Als2cr4, also known as hypothetical protein FLJ33282, is predicted to be a tetraspanin membrane–associated protein that may function in photoreceptor protein trafficking and may contribute to photoreceptor structural integrity. Defects in other tetraspanin proteins lead to photoreceptor cell death and visual loss.
Abstract
Purpose.
The role of Als2cr4 (amyotrophic lateral sclerosis 2 [juvenile] chromosome region, candidate 4; also known as hypothetical protein FLJ33282) in the mouse retina was determined by characterizing the molecular structure, cellular interacting partners, and potential biochemical functions. Previous in situ hybridization and gene expression profiles show that the mRNAs encoding Als2cr4 are abundant in the eye, hippocampus, cerebellum, and olfactory bulb.
Methods.
From predicted antigenic epitopes of Als2cr4, two novel antibodies were developed to examine protein expression and morphologic localization in retinas from light-adapted and dark-adapted mice by immunohistochemistry, immunoblot analysis, and immunoelectron microscopy, and then immunoprecipitation was performed to identify interacting proteins by mass spectroscopy.
Results.
Peptide antibodies with Als2cr4 antigenic epitopes from either the amino- or carboxyl terminus were characterized with Als2cr4 recombinant proteins and peptide competition assays. Als2cr4 is a 45-kDa insoluble protein, highly enriched in retina, and localizes to photoreceptor outer segments, ciliary complex, and horizontal cells in the outer plexiform layer. Immunoelectron microscopy for Als2cr4 verified its expression in the discs of photoreceptor outer segments. Immunoprecipitation and mass spectroscopy identified eight potential interacting partners: vimentin, actin, myosin Va, myosin VI, myosin X, myosin XIV, kinesin 1, Als2cr4, and lamin B-1.
Conclusions.
Als2cr4 is a novel protein, with a probable tetraspanin-like membrane structure, that is localized in photoreceptors and in the postsynaptic outer plexiform layer and that interacts with cytoskeletal proteins. Als2cr4 may be involved in membrane transport between the photoreceptor inner and outer segments and may be a key component in maintaining the structural integrity of the outer segment.
The vertebrate retina is an intricate organ consisting of five distinct types of neurons: photoreceptors, horizontal cells, bipolar cells, amacrine cells, and ganglion cells. These specialized cell types process information in the form of light quanta and relay information to the lateral geniculate nucleus in the brain and ultimately to the primary visual cortex. Photoreceptors are the polarized, light-sensitive cell types responsible for initiating the phototransduction cascade in the mammalian retina and are of two types: rods and cones, with the predominant ones being rods. Although photoreceptors share similar cellular mechanisms for the activation and deactivation of phototransduction, they respond to various intensities of light and show distinction in the wavelengths of light absorbed by the visual pigments.1
Although much is known about the visual process, a better understanding of phototransduction is essential, since many visual impairments develop as a direct result of mutations in the genes encoding the proteins essential for phototransduction.2 One such protein is arrestin 1 (ARR1), which, when defective, leads to a form of retinitis pigmentosa known as Oguchi disease, which has congenital stationary night blindness phenotype.3 Arrestins downregulate activated, phosphorylated G-protein-coupled receptors and include four subtypes: ARR1 and arrestin-4 (ARR4, cone or X-arrestin) and the ubiquitously expressed β-adrenergic arrestin-1 and -2 (ARR2 and -3).4–7
To identify and characterize potential functional partners for ARR4, we screened a mouse retinal cDNA yeast two-hybrid (Y2H) library by using ARR4 as bait (Zuniga FI, et al. IOVS 2007;48:ARVO E-Abstract 601; Zuniga FI, et al. IOVS 2009;50:ARVO E-Abstract 5453). From the list of candidate proteins, Als2cr4 (NM_001037812, hypothetical protein FLJ33282; accession numbers are all GenBank; http://www.ncbi.nlm.nih.gov/Genbank; National Center for Biotechnology Information [NCBI], Bethesda, MD) was further investigated. Als2cr4 was first identified in Mus musculus in 2005 by the Laboratory for Genome Exploration Research Group at the RIKEN Genomic Sciences Centre (Kanagawa, Japan). The official gene symbol is Als2Cr4, corresponding to mouse amyotrophic lateral sclerosis (ALS) 2 (juvenile) chromosome region, candidate 4. As indicated by the name, this gene was in a candidate region for the neurodegenerative disease ALS; however, it was found that defects in ALS2 and its encoded protein, alsin, are responsible for neurotoxicity by the familial ALS-linked mutant Cu/Zn-superoxide dismutase (FSOD1).8,9 In the mouse, Als2cr4 is located on chromosome 1 and consists of two isoforms (1 [NP_001028621] and 2 [NP_001032901]), sharing more than 95% identity. The divergent sequence is located at the extreme amino terminus: amino acids (AA) 1-21. Moreover, Als2Cr4 shares approximately 83% identity with its human ortholog located on 2q33.2. The in situ hybridization (St. Jude's Brain Gene Expression Map www.stjudebgem.org/ provided in the public domain by St. Jude's Research Hospital, Memphis, TN) gene expression patterns show that the mRNA is highly abundant in the mouse eye from embryonic day (E)15 through adulthood. Prominent expression was also noted in the hippocampus, cerebellum, and olfactory bulb from postnatal day (P)7 through adulthood.
Recent technological advances are now available to aid in the study of the proteome of photoreceptors and have led to previously unidentified candidates that cause visual dysfunction. In two retinal proteomic analyses, Als2cr4 was identified in mouse sensory ciliary complex (CC)10 and in a rod outer segment (OS) preparation.11
We explored the structural and biochemical role that this novel protein plays in the retina under different environmental lighting conditions and in vitro cultures by using newly developed antibodies for Als2cr4 with immunoblot (IB) analysis, immunohistochemistry (IHC), and immunoelectron microscopy (IM-EM). In addition, to differentiate between the role of Als2cr4 in rod and cone photoreceptors, we compared retinas from C57Bl/6J (wild-type; WT) mice with those of neural retina leucine zipper knockout mice (Nrl−/−). Nrl is a key transcription factor responsible for the developmental program switch of rods to enhanced S-cone expression.12 Using these reagents for co-immunoprecipitation (IP), we identified interacting proteins through mass spectrometry.
Methods
Animals
All procedures involving mice were approved by the Institute for Laboratory Animal Research and conducted according to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Original breeders for the Nrl−/− and Arr1−/− mice were generated and generously provided by Anand Swaroop (University of Michigan, NEI-NIH)12 and Jeannie Chen (University of Southern California; USC),13 respectively. The Arr4−/− and Arr1−/−Arr4−/− double-knockout mice were generated in our laboratory.14 The WT (C57BL/6J and 129SVJ mixed background) mice were from our breeding colony. All animals were born and maintained in the USC vivarium in the Zilkha Neurogenetic Institute. They were kept under controlled ambient illumination on a 12-hour:12-hour light–dark cycle, with the exception of the Arr1−/− mice, which were reared in constant darkness because of their susceptibility to light-induced degeneration.15
Computer-Generated Primary and Two-Dimensional (2-D) Secondary Structure
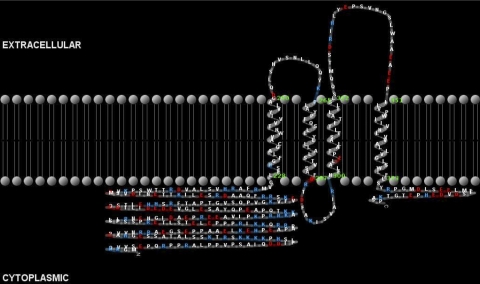
The Als2cr4 (NP_001032901) predicted translated AA sequence was used to identify protein transmembrane helices with version 2 of the TMHMM Server (Transmembrane Helices Markov Model; http://www.cbs.dtu.dk/services/TMHMM-2.0; provided in the public domain by the Center for Biological Sequence Analysis, the Technological University of Denmark, Lyngby, Denmark). These data were input into TMRPres2D software (http://bioinformatics.biol.uoa.gr/TMRPres2D/download.jsp) to create a 2-D model of α-helical or β-barrel transmembrane proteins.
