Abstract
MutLα (MLH1–PMS2) is a latent endonuclease that is activated in a mismatch-, MutSα-, proliferating cell nuclear antigen (PCNA)-, replication factor C (RFC)-, and ATP-dependent manner, with nuclease action directed to the heteroduplex strand that contains a preexisting break. RFC depletion experiments and use of linear DNAs indicate that RFC function in endonuclease activation is limited to PCNA loading. Whereas nicked circular heteroduplex DNA is a good substrate for PCNA loading and for endonuclease activation on the incised strand, covalently closed, relaxed circular DNA is a poor substrate for both reactions. However, covalently closed supercoiled or bubble-containing relaxed heteroduplexes, which do support PCNA loading, also support MutLα activation, but in this case cleavage strand bias is largely abolished. Based on these findings we suggest that PCNA has two roles in MutLα function: The clamp is required for endonuclease activation, an effect that apparently involves interaction of the two proteins, and by virtue of its loading orientation, PCNA determines the strand direction of MutLα incision. These results also provide a potential mechanism for activation of mismatch repair on nonreplicating DNA, an effect that may have implications for the somatic phase of triplet repeat expansion.
Keywords: MutLalpha, genetic instability, cancer, triplet repeats
The best understood function of mismatch repair is its role in correction of replication errors, which requires that repair be directed to a newly synthesized DNA strand. A strand-specific nick or gap is sufficient to direct mismatch repair in extracts of mammalian and Drosophila cells, or in Xenopus egg extracts (1–4), and an obvious possibility is that DNA termini that occur naturally at the replication fork serve as the strand signals that direct the reaction in the eukaryotic cell. Whereas the strand break that directs in vitro repair can be located either 3′ or 5′ to the mismatch, only a single excision activity has been implicated in the eukaryotic reaction. Exonuclease 1 (Exo1), which hydrolyzes duplex DNA with 5′ to 3′ polarity, has been implicated in both yeast and mammalian mismatch repair, and in the latter case has been shown to function in both 3′- and 5′-directed repair events (1–4).
A possible explanation for this polarity paradox has been provided by analysis of a purified system comprised of MutSα (MSH2–MSH6 heterodimer), MutLα (MLH1–PMS2 heterodimer), the replication clamp proliferating cell nuclear antigen (PCNA), the clamp loader replication factor C (RFC), the single-stranded DNA binding protein replication protein A, Exo1 and DNA polymerase δ. Repair and mismatch-provoked excision in this minimal system recapitulate bidirectional features of the extract reaction, and as in extracts, Exo1 supports repair directed by either a 3′ or 5′ strand break (5, 6). This puzzling observation was clarified by the finding that eukaryotic MutLα is a latent endonuclease that is activated in a mismatch-, MutSα- (or MutSβ-), PCNA-, RFC-, and ATP-dependent fashion in a reaction that also depends on presence of a preexisting break in one heteroduplex strand (7–9). Incision by activated MutLα is directed to the heteroduplex strand that contains a preexisting strand break and is biased to the distal side of the mismatch. For a 3′ heteroduplex this results in incision 5′ to the mispair to yield a product in which the mismatch is bracketed by strand breaks. Multiply incised molecules produced in this manner are substrates for MutSα-activated Exo1 (7, 10), which removes the DNA segment spanning the mismatch by 5′ to 3′ hydrolysis.
Although attempts to identify other exonucleases that function in eukaryotic mismatch repair have yielded negative results, eukaryotic cells support a significant level of Exo1-independent mismatch correction that accounts for 10–40% of the repair events that occur in mammalian cells (11, 12). A recent analysis of this effect has led to the suggestion that repair in the absence of Exo1 occurs via strand displacement synthesis by DNA polymerase δ in a reaction that depends on the introduction of secondary strand breaks by MutLα (13).
Although the endonuclease activity of MutLα plays a central role in the current understanding of eukaryotic mismatch correction, the mechanism of endonuclease activation and the factor(s) involved in strand direction of this activity are not known. The study described here addresses involvement of RFC, PCNA, and a DNA strand break in these effects.
Results
The Primary Function of RFC in MutLα Activation Is PCNA Loading.
