Abstract
Apidae is the most speciose and behaviorally diverse family of bees. It includes solitary, eusocial, socially parasitic, and an exceptionally high proportion of cleptoparasitic species. Cleptoparasitic bees, which are brood parasites in the nests of other bees, have long caused problems in resolving the phylogenetic relationships within Apidae based on morphological data because of the tendency for parasites to converge on a suite of traits, making it difficult to differentiate similarity caused by common ancestry from convergence. Here, we resolve the evolutionary history of apid cleptoparasitism by conducting a detailed, comprehensive molecular phylogenetic analysis of all 33 apid tribes (based on 190 species), including representatives from every hypothesized origin of cleptoparasitism. Based on Bayesian ancestral state reconstruction, we show that cleptoparasitism has arisen just four times in Apidae, which is fewer times than previously estimated. Our results indicate that 99% of cleptoparasitic apid bees form a monophyletic group. Divergence time estimates reveal that cleptoparasitism is an ancient behavior in bees that first evolved in the late Cretaceous 95 Mya [95% highest posterior density (HPD) = 87–103]. Our phylogenetic analysis of the Apidae sheds light on the macroevolution of a bee family that is of evolutionary, ecological, and economic importance.
Keywords: ancestral state reconstruction, apidae, divergence dating, kleptoparasitism, molecular phylogeny
Apidae is the largest family of bees, with over 5,600 described species. The family includes the most important managed pollinator (Apis mellifera, the honey bee) and the only bees domesticated by humans for honey production (1). The honey bee is one of the more important model organisms, especially for all aspects of eusociality (2–5). Apid species represent a rich diversity of solitary, social, and parasitic lifestyles, and they pollinate a wide variety of agricultural and native plants. Despite the importance of this group, a robust comparative framework for evolutionary studies on the ecological and behavioral diversity of apid bees is lacking, primarily because of problems caused by the high proportion of cleptoparasitic species (28%) and tribes (50%).
Cleptoparasitism (or kleptoparasitism), which involves the stealing of food or nesting material by one animal from another, is a widespread phenomenon found in many animal groups, including birds (6), bees (7), wasps (8), spiders (9), fish (10), and mammals (11). In bees, cleptoparasitic species do not build or provision their own nests; instead, they enter the nests of other bees and lay their eggs in either closed or open, partially provisioned brood cells (12). In a few cases, the adult female parasite destroys the host egg (13), but more commonly, a specialized larval instar kills the host larva (14). The parasitic larva then consumes the pollen and nectar provisions gathered by the host adult and completes its development before emerging from the host nest. This form of parasitism differs from that found in social parasitic bees (e.g., Psithyrus), in which the female enters the nest of its social host and replaces the queen so that the workers of the colony now rear the parasites offspring (15).
Darwin (16) described cleptoparasitism in bees as being “more remarkable than that of the cuckoo; for these bees have not only their instincts but their structure modified in accordance with their parasitic habits.” Because bee cleptoparasites do not build or provision nests, they lack pollen-collecting structures, and they are often heavily armored relative to pollen-collecting species. Convergent evolution in cleptoparasitic bees has been well-documented (17, 18), making it difficult to differentiate between features that are similar because of shared ancestry versus convergence. In the phylogenetic analysis on which the present classification of Apidae is based (19), characters that were considered a priori to have arisen convergently in cleptoparasites were excluded, effectively biasing the results to a conclusion of multiple, independent parasitic origins.
In this paper, we resolve the phylogenetic relationships of the apid tribes and reconstruct the evolutionary history of cleptoparasitism within apids by conducting a comprehensive phylogenetic analysis of Apidae using molecular data. We estimate the number of independent origins of cleptoparasitism using model-based methods and estimate the antiquity of cleptoparasitism, Apidae, and its major clades using a relaxed fossil-calibrated molecular-clock model. Because our analysis is based exclusively on DNA sequence data, it provides a phylogenetic hypothesis that is independent of possible morphological convergence in the cleptoparasites.
Results and Discussion
Phylogenetic Relationships.
