Abstract
Nanopore sequencing has the potential to become a direct, fast, and inexpensive DNA sequencing technology. The simplest form of nanopore DNA sequencing utilizes the hypothesis that individual nucleotides of single-stranded DNA passing through a nanopore will uniquely modulate an ionic current flowing through the pore, allowing the record of the current to yield the DNA sequence. We demonstrate that the ionic current through the engineered Mycobacterium smegmatis porin A, MspA, has the ability to distinguish all four DNA nucleotides and resolve single-nucleotides in single-stranded DNA when double-stranded DNA temporarily holds the nucleotides in the pore constriction. Passing DNA with a series of double-stranded sections through MspA provides proof of principle of a simple DNA sequencing method using a nanopore. These findings highlight the importance of MspA in the future of nanopore sequencing.
Keywords: bionanotechnology, next generation sequencing, single-molecule, stochastic sensing, protein pore
The information encoded in DNA is of paramount importance to medicine and to the life sciences. The mapping of the human genome is revolutionizing the understanding of genetic disorders and the prediction of disease and will aid in developing therapies as in refs. 1–3. The ability to sequence DNA quickly and inexpensively is essential to individualized medicine and to scientific research and has prompted the development of new sequencing techniques beyond the original Sanger sequencing (4–7). Nanopore DNA sequencing represents one of the approaches being developed to rapidly sequence a human genome for under $1,000 (www.genome.gov/12513210).
In the most elementary form of nanopore DNA sequencing, a nanometer-scale pore provides the sole pathway for an ionic current. Single-stranded DNA (ssDNA) is electrophoretically driven through the pore, and as the ssDNA passes through, it reduces the ionic current through the pore. If each passing nucleotide yields a characteristic residual ionic current then the record of the current will correspond to the DNA sequence. This simple and reagent-free sequencing technique holds the promise to inexpensively read long lengths of DNA molecules at intrinsically fast rates (8). Due to its inherently small size, this system is amenable to parallelization (9).
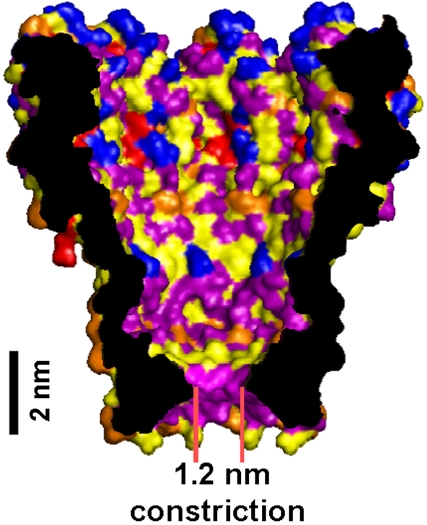
Lately, nanopore sequencing techniques have progressed substantially. This progress and the remaining challenges in nanopore DNA sequencing are summarized in a review article by Branton et al. (8). While nanotechnology usually involves materials such as Si and SiN, nanopore DNA sequencing first evolved using the well-studied protein porin α-hemolysin (10). In contrast to pores made from inorganic materials (11, 12), protein pores can be easily modified and produced with repeatable subnanometer accuracy. Stoddart (13) and Purnell (14) demonstrated that several locations within the beta barrel of α-hemolysin exhibit nucleotide-specific sensitivity with immobilized ssDNA (13). However, α-hemolysin’s 5 nm-long cylindrical beta barrel presents a structural limitation that dilutes the ion current specific to individual nucleotides and yields small current differences between the nucleotides. This structural drawback is overcome by the channel protein Mycobacterium smegmatis porin A (MspA) (15). MspA is an octameric protein with a single pore of dimensions (16) ideal for nanopore sequencing (Fig. 1). We hypothesized that the single constriction of MspA, with a diameter of ∼1.2 nm and a length of ∼0.5 nm, could yield well-resolved current signatures for each nucleotide. Initially, the negative charges at the constriction of wild-type MspA prevented ssDNA translocation. We used site-directed mutagenesis to remove three negatively charged aspartic acids within the constriction (positions 90, 91, and 93) and replaced them with neutral asparagines. The resulting mutant MspA, called M1-NNN-MspA, allowed ssDNA translocation (15).
Fig. 1.
Crystal structure of MspA. The cross-sectional view through M1-NNN-MspA’s structure using a space-filling model displays the classes of amino acids: Red are positively charged; blue are negatively charged; purple are polar; yellow are hydrophobic-aliphatic; orange are hydrophobic-aromatic. The single constriction, with dimensions similar to that of a single-nucleotide, makes MspA a good candidate for nanopore DNA sequencing techniques.