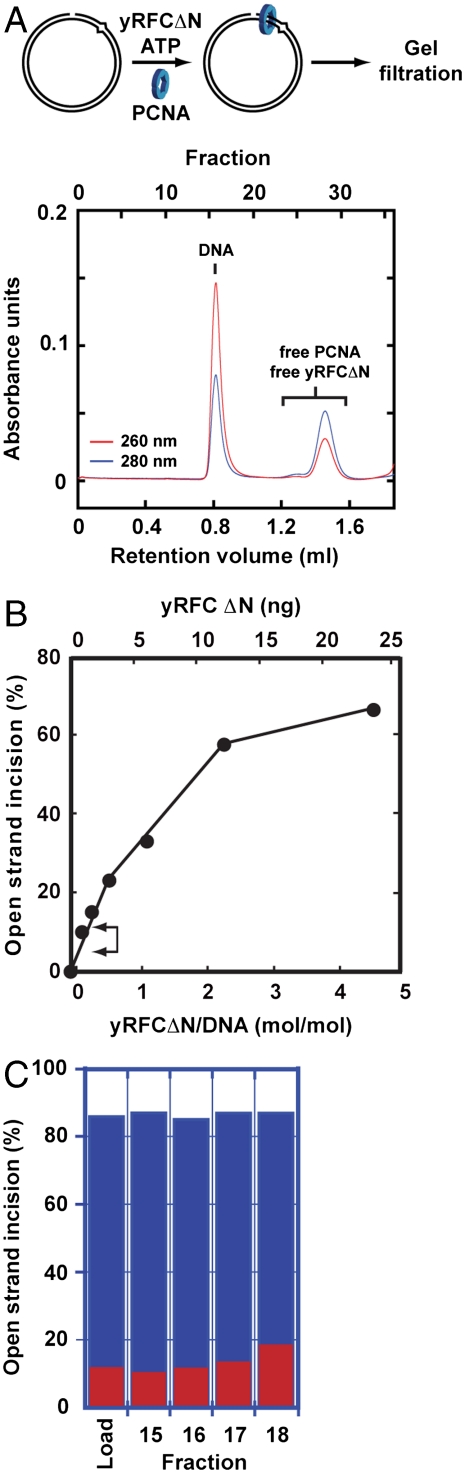
At physiological ionic strength in the presence of Mg2+, MutLα endonuclease activation requires a mismatch, MutSα (or MutSβ), PCNA, RFC, and a strand break (7–9). MutSα/MutSβ is required for mismatch recognition, but the individual roles of PCNA and RFC in this reaction have not been defined. PCNA interaction with MutLα has been demonstrated (5, 14, 15), suggesting that this interaction may play a role in endonuclease activation (8). However, it is unclear whether RFC function is restricted to clamp loading. We have addressed this issue using a set of near homogeneous proteins (Fig. S1). PCNA was loaded onto 3′-nicked G–T heteroduplex DNA and Superdex 200 gel filtration employed to resolve PCNA–DNA complexes from unbound protein. This procedure was facilitated by use of yeast RFCΔN (yRFCΔN; this designation will be used below to distinguish this activity from human RFC), which lacks the ligase homology domain of the large subunit and displays reduced DNA affinity (16), but nevertheless efficiently loads human PCNA onto nicked DNA and is functional in reconstituted human mismatch-provoked excision (5).
As shown in Fig. 1A and Fig. S2, the bulk of the DNA eluted from Superdex 200 in the column void volume in fractions 15–18, well separated from free yRFCΔN and PCNA, which eluted in fractions 25–31. Western blot analysis of column fractions (Fig. S2B) demonstrated that PCNA coeluted with DNA in fractions 15–18 with an efficiency of clamp loading of about 4 PCNA trimers per heteroduplex. However, these fractions contained only trace amounts of yRFCΔN, levels sufficient to support MutLα activation at only 5–11% of the maximal level (Fig. 1B, bracket). Neverthless, supplementation of samples of these fractions with MutSα and MutLα resulted in robust endonuclease activation (> 70% cleavage, Fig. 1C), with incision directed to nicked heteroduplex strand (Fig. S2A). These findings suggest that yRFCΔN function in MutLα activation is restricted to clamp loading.
Fig. 1.
RFC function in MutLα activation is limited to PCNA loading. (A) Human PCNA was loaded onto 3′ G-T heteroduplex DNA using yRFC∆N, and reaction products subjected to Superdex 200 gel filtration (Materials and Methods). DNA eluted in fractions 15–18 and unbound protein in fractions 25–31. (B) The yRFC∆N dependence of MutLα endonuclease activation was determined using 24 fmol 3′ G–T heteroduplex DNA as described in Materials and Methods. Values shown are corrected for a modest background level of random strand breaks in the heteroduplex preparation used. The bracket indicates the activity range supported by the yRFC∆N levels present in 10 μL of Superdex 200 fractions 15–18 as estimated by Western blot (Fig. S2B). (C) Ten μL of the indicated Superdex 200 fractions were assayed for strand-directed endonuclease activity in the absence (red bars) or presence of MutSα and MutLα [blue bars (Materials and Methods), mismatch-dependent conditions)]. Background strand breaks in the heteroduplex preparation used were comparable to those observed when MutSα and MutLα were omitted from the incubation (red bars). Raw data for this analysis is shown in Fig. S2A.