Applying a range of analytical methods to our dataset resulted in a well-resolved phylogeny (Fig. 1). Our phylogenetic hypothesis based on an extensive Bayesian analysis (Fig. S1) of the dataset was almost identical to that based on maximum likelihood (ML) (Fig. S2) and mostly congruent with that based on parsimony (Fig. S3). Differences in phylogenetic relationships among the three analytical methods do not impact our inferences about the evolutionary history of cleptoparasitism.
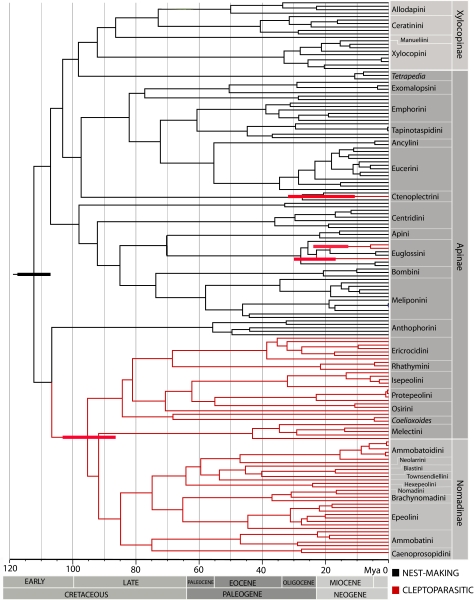
Fig. 1.
Maximum clade credibility tree of the Bayesian phylogenetic analysis of Apidae with a relaxed fossil-calibrated molecular-clock model. Cleptoparasitism is traced in red on the tree. Red bars indicate the 95% HPD on the estimated age of the cleptoparasitic lineages, and the black bar indicates the 95% HPD of the estimated age of Apidae. Outgroup taxa used in the analysis have been removed.
Our phylogenetic results imply significant changes to the current understanding of apid higher-level relationships. This is mostly (but not exclusively) because of the formation of the large cleptoparasitic clade comprised of the subfamily Nomadinae and most of the cleptoparasitic tribes previously placed in the subfamily Apinae (including the tribes Melectini, Ericrocidini, Protepeolini, Isepeolini, Osirini, and Rhathymini). The only cleptoparasitic apids not included within this group are the cleptoparasitic orchid bees Aglae and Exaerete (Euglossini) and Ctenoplectrina (Ctenoplectrini). The large cleptoparasitic clade, which includes over 99% of parasitic apid species, has a posterior probability (PP) of 100, ML bootstrap proportion of 98, and parsimony bootstrap proportion of 42.
Recovery of the large cleptoparasitic clade is a surprising result but is supported to some extent by a recent morphological analysis of the family Apidae. Straka and Bogusch (20), in an analysis based only on larval characters, obtained a tree in which many of the cleptoparasitic Apinae formed a monophyletic group. However, the details of their topology differ substantially from our tree. Their tree recovered a monophyletic group of cleptoparasitic Apinae, but it excluded the Nomadinae and Coelioxoides. In addition, their tree was relatively poorly supported (based on bootstrap proportions), and the Apidae had to be constrained to form a monophyletic group, making it difficult to conclusively resolve the evolutionary history of cleptoparasitism.
We recover a monophyletic Xylocopinae and Nomadinae and find that Osirini, Protepeolini, and Isepeolini should not be placed within Nomadinae. Previous morphological studies have strongly supported monophyly of Nomadinae (19, 21). However, it has been unclear whether Osirini, Protepeolini, and Isepeolini (three cleptoparasitic tribes of Apinae) form a monophyletic group with Nomadinae or whether they belong to Apinae (21). In our analysis, they are part of the large cleptoparasitic clade. The three main lineages of Osirini (22) do not form a monophyletic group; instead, Osiris and Epeoloides group with Protepeolini and Isepeolini, whereas Parepeolus groups with Coelioxoides (Tetrapediini). Tetrapediini, a tribe of Apinae, consists of the cleptoparasitic genus Coelioxoides and its host, Tetrapedia (14). We do not recover the monophyly of Tetrapediini or place it within the Apinae. Instead, Coelioxoides is part of the large cleptoparasitic clade and Tetrapedia comes out sister to Xylocopinae. Both Tetrapedia and Xylocopinae nest in dead plant material, whereas most other solitary apids nest in the ground.
