Abstract
The human mitochondrial transcription machinery generates the primers required for initiation of leading-strand DNA replication. According to one model, the 3′ end of the primer is defined by transcription termination at conserved sequence block II (CSB II) in the mitochondrial DNA control region. We here demonstrate that this site-specific termination event is caused by G-quadruplex structures formed in nascent RNA upon transcription of CSB II. We also demonstrate that a poly-dT stretch downstream of CSB II has a modest stimulatory effect on the termination efficiency. The mechanism is reminiscent of Rho-independent transcription termination in prokaryotes, with the exception that a G-quadruplex structure replaces the hairpin loop formed in bacterial mRNA during transcription of terminator sequences.
Keywords: mitochondrion, RNA polymerase
Human mitochondria contain their own genetic material called mitochondrial DNA (mtDNA), a double-stranded circular molecule encoding 13 subunits of the respiratory chain, 22 tRNAs, and 2 rRNAs. The two strands are referred to as the heavy and light strand based on their buoyant density in cesium chloride gradients (1). Transcription from the light strand promoter (LSP) and heavy strand promoter is polycistronic and produces near genome-length transcripts, which are processed to release the individual RNA molecules (2, 3). MtDNA contains few noncoding sequences, with the exception of the control region, which harbors DNA elements required for initiation of transcription and DNA replication (Fig. 1A).
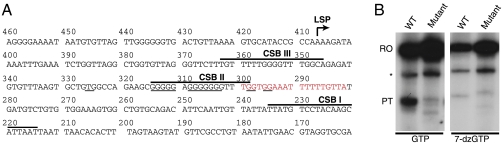
Fig. 1.
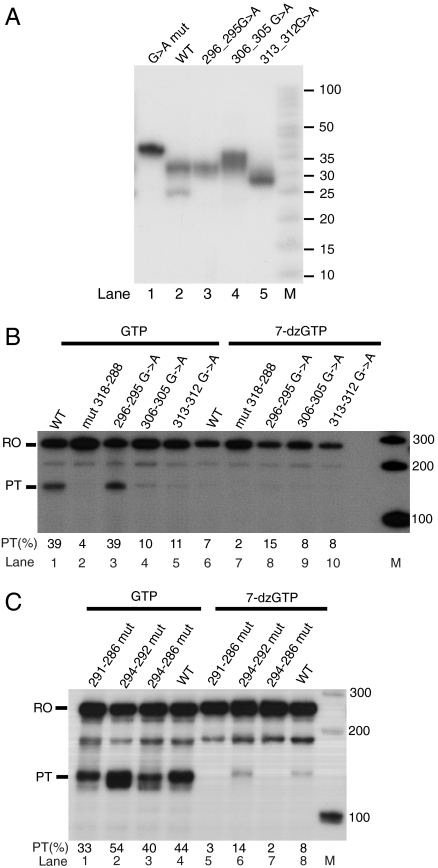
Premature termination of transcription is reduced in 7-deaza-GTP. (A) Partial sequence of the mtDNA control region. The LSP is denoted by a bent arrow. Positions of premature transcription termination (nucleotides 282–300) are shown in red. Conserved sequence blocks I–III are marked with lines above the sequence, and the guanines with potential to contribute to one of the predicted G-quadruplexes at CSB II are underlined. (B) Products of in vitro transcription on WT and mutant 318–288 CSB II template with GTP or 7-deaza-GTP (7-dzGTP) were separated on a 6% PAGE gel. RO, full-length runoff transcript; PT, preterminated transcript; asterisk denotes a minor product that was unaffected in 7-deaza-GTP and acts as an internal control.