A common challenge to all nanopore sequencing approaches is that the DNA translocation is rapid, with speeds > 10 nt/μs (11) in solid state pores and > 1 nt/μs in α-hemolysin (17, 18) and MspA (15). While MspA may offer large current distinctions for individual nucleotides, the current levels are too short to be resolved. To explore the nucleotide discrimination and resolution possible with M1-NNN-MspA, we used DNA with double-stranded sections of DNA that are too thick to pass through MspA, similar to studies with α-hemolysin (19, 20). The double-stranded section causes the translocation to halt while it holds the single-stranded section of interest within the pore’s constriction. We changed the composition of the single-stranded section to examine how the nucleotides modulate the residual ion current.
With duplexes halting translocation, we found that the residual current through M1-NNN-MspA can distinguish the nucleotides adenine (dA), cytosine (dC), thymine (dT), and guanine (dG) with differences in conductivity on the order of 0.1 nS. Furthermore, we found that with the residual current within MspA one can discriminate individual nucleotides within a homopolymer background.
We extend the concept of using one double strand to pause DNA movement, to using multiple double-stranded sections. With this extension, we present proof of principle of a previously undescribed sequencing method that would use DNA that has double-stranded sections between each nucleotide of the DNA to be sequenced.
Results and Discussion
Nucleotide Identification by MspA.
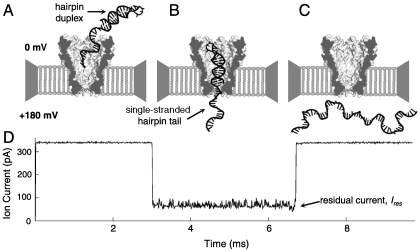
We conducted translocation experiments with DNA that forms a 14-base pair hairpin duplex and has a 50-nucleotide ssDNA “tail” (Table S1). When a voltage is applied across the pore, the long single-stranded tail facilitates capture and insertion of the DNA into the pore’s constriction (Fig. 2). At a driving voltage of 180 mV, the hairpin duplex dissociates after ∼10 ms. During the time that the hairpin tail is held, the measured residual ionic current (Ires) depends strongly on the composition of the ssDNA section residing in the confining constriction of the pore. Once the duplex dissociates, the DNA completes translocation to the lower potential chamber at speeds faster than 1 nt/μs (15).
Fig. 2.
DNA translocation through the nanopore MspA. The cartoon depicts DNA translocation through MspA and the resulting residual current. (A) The positive voltage attracts the negatively charged hairpin DNA into the pore. (B) The DNA threads through the pore until the wider hairpin duplex prevents further translocation. (C) After a few milliseconds the hairpin dissociates allowing for complete translocation. (D) The resulting current trace associated with the above cartoon shows that the hairpin DNA present in the pore allows a residual current, Ires, until the hairpin duplex dissociates.
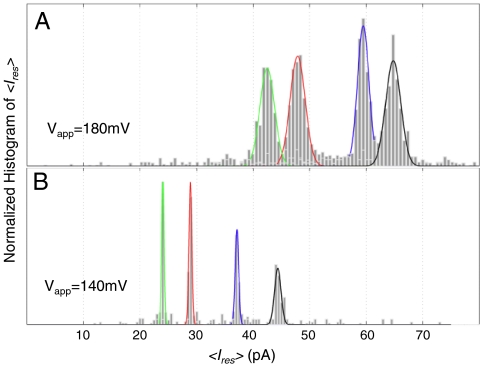
First, we determined the characteristic residual currents associated with the four bases by using the “homopolymer” DNA hairpin tails (dA)50, (dC)50, (dT)50, and (dG)3 (dA)47 held in M1-NNN-MspA. Note that we used (dG)3 (dA)47, instead of (dG)50, because of G-tetrad formation. For each polynucleotide tail, the histograms of the average residual current reveal unique values at an applied voltage of 180 mV (Fig. 3). The Gaussian mean (μ) and half-width (σ) of the residual ion currents caused by (dA)50 tails was IdA = 65.5 ± 1.5 pA (μ ± σ, averaged over N = 7 experiments with different pores, n = 3257 total translocations). The tails (dG)3 (dA)47, (dC)50, and (dT)50 yield residual ion currents of IdG = 59.4 ± 1.2 pA (N = 5, n = 2938), IdC = 48.4 ± 1.1 pA (N = 7, n = 1830), and IdT = 41.9 ± 1.2 pA (N = 4, n = 2407), respectively. At a lower voltage of 140 mV, we saw both narrower Gaussian widths and reduced separations within the distributions, with IdA = 43.6 ± 0.4 pA (n = 117), IdG = 37.5 ± 0.6 pA (n = 93), IdC = 29.2 ± 0.3 pA (n = 87), and IdT = 24.4 ± 0.5 pA (n = 169). The bulkier purines, dA and dG, have greater residual currents than the pyrimidines, dT and dC, indicating that steric restriction is not the primary determinant of residual current values, as has been noticed in α-hemolysin (21).