Evolution of Cleptoparasitism.
If we use parsimony to reconstruct the evolutionary history of cleptoparasitism in Apidae, it is clear that cleptoparasitism evolved one time in the common ancestor of the large cleptoparasitic clade and one time in the common ancestor of Ctenoplectrina. It is unclear, however, if cleptoparasitism evolved one time in orchid bees in the common ancestor of Exaerete, Algae, and Euglossa, with a reversal to nest making in the Euglossa, or if it evolved two times independently in Exaerete and Aglae (Fig. S4).
To resolve the ambiguity found within orchid bees, we used model-based methods implemented in the program BayesTraits v.1.0 (23). These methods incorporate branch-length information, transition rates, and uncertainty in tree topology into the ancestral state reconstruction. Analysis based on ML supports four independent origins with no reversals. The rate of transition from cleptoparasitism to nest making was found to be 0 in the ML analysis. Results based on Bayesian methods also support the hypothesis of four independent origins with no reversals (Fig. 1 and Fig. S5). In this analysis, the rate of transition from nest making to cleptoparasitism was 1.31 ± 0.22, and the rate of transition from cleptoparasitism to nest making was 0.05 ± 0.05. The common ancestor of Exaerete, Algae, and Euglossa is reconstructed as being nest making with a probability of 0.95. The common ancestor of the large cleptoparasitic clade and Ctenoplectrina is reconstructed as being cleptoparasitic with a probability of 1.0. We used the Bayes Factor test to determine if the dataset significantly supports four independent origins over three origins with one reversal. The Bayes Factor was 8.04 ± 1.2 (Fig. S6), which indicates strong support for the hypothesis of four independent origins of cleptoparasitism (24). Although previous studies of orchid bee generic relationships based on morphological (25–27) and molecular (28, 29) data have provided conflicting results, no previous studies hypothesize a sister group relationship between the two cleptoparasitic genera Exaerete and Aglae. Previous studies are, therefore, consistent with our hypothesis of dual origins of cleptoparasitism in orchid bees.
Our analysis reduces the presumed number of independent origins of cleptoparasitism from 11 (30, 31) (estimate based on the prevailing concept for the phylogeny of apids) to just 4 [Straka and Bogusch (20) estimated six origins but see comments above]. The observation that cleptoparasitism evolved relatively few times in Apidae is in striking contrast to another bee family, Halictidae, where cleptoparasitism is estimated to have arisen nine times from nest-making ancestors (31, 32). Our results establish, based on a comprehensive analysis of the data, that cleptoparasitism evolved relatively few times in Apidae.
Divergence Time Estimates and the Antiquity of Cleptoparasitism.
To estimate the antiquity of cleptoparasitism, we used a fossil-calibrated relaxed molecular-clock model (33) as implemented in Beast v1.4.8 (34). We applied lognormal distributed prior age estimates with a minimum age constraint on 10 nodes using information from the fossil record. Varying the prior age set on the root node affected the estimated divergence times of nodes close to the root of the tree but did not have an effect on nodes near the tips of the tree. Because bees are thought to have originated in the mid-Cretaceous about 120–125 Mya (35), we present results based on the analysis in which the root node of the bees was set to 120 Mya (Fig. 1).
Cleptoparasitism seems to be an ancient behavior in apid bees. It first evolved 95 Mya [95% highest posterior density (HPD) = 87–103] in the common ancestor of the Nomadinae and cleptoparasitic Apinae (i.e., the large cleptoparasitic clade). The other three origins of cleptoparasitism are much more recent: 21 Mya (95% HPD = 11–32) in the Ctenoplectrini, 23 Mya (95% HPD = 17–30) in Aglae, and sometime after Exaerete diverged from Euglossa 19 Mya (95% HPD = 13–24). Before this analysis, the oldest evidence of cleptoparasitism in bees is the extinct Protomelecta brevipennis Cockerell (1908) from the Florissant Formation of Colorado dated to the Eocene/Oligocene boundary (36). The phylogenetic affinities of this fossil are unclear, but it seems to be closely related to Melectini (37).