Antibody Production
The mouse Als2cr4 AA sequence was assessed for hydrophilicity, antigenicity, surface accessibility, and predicted secondary structure by using Invitrogen's open source PeptideSelect Online Designer software (http://peptideselect.invitrogen.com/peptide/; Invitrogen, Carlsbad, CA). Candidate sequences were subjected to BLAST queries to circumvent cross-reactivity with other proteins (www.ncbi.nlm.nih.gov/blast/ provided in the public domain by NCBI, Bethesda, MD). The Zymed Laboratory (Invitrogen) developed two affinity-purified polyclonal antibodies (pAbs) with predicted antigenic epitopes (FLJ-Frederick and Dorie Miller [FLJ-FM] AA 76-88 EAPAPRRQKKIRP); and (FLJ-Thomas and Laurene Gray [FLJ-LG] AA 390-403 DVDEHPETGTKASP) to the amino- and carboxyl-terminal domains, respectively.
Immunoblot Analysis
The immunoblot analysis using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was modified from previously published protocols.16 Briefly, mice were kept in darkness for 24 hours, exposed to room light for 10 minutes, and killed or dark adapted (DA) overnight, killed in the dark, and the retinas dissected under infrared (IR) light. The retinas were homogenized (Polytron PT1200; Kinematica AG, Lucerne, Switzerland) in NP-40 lysis buffer (50 mM Tris-Cl [pH 7.4], 250 mM NaCl, 5 mM EDTA, 1% NP-40, and 0.02% NaN3 plus protease inhibitor cocktail; Halt Protease/Phosphatase Inhibitor Cocktail; Thermo Scientific, Rockford, IL) and centrifuged at 13,000 rpm for 15 minutes at 4°C. The pellet fractions were further disrupted (Microson sonicator, Misonex, NY) in minimum CHAPS lysis buffer (50 mM Tris-Cl [pH 7.6], 150 mM NaCl, 10 mM CHAPS, and 0.02% NaN3 plus protease/phosphatase inhibitor cocktail) and centrifuged as just described. The cell lysates were pooled, and 20 μg of retinal homogenate was mixed with Laemmli buffer, boiled for 10 minutes, and resolved on 10% to 12% SDS-PAGE. Immobilized proteins were stained with Coomassie blue or transferred onto PVDF membrane (Immobilon-P PVDF; Millipore, Billerica, MA) and reversibly stained (MemCode Reversible stain; Thermo Scientific). The membranes were blocked in 5% milk for 30 minutes at room temperature (RT), incubated with primary antibody overnight, labeled with horseradish peroxidase (HRP)–conjugated secondary antibody (Bio-Rad, Hercules, CA), and visualized by enhanced chemiluminescence (ECL) detection. Antibodies were used at the following concentrations: FLJ-FM pAb, 1:1,000; FLJ-LG pAb, 1:1,000; anti-GAPDH mAb (G8795; Sigma-Aldrich, St. Louis, MO), 1:5,000; anti-Arr1 mAb (D9F2), 1:50,000; anti-Arr4 rabbit pAb (mCAR-LUMIj), 1:1,000; anti-myosin Va pAb (sc-17707; Santa Cruz Biotechnology, Santa Cruz, CA), 1:375; and anti-myosin VI pAb (sc-23,68; Santa Cruz), 1:1,500. The secondary antibodies goat anti-rabbit HRP-conjugated IgG (170-6515; Bio-Rad) and goat anti-mouse HRP-conjugated IgG (172-1011; Bio-Rad) were used at 1:10,000.
Peptide Competition Assay
Samples were prepared as just stated, resolved on 12% SDS-PAGE, transferred onto PVDF membrane, reversibly stained (MemCode Reversible Protein stain; Thermo Scientific), and cut into equal halves. Als2cr4 antibody (FLJ-FM pAb) was preadsorbed with excess FLJ-FM peptide in 5% milk overnight at 4°C with gentle agitation. The membranes were probed with FLJ-FM pAb or preadsorbed FLJ-FM pAb (+ peptide) overnight, followed by secondary goat anti-rabbit HRP-conjugated antibody for ECL detection.
Immunoprecipitation
Retinal lysates from WT or Nrl−/− mice were prepared in either room light or in total darkness, as stated earlier. Each procedure was conducted in its respective lighting condition throughout the experiment. Magnetic beads (Dynal; Invitrogen) were prepared and incubated with retinal lysate for 2 hours at 4°C with end-over-end shaking. The precleared supernatant was collected and incubated with 10 μg of pAb FLJ-FM or a nonspecific antibody at 4°C overnight. Magnetic protein-G beads were added and incubated for 2 hours at 4°C with shaking. The beads were gently washed three times with 1× PBS before 5× SDS sample loading buffer (2× final concentration) was added and the mixture heated at 95°C for 10 minutes. The precipitated proteins were resolved on 10% SDS-PAGE and either stained with Coomassie blue for mass spectra analysis or transferred to PVDF membranes and subjected to IB analysis.
Mass Spectrometry
Selected fragments from the IP identified by pAb FLJ-FM were excised from the Coomassie blue–stained gel; in-gel trypsin digestion was performed and tryptic fragments were analyzed by liquid chromatography with tandem mass spectrometry detection (LC-MS/MS).17 A portion of the digested peptide mixture was also analyzed with a custom-built capillary liquid chromatography system18 interfaced directly to an ion-trap mass spectrometer (Thermo-Finnigan LCQ; San Jose, CA) as described previously.19 Full mass range spectra, high-resolution zoom scan spectra, and fragment ion (MS/MS) spectra were collected with the automated, data-dependent acquisition functions of the data system. Scaffold (version Scaffold_2_04_01; Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 50.0% probability, as specified by the peptide prophet algorithm.20 Protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least five identified peptides. Protein probabilities were assigned by the protein prophet algorithm.21
Immunohistochemistry
One-month-old mice were DA for 24 hours, exposed to room light for 1 hour and euthanatized or kept in total darkness where they were killed and the eyecups dissected under infrared light.22 The eyecups were fixed in 4% paraformaldehyde (PFA) for 2 hours at 4°C, incubated in 30% sucrose overnight, embedded in optimal cutting temperature (OCT; Sakura Finetek, Torrance, CA) embedding medium, and cryosectioned (model CM 1800; Leica-Jung, Wetzlar, Germany) at 7 μm through the optic nerve head; the slides were stored at −80°C. Tissue slides were thawed for 30 minutes at RT, washed two times in 1× PBS-Tween for 5 minutes, and blocked for 30 minutes with blocking buffer (10% normal goat serum, 0.2% Triton X-100, in 1× PBS) in a humidified chamber. The sections were incubated with primary antibody overnight at 4°C in antibody dilution buffer (2% goat serum, 0.2% Triton X-100, in 1× PBS) with gentle agitation. The day after, the slides were washed three times in 1× PBS-Tween for 15 minutes followed by direct protein labeling with Alexa Fluor-conjugated secondary antibodies (Invitrogen), and the nuclei were counterstained (TO-PRO-3, T3605; Invitrogen) in antibody dilution buffer. The slides were washed and vacuum dried before mounting medium was added for immunofluorescence (Vector Laboratories, Burlingame, CA) with nail polish to eliminate evaporation. The immunofluorescence from the retinal sections were visualized and photographed with a confocal laser scanning microscope (Carl Zeiss Meditec, Dublin, CA) at 63× magnification. Antibodies were used at the following concentrations: FLJ-FM pAb, 1:1,000; anti-calbindin-D-28K mAb (C9848; Sigma-Aldrich), 1:2,000; anti-Arr1 mAb (D9F2), 1:20,000; anti-Arr4 pAb (Chicken LUMIj), 1:2,500; secondary antibodies: Alexa Fluor-conjugated IgG, 1:500; and nuclear stain TO-PRO-3, 1:2,500.