Mitochondrial DNA replication initiates at two major sites, the origins of light and heavy (OriH) strand replication. The mitochondrial RNA polymerase generates the RNA primers required for initiation of DNA synthesis at both these sites (4–7). Primers required for OriH activation are produced by transcription initiated at LSP, and there must thus exist a mechanism that enables the transcription machinery to switch from genome-length transcription to primer formation. One model suggests that the primers are formed by cleavage of the primary LSP transcript by the mitochondrial RNA processing endonuclease (8, 9), but this idea has been questioned, because at least human mitochondrial RNA processing endonuclease is primarily localized to the nucleolus, where it is required for rRNA processing (10). Some years ago, we suggested an alternative model for primer formation at OriH. Using a reconstituted in vitro transcription system containing the mitochondrial RNA polymerase (POLRMT) and mitochondrial transcription factors A (TFAM) and B2 (TFB2M), we observed that a large fraction (up to 65%) of the transcription events initiated at LSP were prematurely terminated at a conserved DNA element within the control region, denoted conserved sequence block II (CSB II). The 3′ ends of these terminated transcription products coincided with the RNA-to-DNA transition sites mapped in vivo (5). The mechanism behind the observed transcription termination was not elucidated, but mutational analysis demonstrated that sequence elements within CSB II were of essential importance (4, 5). CSB II could also stimulate transcription termination by the structurally related T7 RNA polymerase, and the effect was therefore not due to a specific CSB II–POLRMT interaction. Instead, it appeared likely that the primary DNA sequence, and probably also structural features of CSB II, caused transcription termination.
Four-stranded G-quadruplex or G4 nucleic acid structures form in guanine-rich sequences through the stacking of planar G tetrads or quartets (11, 12). Such structures are very stable due to G-G hydrogen bonding within each quartet and are further stabilized by monovalent cations that fit into the central cavity (12–14). G-rich sequences occur in the human genome at telomeres, ribosomal DNA, immunoglobulin switch regions, and in minisatellites, and they are also enriched in promoter regions, with more than 40% of promoters containing a sequence motif with potential to form G-quadruplexes (15–17). This enrichment to promoters, as well as the finding that potential to form G-quadruplexes correlates with gene function (18) implies a role for G-quadruplexes in the regulation of global gene expression. G-quadruplexes in the nontranscribed DNA strand can also form a physical obstacle for a progressing RNA polymerase (19). G-quadruplexes may also form in RNA, and previous work has linked these structures to termination of transcription, even if the mechanisms of G-quadruplex-induced transcription termination remains unresolved (20, 21).
In this work, we investigate the molecular mechanisms of transcription termination and primer formation at CSB II. We find that the CSB II sequence has a strong potential to form G-quadruplexes in RNA and that the presence of G-quadruplexes is required for primer formation. Based on our findings, we suggest a model for mitochondrial transcription termination, which is partially reminiscent of Rho-independent transcription termination in bacteria.
Results
Conserved Sequence Block II Has the Potential to Form G-Quadruplex Structures.
The majority of transcripts initiated from LSP are terminated between nucleotide positions 300–282 immediately downstream of CSB II by an unknown mechanism, and deletion of this DNA region abolishes transcription termination (5). Please note that the numbering of positions in the circular mtDNA is fixed and follows the orientation of the mtDNA light strand (Fig. 1A).
Human CSB II has a distinctive sequence containing a stretch of 12 guanines interrupted by a single adenine. The abundance of guanines in the CSB II region prompted us to search for sequences that may fold into G-quadruplexes using the software “QGRS mapper” (22). The analysis demonstrated that most of the Gs between nucleotide positions 325–295 in the CSB II region (underlined in Fig. 1A) can contribute to one of the several G-quadruplexes predicted to form in this region.
Pretermination of LSP Transcripts at CSB II Involves G-Quadruplex RNA.