Despite the antiquity of cleptoparasitism in apid bees (∼95 Mya for the Nomadinae + clepto Apinae), we do not see any reversals back to nest making. Examination of clade ages and relationships within orchid bees provides the intuitively appealing result that the cleptoparasitic genera (Aglae and Exaerete) evolved after their pollen-collecting hosts (Eulaema and Eulaema/Eufriesia, respectively). Previous studies of orchid bee generic relationships using molecular data would suggest the contrary: that cleptoparasites evolved before the first appearance of their hosts (28, 29). This is a less parsimonious hypothesis, because it requires the origin and subsequent extinction of pollen-collecting stem lineages, host switches, or a reversal from cleptoparasitism back to nest making. A reversal in nesting habit would require females to reevolve the structures associated with foraging for pollen and nest making (such as the corbicula), which is unlikely (19, 28, 29).
Concluding Remarks.
Our phylogenetic hypothesis indicates that most cleptoparasitic apid bees form a monophyletic group and therefore, stem from a single, ancient origin of cleptoparasitism. We find two origins of cleptoparasitism in the orchid bees and one in the tribe Ctenoplectrini. The large cleptoparasitic clade formed by the Nomadinae and most cleptoparasitic Apinae renders the subfamily Apinae paraphyletic and indicates that major changes are needed to the higher-level classification of Apidae. Divergence time estimates using a relaxed fossil-calibrated molecular-clock model reveal that cleptoparasitism is an ancient behavior in apid bees that first evolved ∼95 Mya, which is ∼60 My earlier than the appearance of the first cleptoparasitic bee in the fossil record.
Our molecular phylogeny is broadly supported, although some nodes in the tree would benefit from additional data and analyses. We also suggest that morphology should be reinvestigated to see if synapomorphies can be found supporting the clades suggested in this tree. By providing a well-supported hypothesis of the relationships among apid tribes, we have laid the foundation for future work on fundamental evolutionary questions, such as the study of host–parasite coevolution and the ecological conditions under which cleptoparasitism evolves. Furthermore, our divergence time estimates provide a framework within which the biogeography of Apidae can now be more thoroughly investigated.
Materials and Methods
Taxon and Gene Sampling.
We sampled representatives from all 33 currently recognized tribes (31) of Apidae. We also included all apid genera for which we had access to high-quality molecular material (106/209 genera). An effort was made to choose taxa representing the morphological diversity found within tribes for a total of 160 ingroup exemplars. As outgroups, representatives from all currently recognized tribes of the family Megachilidae, which has been well-supported as the sister clade to the Apidae (19, 38–40), were included in addition to taxa from all three melittid subfamilies hypothesized to be members of the earliest diverging lineages of bees (39, 40). This totaled 30 outgroup taxa. We did not include representatives from all bee families as outgroups to facilitate alignment of the ribosomal genes and to keep the number of taxa included to a manageable size. Outgroup selection can have an effect on the rooting of the ingroup, but we are confident that we have correctly recovered the apid root node based on preliminary analyses that included representatives of all bee families but a limited number of apid taxa. The entire dataset includes 190 taxa. Voucher and locality information for all taxa in this study are provided in Table S1.
Most sequences used in the study are previously unpublished sequences. Previously published sequences were downloaded from GenBank. All new sequences were obtained after standard PCR and sequencing protocols (41). The dataset consists of sequences of two nuclear ribosomal genes (18S and 28S) and five nuclear protein-coding genes [wingless, pol II, long-wavelength rhodopsin (opsin), Nak, and EF-1α]. Primer pairs and PCR conditions for all genes are listed in Table S2, and GenBank accession numbers are in Table S3.
All genes were separately aligned in the Lasergene DNAStar (42) software package using ClustalW. Alignments for 28S and 18S were subsequently adjusted by referring to the secondary structure of these genes proposed for A. mellifera (43). Regions that could not be aligned with confidence were excluded from the analysis (including introns of opsin and EF-1α).
Phylogenetic Inference.
Phylogenies were inferred using parsimony, ML, and Bayesian methods. Selection of best-fit models of nucleotide substitution for each data partition used in a Bayesian or ML analysis was based on the Akaike (AIC) and Bayesian Information Criteria (BIC) as implemented in JModelTest v.0.1.1 (44). Several different partitioning schemes were explored, but results presented herein are based on a matrix in which the two ribosomal genes were placed in one partition and the protein-coding genes were combined together and partitioned by codon position. Detailed information on the implementation of all phylogenetic methods is found in SI Materials and Methods.