Immunoelectron Microscopy
One-month-old WT mice were light-adapted (LA) or DA and euthanatized, and the eyes were enucleated. The eyecups were fixed in 4% PFA, washed three times (10 minutes) in fixative buffer, and dehydrated in graded ethanol (30%–100%). Eyecups were then infiltrated with various ratios of resin (LR White Medium Grade, EMS, Hatfield, PA) to ethanol, embedded in fresh resin in gelatin capsules, and polymerized under UV light for 48 hours at 4°C. Sections were cut on a microtome (Ultracut E Ultramicrotome; Reichert, Vienna, Austria) with a diamond knife and placed on uncoated 75/300-mesh nickel grids. Grids were incubated in 0.02 M glycine in 1× PBS for 15 minutes, washed four times in PBS, blocked (5% NGS, 1% BSA in PBS) at RT for 15 minutes, and incubated overnight at 4°C in primary antibody. Sections were washed five times in PBS and incubated in goat anti-rabbit 10-nm colloidal gold (1:50) for 2 hours at RT. The grids were washed three times in PBS, washed two times for 3 minutes each in distilled water (DW), fixed for 5 minutes with 1% glutaraldehyde in DW, washed 30 minutes in 5 drops of DW, and stained with aqueous 2% uranyl acetate in darkness. This procedure was followed by a wash in DW, staining for 15 minutes with Reynolds' lead citrate at RT and a wash in DW. The grids were allowed to dry and fumed with aqueous 2% osmium tetroxide for 30 minutes at RT. The sections were viewed with an electron microscope (100CX; JEOL, Tokyo, Japan), as previously described.23,24
cDNA Constructs for Als2cr4
The cDNA encoding mouse Als2cr4 was obtained from total RNA of retinas from Nrl−/− mice by using a first-strand cDNA synthesis kit (SuperScript III; (Invitrogen). The complete predicted coding region of Als2cr4 (NM_001037812) was amplified by polymerase chain reaction (PCR) technology with sense, 5′-ATGGGGAAGAAGCAGGTGGT-3′, and antisense, 5′-CTATGGAGAGGCTTTGGTT-3′, primer pairs. The amplified product was purified and then subcloned into the plasmid vectors pTrcHis-TOPO and pcDNA4/HisMax-TOPO (Invitrogen). The integrity and nucleotide sequence of the constructs was confirmed by automated DNA nucleotide sequencing.
Cell Culture
HEK293 (ATCC, Manassas, VA) cells were grown in minimum essential medium (MEM; ATCC), supplemented with 10% bovine serum albumin (BSA) and penicillin/streptomycin antibiotics in 5% CO2 at 37°C until confluent. The cells were plated on four-well culture slides (BD Falcon, Lincoln Park, NJ) at an initial density of 50,000 cells/chamber and grown for 24 hours. Plasmid cDNAs were transfected into HEK293 cells (FugeneHD Transfection reagent; Roche, Mannheim Germany) as suggested in the manufacturer's protocol. The HEK293 cells were transfected with either pcDNA4-Als2cr4-HisMax or empty vector and allowed to incubate for 24 hours before they were washed with ice-cold PBS and fixed in 4% PFA for 15 minutes at 4°C. The cells were blocked with 10% goat serum and 0.2% Triton X-100 in 1× PBS for 30 minutes at RT, followed by incubation for 1 hour at RT with primary antibody (diluted in 2% goat serum and 0.2% Triton X-100 in 1× PBS). The cells were washed three times in ice-cold PBS and then incubated with the appropriate secondary antibody before being mounted with coverslips and viewed with a laser scanning confocal microscope (Carl Zeiss Meditec) at 63× magnification. The antibodies were used at the following concentrations: FLJ-FM pAb, 1:1,000; anti-6xHis mAb (H1029; Sigma-Aldrich, MO), 1:3,000; and secondary antibody Alexa Fluor-conjugated IgG, 1:500.
Results
Primary and Secondary 2-D Protein Structure of Als2cr4
To elucidate the predicted structure of Als2cr4, the translated AA sequence was subjected to a functional domain query. The NCBI conserved-domain database identified a multidomain conservation between the Als2cr4 isoform 1 and DNA polymerase III subunits gamma and tau.25 The protein coding sequence of Als2cr4 isoform 2 is composed of 1212 nucleotides (nt 74-1285), and the translational product consists of 403 AAs (NP_001032901), resulting in a predicted translated protein of molecular mass of ∼45 kDa. Furthermore, the translated coding sequence of mouse Als2cr4 (isoform 2) was aligned with various orthologs (Supplementary Fig. S1, http://www.iovs.org/cgi/content/full/51/9/4407/DC1) and resulted in the following percent identities: 98% (Mus musculus isoform 1), 81% (Homo sapiens isoform a), 82% (Homo sapiens isoform b), 82% (Macaca mulatta), 79% (Bos taurus), and 49% (Danio rerio).
Computer modeling studies (Fig. 1) predict that the Als2cr4-translated protein contains four transmembrane helices (AAs 228-250, 265-287, 300-322, and 351-373), along with a lengthy 226-AA amino terminus and a short 30-AA carboxyl terminal tail, both predicted to be in the cytoplasmic space. In addition to these structural motifs, Als2cr4 contains a short (15 AA) loop linking transmembrane helices 1 and 2, as well as a longer (29-AA) loop linking helices 3 and 4, both in the extracellular space. An additional 13-AA loop is present in the cytoplasmic space linking helices 2 and 3. Moreover, Als2cr4 is predicted to have a tetratricopeptide motif present in the long (160-192-AA) tail. Tetratricopeptides are structural motifs that form scaffolds to mediate protein–protein interaction.26 The prediction that Als2cr4 is a structural membrane protein agrees with our biochemical and morphologic analyses, as we observed Als2cr4 to be present exclusively in the insoluble retina fraction (Supplementary Fig. S2) and to be embedded within the membrane discs of mouse photoreceptor OS.
Figure 1.
Computer-modeled 2-D structure. An Als2cr4-translated AA sequence was used to identify transmembrane helices with version 2 of the TMHMM Server. The data were input into TMRPres2D software to create a 2-D model of α-helical or β-barrel transmembrane proteins. Four potential transmembrane domains were identified, in addition to a long amino-terminal tail (226 AAs) and a short carboxyl-terminal tail (30 AAs), which are predicted to reside within the cytoplasm.