Fig. 3.
Example histograms of the averaged residual ion currents, 〈Ires〉 are shown for different “hompolymer” single-stranded tails of a 14 base pair hairpin (hp). Data were taken at (A) 180 mV and (B) 140 mV. The translocations included in the above histograms have durations longer than 1 millisecond and reveal distinguishable and well-resolved current levels. We give the average of the fitted Gaussian mean of a number of experiments in the main text. There were at least four experimental repetitions with each of the above hairpin DNA. The reduction in widths at the 140 mV is due to increased time averaging because the dissociation times are nearly 30 × longer than the dissociation times at 180 mV. Additional information may be found in Table S1.
The residual currents due to the different homopolymer hairpin tails are well-separated and well-resolved. For example, the residual ion current difference between poly-dA and poly-dC is IdA-IdC = 14.4 ± 0.5 pA in 1 M KCl using a 140 mV driving voltage. Comparable hairpin DNA experiments conducted with DNA hairpins in WT- α-hemolysin at 170 mV yielded less resolved current differences of IdA-IdC = -8.6 ± 4.0 pA with 0.5 M KCl (21). Even smaller current differences are found when ssDNA is held by streptavidin in α-hemolysin for long durations at 120 mV, yielding IdA-IdC = -3.3 pA ± 0.4 pA in 1.0 M KCl (22). MspA provides a minimum 3.5-times enhanced separation of nucleotide-specific currents in comparison to α-hemolysin.
Region of Sensitivity.
Next, we searched for the nucleotide location within the hairpin tail that influences the residual current. For this we used a series of hairpin DNA with (dC)4 sections at various positions in an otherwise poly-dA tail. When the (dC)4 was adjacent to the hairpin duplex, the currents were identical to IdC. When the (dC)4 section was located more than three bases away from the hairpin duplex, the residual current was undifferentiated from IdA (Table S1). To test the apparent importance of the first three nucleotides adjacent to the hairpin, we used tails with either (dA)3 or (dC)3 followed by two different random heteropolymers. We found the residual currents to be independent of the heteromeric section and to be indistinguishable from IdA or IdC, respectively (Table S1). Given MspA’s geometry, the hairpin duplex is expected to reside near the constriction. Because fewer than four nucleotides influence the residual current, the nucleotides within and near the constriction of MspA govern the residual currents.
Single Nucleotide Recognition.
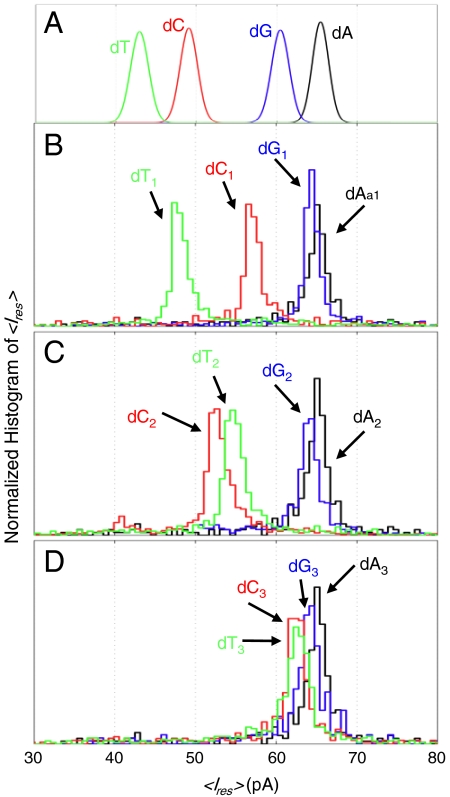
Recognition and identification of individual nucleotide sites is required for nanopore sequencing. We examined MspA’s sensitivity to individual nucleotides by making single nucleotide substitutions in the ssDNA hairpin tail. The substitution of a single nucleotide, dN in an otherwise poly-dA hairpin tail, in the first three positions as counted from the duplex, x = 1,2,3, and is denoted dNx. For example, a dC at the first position after the duplex (x = 1) is called dC1. Fig. 4 displays the histograms of the averaged Ires. With a dC2 substitution, Ires is close to the current associated with poly-dC, IdC. For a dC1 substitution, the measured ion currents were between the ion currents found using homopolymers, IdA and IdC. We found that substitutions of dT1, dT2 and 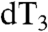 cause an ion current between IdA and IdT, with the current for the dT1 substitution nearest to IdT. A single dG within poly-dA does not appear to modulate the current appreciably from the current of a pure poly-dA, as might be expected from the relative closeness of IdA and IdG. The residual current tends toward IdA as the heteronucleotide substitution is placed further from the duplex, giving additional evidence that the residual current signature is primarily due to the first two nucleotides after the duplex, and partly due the third nucleotide. See Table S1 for additional information on these hairpin tails and their associated residual current values.