Ancestral State Reconstruction of Cleptoparasitism.
All terminals in the tree were coded for a behavioral character, based on information in the literature, consisting of two states: cleptoparasitic and nest making (Table S4). Parsimony methods were used to reconstruct the evolution of cleptoparasitism in apid bees on the Bayesian maximum clade credibility tree using MacClade v.4.0 (45).
To take phylogenetic uncertainty, branch lengths, and relative rates of gains and losses into account, Bayesian ancestral-state reconstruction methods were implemented in the program BayesTraits v.1.0 (23). We used a random sample of 10,000 trees from the Bayesian analysis of the phylogeny. We initially ran a likelihood analysis to get reasonable starting values (priors) on the transition rate from one character state to another for the Bayesian MCMC analysis, as recommended in the manual (23). Both transition rates were below 1.3. We, therefore, used a reversible jump model with priors obtained from an exponential prior seeded from a uniform on the interval 0–5 (hyperprior). A reversible-jump model automatically finds the posterior distribution of models of evolution for the data (46). In this case, it would determine if a model where the rate of transition from nest making to cleptoparasitic was the same as the rate of transition from cleptoparasitic to nest making or if a model with these two transition rates being different fit the data better. Using a hyperprior allowed us to choose an exponential distribution for the prior and let the program estimate this prior from the data using a uniform hyperprior to seed the prior (23). We ran the analysis several times for 2 million generations and discarded the first 1 million as burn-in. Examination of the output files indicated that our burn-in was sufficient, because parameter estimates were stable over the postburn-in period.
To test whether there was significant support for four independent origins of cleptoparasitism instead of just three, we ran the MCMC analysis 100 times with the common ancestor of Euglossa, Exaerete, and Aglae fixed alternatively as cleptoparasitic and nest making. We compared the log of the harmonic means obtained for each of the 100 replicates. Two times the difference in the harmonic mean of the log-likelihood scores represents the Bayes Factor. Values above 0 are considered positive support for the hypothesis, and values above 6 are considered strong support (24).
Estimating Divergence Times.
We used a Bayesian phylogenetic relaxed molecular-clock model (33) with multiple calibration points to estimate divergence times in the program BEAST v1.4.8 (34). We applied a GTR+I+G model as in the phylogenetic analysis described above. Branch rates were estimated with an uncorrelated relaxed clock model in which the rate at each branch was drawn from an underlying log-normal distribution. This allowed for the rate of evolution to vary among the branches of the tree with no a priori correlation between a lineage's rate and that of its ancestor. Parameters were unlinked across partitions. The Yule tree prior was used, which assumes a constant per lineage selection rate as recommended in the manual for species-level phylogenies. We randomly selected a starting tree from the posterior distribution of trees from the MrBayes analysis.
The tree was time-calibrated by setting priors on the ages of 10 internal nodes and the root of the tree. Age estimates were based on paleontological evidence. Uncertainty in the age of the calibration points was incorporated into the analysis by assuming that the probability of the node being a certain age follows a lognormal distribution with a rigid minimum bound. This required us to specify a mean, SD, and rigid lower bound for the age of each calibration point. Selection of the values for these parameters is somewhat subjective, but we outline the reasoning behind each of our choices for every calibration point in SI Materials and Methods. Applying a lognormal distribution to our age estimates allows us to assume that the actual divergence event took place some time before the earliest appearance of fossil evidence but that the age of the node is more likely to be close to the age of the oldest known fossil and less likely to be significantly older. Fossils can only provide minimum age estimates, and their appearance must postdate the origin of the clade to which they belong. By how much the appearance of a clade predates the age of the first fossil is unclear. We, therefore, made sure that the 95% probability included the oldest reasonable age for the clade. For our more basally positioned calibration points, that meant including the start of the Cretaceous (145 Mya). Angiosperms are thought to have originated ∼130 Mya, shortly before the origin of the eudicots ∼125 Mya (47–49). Bees are mostly dependent on floral resources from eudicots, and therefore, it would be very unlikely for bees to predate the origin of eudicots or angiosperms. Furthermore, it is estimated that the crabronid–bee divergence took place ∼120 Mya (35).