IB Analysis of Als2cr4
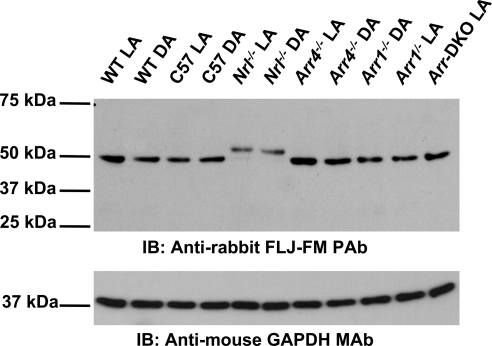
To confirm the expression of this newly identified protein, we generated polyclonal antibodies to two unique antigenic epitope domains: FLJ-FM AA 76-88 and FLJ-LG AA 390-403. Antibody specificity was assessed by peptide competition assays (Supplementary Fig. S2) and detection of Als2cr4 recombinant proteins (data not shown). In Supplementary Figure S2, FLJ-FM antibody specificity was abolished by using excess FLJ-FM immunizing peptide. Next, Als2cr4 was tested for differential expression in visual knockout mice (Arr1−/−, Arr4−/−, Arr1−/−Arr4−/− [Arr double knockout, Arr-DKO], and Nrl−/−) compared with control WT and also differences in expression in LA and DA conditions. In control WT retina, IB analysis (Fig. 2) of retinal lysates showed that Als2cr4 is expressed ubiquitously as a 45-kDa protein. Additional experiments with protein homogenates from the Nrl−/− retina produced a doublet of 45 and 52 kDa, respectively, suggesting that Als2cr4 isoform 1 is a cone-enriched isoform below detectable levels in WT retina that contains 3% to 5% cones.27 In addition, the mice from DA environment compared with LA have similar Als2cr4 protein expression levels (Fig. 2). Furthermore, no differences in Als2cr4 expression patterns were observed on either IB analysis (Fig. 2) or immunofluorescent localization (data not shown) when either Arr1 or Arr4 was knocked out. Its absence may be explained by brief interaction between these proteins or by the use of nonoptimal conditions.
Figure 2.
IB analysis of Als2cr4 protein expression. Each lane contains 20 μg of total retinal lysate from P30 mice. Control WT, Nrl−/−, Arr4−/−, Arr1−/− or Arr-DKO mice were light or dark adapted and killed, and retinal lysates were prepared. The proteins were resolved on 10% SDS-PAGE, transferred onto PVDF membrane, probed with the pAb FLJ-LG followed by the secondary antibody, goat anti-rabbit HRP-conjugated IgG, and processed for ECL. As a loading control, the membrane was probed with the mAb GAPDH and the appropriate secondary antibody.
Cellular and Subcellular Immunohistochemical Localization of Als2cr4
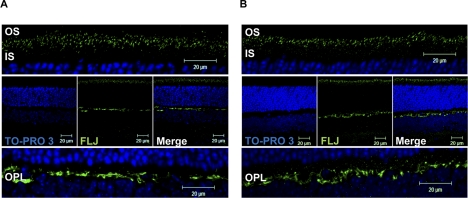
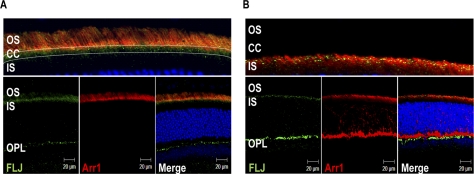
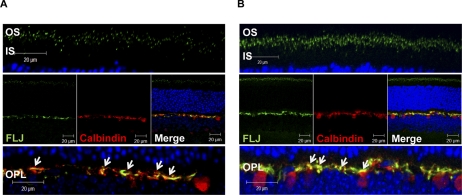
Our immunofluorescence analysis revealed that Als2cr4 is expressed in both rods (Figs. 3, 4, 5) and Nrl−/− cones (Supplementary Fig. S3). This observation is in agreement with our IB analysis showing Als2cr4 expression in WT and Nrl−/− mice (Fig. 2). As shown in Figures 3, 4, and 5 with immunohistochemical localization, Als2cr4 localized predominantly to photoreceptor OS, CC, and the outer plexiform layer (OPL). Dual immunohistochemical labeling experiments with a well-characterized monoclonal D9F2 antibody to Arr128 and pAb-FLJ-FM to Als2cr4 (Fig. 4A) showed that in light, Arr1 is localized to the rod and cone OS, whereas Als2cr4 is present both in the photoreceptor OS and in the CC. A higher magnification of the photoreceptor layer in Figure 4A (top) clearly shows the limits of the OS, and dotted white lines indicate the CC where Als2cr4 staining was also observed. The Als2cr4 immunostaining pattern appeared punctate in the photoreceptor layer and uniform in the OPL. We observed a similar immunologic staining pattern in the DA (Figs. 3B, 4B) retinas. Figure 4B shows that in DA WT retina, Arr1 and Als2cr4 do not co-localize in either rod spherules or cone pedicles, indicating that Als2cr4 expression is postsynaptic. Furthermore, dual immunohistochemical stain labeling experiments (Fig. 5) demonstrated that Als2cr4 co-localizes with calbindin, a well-characterized marker used to identify horizontal cells. At present, however, we are not certain whether expression of Als2cr4 is limited to horizontal cell axons, dendrites, or both.
Figure 3.
Als2cr4 immunohistochemical localization. P30 WT mice were either (A) light or (B) dark adapted and killed. The retina sections (7 μm) were incubated with pAb FLJ-FM followed by incubation with Alexa Fluor-488 (Als2cr4) goat anti-rabbit IgG. The nuclei were stained with TO-PRO3 iodide. Top: magnified region of the photoreceptor layer. Middle: view of the retina at ×63 magnification. Bottom: magnified region of the OPL. Results demonstrate a similar staining pattern for Als2cr4 regardless of lighting condition. Punctate staining was observed in the photoreceptor layer, whereas staining in the OPL was more uniform. Magnification, ×63.
Figure 4.
Dual IHC of Als2cr4 and Arr1 in retinas from light- or dark-adapted mice. P30 WT mice were either (A) light- or (B) dark-adapted and killed. The retina sections were dual stained with mAb D9F2 for Arr1 and pAb FLJ-FM for Als2cr4. Secondary antibodies were Alexa Fluor-568 goat anti-mouse IgG (Arr1) and Alexa Fluor-488 goat anti-rabbit IgG (Als2cr4). Nuclei were stained with TO-PRO3 iodide. (A, top) A magnified region of the merged photoreceptor layer in the bottom panel demonstrates that Als2cr4 was present in the connecting cilia. After 1 hour of light exposure, Arr1 localization was exclusive to the OS, whereas Als2cr4 was present in both the OS and connecting cilia. (B) Al2cr4 was localized in the postsynaptic OPL, with no co-localization of Arr1 with Als2cr4 in rod spherules or cone pedicles. Magnification, ×63.
Figure 5.