cause an ion current between IdA and IdT, with the current for the dT1 substitution nearest to IdT. A single dG within poly-dA does not appear to modulate the current appreciably from the current of a pure poly-dA, as might be expected from the relative closeness of IdA and IdG. The residual current tends toward IdA as the heteronucleotide substitution is placed further from the duplex, giving additional evidence that the residual current signature is primarily due to the first two nucleotides after the duplex, and partly due the third nucleotide. See Table S1 for additional information on these hairpin tails and their associated residual current values.
Fig. 4.
Residual current histograms due to single nucleotide substitutions in an otherwise poly-dA hairpin tail. (A) For comparison purposes the top panel summarizes the averaged Gaussian mean and width of Ires of the homopolymer hairpin tails at 180 mV (with fit values in text). Colors of black, blue, red, and green are used to help separate effects on Ires due to dA, dG, dC, and dT, respectively. The residual current markedly changes with the position, x, of a single nucleotide dNx, within an otherwise poly-dA homopolymer hairpin (hp) tail. (B) When the nucleotide substitution is adjacent to the double-stranded terminus, x = 1, the residual current deviates to resemble the hompolymer values associated with the substituted nucleotide. The dT1 substitution most closely resembles IdT. (C) At x = 2, the nucleotide substitution also causes residual current is closer to the homopolymer associated with the substituted nucleotide. The dC2 substitution is closest to IdC. (d) With any substitution at x = 3, Ires is only slightly different fromIdA, suggesting that MspA is primarily sensitive to the two nt after the hairpin duplex. A dGx substitution at x = 1, 2, or 3, does not significantly influence the current, as may be expected given the relative closeness of IdG and IdA.
The homopolymer background may influence the effect a heteronucleotide substitution has on Ires (14). We examined this effect by using a dA substitution in a poly-dC background (Fig. S1). The residual current was affected by these substitutions at x = 1,2,3, but the current differences from IdC were not as large as the influence of dCx substitutions in poly-dA. Similarly, we substituted a single dA in the background of poly-dT and did not find as consistent of an influence on Ires when compared to a dTx substitution in a background of poly-dA. These observations are not well described by a resistor model associated with each nucleotide substitution. However, the observed asymmetry may be qualitatively understood with rate limits to ion transport caused by energetic barriers (See SI Text).
These results indicate that the short constriction zone of MspA is indeed responsible for nucleotide identification. Compared to α-hemolysin, MspA produces a larger and more focused ion current density in its constriction zone. The length along the constriction where the current is most sensitive to nucleotide identity is about the length of two nucleotides. It is possible that nucleotide specificity and spatial separation may be enhanced with additional mutations to MspA, especially because the data presented were taken using M1-NNN-MspA, the first MspA mutant to allow DNA translocation. Because of its importance to the residual current, the constriction will likely be a very promising location for site-mutations.
Effect of the Hairpin Terminus.
Because the hairpin duplex rests near MspA’s constriction, it is possible that it also affects the ion current. To explore this, we investigated hairpin DNA with various duplex lengths but with the same terminal bases. The measured currents were found to be weakly dependent on the hairpin duplex length, with the longer duplex lengths inducing lower Ires than shorter duplex lengths (Table S2). In further experiments, we kept the original 14 bp duplex and varied only the terminal bp. We found that the residual current is strongly dependent on this terminal bp (Table S2), altering Ires by up to ∼20%. In order to compare the influence of the hairpin’s tail, the experiments presented in previous sections were acquired with the same 14 bp duplex.
Duplex Interrupted (DI) Nanopore Sequencing.