Bees are thought to have originated in the mid-Cretaceous about 120–125 Mya (35). We, therefore, ran two analyses of 100 million generations with the root node set to 120 Mya and the calibration points set with a lognormal distribution as describe above. We also ran additional shorter analyses with the age of the root node sampled from a normal distribution with a mean of 145.5, 130.0, 120.0, 110.0, 100.0, and 90.0 ± 1.0 Mya to examine the effect of different root-node ages on the age estimates of internal nodes. We also ran analyses for all of these different root-node ages with the internal calibration-point age priors changed to be sampled from a uniform distribution with a specified minimum and maximum age. In all cases, the start of the Cretaceous was used as an absolute maximum (145.5 Mya), and the youngest age assigned to a fossil was used as the minimum age for the group. We did five independent runs of 10 million generations for each of these exploratory analyses. Results of the five independent runs were combined together, giving us a total of 50 million generations. Applying a uniform distribution instead of a lognormal distribution to the prior age estimates of the calibration points did not substantially alter our divergence time estimates, and therefore, we do not present those results.
Supplementary Material
Acknowledgments
We thank Drs. Jerome G. Rozen, Jr (American Museum of Natural History, New York), Laurence Packer (York University, Toronto), Eduardo Almeida (Cornell University, Ithaca, NY), Robert Minckley (University of Rochester, Rochester, NY), John Neff (Central Texas Melittological Institute, Austin, TX), Christophe Praz (ETH Zürich, Zurich), John Ascher (American Museum of Natural History, New York), David L. Wagner (The University of Connecticut, Storrs, CT), Hanno Schaefer (Ludwig-Maximilians-Universität München, Munich), Jan Batelka (Charles University in Prague, Prague), Ivan Cepicka (Charles University in Prague, Prague), and Santiago Ramírez (Harvard University, Cambridge, MA) for specimens, A. Kawakita (Kyoto University, Kyoto) for sequence data, A. Agrawal, J. Liebherr, and S. Campbell for comments on the manuscript, and W.T. Wcislo and two anonymous reviewers for improvements to the manuscript. This work was supported by National Science Foundation Grants DEB 0709956 (to S.C.) and DEB 0814544 (to B.N.D.) and institutional support through Grant MSM0021620828 (to J.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Database deposition: The sequences reported in this paper have been deposited in the GenBank database (accession numbers are given in Table S3).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006299107/-/DCSupplemental.
References
- 1.Buchmann S, Repplier B. Letters from the Hive: An Intimate History of Bees, Honey, and Humankind. New York: Bantam Dell; 2005. [Google Scholar]
- 2.Seeley TD. The Wisdom of the Hive: The Social Physiology of Honey Bee Colonies. Cambridge, MA: Harvard Univ Press; 1995. [Google Scholar]
- 3.Frisch KV. The Dance Language and Orientation of the Bees. Cambridge, MA: Harvard Univ Press; 1967. [Google Scholar]
- 4.Ben-Shahar Y, Robichon A, Sokolowski MB, Robinson GE. Influence of gene action across different time scales on behavior. Science. 2002;296:741–744. doi: 10.1126/science.1069911. [DOI] [PubMed] [Google Scholar]
- 5.Honeybee Genome Sequencing Consortium Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brockmann HJ, Barnard CJ. Kleptoparasitism in birds. Anim Behav. 1979;27:487–514. [Google Scholar]
- 7.Rozen JG, Jr, Kamel SM. Investigations on the biologies and immature stages of the cleptoparasitic bee genera Radoszkowskiana and Coelioxys and their Megachile hosts (Hymenoptera: Apoidea: Megachilidae: Megachilini) Am Mus Novit. 2007;3573:1–43. [Google Scholar]