Dual IHC localization of Als2cr4 and calbindin in horizontal cells. P30 WT mice were either (A) light- or (B) dark-adapted and killed. The retinal sections were dual stained with mAb calbindin and pAb FLJ-FM. Secondary antibodies were Alexa Fluor-568 goat anti-mouse IgG (Calbindin) and Alexa Fluor-488 goat anti-rabbit IgG (Als2cr4). The nuclei were stained with TO-PRO3 iodide. Top: magnified region of the photoreceptor layer. Middle: view of the retina at ×63 magnification. Bottom: magnified region of the OPL. White arrows denote dual immunohistochemical localization of Als2cr4 with calbindin at the OPL. Magnification, ×63.
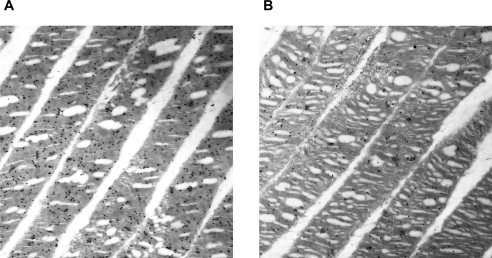
In other experiments, the photoreceptor ultrastructure was examined by IM-EM to visualize the subcellular location of Als2cr4. Figure 6 shows data from LA (Fig. 6A) and DA (Fig. 6B) WT retinas. IM-EM with pAb FLJ-FM shows colloidal gold particles present in the discs of photoreceptor OS. Figure 6 shows rod OS; however, we observed that cone OS were also labeled in other micrographs (data not shown). In addition, IM-EM results suggested that more Als2cr4 is expressed in LA (Fig. 6A) than in DA (Fig. 6B) retinas; however, IB (Fig. 2) and IHC experiments (Figs. 3, 4, 5) revealed comparable protein expression levels in both lighting conditions. These experiments should be replicated and the results analyzed statistically. With regard to the presence of Als2cr4 in connecting cilia, which has been reported in the CC proteome10 and which we showed by IHC in mouse retinas (Fig. 4), more studies are warranted.
Figure 6.
Immuno-EM analysis of Als2cr4 in WT retinas. Retinas from either (A) light- or (B) dark-adapted mice were identified with pAb FLJ-FM and secondary goat anti-rabbit-10 nm colloidal gold antibody particles. (A, B) Rod OS. IM-EM shows Als2cr4 embedded within the discs of photoreceptor OS. Magnification, ×20,000.
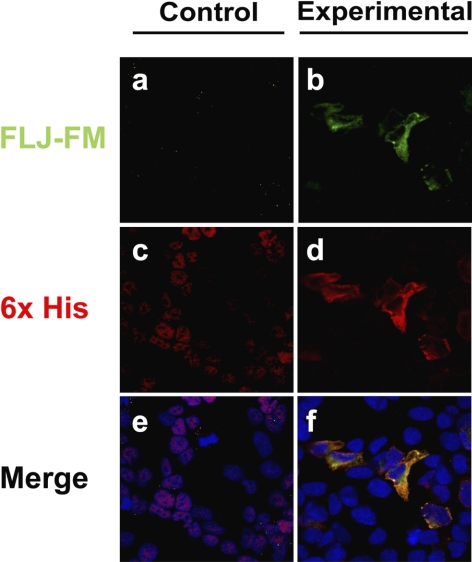
Subsequently, immunohistochemical localization of exogenous Als2cr4 expression and localization in cultured cells was examined. The 6xHis-tagged cDNA for Als2cr4 was transiently transfected into HEK293 cells to study potential morphologic and physiological effects that were due to introduction of the tagged protein, as well as to determine where the subcellular recombinant protein is expressed. The Als2cr4 expression pattern was visualized by dual-IHC labeling with pAb FLJ-FM (green) and 6xHis mAb (red). In Figures 7b, 7d, and 7f, the transfected cells show poly-histidine-tagged Als2cr4 recruitment to the cytoplasm and plasma membrane. This observation correlates with our findings that Als2cr4 was present only in the pellet fractions of retinal homogenates and not in the soluble fractions (Supplementary Fig. S2). Furthermore, transfection of 6xHis-Als2cr4 did not disrupt or change cellular morphology. Last, to resolve whether Als2cr4 is recruited to the plasma membrane because of its primary AA sequence or whether the localization is an artifact of the epitope tag, a control pCDNA4/HisMax without a cDNA insert was transfected into the cells. The data confirm (Figs. 7a, 7c, 7e) that expression of pCDNA4/HisMax without the recombinant Als2cr4 insert was limited exclusively to the nucleus, thereby substantiating that the 6xHis-tag alone would not drive expression of the His tag control to the cytoplasm or the plasma membrane.
Figure 7.
Membrane localization of Als2cr4 transfected into HEK293 cells. Confocal microscopy images of 6xHis tagged-Als2cr4, transiently transfected into HEK293 cells. Als2cr4 subcellular localization was identified by immunohistochemical labeling with antibodies recognizing Als2cr4 with pAb FLJ-FM (green; a, b) or the His-tag with mAb 6xHis (red; c, d). Nucleic acids in the nuclei were labeled with TO-PRO 3 iodide. Cells transfected with a recombinant Als2cr4 cDNA construct expressed the recombinant Als2cr4 protein, which is localized to the cytoplasm and the plasma membrane, (f) whereas those transfected with no recombinant cDNA inserted into the pCDNA4/HisMax vector (e) exhibited no Als2cr4 expression, and the His-tag was limited to the nucleus. Labeling patterns of Als2cr4 pAb (b) and 6xHis mAb (d) displayed co-localization in the plasma membrane (f), thereby confirming Al2cr4 antibody specificity.
IP Showing Potential Interacting Partners of Als2cr4
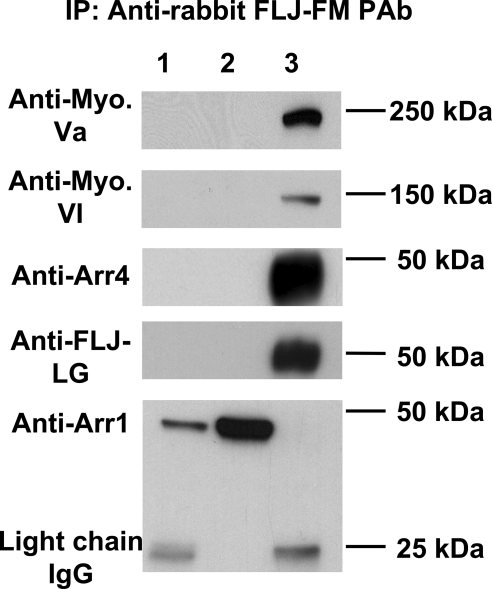
IP experiments were performed29,30 to identify potential binding partners for Als2cr4 in response to LA and DA conditions. Supplementary Figure S4 shows DA IP studies that were analyzed from stained gels with either a control IgG or Als2cr4-specific pAb-FLJ-FM. Lane 1 represents the precleared supernatant fraction, and lane 2 shows the proteins eluted after IP with pAb FLJ-FM (Supplementary Table S1, http://www.iovs.org/cgi/content/full/51/9/4407/DC1). Eight bands were excised and analyzed by mass spectra analysis (Table 1). Of the 15 peptides identified in the eight bands, most corresponded to cytoskeletal components. In contrast, the only bands detected in the control IP fraction were IgG light and heavy chains. Figure 8 shows IB analysis with specific antibodies verifying the direct interaction of Als2cr4 with myosin Va, myosin VI, and Arr4. Moreover, the observation that Als2cr4 immunoprecipitates Arr4 independently confirms the protein–protein interaction previously observed by the retinal yeast two-hybrid screen with cone arrestin used as the bait. Further experimental work is ongoing to determine the biological significance of this interaction. In addition, Als2cr4, the positive control, is clearly shown to be immunoprecipitated with FLJ-FM pAb and detected by the second Als2cr4 antibody FLJ-LG pAb; and Arr1, a negative control in this experiment, verifies that Arr1 binds nonspecifically to the protein G beads.