The ion current is uniquely sensitive to single-stranded DNA nucleotides in MspA’s constriction, one of the necessary requirements of nanopore sequencing. While the speed of unimpeded ssDNA translocation is still too fast to utilize this sensitivity, we can control the translocation speed of DNA using duplex regions. In the following section we describe a DNA sequencing method using double-stranded DNA sections to slow the translocation of DNA. With an existing biochemical conversion processes, short sections of double-stranded DNA can be effectively placed between the nucleotides of an analyte DNA. When this converted DNA is driven through MspA, each duplex section sequentially halts the translocation. As the nucleotide of the analyte DNA is held in the confining aperture by the duplex section, the residual ion current can identify the analyte nucleotide. After one DNA duplex dissociates, the DNA quickly advances until the next DNA duplex pauses the translocation, allowing the next analyte nucleotide to be determined. Such a method, which we term Duplex Interrupted (DI) nanopore sequencing, may ultimately yield a fast and sequential read of an analyte DNA using a nanopore.
Conversion of DNA.
DI sequencing depends on modifying the analyte DNA to have double-stranded sections between each nucleotide. Such DNA conversion schemes were initially developed in hopes of sequencing freely translocating DNA that is expanded into many nucleotides of the same type to produce sufficiently long current signatures. This conversion is typically accomplished using cyclic application of DNA restriction and ligation enzymes. Meller et al. postulated an optical-nanopore sequencing strategy with each nucleotide converted into a specific binary code made of two 12-mer oligos using such a DNA conversion scheme (23–25). An automated, massively parallel process (26, 27) requires ∼24 h for the conversion of a complete human genome into a DNA mixture consisting of fragments, each corresponding to a 24 bp segment of the original genome (8). The DNA conversion required for DI sequencing is less demanding because each inserted DNA could be identical and independent of the analyte nucleotides. Work is currently under way to develop inexpensive, low-error conversion of long segments of the original genome with reduced conversion time (25). Further reduction in conversion cost and speed may be attained through massive parallelization, comparable to sequencing-by-ligation technologies that rely on ligation reactions (4).
DI Sequencing Proof of Principle Experiments.
To explore the feasibility of DI sequencing, we used synthesized DNA where each nucleotide of a hypothesized analyte DNA is followed by a 14 bp duplex region. The duplex sections had sequences identical to those of hairpins examined above and were formed by annealing complementary oligo nucleotides. Instead of using single-nucleotides between each duplex section, we chose to use trinucleotides to easily compare the ion currents to our well-characterized homopolymer hairpin experiments. A poly-dA tail was added to the 3′ end of the synthesized sequence to initiate DNA threading into MspA. For example, the analyte sequence 3′-ATGC-5′ would be converted to the analyzable DI sequence 5′duplex-CCC-duplex-GGA- duplex-TTT-duplex-AAA-dA32 3′. These synthesized sequences containing the trinucleotide regions could also be the product of DNA conversion (26).
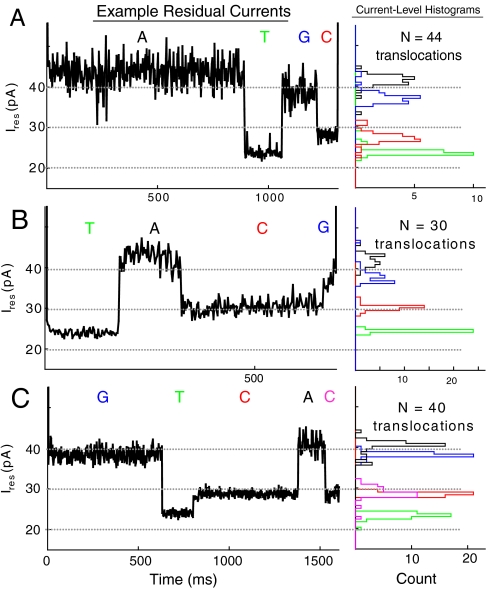
Using M1-NNN-MspA, we examined the DNA constructs for the analyte sequences 3′-ATGC-5′ and 3′-TACG-5′, both containing all four nucleotides. We observed successive discrete steps in the ion current for synthesized sequences with residual currents shown in Fig. 5 A and B. Each level was consistent with one of the levels observed in the homopolymer hairpin experiments. Using an edge detection algorithm (SI Text) on the translocations that had an average current of < 25% of the open pore current, we found that ∼4% of the translocations exhibited all four current levels, IdA, IdC, IdT and IdG in the anticipated order. In Fig. 5 A and B we present four-level sample traces recorded at 140 mV together with histograms of the average current of the levels found for many translocations. Additional data for DI sequencing at other voltages is presented in Figs. S2, S3, and S4. A large fraction of the translocations exhibited three or fewer distinct levels (Tables S3, S4, and S5). The levels found in these translocations also contained the homopolymer current levels with ordering consistent with the analyte sequence but with one or more levels missing. We believe that the translocations with fewer than four levels are due to incompletely annealed duplex regions or level durations that were too short (< 1 ms) to be properly identified.