- 8.Rosenheim JA. Host location and exploitation by the cleptoparasitic wasp Argochrysis armilla: The role of learning (Hymenoptera: Chrysididae) Behav Ecol Sociobiol. 1987;21:401–406. [Google Scholar]
- 9.Vollrath F. Behaviour of the kleptoparasitic spider Argyrodes elevatus (Araneae, theridiidae) Anim Behav. 1979;27:515–521. [Google Scholar]
- 10.Hamilton IM, Dill LM. The use of territorial gardening versus kleptoparasitism by a subtropical reef fish (Kyphosus cornelii) is influenced by territory defendability. Behav Ecol. 2003;14:561–568. [Google Scholar]
- 11.Honer OP, Wachter B, East ML, Hofer H. The response of spotted hyaenas to long-term changes in prey populations: Functional response and interspecific kleptoparasitism. J Anim Ecol. 2002;71:236–246. [Google Scholar]
- 12.Rozen JG., Jr Eggs, ovariole numbers, and modes of parasitism of cleptoparasitic bees, with emphasis on neotropical species (Hymenoptera: Apoidea) Am Mus Novit. 2003;3413:1–36. [Google Scholar]
- 13.Garófalo CA, Rozen JG., Jr Parasitic behavior of Exaerete smaragdina with descriptions of its mature oocyte and larval instars (Hymenoptera: Apidae: Euglossini) Am Mus Novit. 2001;3349:1–26. [Google Scholar]
- 14.Alves-dos-Santos I, Melo GAR, Rozen JG., Jr Biology and immature stages of the bee tribe Tetrapediini (Hymenoptera: Apidae) Am Mus Novit. 2002;3377:1–45. [Google Scholar]
- 15.Bohart GE. The Evolution of Parasitism Among Bees. Logan, UT: Utah State University; 1970. [Google Scholar]
- 16.Darwin CR. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. 4th Ed. London: J. Murray; 1866. [PMC free article] [PubMed] [Google Scholar]
- 17.Michener CD. The parasitic groups of Halictidae (Hymenoptera, Apoidea) Univ Kansas Sci Bull. 1978;51:291–339. [Google Scholar]
- 18.Wcislo WT. Transvestism hypothesis: a cross-sex source of morphological variation for the evolution of parasitism among sweat bees (Hymenoptera: Halictidae)? Ann Entomol Soc Am. 1999;92:239–242. [Google Scholar]
- 19.Roig-Alsina A, Michener CD. Studies of the phylogeny and classification of long-tongued bees (Hymenoptera: Apoidea) Univ Kansas Sci Bull. 1993;55:124–162. [Google Scholar]
- 20.Straka J, Bogusch P. Phylogeny of the bees of the family Apidae based on larval characters with focus on the origin of cleptoparasitism (Hymenoptera: Apiformes) Syst Entomol. 2007;32:700–711. [Google Scholar]
- 21.Alexander B. A cladistic analysis of the nomadine bees (Hymenoptera: Apoidea) Syst Entomol. 1990;15:121–152. [Google Scholar]
- 22.Roig-Alsina A. The tribe Osirini, its scope, classification, and revisions of the genera Parepeolus and Osirinus (Hymenoptera, Apoidea, Anthophoridae) Univ Kansas Sci Bull. 1989;54:1–23. [Google Scholar]
- 23.Pagel M, Meade A, Barker D. Bayesian estimation of ancestral character states on phylogenies. Syst Biol. 2004;53:673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- 24.Kass RE, Raftery AE. Bayes factors. J Am Stat Assoc. 1995;90:773–795. [Google Scholar]
- 25.Kimsey LS. Generic relationships within the Euglossini (Hymenoptera: Apidae) Syst Entomol. 1987;12:63–72. [Google Scholar]
- 26.Michener CD. Classification of the Apidae (Hymenoptera) Univ Kansas Sci Bull. 1990;54:75–164. [Google Scholar]
- 27.Engel MS. The first fossil Euglossa and phylogeny of the orchid bees (Hymenoptera: Apidae; Euglossini) Am Mus Novit. 1999;3272:1–14. [Google Scholar]