Further support for this idea was provided by analysis of MutLα endonuclease on linear homoduplex DNA. We have previously shown that constitutive endonuclease activity can be demonstrated in MutLα preparations provided the ionic strength is low and Mn2+ is substituted for Mg2+. The Mn2+-dependent activity does not respond to MutSα, but is strongly stimulated on the incised strand of nicked circular DNA by PCNA and RFC, and both proteins are required for this effect (7, 8). Fig. 2A demonstrates that incision of a linear 202 bp duplex by the Mn+2-dependent nuclease is also dramatically activated in the presence of yRFCΔN and PCNA (compare lanes 1 and 3), but in contrast to results with nicked circular DNA, PCNA alone is sufficient to account for the MutLα activation observed (compare lanes 2 and 3). The RFC independence of this linear DNA effect is reminiscent of RFC-independent PCNA activation of DNA polymerase δ on linear template-primers (17), and provides additional support for the idea that RFC function in MutLα endonuclease activation is restricted to loading of PCNA onto DNA to which it is otherwise topologically inaccessible. The fact that PCNA is sufficient to activate MutLα in this system indicates that interaction of the two proteins, presumably while DNA-bound, is required for this effect. Consistent with this idea is the finding that PCNA activation of MutLα in this two-protein system is blocked by a peptide containing the p21 PCNA interaction motif, which interferes with a number of PCNA-dependent processes (18), but not by a scrambled sequence peptide (Fig. S3A).
Fig. 2.
Endonuclease activity on linear DNA requires PCNA but not RFC. Mismatch-independent MutLα endonuclease activity on 202 bp linear homoduplex DNA as described in Materials and Methods in the presence of reaction components as indicated. (A) Reactions contained 30 mM KCl, 0.5 mM ATP, 1 mM MnSO4, and protein components as indicated. (B) Reactions contained 60 mM KCl, 1.5 mM ATP, 5 mM MgCl2, and indicated proteins. ATP was omitted from the reaction in lane 8 and EDTA substituted for MgCl2 in lane 9. Reactions shown in lanes 11–13 were performed as for lane 8 except that final KCl concentration was varied as shown.
Mismatch- and RFC-Independent but PCNA- and MutSα-Dependent Activation of MutLα.
The simplified Mn2+-dependent mismatch-independent reaction described above has partially clarified PCNA function in MutLα activation. By manipulating reaction components, we have also sought conditions where MutLα activation depends on both PCNA and MutSα, but is independent of RFC and a mismatched base pair. As shown in Fig. 2B (lanes 1–10), a neutral pH buffer containing 60 mM KCl and 5 mM MgCl2 is sufficient in this regard. Under these conditions, MutLα activation requires MutSα, PCNA, ATP, and a divalent cation, but is independent of RFC and mismatch. Inasmuch as PCNA–MutSα interaction is not necessary for MutLα activation (19), we infer that the MutSα requirement for this reduced-specificity reaction reflects a requirement for MutSα–MutLα interaction. Endonuclease activation in this manner was abolished when the KCl concentration was increased to physiological levels that are employed for study of mismatch-dependent activation of MutLα (120–125 mM; Fig. 2B [lanes 11–13, (7)].
The observed distributions of cleavage sites within the 202 bp homoduplex under MutSα-independent and MutSα-dependent conditions were similar, and in both cases were nonrandom (Figs. 2 A and B). Furthermore, the distribution of incision sites relative to labeled 5′-termini differed for the two strands of the homoduplex (Fig. S3B), suggesting that MutLα incision responds to some extent to sequence context.
Loaded PCNA on a Supercoiled Heteroduplex Is Sufficient to Activate MutLα Nuclease in a MutSα-Dependent Manner, but Incision Occurs Without Strand Bias.
At physiological ionic strength in the presence of Mg2+, activation of MutLα endonuclease requires a mismatch, MutSα, PCNA, RFC, and depends on the presence of a single strand interruption within the heteroduplex (7, 8). In addition to its involvement in MutLα activation, the strand break also determines the strand-specificity of incision by the activated endonuclease. One probable function of the strand discontinuity is provision of a loading site for PCNA (20, 21), and it has been suggested that loaded PCNA may function as an effector for communication between the strand break and mismatch for the purpose of strand discrimination (8, 14). To evaluate potential involvement of PCNA in strand discrimination, we have sought conditions that support PCNA loading onto closed circular DNA.