Fig. 5.
Demonstration of duplex interrupted (DI) nanopore sequencing using MspA. We use synthesized DNA simulating analyte DNA converted to have duplexes between information carrying nucleotides. Each of these duplexes must be sequentially melted as the DNA is pulled through the pore, enabling the residual current to determine the sequence. We provide proof of principle of this technique using DNA chosen to represent different analyte sequences (A) 3′-ATGC-5′, (B) 3′-TACG-5′, and (C) the “blind” sequence determined to be 3′-GTCAC-5′. We display example traces of residual currents (left). Each step in the residual current is representative of three nucleotides within MspA’s constriction held by a 14 bp DNA duplex. For each of these steps we generate a histogram of each level (right) for N translocations. These results are generated from three or more experiments at 140 mV. At higher voltages, the number of translocations increases (Tables S3, S4, and S5), but the level specificity decreases (Figs. S2, S3, and S4) due to reduced time averaging.
With our edge detection algorithm tuned by analyzing the DI sequences 3′-ATGC-5′ and 3′-TACG-5′, we conducted a blind test with synthesized DNA derived from a short sequence of unknown composition and length. We found ∼3% of the translocations to have five levels in the current traces corresponding to 3′-GTCAC-5′, which was later confirmed to be the unknown sequence. Fig. 5C displays an example current trace from the blind test. These DI experiments demonstrate that sequence information can be extracted from DNA molecules serially passing through a nanopore.
Nanopore sequencing must be able to distinguish repeat nucleotides using the residual current. With DI sequencing in MspA, this requirement may be accomplished by using a “fifth level” that marks the progression to the next analyte nucleotide. The fifth level is made by partitioning the interrupting-duplex with two separate complimentary oligos. The resulting first duplex produces the levels specific to analyte nucleotides and the second duplex produces a distinct level. The distinct fifth level can be made by choosing the second duplex to have terminal 5′ (dC) (Table S2) that yields a residual current higher than the residual current of a 5′ that is (dG) (Table S1). With this choice, the residual current would toggle between ∼77 pA and the analyte nucleotide-specific currents between 42 pA and 66 pA using 180 mV voltage. The fifth level would separate the current level of every analyte nucleotide and would allow nucleotide repeats of any length to be read.
While an individual translocation may indicate the sequence, missed bases require the statistics of multiple translocations to enhance the sequencing fidelity. Statistical analysis of current-level durations can provide additional information about sequence and missed nucleotides, but this will be the subject of a subsequent publication.
The experiments and data presented here are intended to be proof of principle of a previously undescribed sequencing technique. There is potential to optimize both the speed and sensitivity of DI sequencing by altering operating parameters such as pH and ionic strength of the buffer, using duplex-binding reagents, improving oligo annealing, and using more sophisticated data analysis techniques (28). In comparison to previously proposed sequencing methods involving converted DNA (19, 29), DI sequencing does not require additional hardware such as fluorescence detection or conversion to binary codes. While DI sequencing is a promising step forward, the realization of MspA-based nanopore sequencing using unmodified DNA remains the primary goal of our research.
Conclusions
Our experiments demonstrate that the protein pore MspA has great potential to advance nanopore DNA sequencing. MspA’s short and narrow constriction can be used to discriminate individual nucleotides of ssDNA while duplex DNA sections pause the translocation. Using this concept, we provide proof of principle for a nanopore DNA sequencing technique that would use DNA converted to have double-stranded sections between analyte nucleotides.
Materials and Methods
Biological Material.
MspA mutant porin M1-NNN-MspA was selectively prepared and extracted from M. smegmatis (15). DNA was synthesized by Integrated DNA Technologies with no additional purification for hairpin DNA, or with PAGE purification for some of the DNA used in DI experiments. We used DNA concentrations of ∼10 μM to 100 μM. To prevent self-dimerization, we prepared hairpin DNA by heating it to 90 °C for 1 min, cooling in a -8 °C freezer for an additional minute, and then returning it to room temperature before use.