- 28.Michel-Salzat A, Cameron SA, Oliveira ML. Phylogeny of the orchid bees (Hymenoptera: Apinae: Euglossini): DNA and morphology yield equivalent patterns. Mol Phylogenet Evol. 2004;32:309–323. doi: 10.1016/j.ympev.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 29.Ramírez SR, Roubik DW, Skov C, Pierce NE. Phylogeny, diversification patterns and historical biogeography of euglossine orchid bees (Hymenoptera: Apidae) Biol J Linn Soc Lond. 2010;100:552–572. [Google Scholar]
- 30.Rozen JG., Jr . 2000. Anais do IV Encontro Sobre Abelhas, eds Bitondi MMG, Hartfelder K, et al. (Ribeirão Preto, Brazil), pp. 204–210. [Google Scholar]
- 31.Michener CD. The Bees of the World. Baltimore: Johns Hopkins Univ Press; 2007. p. 953. [Google Scholar]
- 32.Danforth BN, et al. Phylogeny of Halictidae with an emphasis on endemic African Halictinae. Apidologie. 2008;39:86–101. [Google Scholar]
- 33.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grimaldi DA, Engel MS. Evolution of the Insects. New York: Cambridge University Press; 2005. [Google Scholar]
- 36.Evanoff E, McIntosh WC, Murphey PC. 2001. Fossil flora and stratigraphy of the Florissant Formation, Colorado, eds Evanoff E, Gregory-Wodzicki KM, and Johnson KR (Proceedings of the Denver Museum of Nature & Science, ser 4, no. 1), pp. 1–16. [Google Scholar]
- 37.Engel MS. Notes on a megachiline bee (Hymenoptera: Megachilidae) from the Miocene of Idaho. Trans Kans Acad Sci. 2004;107:97–100. [Google Scholar]
- 38.Alexander BA, Michener CD. Phylogenetic studies of the families of short-tongued bees (Hymenoptera: Apoidea) Univ Kansas Sci Bull. 1995;55:377–424. [Google Scholar]
- 39.Danforth BN, Fang J, Sipes S. Analysis of family-level relationships in bees (Hymenoptera: Apiformes) using 28S and two previously unexplored nuclear genes: CAD and RNA polymerase II. Mol Phylogenet Evol. 2006;39:358–372. doi: 10.1016/j.ympev.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 40.Danforth BN, Sipes S, Fang J, Brady SG. The history of early bee diversification based on five genes plus morphology. Proc Natl Acad Sci USA. 2006;103:15118–15123. doi: 10.1073/pnas.0604033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Danforth BN, Sauquet H, Packer L. Phylogeny of the bee genus Halictus (Hymenoptera: Halictidae) based on parsimony and likelihood analyses of nuclear EF-1α sequence data. Mol Phylogenet Evol. 1999;13:605–618. doi: 10.1006/mpev.1999.0670. [DOI] [PubMed] [Google Scholar]
- 42.DNASTAR . Madison, WI: DNASTAR, Inc.; 1999. MEGALIGN 4.00. [Google Scholar]
- 43.Gillespie JJ, Johnston JS, Cannone JJ, Gutell RR. Characteristics of the nuclear (18S, 5.8S, 28S and 5S) and mitochondrial (12S and 16S) rRNA genes of Apis mellifera (Insecta: Hymenoptera): Structure, organization, and retrotransposable elements. Insect Mol Biol. 2006;15:657–686. doi: 10.1111/j.1365-2583.2006.00689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Posada D. jModelTest: Phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 45.Maddison DR, Maddison WP. MacClade 4.0: Analysis of phylogeny and character evolution. 2000 doi: 10.1159/000156416. [DOI] [PubMed] [Google Scholar]
- 46.Pagel M, Meade A. Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov chain Monte Carlo. Am Nat. 2006;167:808–825. doi: 10.1086/503444. [DOI] [PubMed] [Google Scholar]
- 47.Davies TJ, et al. Darwin's abominable mystery: Insights from a supertree of the angiosperms. Proc Natl Acad Sci USA. 2004;101:1904–1909. doi: 10.1073/pnas.0308127100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soltis PS, Soltis DE. The origin and diversification of angiosperms. Am J Bot. 2004;91:1614–1626. doi: 10.3732/ajb.91.10.1614. [DOI] [PubMed] [Google Scholar]
- 49.Magallón S, Castillo A. Angiosperm diversification through time. Am J Bot. 2009;96:349–365. doi: 10.3732/ajb.0800060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.