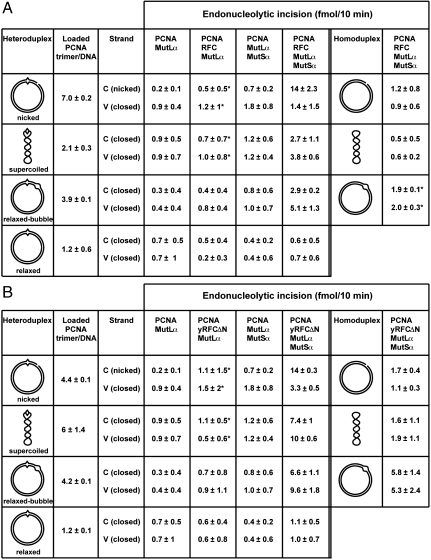
RFC has been shown to load PCNA onto closed circular DNA at low ionic strength, with negatively supertwisted molecules being better substrates than relaxed circles (20), and similar results have been obtained with the Escherichia coli β clamp and the γ complex clamp loader (22). It has been suggested that loading onto supertwisted DNA is due to transient helix opening driven by the free energy of supercoiling and the consequent production of double strand-single strand junctions that serve as sites for clamp assembly (22). To evaluate PCNA loading onto circular heteroduplex DNAs, we utilized a PCNA variant that can be 32P-labeled coupled with gel filtration assay to score loading of clamps onto the helix (refs. 20 and 23 and Fig. S4). As summarized in the second column of Fig. 3 A and B, RFC and yRFCΔN are capable of loading PCNA onto supercoiled heteroduplex DNA at physiological salt concentration (125 mM NaCl, 5 mM MgCl2). In fact, the efficiency of PCNA loading by yRFCΔN onto a supercoiled G–T heteroduplex (6 clamps/DNA) is similar to that observed with the nicked substrate (4 clamps/DNA). As expected, human RFC also efficiently loaded PCNA onto nicked DNA (7 clamps/DNA), but was less effective than yRFCΔN with the supercoiled heteroduplex (2 clamps/DNA). Relaxed closed circular heteroduplex was the weakest substrate for clamp loading under these conditions (1 clamp per heteroduplex). Our results with supercoiled DNA differ somewhat from those of Podust et al. (20), who found that RFC loading of PCNA onto supercoils is greatly reduced at physiological salt concentration. We attribute this difference to the fact that the supertwisted heteroduplexes used in our studies were prepared to have approximately twice the superhelical density of plasmid DNA isolated from E. coli (Materials and Methods) to facilitate helix opening at elevated ionic strength.
Fig. 3.
Strand break and DNA topology dependence of PCNA loading and MutLα activation. Efficiency of PCNA loading onto nicked, supercoiled (σ = -0.12), relaxed bubble, and relaxed G–T heteroduplexes was determined by gel filtration assay (Materials and Methods and Fig. S4). Loading values shown are corrected for that observed in the absence of ATP (0.5 PCNA trimer/DNA). Clamp loading utilized native human RFC (A) or yRFCΔN (B). For supercoiled DNA, loading values are the mean of 3 determinations (± 1 standard deviation); for other DNAs, values are the average of 2 determinations, with the variation shown corresponding to the range observed. MutLα endonucleolytic incision of complementary (C) and viral (V) strands of these heteroduplexes (or A–T homoduplex controls) was determined in the presence of the indicated proteins and quantified as described under Materials and Methods, and corrected for presence of background nicks in DNA preparations as described in Fig. 1. Incision values indicated with an asterisk are the average of 2 determinations, with the range observed indicated. Other values are the mean of 3 to 6 determinations ± one standard deviation.
We thus asked whether the ability of yRFCΔN and RFC to load PCNA onto negatively supertwisted heteroduplex DNA bypasses the requirement for a strand break in MutLα activation. Fig. 3 (columns 7 and 9) summarizes the efficiencies of MutLα activation for several G–T heteroduplex and control A–T homoduplex DNAs as a function of strand break status and tertiary conformation. As observed previously (7), MutLα incision of nicked heteroduplex DNA requires a mismatch, MutSα, PCNA, and RFC (or yRFCΔN), and is strongly biased to the heteroduplex strand that contains a preexisting break. However, an otherwise identical relaxed closed circular heteroduplex was found to be inert as substrate, with levels of incision similar to those obtained with nicked or supercoiled homoduplex controls. By contrast, the negatively supercoiled G–T heteroduplex does support mismatch-dependent MutLα endonuclease activation with the same protein requirements as those for a nicked heteroduplex, but incision of the superhelical substrate occurs with minimal strand bias (Fig. 3 A and B). The implications of this finding will be considered below.
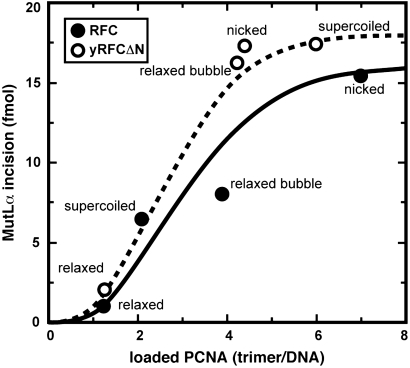
Although RFC and yRFCΔN support comparable levels of MutLα activation on nicked heteroduplex DNA, yRFCΔN was consistently found to provide higher levels of activation with a superhelical heteroduplex (Fig. 3 A and B), an effect that correlates with the differential ability of the two activities to load PCNA onto negatively supertwisted DNA. This effect and an empirical comparison of the efficiency of MutLα activation as a function of the extent of PCNA loading by RFC and yRFCΔN are illustrated in Fig. 4. For a given extent of PCNA loading, reactions containing native human RFC resulted in a somewhat reduced level of MutLα activation as compared to that observed with yRFCΔN. Although we have not established the basis for this effect, it is known that native RFC avidly binds to DNA, an effect that is abrogated by deletion of the ligase homology domain of the large subunit as in yRFCΔN (16). It is possible that the modest activity reduction observed with native RFC is the result of inhibition by the DNA-bound protein.
