopting a typical three-helical junction: typical Helices I and III in the 5′ and 3′ surrounding regions (Fig. 1) and the 5′-RUH-3′ cleavage site (R is a purine, and H can be A, C, or U) between Helix I and Helix III had to be found. An extra point of validation was the presence and nature of loops 1 and 2, which should establish the tertiary interaction required for in vivo ribozymatic activity (de la Peña et al. 2003; Khvorova et al. 2003); although due to the known heterogeneity among these interactions (de la Peña et al. 2009), a direct confirmation could not be always found. Obtained targets fulfilling the requirements were employed as new seeds for BLAST searches that were again manually inspected and selected for further analysis.
Analysis of the data
Genome servers employed for the analysis of the data and mapping of the ribozymes were ENSEMBL (www.ensembl.org), University of California Santa Cruz Genome Bioinformatics (genome.ucsc.edu), Wellcome Trust Sanger Institute (www.sanger.ac.uk), and DOE Joint Genome Institute (www.jgi.doe.gov) sites. The sequence alignments and alignment figures were done using Clustal X (Larkin et al. 2007). RNA secondary structure predictions were performed through the mFOLD server (Zuker 2003).
In vitro transcription
Cis-acting hammerheads were synthesized by in vitro transcription of XbaI-linearized plasmids containing the corresponding cDNA inserts immediately preceded and followed by the promotor of T7 RNA polymerase and the XbaI site, respectively. Transcription reactions (20 μL) contained the following: 40 mM Tris-HCl (pH 8); 6 mM MgCl2; 2 mM spermidine; 0.5 mg/mL RNase-free bovine serum albumin; 0.1% Triton X-100; 10 mM dithiothreitol; 1 mM each of ATP, CTP, and GTP; 0.1 mM UTP plus 0.5 μCi/μL (α-32P)UTP; 2 U/μL of human placental ribonuclease inhibitor; 20 ng/μL of plasmid DNA; 4 U/μL of T7 RNA polymerase; and 0.1–1 mM of the blocking deoxyoligonucleotide. Blocking deoxyoligonucleotide for D. reducens HHR was 5′-TTCCTGGACTCATCAGTGGGAGGG-3′. After incubation for 1 h at 37°C, products were fractionated by PAGE in 15% gels with 8 M urea, and the uncleaved primary transcripts were eluted by crushing the gel pieces and extracting them with phenol saturated with buffer (10 mM Tris-HCl at pH 7.5, 1 mM EDTA, 0.1% SDS), recovered by ethanol precipitation, and resuspended in deionized and sterile water.
Cis cleavage kinetics under protein-free conditions
For determining the cleaving rate constants, uncleaved primary transcripts (from 1 nM–1 μM) were incubated in 20 μL of 50 mM PIPES-NaOH (pH 6.5) for 1 min at 95°C and slowly cooled for 15 min to 25°C. After taking a zero-time aliquot, self-cleavage reactions were triggered by adding MgCl2 to 1 mM. Aliquots were removed at appropriate time intervals and quenched with a fivefold excess of stop solution at 0°C. The substrates and cleavage products were separated by PAGE in 15% denaturing gels. The product fraction at different times, Ft, was determined by quantitative scanning of the corresponding gel bands and fitted to the equation Ft = Fo + F∞(1 − e–kt), where Fo and F∞ are the product fractions at zero time and at the reaction endpoint, respectively, and k is the first-order rate constant of cleavage (kobs).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank S. Delgado and R. Flores for advice and critical reading of the manuscript. This work was supported by Ministerio de Educación y Ciencia of Spain (Ramón y Cajal contract and BFU2008-03154 to M.d.l.P.) and Generalitat Valenciana (GV06/206 to M.d.l.P.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2130310.