Table 1.
Retinal Proteins Identified by Mass Spectrometry from Immunoprecipitation with Als4cr2 pAb FLJ-FM
| Band No. | NCBI Accession No. | Protein ID | Mass (kDa) | Peptides (n) | Coverage (%) |
|---|---|---|---|---|---|
| 2* | NP_780469 | Myosin X | 233 | 18 | 9 |
| 2* | NP_082297 | Myosin XIV | 229 | 10 | 6 |
| 3 | NP_034994 | Myosin Va | 215 | 18 | 11 |
| 4 | NP_001034635 | Myosin VI | 146 | 18 | 18 |
| 5 | Q61768 | Kinesin-1 | 110 | 7 | 9 |
| 6 | NP_034851 | Lamin-B1 | 67 | 5 | 9 |
| 7* | NP_035831 | Vimentin | 54 | 34 | 59 |
| 7* | NP_001032901 | Als2cr4 | 45 | 6 | 16 |
| 8* | NP_031419 | Beta-actin | 42 | 16 | 47 |
Summary of the eight proteins isolated by immunoprecipitation with the FLJ-FM pAb and identified by mass spectrometry, their predicted molecular mass, the number of peptides identified for each protein, and the percentage of the protein covered.
Bands that showed protein grouping ambiguity.
Figure 8.
IP with Als2cr4 and IB analysis verified interacting partners. The retinas of LA WT mice were used to immunoprecipitate novel Als2cr4-interacting partners. Individual membranes were probed with the indicated antibody (left) followed by secondary goat anti-rabbit HRP-conjugated antibody for ECL detection. Lane 1: experimental precleared beads; lane 2: experimental precleared supernatants; and lane 3: eluted antigens immunoprecipitated with 10 μg of FLJ-FM pAb. Results showed that Als2cr4 specifically immunoprecipitated myosin Va, myosin VI, and Arr4. The positive control showed that Als2cr4 was present in the eluted fraction (lane 3), and the negative control showed that Arr1 bound nonspecifically to the beads, was present in the supernatants, and did not interact with Als2cr4. Molecular mass markers (kilodaltons) are identified on the right. Bottom: light chain IgG.
Discussion
Initially, visual Arr4 was used as a bait to screen a yeast two-hybrid Nrl−/− retinal cDNA library, and one of the candidates for protein–protein interaction was a novel hypothetical protein known as Als2cr4. Because of its high mRNA expression in the eye and the observation that Als2cr4 is originally a candidate gene for ALS, we explored the potential physiological relevance of this novel protein. In parallel to our work, researchers in a recent study tested the functionality of Als2cr4 and, in zebrafish knockdown studies with morpholino technology, demonstrated an “eyes absent” phenotype (The Zebrafish Model Organism Database, www.zfin.org).
Photoreceptor studies with confocal IHC and IM-EM revealed that Als2cr4 is targeted to the membrane and is concentrated in the rod OS. These data correlate well with our IB analysis (Fig. 2) showing lower Als2cr4 protein levels in retinas from “all cone” Nrl−/− mice. Moreover, Als2cr4 appeared to be embedded in the outer segment discs; however, whether it is present in the photoreceptor disc rim like peripherin/rds or in the flat lamellar disc region where opsins are localized is not known. These morphologic studies suggested that Als2cr4 contributes to either phototransduction protein trafficking or participates as a structural membrane protein, in agreement with previous proteomic observations.10,11 In ongoing experiments, we are examining Als2cr4 within the CC and the horizontal cells of the OPL.
Using computer modeling software, we predicted that Als2cr4 is related to the tetraspanin superfamily of proteins containing four transmembrane-spanning domains, two cytoplasmic tails, two extracellular loops, and one intracellular loop. However, Als2cr4 has major differences with canonical tetraspanins: mainly, the presence of a shorter extracellular loop (29 AA vs. 100 AA) and the existence of a large amino-terminal tail (226 AA) compared with the relatively short amino-terminal tail of other tetraspanins. In addition, tetraspanins contain a characteristic four to six conserved cysteine residues in the large outer loop that Als2cr4 does not contain.31
In retinal disease, the most prominent members of the tetraspanin family are peripherin/rds32,33 and ROM-1.34 These proteins participate in outer segment morphogenesis by providing a structural basis whereby new discs and lamellae are continually added to the photoreceptor outer segment basal region.32
The outer segment renewal is a critical process as photoreceptors undergo daily phagocytosis of apical discs and lamellae by retinal pigment epithelium. Moreover, forms of retinitis pigmentosa, macular degeneration, and macular dystrophy are linked to defective genes encoding peripherin/rds, ROM-1, and prominin (Prom1).35 In mouse models, Rds mutants fail to form rod OS,36 whereas in cones the phenotype is less severe with formation of enlarged and morphologically disorganized cone OS.24 In comparable studies, loss of ROM-1 leads to engorged discs and disordered rod OS.37 Furthermore, loss of Prom1 is associated with overgrown and misoriented disc membranes.38 As is evident from these studies, the loss of structural proteins has devastating effects on photoreceptor morphology and leads to loss of photoreceptors. Because Als2cr4 shares structural similarities, has a comparable immunostaining pattern,24,34 and is embedded in the discs, we hypothesize that, similar to peripherin/rds and ROM-1, this structural protein contributes to maintaining the structural integrity of photoreceptors.
To further decipher the protein biochemistry for Als2cr4, we looked at its interacting partners in the DA Nrl−/− retina with co-IP experiments with pAb FLJ-FM (Supplementary Fig. S4, Supplementary Table S1). In total, eight bands were excised, subjected to mass spectra analysis, and identified: vimentin, actin, myosin Va, myosin VI, myosin X, myosin XIV, kinesin 1, Als2cr4, and lamin B-1 (Table 1). The cytoskeleton has been implicated in many aspects of vision ranging from structural maintenance of photoreceptors to protein trafficking between the inner segment (IS) and OS and for active transport in the connecting cilium.39 Cilia are hairlike protrusions extending from the surface of cells involved in cellular motility or, in the case of primary cilia, specialized functions including sight, smell, and hearing. Ciliopathy is a disease state in which mutations are located in protein coding regions and are involved in cilia formation or protein transport. Primary ciliary dyskinesia, kidney diseases, respiratory diseases, and retinal degenerations are all examples of ciliopathies. In addition, mounting evidence links mutations in genes responsible for photoreceptor morphogenesis and intraflagellar transport (IFT) with photoreceptor degeneration.40–46 These diseases are usually accompanied by other clinical symptoms due to global detrimental effects on primary cilia and are syndromic.39
Like all ciliated cells, photoreceptor OS lack the organelles for protein synthesis and are thus dependent on protein trafficking. To date, there is an ongoing debate as to the major mode of shuttling signaling proteins between the inner and outer segment subcellular compartments. Some experts argue for diffusion,47 whereas others support active transport mediated by molecular motors along actin filaments or microtubules.48 Because of the abundance of cytoskeletal components interacting with Als2cr4 (Table 1, Fig. 8), and in particular molecular motors, we propose that Als2cr4 is actively transported to its destination by members of the kinesin and/or myosin families.