Fig. 4.
MutLα activation as a function of extent of PCNA loading. Extents of MutLα activation are plotted against efficiencies of PCNA loading by RFC and yRFCΔN for the heteroduplexes shown in Fig. 3. Lines shown are empirical fits.
Relaxed, Bubble-Containing Heteroduplex DNA Supports PCNA Loading and MutLα Activation, but Incision Occurs Without Strand Bias.
Clamp loading onto supercoiled DNA has been postulated to occur at transient double strand-single strand junctions that result from helix breathing events driven by the free energy of supercoiling (22). To directly test whether double strand-single strand junctions within a covalently continuous duplex can provide PCNA loading sites, we prepared a relaxed closed circular G–T heteroduplex that contains a 10 bp bubble comprised of a d(T)10 sequence within each strand. Unlike simple relaxed circular heteroduplex DNA, the bubble-containing heteroduplex does support efficient PCNA loading by both RFC and yRFCΔN (Fig. 3 and Fig. S4). Furthermore, like supercoiled heteroduplex DNA, the bubble-containing relaxed substrate supports mismatch-, MutSα-, PCNA- and RFC/yRFCΔN-dependent activation of MutLα endonuclease (Fig. 3). As in the case of supercoiled substrate, incision of the bubble-containing relaxed heteroduplex occurs with limited strand bias.
Bubble-containing A–T control DNA was subject to incision at 50 to 70% the efficiency observed with the bubble-containing G–T heteroduplex, and this occurred without significant strand bias. Because incision in this manner was MutSα-dependent (Fig. S5), we attribute this effect to bubble recognition by the MSH2–MSH6 heterodimer, a finding consistent with the demonstration that MutSα is capable of recognizing heterologies involving multiple unpaired nucleotides (1).
Discussion
This study addresses the functions of RFC, PCNA, and the heteroduplex strand break in the activation and strand direction of the MutLα endonuclease. Our results strongly suggest that RFC function in this reaction is restricted to PCNA loading and that MutLα–clamp interaction is required for endonuclease activation. We have also found that the requirement for a strand break in MutLα activation can be bypassed by DNA structural anomalies within a covalently closed circular heteroduplex that provide sites for PCNA loading. In contrast to MutLα action on nicked circular heteroduplex DNA, where incision is directed to the strand containing a preexisting break, incision of these closed circular substrates targets both DNA strands.
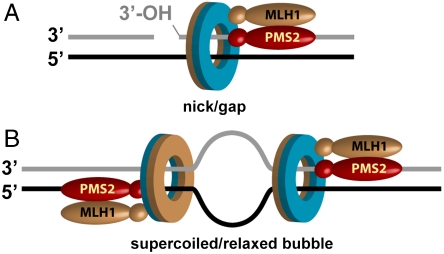
These findings, coupled with a previous analysis of mismatch-independent MutLα incision of nicked circular homoduplex DNA in the presence of low salt and Mn2+, have led us to the model for MutLα action shown in Fig. 5. In the presence of Mn2+ but in the absence of other proteins, MutLα incises the nicked and continuous strands of nicked circular DNA at the same rate (8). Although without effect on the rate of incision of the continuous strand, supplementation with both RFC and PCNA dramatically activates Mn2+-dependent incision of the strand with a preexisting nick. Based on the results described here, we attribute this effect to PCNA loading. The β and PCNA sliding clamps are loaded at a 3′ double strand-single strand junction (24). The two faces of each of these clamps are nonequivalent (18, 25), and available evidence indicates that they are loaded onto DNA with a specific orientation relative to that of the 3′ double strand-single strand junction (refs. 26 and 27 and Fig. 5A). We thus propose that the asymmetry of the loaded clamp is manifested in the PCNA–MutLα complex, and that this asymmetry directs the strand-specificity of MutLα action. Interaction of MutLα with a particular face of the clamp is consistent with our finding that PCNA activation of MutLα in the presence of Mn2+ is blocked by p21 peptide (Fig. S3A), which preferentially interacts with one side of the clamp (18). Because PCNA can diffuse along DNA (28), the asymmetry conferred during clamp loading could be carried along the helix to dictate the strand-specificity of MutLα action elsewhere on the DNA.
Fig. 5.
Model for PCNA involvement in the strand direction of MutLα endonuclease. As indicated by blue and tan colors, the two faces of PCNA are nonequivalent, and the clamp is loaded with a unique orientation relative to the 3′ double strand-single strand junction at a nick or gap (A). A transient (22) or artificial bubble (this study) within a covalently closed circular DNA also supports clamp loading. Because the double strand-single strand junctions at the two ends of a bubble are rotationally symmetric, such structures must support PCNA loading in either orientation (B). We suggest that the MutLα–PCNA complex is characterized by intrinsic asymmetry and that this asymmetry coupled with PCNA loading orientation dictates the strand specificity of incision by the MutLα endonuclease active site within the PMS2 subunit.