Hairpin DNA sequences examining MspA’s nucleotide sensitivity had the same 14 base duplex region and 6 nt loop. 5′GCTGGCTCTGTTGC TCTCTC GCAACAGAGCCAGC 〈tail〉 3′. The underlines indicate duplex formation between complementary bases. The hairpin tail sequences are presented in Tables S1 and S2. If residual currents were sufficiently similar to other DNA strands we ran experiments with similar concentration of either poly-dA or poly-dC to provide a residual current calibration. This calibration reduced minor experimental variation in current levels due to Nernst-potentials and buffer evaporation.
The DNA used in DI sequencing were as follows: 3′ATGC5′: 5′ GCAACAGAGCCAGC CCC GCAACAGAGCCAGC GGA GCAACAGAGCCAGC TTT GCAACAGAGCCAGC AAA A32 3′ 3′TACG5′: GCAACAGAGCCAGC GGA GCAACAGAGCCAGC CCC GCAACAGAGCCAGC AAA GCAACAGAGCCAGC TTT A32 3′ BLIND: 5′ GCAACAGAGCCAGC CCC GCAACAGAGCCAGC AAA GCAACAGAGCCAGC CCC GCAACAGAGCCAGC TTT GCAACAGAGCCAGC GGA A15 3′.
The underlined regions formed duplexes with oligonucleotides of sequence 5′ GCTGGCTCTGTTGC 3′. The oligonucleotides and synthesized DI DNA were combined in a molar ratio > 32∶1, annealed by heating to 95 °C for 5 min and then gradually cooled to 23 ± 1 °C.
Single-Pore Experiments.
Pores were established with previously described methods (15). Briefly, lipid bilayers were formed from either 1,2-diphytanoyl-sn-glycerol-3-phosphocholine, 1,2-diphytanoyl-sn-glycero-3-phosphate (Avanti Polar Lipids) or from equal mixtures thereof. The bilayer spanned a horizontal ∼20 micron diameter aperture in Teflon. M1-NNN-MspA was added to the grounded side of the bilayer at a concentration of ∼2.5 ng/ml. An Axopatch-1B or 200B patch clamp amplifier (Axon Instruments) applied a voltage across the bilayer and measured the ionic currents. The analog signal was low-pass filtered at 10, 50 or 100 kHz with a 4-pole Bessel filter and was then digitized at five times the low-pass filter frequency. Data acquisition was controlled with custom software written in LabWindows/CVI (National Instruments). For display purposes, the residual current traces were digitally filtered at 2 kHz. All experiments were performed at 23 ± 1 °C in 1 M KCl, 10 mM HEPES/KOH buffered at pH 8. Data was analyzed with custom software written in Matlab (The Mathworks) (see SI Text).
Supplementary Material
Acknowledgments.
We thank Kyle Langford, Risa Wong, Boyan Penkov, and Meredith Brown for help in gathering the data and Mark Troll for stimulating discussions and for obtaining the blind DI sequence. This work was supported by the National Institutes of Health, National Human Genome Research Institute $1,000 Genome Program Grant 5R21HG004145 and R01H6005115.
Footnotes
Conflict of interest statement: The authors declare a conflict of interest (such as defined by PNAS policy). J.H.G., M.N., T.Z.B., and M.P. have filed a patent on the use of MspA for single-molecule analysis. I.M.D., M.D.C, and J.H.G. have filed a provisional patent on the sequencing strategy presented herein.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001831107/-/DCSupplemental.