Investigation as to the etiology of Usher 1B syndrome,49 which is the most frequent disease causing combined deafness and blindness in humans, identified defects in the myosin VII gene. The absence of the molecular motor myosin VII is deleterious, as it plays a role in transporting rhodopsin and other proteins essential for normal phototransduction. The cytoskeleton plays an essential role in the photoreceptor's maintenance by providing structure in the form of actin filaments and microtubules forming the axoneme. A further example is the loss of the protein retinitis pigmentosa 1 (RP1) that binds to and stabilizes microtubules and is involved in the regulation of axonemal length and stability.40 The cytoskeleton also plays an indispensable role in transport mechanisms, as is evident by the conditional knockout of kinesin (Kif3a), which results in the mislocalization of opsin, Arr1, and peripherin to the inner segment,50 and the IFT88 knockout mouse,51 which exhibits uncharacteristic stacking of disc membranes reminiscent of ROM-1 loss in rods or peripherin/rds loss in cones. In addition to irregular morphology, loss of IFT88 also leads to an accumulation of opsin in the outer segment and, ultimately, to cytotoxicity.
In conclusion, we have for the first time, characterized the novel protein Als2cr4 in the retina and proposed that it has a 2-D structure consisting of four transmembrane domains. We also discovered this structural protein is embedded in the photoreceptor outer segment discs and in the postsynaptic OPL in horizontal cell axons. Furthermore, we identified cytoskeletal interacting partners and hypothesize that Als2cr4 functions in membrane trafficking and contributes to maintaining photoreceptor structure. Although additional experiments are essential for resolving the question of the function of Als2cr4 in the mouse retina, the recent morpholino knockdown experiment using the Als2cr4 orthologue (XP_685343) to demonstrate a significant phenotype with “failure to develop the eye” (The Zebrafish Model Organism Database www.zfin.org, NCBI gene ID Q66IE4) suggests that our hypothesis is valid. Future experiments will determine whether Als2cr4 functions analogously to what is observed in zebrafish retina as an early embryonic component that is indispensable for normal mouse eye development and for maintaining the photoreceptor structural integrity.
Supplementary Material
Acknowledgments
The authors thank Mary D. Allen for generous and continuous support; Jeannie Chen for the Arr1−/− mice; Anand Swaroop and Jose Luis Linares for providing Nrl−/− mice and Nrl−/− cDNA library for the yeast two-hybrid screen; and Bruce Brown, Xuemei Zhu, and Jeffrey Raskin for their technical and scientific expertise; Barbara Nagel for her immunoelectron microscopy; India Jane Bradley for her help with immunohistochemistry and confocal microscopy; and members of the Mary D. Allen Laboratory, including Shun-Ping Huang, Rosanne Yetemian, Leng-Ying Chen, and Guey Shuang Wu.
Footnotes
Supported by National Institutes of Health EY015851 (CMC), NIH 1F31GM079910 (FIZ, CMC), NIH EY03040 (NEI Core, Doheny Eye Institute), Research to Prevent Blindness (DEI), Charles Frederick and Dorie Miller, Thomas and Laurene Gray (Tony Gray Foundation), William Hansen Sandberg Memorial Foundation (FIZ, CMC), and the Mary D. Allen Foundation (CMC). CMC is the Mary D. Allen Chair in Vision Research, Doheny Eye Institute and a recipient of a Research to Prevent Blindness Senior Scientific Investigator Award.
Disclosure: F.I. Zuniga, None; C.M. Craft, None
References
- 1.Palczewski K, Saari JC. Activation and inactivation steps in the visual transduction pathway. Curr Opin Neurobiol. 1997;7:500–504 [DOI] [PubMed] [Google Scholar]
- 2.Mendez A, Burns ME, Sokal I, et al. Role of guanylate cyclase-activating proteins (GCAPs) in setting the flash sensitivity of rod photoreceptors. Proc Natl Acad Sci U S A. 2001;98:9948–9953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuchs S, Nakazawa M, Maw M, Tamai M, Oguchi Y, Gal A. A homozygous 1-base pair deletion in the arrestin gene is a frequent cause of Oguchi disease in Japanese. Nat Genet. 1995;10:360–362 [DOI] [PubMed] [Google Scholar]
- 4.Craft CM, Whitmore DH, Wiechmann AF. Cone arrestin identified by targeting expression of a functional family. J Biol Chem. 1994;269:4613–4619 [PubMed] [Google Scholar]
- 5.Murakami A, Yajima T, Sakuma H, McLaren MJ, Inana G. X-arrestin: a new retinal arrestin mapping to the X chromosome. FEBS Lett. 1993;334:203–209 [DOI] [PubMed] [Google Scholar]
- 6.Lefkowitz RJ, Inglese J, Koch WJ, Pitcher J, Attramadal H, Caron MG. G-protein-coupled receptors: regulatory role of receptor kinases and arrestin proteins. Cold Spring Harb Symp Quant Biol. 1992;57:127–133 [DOI] [PubMed] [Google Scholar]
- 7.Gurevich VV, Dion SB, Onorato JJ, et al. Arrestin interactions with G protein-coupled receptors: direct binding studies of wild type and mutant arrestins with rhodopsin, beta 2- adrenergic, and m2 muscarinic cholinergic receptors. J Biol Chem. 1995;270:720–731 [DOI] [PubMed] [Google Scholar]
- 8.Kanekura K, Hashimoto Y, Niikura T, Aiso S, Matsuoka M, Nishimoto I. Alsin, the product of ALS2 gene, suppresses SOD1 mutant neurotoxicity through RhoGEF domain by interacting with SOD1 mutants. J Biol Chem. 2004;279:19247–19256 [DOI] [PubMed] [Google Scholar]
- 9.Matsuoka M, Nishimoto I. Anti-ALS activity of alsin, the product of the ALS2 gene, and activity-dependent neurotrophic factor. Neurodegener Dis. 2005;2:135–138 [DOI] [PubMed] [Google Scholar]
- 10.Liu Q, Tan G, Levenkova N, et al. The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics. 2007;6:1299–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwok MC, Holopainen JM, Molday LL, Foster LJ, Molday RS. Proteomics of photoreceptor outer segments identifies a subset of SNARE and Rab proteins implicated in membrane vesicle trafficking and fusion. Mol Cell Proteomics. 2008;7:1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mears AJ, Kondo M, Swain PK, et al. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447–452 [DOI] [PubMed] [Google Scholar]
- 13.Xu J, Dodd RL, Makino CL, Simon MI, Baylor DA, Chen J. Prolonged photoresponses in transgenic mouse rods lacking arrestin. Nature. 1997;389:505–509 [DOI] [PubMed] [Google Scholar]
- 14.Nikonov SS, Brown BM, Davis JA, et al. Mouse cones require an arrestin for normal inactivation of phototransduction. Neuron. 2008;59:462–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Simon MI, Matthes MT, Yasumura D, LaVail MM. Increased susceptibility to light damage in an arrestin knockout mouse model of Oguchi disease (stationary night blindness). Invest Ophthalmol Vis Sci. 1999;40:2978–2982 [PubMed] [Google Scholar]