This model accounts for the mismatch-dependent results we have obtained with supercoiled DNA and relaxed bubble-containing heteroduplexes. Clamp loading onto negatively supertwisted DNA has been postulated to occur at double strand-single strand junctions that result from transient helix opening (22). We have shown that a short region of open helix within a relaxed circular duplex does indeed provide a target for PCNA loading. An open region of helix has two double strand-single strand junctions, and these are rotationally symmetric, implying that such structures must support the loading of PCNA in both orientations relative to the absolute orientation of the duplex (Fig. 5B). Consequently, PCNA loaded onto supercoiled or a relaxed bubble-containing heteroduplex would be expected to activate MutLα cleavage of both DNA strands, which is what we have observed.
The process of MutLα endonuclease activation displays strong apparent cooperativity with respect to the number of PCNA clamps present on the circular heteroduplexes used in this study (Fig. 4). There are two types of explanation for this observation. Activation of the endonuclease could involve MutLα interaction with several clamps or the assembly of a higher order complex involving multiple molecules of both MutLα and PCNA. We regard such mechanisms as unlikely because the dependence of endonuclease activation on MutLα concentration is not cooperative (7). A second possibility is that the PCNA cooperativity observed is an apparent effect that is not actually manifested at the level of the mechanism. For example, whereas MutLα is recruited by MutSα to the vicinity of the mismatch (29), loaded PCNA is free to diffuse along the helix (28). The loading of multiple clamps onto the 6,400 bp heteroduplex used in our experiments may thus be required simply to achieve a local PCNA concentration that is sufficient to activate MutLα. We favor this sort of explanation for the cooperativity observed in Fig. 4.
In addition to clarifying the nature of MutLα activation, the findings described here may have implications for the expansion of (CAG)n∶(CTG)n triplet repeat sequences, the cause of a number of neurodegenerative diseases (30, 31). Somatic expansion, which depends on both MutSβ and MutLα has been attributed to processing of strand slippage products, with the probability of such slippage events increasing with repeat number (31, 32). However, such expansions have been shown to occur in postmitotic neurons (33) in the absence of DNA replication. This finding is puzzling because previous work on both bacterial and eukaryotic mismatch correction has indicated a requirement for a strand break to initiate excision repair (1–4). We have shown here that DNA strand break involvement in the human repair reaction is restricted to provision of a loading site for PCNA, and that the strand break requirement can be bypassed if an alternate clamp loading site is available; e.g. a small bubble within a covalently continuous heteroduplex. It is thus plausible that extruded (CAG)x and/or (CTG)x elements resulting from strand slippage within a nonreplicating (CAG)n∶(CTG)n repeat could provide not only a MutSβ-recognizable lesion, but a PCNA loading site as well, which would lead to functional activation of the repair system on nonreplicating DNA. We are pursuing this possibility.
Materials and Methods
Proteins and DNAs.
Proteins, DNAs, and Western blot procedures used in this study are described in SI Text.
PCNA Loading Assay.
PK-PCNA was radiolabeled at 30 °C for 30 min in 30 μL reactions containing 20 μg PK-PCNA, 20 mM Tris-HCl pH 7.5, 60 μCi [γ32P]ATP (6000 Ci/mmol), 13 μM cold ATP, 10 mM MgCl2, 17 mM NaCl, 0.4 mM DTT and 500 units of cAMP-dependent protein kinase catalytic subunit (New England Biolabs) (20, 23). Free ATP was removed by filtration through a Quick Spin Sephadex G-50 column (Roche). Specific activities of preparations ranged from 1 × 105 to 3 × 105 cpm/pmol trimer. [32P]PK-PCNA loading reactions (20 μL) contained 20 mM Tris-HCl, pH 7.6, 125 mM NaCl, 5 mM MgCl2, 1.5 mM ATP, 1 mM glutathione, 0.05 mg/mL BSA, 100–140 fmol DNA, 2100 fmol [32P]PK–PCNA homotrimer, 620 fmol RFC or 820 fmol yRFCΔN. After incubation at 37 °C for 3 min, the reaction was chilled on ice and 10 μL was loaded onto a 3.2 × 300 mm Superdex 200 column (GE Healthcare) equilibrated with 20 mM Tris-HCl, pH 7.6, 125 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 1 mM DTT at 95 μL/ min at 4 °C. The column eluate was monitored at 260 and 280 nm. Extent of [32P]PK–PCNA loading was determined by liquid scintillation counting.