REFERENCES
- Chi YI, Martick M, Lares M, Kim R, Scott WG, Kim SH 2008. Capturing hammerhead ribozyme structures in action by modulating general base catalysis. PLoS Biol 6: e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daròs JA, Flores R 1995. Identification of a retroviroid-like element from plants. Proc Natl Acad Sci 92: 6856–6860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Peña M, Flores R 2001. An extra nucleotide in the consensus catalytic core of a viroid hammerhead ribozyme: Implications for the design of more efficient ribozymes. J Biol Chem 276: 34586–34593 [DOI] [PubMed] [Google Scholar]
- de la Peña M, García-Robles I 2010. Intronic hammerhead ribozymes are ultraconserved in the human genome. EMBO Rep doi: 10.1038/embor.2010.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Peña M, Gago S, Flores R 2003. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J 22: 5561–5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Peña M, Dufour D, Gallego J 2009. Three-way RNA junctions with remote tertiary contacts: A recurrent and highly versatile fold. RNA 15: 1949–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour D, de la Peña M, Gago S, Flores R, Gallego J 2009. Structure–function analysis of the ribozymes of chrysanthemum chlorotic mottle viroid: A loop–loop interaction motif conserved in most natural hammerheads. Nucleic Acids Res 37: 368–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein LM, Gall JG 1987. Self-cleaving transcripts of satellite DNA from the newt. Cell 48: 535–543 [DOI] [PubMed] [Google Scholar]
- Ferbeyre G, Smith JM, Cedergren R 1998. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol Cell Biol 18: 3880–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferbeyre G, Bourdeau V, Pageau M, Miramontes P, Cedergren R 2000. Distribution of hammerhead and hammerhead-like RNA motifs through the GenBank. Genome Res 10: 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores R, Delgado S, Gas ME, Carbonell A, Molina D, Gago S, de la Peña M 2004. Viroids: The minimal noncoding RNAs with autonomous replication. FEBS Lett 567: 42–48 [DOI] [PubMed] [Google Scholar]
- Forster AC, Davies C, Sheldon CC, Jeffries AC, Symons RH 1988. Self-cleaving viroid and newt RNAs may only be active as dimers. Nature 334: 265–267 [DOI] [PubMed] [Google Scholar]
- Gilbert W 1986. Origin of life: The RNA world. Nature 319: 618 [Google Scholar]
- Hajimorad MR, Ghabrial SA, Roossinck MJ 2009. De novo emergence of a novel satellite RNA of cucumber mosaic virus following serial passages of the virus derived from RNA transcripts. Arch Virol 154: 137–140 [DOI] [PubMed] [Google Scholar]
- Hammann H, Westhof E 2007. Searching genomes for ribozymes and riboswitches. Genome Biol 8: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR 1982. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31: 147–157 [DOI] [PubMed] [Google Scholar]
- Khvorova A, Lescoute A, Westhof E, Jayasena SD 2003. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol 10: 708–712 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lee KB, De Backer P, Aono T, Liu CT, Suzuki S, Suzuki T, Kaneko T, Yamada M, Tabata S, Kupfer DM, et al. 2008. The genome of the versatile nitrogen fixer Azorhizobium caulinodans ORS571. BMC Genomics 9: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martick M, Horan LH, Noller HF, Scott WG 2008. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature 454: 899–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevin KP, Finneran KT, Lovley DR 2003. Microorganisms associated with uranium bioremediation in a high-salinity subsurface sediment. Appl Environ Microbiol 69: 3672–3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G 1986. Autolytic processing of dimeric plant virus satellite RNA. Science 231: 1577–1580 [DOI] [PubMed] [Google Scholar]
- Prust C, Hoffmeister M, Liesegang H, Wiezer A, Fricke WF, Ehrenreich A, Gottschalk G, Deppenmeier U 2005. Complete genome sequence of the acetic acid bacterium Gluconobacter oxydans. Nat Biotechnol 23: 195–200 [DOI] [PubMed] [Google Scholar]
- Przybilski R, Gräf S, Lescoute A, Nellen W, Westhof E, Steger G, Hammann C 2005. Functional hammerhead ribozymes naturally encoded in the genome of Arabidopsis thaliana. Plant Cell 17: 1877–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, et al. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317: 86–94 [DOI] [PubMed] [Google Scholar]
- Richards EJ, Ausubel FM 1988. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell 53: 127–136 [DOI] [PubMed] [Google Scholar]
- Rojas AA, Vazquez-Tello A, Ferbeyre G, Venanzetti F, Bachmann L, Paquin B, Sbordoni V, Cedergren R 2000. Hammerhead-mediated processing of satellite pDo500 family transcripts from Dolichopoda cave crickets. Nucleic Acids Res 28: 4037–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, et al. 2007. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol 5: e77 doi: 10.1371/journal.pbio.0050077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi-Ashtiani K, Szostak JW 2001. In vitro evolution suggests multiple origins for the hammerhead ribozyme. Nature 414: 82–84 [DOI] [PubMed] [Google Scholar]
- Salehi-Ashtiani K, Lupták A, Litovchick A, Szostak JW 2006. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science 313: 1788–1792 [DOI] [PubMed] [Google Scholar]
- Tebo BM, Obraztsova A 1998. Sulfate-reducing bacterium grows with Cr(VI), U(VI), Mn(IV), and Fe(III) as electron acceptors. FEMS Microbiol Lett 162: 193–198 [Google Scholar]
- Webb CT, Riccitelli NJ, Ruminski DJ, Lupták A 2009. Widespread occurrence of self-cleaving ribozymes. Science 326: 953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichenrieder O, Wild K, Strub K, Cusack S 2000. Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature 408: 167–173 [DOI] [PubMed] [Google Scholar]
- Zuker M 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]