References
- 1.Goldstein DB. Common Genetic Variation and Human Traits. N Engl J Med. 2009;360(17):1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 2.Hirschhorn JN. Genomewide association studies—Illuminating biologic pathways. N Engl J Med. 2009;360(17):1699–1701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- 3.Morris JR, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462(7275):886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 4.Shendure JA, Porreca GJ, Church GM. Overview of DNA sequencing strategies. In: Ausubel Frederick M, et al., editors. Current Protocols in Molecular Biology. New York: Wiley Interscience; 2008. Chapter 7. [DOI] [PubMed] [Google Scholar]
- 5.Fuller CW, et al. The challenges of sequencing by synthesis. Nat Biotechnol. 2009;27(11):1013–1023. doi: 10.1038/nbt.1585. [DOI] [PubMed] [Google Scholar]
- 6.Drmanac R, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2009;327(5961):78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 7.Bentley DR. Whole-genome re-sequencing. Curr Opin Genet Dev. 2006;16(6):545–552. doi: 10.1016/j.gde.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 8.Branton D, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26(10):1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osaki T, Suzuki H, Le Pioufle B, Takeuchi S. Multichannel simultaneous measurements of single-molecule translocation in alpha-hemolysin nanopore array. Anal Chem. 2009:105–114. doi: 10.1021/ac901732z. [DOI] [PubMed] [Google Scholar]
- 10.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA. 1996;93(24):13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Gershow M, Stein D, Brandin E, Golovchenko JA. DNA molecules and configurations in a solid-state nanopore microscope. Nat Mater. 2003;2(9):611–615. doi: 10.1038/nmat965. [DOI] [PubMed] [Google Scholar]
- 12.van den Hout M, et al. Controlling nanopore size, shape and stability. Nanotechnology. 2010;21(115304):115304. doi: 10.1088/0957-4484/21/11/115304. [DOI] [PubMed] [Google Scholar]
- 13.Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H. Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc Natl Acad Sci USA. 2009;106(19):7702–7707. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purnell RF, Schmidt JJ. Discrimination of single base substitutions in a DNA strand immobilized in a biological nanopore. ACS Nano. 2009;3(9):2533–2538. doi: 10.1021/nn900441x. [DOI] [PubMed] [Google Scholar]
- 15.Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. Single-molecule DNA detection with an engineered MspA protein nanopore. Proc Natl Acad Sci USA. 2008;105(52):20647–20652. doi: 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faller M, Niederweis M, Schulz GE. The structure of a mycobacterial outer-membrane channel. Science. 2004;303(5661):1189–1192. doi: 10.1126/science.1094114. [DOI] [PubMed] [Google Scholar]
- 17.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys J. 1999;77(6):3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Rapid nanopore discrimination between single polynucleotide molecules. Proc Natl Acad Sci USA. 2000;97(3):1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sauer-Budge AF, Nyamwanda JA, Lubensky DK, Branton D. Unzipping kinetics of double-stranded DNA in a nanopore. Phys Rev Lett. 2003;90(23):238101. doi: 10.1103/PhysRevLett.90.238101. [DOI] [PubMed] [Google Scholar]
- 20.Vercoutere W, et al. Rapid discrimination among individual DNA hairpin molecules at single-nucleotide resolution using an ion channel. Nat Biotechnol. 2001;19(3):248–252. doi: 10.1038/85696. [DOI] [PubMed] [Google Scholar]
- 21.Ashkenasy N, Sanchez-Quesada J, Bayley H, Ghadiri MR. Recognizing a single base in an individual DNA strand: A step toward DNA sequencing in nanopores. Angew Chem Int Edit. 2005;44(9):1401–1404. doi: 10.1002/anie.200462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purnell RF, Mehta KK, Schmidt JJ. Nucleotide identification and orientation discrimination of DNA homopolymers immobilized in a protein nanopore. Nano Lett. 2008;8(9):3029–3034. doi: 10.1021/nl802312f. [DOI] [PubMed] [Google Scholar]
- 23.Lee J, Meller A. Rapid DNA sequencing by direct nanoscale reading of nucleotide bases on individual DNA chains. In: Mitchelson K, editor. New High Throughput Technologies for DNA Sequencing and Genomics. Oxford, UK: Elsevier; 2007. pp. 245–264. (Vol Perspectives in Bioanalysis). [Google Scholar]
- 24.Soni GV, Meller A. Progress toward ultrafast DNA sequencing using solid-state nanopores. Clin Chem. 2007;53(11):1996–2001. doi: 10.1373/clinchem.2007.091231. [DOI] [PubMed] [Google Scholar]
- 25.McNally B, Yu Z, Sun Y, Weng Z, Meller A. Optical recognition of converted DNA nucleotides for single-molecule DNA sequencing using nanopore arrays. Nano Lett. 2010 doi: 10.1021/nl1012147. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lexow P. Sequencing method using magnifying tags. WO 2000/39333. Int Patent. 2004
- 27.Lexow P. Method for producing and amplified polynucleotide sequence. WO 2006/092582. Int Patent. 2006
- 28.Churbanov A, Baribault C, Winters-Hilt S. Duration learning for analysis of nanopore ionic current blockades. BMC Bioinformatics. 2007;8(Suppl 7):S14. doi: 10.1186/1471-2105-8-S7-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim YR, Li CM, Wang Q, Chen P. Detecting translocation of individual single stranded DNA homopolymers through a fabricated nanopore chip. Front Biosci. 2007;12:2978–2983. doi: 10.2741/2287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.