Preparative PCNA loading reactions (70 μL) for isolation of PCNA–DNA complexes contained 20 mM Tris-HCl, pH 7.6, 125 mM NaCl, 5 mM MgCl2, 1.5 mM ATP, 1 mM glutathione, 336 fmol nicked circular 3′ G-T heteroduplex DNA, 10 pmol PCNA trimer and 1.9 pmol yRFCΔN. After incubation at 37 °C for 3 min, reactions were chilled on ice. Two-5 μL samples were removed, adjusted to 10 μL with 20 mM Tris-HCl, pH 7.6, 125 mM NaCl, 5 mM MgCl2, 1 mM EDTA and 1 mM DTT, and scored for endonuclease activity as described below in 20 μL reactions in the absence or presence of 390 fmol MutSα and 460 fmol MutLα. Fifty μL of the remainder was loaded onto a Superdex 200 column and chromatographed as described above and 52 μL fractions collected. Twenty μL samples were analyzed by immunoblot for presence of yRFCΔN and PCNA (SI Text). Ten μL samples of fractions 14–18 containing DNA were supplemented with 10 μL of 20 mM Tris-HCl, pH 7.6, 125 mM NaCl, 5 mM MgCl2, 3 mM ATP, 2 mM glutathione, 0.1 mg/mL BSA, 8% vol/vol glycerol containing 390 fmol MutSα, and 460 fmol MutLα. After incubation for an additional 10 min at 37 °C, reactions were stopped with 5 μL of 0.3 M NaOH, 0.1 M EDTA and analyzed by southern hybridization as described below.
MutLα Endonuclease Assays.
Mismatch-dependent MutLα endonuclease activation assay was performed by a modification of that used previously (7). Reactions (10 μL) contained 20 mM Tris-HCl, pH 7.6, 125 mM KCl, 5 mM MgCl2, 1.5 mM ATP, 1 mM glutathione, 0.05 mg/mL BSA, 4% vol/vol glycerol, 24 fmol DNA as indicated, 580 fmol PCNA trimer, and unless indicated otherwise, 100 fmol RFC or 136 fmol yRFCΔN. After 10 min at 37 °C, 10 μL of a solution containing 390 fmol MutSα, and 460 fmol MutLα in the same buffer were added. After incubation for an additional 10 min, reactions were terminated by addition of 40 μL of 30 mM EDTA, 180 μg/mL proteinase K and 0.4 mg/mL glycogen. After 20 min at 55 °C, reactions were phenol extracted, and DNA ethanol precipitated and digested with ClaI. Alternatively, reactions were terminated with 5 μL of 0.3 M NaOH, 0.1 M EDTA. DNA products were resolved by electrophoresis through alkaline agarose, transferred to Hybond membrane, and the two strands detected by indirect end labeling with 32P-labeled oligonucleotides V5504, V2505 (for DNAs digested with ClaI) or C2526, corresponding to f1 viral strand coordinates 5504–5523 (d(ATCATTAAGCGCGGCGGGTG)) or 2505–2526, or complementary strand coordinates 2505–2526, respectively (5).
Mismatch-independent MutLα endonuclease activity was determined in 10 μL reactions containing 20 mM Tris-HCl, pH 7.6, 100 fmol 200 or 202 bp homoduplex DNA (5′-32P labeled on one strand) in the presence of salt, ATP, divalent cation, MutSα (780 fmol), MutLα (1,700 fmol), PCNA (2,300 fmol, trimer) and yRFCΔN (470 fmol) as indicated. After incubation at 37 °C for 20 min, reactions were stopped with 90% formamide and products resolved by electrophoresis through 8% polyacrylamide gels containing 8 M urea in 45 mM Tris-borate, 1 mM EDTA, pH 8.0, followed by phosphorimaging.
Supplementary Material
Acknowledgments.
We thank Bruce Stillman, Peter Burgers, and Mike O’Donnell for expression plasmids, Hongbing Shao for chromatography assistance, Elisabeth Penland for tissue culture support, and Tao-shih Hsieh for suggesting the bubble heteroduplex experiment. This work was supported in part by National Institutes of Health Grants R01 GM45190 and P01 CA092584. P.M. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010662107/-/DCSupplemental.