- 16.Zhang Y, Li A, Zhu X, Wong CH, Brown B, Craft CM. Cone arrestin expression and induction in retinoblastoma cells. In: Anderson RE, LaVail MM, Hollyfield JG. eds. Proceedings of the Ninth International Symposium on Retinal Degeneration New York: Kluwer Academic/Plenum Publishers; 2001:309–317 [Google Scholar]
- 17.Hellman U, Wernstedt C, Gonez J, Heldin CH. Improvement of an “In-Gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal Biochem. 1995;224:451–455 [DOI] [PubMed] [Google Scholar]
- 18.Davis MT, Stahl DC, Hefta SA, Lee TD. A microscale electrospray interface for on-line, capillary liquid chromatography/tandem mass spectrometry of complex peptide mixtures. Anal Chem. 1995;67:4549–4556 [DOI] [PubMed] [Google Scholar]
- 19.Davis MT, Lee TD. Rapid protein identification using a microscale electrospray LC/MS system on an ion trap mass spectrometer. J Am Soc Mass Spectrom. 1998;9:194–201 [DOI] [PubMed] [Google Scholar]
- 20.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392 [DOI] [PubMed] [Google Scholar]
- 21.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658 [DOI] [PubMed] [Google Scholar]
- 22.Zhu X, Li A, Brown B, Weiss ER, Osawa S, Craft CM. Mouse cone arrestin expression pattern: light induced translocation in cone photoreceptors. Mol Vis. 2002;8:462–471 [PubMed] [Google Scholar]
- 23.Fliesler SJ, Peachey NS, Richards MJ, Nagel BA, Vaughan DK. Retinal degeneration in a rodent model of Smith-Lemli-Opitz syndrome: electrophysiologic, biochemical, and morphologic features. Arch Ophthalmol. 2004;122:1190–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farjo R, Skaggs JS, Nagel BA, et al. Retention of function without normal disc morphogenesis occurs in cone but not rod photoreceptors. J Cell Biol. 2006;173:59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jarvis TC, Beaudry AA, Bullard JM, Janjic N, McHenry CS. Reconstitution of a minimal DNA replicase from Pseudomonas aeruginosa and stimulation by non-cognate auxiliary factors. J Biol Chem. 2005;280:7890–7900 [DOI] [PubMed] [Google Scholar]
- 26.Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;21:932–939 [DOI] [PubMed] [Google Scholar]
- 27.Carter-Dawson LD, LaVail MM, Sidman RL. Differential effect of rd mutation on rods and cones in mouse retina. Invest Ophthalmol Vis Sci. 1978;17:489–498 [PubMed] [Google Scholar]
- 28.Brown BM, Ramirez T, Rife L, Craft CM. Visual arrestin 1 contributes to cone photoreceptor survival and light-adaptation. Invest Ophthalmol Vis Sci. 2010;51:2372–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu X, Craft CM. Modulation of CRX transactivation activity by phosducin isoforms. Mol Cell Biol. 2000;20:5216–5226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu X, Brown B, Li A, Mears AJ, Swaroop A, Craft CM. GRK1-dependent phosphorylation of S and M opsins and their binding to cone arrestin during cone phototransduction in the mouse retina. J Neurosci. 2003;23:6152–6160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stipp CS, Kolesnikova TV, Hemler ME. Functional domains in tetraspanin proteins. Trends Biochem Sci. 2003;28:106–112 [DOI] [PubMed] [Google Scholar]
- 32.Goldberg AF. Role of peripherin/rds in vertebrate photoreceptor architecture and inherited retinal degenerations. Int Rev Cytol. 2006;253:131–175 [DOI] [PubMed] [Google Scholar]
- 33.Travis GH, Brennan MB, Danielson PE, Kozak CA, Sutcliffe JG. Identification of a photoreceptor-specific mRNA encoded by the gene responsible for retinal degeneration slow (rds). Nature. 1989;338:70–73 [DOI] [PubMed] [Google Scholar]
- 34.Boesze-Battaglia K, Stefano FP, Fitzgerald C, Muller-Weeks S. ROM-1 potentiates photoreceptor specific membrane fusion processes. Exp Eye Res. 2007;84:22–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kennedy B, Malicki J. What drives cell morphogenesis: a look inside the vertebrate photoreceptor. Dev Dyn. 2009;238:2115–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanyal S, Jansen HG. Absence of receptor outer segments in the retina of rds mutant mice. Neurosci Lett. 1981;21:23–26 [DOI] [PubMed] [Google Scholar]
- 37.Clarke G, Goldberg AF, Vidgen D, et al. Rom-1 is required for rod photoreceptor viability and the regulation of disk morphogenesis. Nat Genet. 2000;25:67–73 [DOI] [PubMed] [Google Scholar]
- 38.Yang Z, Chen Y, Lillo C, et al. Mutant prominin 1 found in patients with macular degeneration disrupts photoreceptor disk morphogenesis in mice. J Clin Invest. 2008;118:2908–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramamurthy V, Cayouette M. Development and disease of the photoreceptor cilium. Clin Genet. 2009;76:137–145 [DOI] [PubMed] [Google Scholar]
- 40.Liu Q, Zuo J, Pierce EA. The retinitis pigmentosa 1 protein is a photoreceptor microtubule-associated protein. J Neurosci. 2004;24:6427–6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Connell G, Bascom R, Molday L, Reid D, McInnes RR, Molday RS. Photoreceptor peripherin is the normal product of the gene responsible for retinal degeneration in the rds mouse. Proc Natl Acad Sci U S A. 1991;88:723–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams NA, Awadein A, Toma HS. The retinal ciliopathies. Ophthalmic Genet. 2007;28:113–125 [DOI] [PubMed] [Google Scholar]
- 43.Moore A, Escudier E, Roger G, et al. RPGR is mutated in patients with a complex X linked phenotype combining primary ciliary dyskinesia and retinitis pigmentosa. J Med Genet. 2006;43:326–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams DS. Usher syndrome: animal models, retinal function of Usher proteins, and prospects for gene therapy. Vision Res. 2008;48:433–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blacque OE, Leroux MR. Bardet-Biedl syndrome: an emerging pathomechanism of intracellular transport. Cell Mol Life Sci. 2006;63:2145–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leroux MR. Taking vesicular transport to the cilium. Cell. 2007;129:1041–1043 [DOI] [PubMed] [Google Scholar]
- 47.Calvert PD, Strissel KJ, Schiesser WE, Pugh EN, Jr, Arshavsky VY. Light-driven translocation of signaling proteins in vertebrate photoreceptors. Trends Cell Biol. 2006;16:560–568 [DOI] [PubMed] [Google Scholar]
- 48.Reidel B, Goldmann T, Giessl A, Wolfrum U. The translocation of signaling molecules in dark adapting mammalian rod photoreceptor cells is dependent on the cytoskeleton. Cell Motil Cytoskeleton. 2008;65:785–800 [DOI] [PubMed] [Google Scholar]
- 49.Weil D, Blanchard S, Kaplan J, et al. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature. 1995;374:60–61 [DOI] [PubMed] [Google Scholar]
- 50.Marszalek JR, Liu X, Roberts EA, et al. Genetic evidence for selective transport of opsin and arrestin by kinesin-II in mammalian photoreceptors. Cell. 2000;102:175–187 [DOI] [PubMed] [Google Scholar]
- 51.Pazour GJ, Baker SA, Deane JA, et al. The intraflagellar transport protein, IFT88, is essential for vertebrate photoreceptor assembly and maintenance. J Cell Biol. 2002;157:103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.