References
- 1.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 2.Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 4.Hsieh P, Yamane K. DNA mismatch repair: Molecular mechanism, cancer, and ageing. Mech Ageing Dev. 2008;129:391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dzantiev L, et al. A defined human system that supports bidirectional mismatch-provoked excision. Mol Cell. 2004;15:31–41. doi: 10.1016/j.molcel.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 6.Constantin N, Dzantiev L, Kadyrov FA, Modrich P. Human mismatch repair: Reconstitution of a nick-directed bidirectional reaction. J Biol Chem. 2005;280:39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 8.Kadyrov FA, et al. Saccharomyces cerevisiae MutLα is a mismatch repair endonuclease. J Biol Chem. 2007;282:37181–37190. doi: 10.1074/jbc.M707617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iyer RR, et al. MutLalpha and proliferating cell nuclear antigen share binding sites on MutSbeta. J Biol Chem. 2010;285:11730–11739. doi: 10.1074/jbc.M110.104125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol Cell. 2003;12:1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- 11.Genschel J, Bazemore LR, Modrich P. Human exonuclease I is required for 5′ and 3′ mismatch repair. J Biol Chem. 2002;277:13302–13311. doi: 10.1074/jbc.M111854200. [DOI] [PubMed] [Google Scholar]
- 12.Wei K, et al. Inactivation of Exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003;17:603–614. doi: 10.1101/gad.1060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadyrov FA, et al. A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc Natl Acad Sci USA. 2009;106:8495–8500. doi: 10.1073/pnas.0903654106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Umar A, et al. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 15.Lee SD, Alani E. Analysis of interactions between mismatch repair initiation factors and the replication processivity factor PCNA. J Mol Biol. 2006;355:175–184. doi: 10.1016/j.jmb.2005.10.059. [DOI] [PubMed] [Google Scholar]
- 16.Gomes XV, Gary SL, Burgers PM. Overproduction in Escherichia coli and characterization of yeast replication factor C lacking the ligase homology domain. J Biol Chem. 2000;275:14541–14549. doi: 10.1074/jbc.275.19.14541. [DOI] [PubMed] [Google Scholar]
- 17.Podust VN, Chang LS, Ott R, Dianov GL, Fanning E. Reconstitution of human DNA polymerase δ using recombinant baculoviruses: The p12 subunit potentiates DNA polymerizing activity of the four-subunit enzyme. J Biol Chem. 2002;277:3894–3901. doi: 10.1074/jbc.M109684200. [DOI] [PubMed] [Google Scholar]
- 18.Gulbis JM, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- 19.Iyer RR, et al. The MutSalpha-proliferating cell nuclear antigen interaction in human DNA mismatch repair. J Biol Chem. 2008;283:13310–13319. doi: 10.1074/jbc.M800606200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Podust LM, Podust VN, Sogo JM, Hubscher U. Mammalian DNA polymerase auxiliary proteins: analysis of replication factor C-catalyzed proliferating cell nuclear antigen loading onto circular double-stranded DNA. Mol Cell Biol. 1995;15:3072–3081. doi: 10.1128/mcb.15.6.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai J, et al. Reconstitution of human replication factor C from its five subunits in baculovirus-infected insect cells. Proc Natl Acad Sci USA. 1996;93:12896–12901. doi: 10.1073/pnas.93.23.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao N, Leu FP, Anjelkovic J, Turner J, O’Donnell M. DNA structure requirements for the Escherichia coli gamma complex clamp loader and DNA polymerase III holoenzyme. J Biol Chem. 2000;275:11440–11450. doi: 10.1074/jbc.275.15.11440. [DOI] [PubMed] [Google Scholar]
- 23.Kelman Z, Naktinis V, O’Donnell M. Radiolabeling of proteins for biochemical studies. Method Enzymol. 1995;262:430–442. doi: 10.1016/0076-6879(95)62034-6. [DOI] [PubMed] [Google Scholar]
- 24.Yao N, Hurwitz J, O’Donnell M. Dynamics of beta and proliferating cell nuclear antigen sliding clamps in traversing DNA secondary structure. J Biol Chem. 2000;275:1421–1432. doi: 10.1074/jbc.275.2.1421. [DOI] [PubMed] [Google Scholar]
- 25.Kong XP, Onrust R, O’Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: A sliding DNA clamp. Cell. 1992;69:425–437. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- 26.Bowman GD, O’Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp–clamp loader complex. Nature. 2004;429:724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- 27.Georgescu RE, et al. Structure of a sliding clamp on DNA. Cell. 2008;132:43–54. doi: 10.1016/j.cell.2007.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao N, et al. Clamp loading, unloading and intrinsic stability of the PCNA, beta and gp45 sliding clamps of human, E. coli and T4 replicases. Genes Cells. 1996;1:101–113. doi: 10.1046/j.1365-2443.1996.07007.x. [DOI] [PubMed] [Google Scholar]
- 29.Blackwell LJ, Wang S, Modrich P. DNA chain length dependence of formation and dynamics of hMutSa–hMutLa–heteroduplex complexes. J Biol Chem. 2001;276:33233–33240. doi: 10.1074/jbc.M105076200. [DOI] [PubMed] [Google Scholar]
- 30.Brouwer JR, Willemsen R, Oostra BA. Microsatellite repeat instability and neurological disease. Bioessays. 2009;31:71–83. doi: 10.1002/bies.080122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol. 2010;11:165–170. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- 32.Gomes-Pereira M, Fortune MT, Ingram L, McAbney JP, Monckton DG. Pms2 is a genetic enhancer of trinucleotide CAG. CTG repeat somatic mosaicism: Implications for the mechanism of triplet repeat expansion. Hum Mol Genet. 2004;13:1815–1825. doi: 10.1093/hmg/ddh186. [DOI] [PubMed] [Google Scholar]
- 33.Gonitel R, et al. DNA instability in postmitotic neurons. Proc Natl Acad Sci USA. 2008;105:3467–3472. doi: 10.1073/pnas.0800048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.