We used an in vitro transcription system with purified POLRMT, TFAM, and TFB2M to examine the potential role of G-quadruplexes in premature termination of LSP transcripts (23). Because termination occurs just downstream of the CSB II sequence (5), and not before, as one would expect if an obstruction in the template at CSB II would block polymerase passage, we predicted that the terminating structure would involve RNA. We decided to use 7-deaza-GTP, a GTP analogue that lacks one of the atoms (N7) required for G-G hydrogen bonding in G quartets and thus disrupts four of the eight hydrogen bonds within each G quartet, to investigate this possibility. Incorporation of 7-deaza-GTP in the RNA strand has previously been used as a means of preventing G-quadruplex formation (21). We performed in vitro transcription on a linearized template plasmid containing nucleotides 477–1 of human mtDNA (LSP transcription starts at position 407) and compared the outcome in the presence of GTP or 7-deaza-GTP. In the presence of GTP, more than 40% of the transcripts initiated at LSP were prematurely terminated at CSB II (Fig. 1B). In contrast, transcription on a template with nucleotides 318–288 mutated to prevent G-quadruplex formation produced significantly reduced levels of prematurely terminated transcripts (below 10%). When GTP was substituted with 7-deaza-GTP, CSB II-dependent transcription termination was dramatically decreased, suggesting that G-quadruplexes are involved in the regulation of termination. As expected, transcription from the template mutated at positions 318–288 was not affected by 7-deaza-GTP. The intensity of another, minor transcription product (indicated with an asterisk in Fig. 1B) remained unchanged relative to the full-length runoff product, thus providing an internal control.
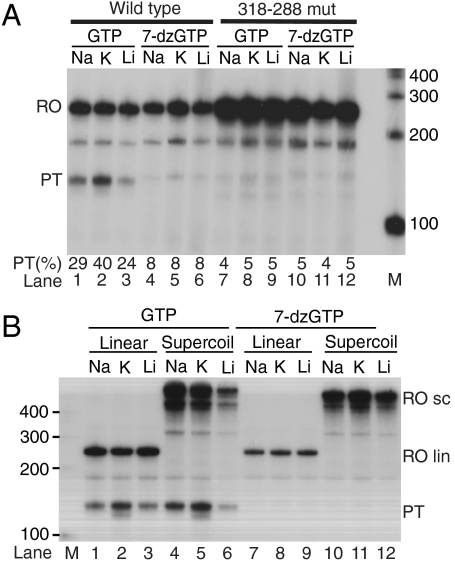
Monovalent cations, especially K+, are known to stabilize G-quadruplexes (11, 13), and their presence in reaction buffers should therefore increase the fraction of prematurely terminated transcripts observed in our system. To test this hypothesis, we performed in vitro transcription on the wild-type CSB II template with Na+, K+, or Li+ added to the reaction buffer. Termination at CSB II was more efficient in the presence of K+ (Fig. 2A, lanes 1–3), as would be expected for a G-quadruplex-dependent event. However, we observed robust CSB II-dependent transcription termination in all of the conditions tested, suggesting that even the low level of monovalent cations deriving from the protein preparations and other sources was enough to support formation of G-quadruplex structures. Substitution of GTP with 7-deaza-GTP dramatically decreased premature termination to below 10% in all reactions (Fig. 2A, lanes 4–6). The efficiency of transcription pretermination using the 318–288 mutant template was less than 10% irrespective of ionic conditions and was unaffected by 7-deaza-GTP (Fig. 2A, lanes 7–12). The stabilizing effect of K+ taken together with the decreased termination in 7-deaza-GTP strongly suggests that premature transcription termination at CSB II is dependent on G-quadruplexes formed in the RNA transcript.
Fig. 2.
The effect of ionic conditions and template topology on premature transcription termination. (A) Wt (lanes 1–6) and mutant (lanes 7–12) templates were transcribed in the presence of GTP (lanes 1–3, 7–9) or 7-deaza-GTP (lanes 4–6, 10–12) and 35 mM of added NaCl, KCl, or LiCl. PT(%), percentage of pretermination; M, marker lane. (B) In vitro transcription on SspI-linearized (lanes 1–3 and 7–9) or supercoiled (lanes 4–6, 10–12) template. A final concentration of 35 mM of the indicated ion was added, and reactions contained either GTP or 7-deaza-GTP. PT, preterminated transcript; RO lin, runoff transcript from linearized template (∼240 nts); RO sc, runoff transcripts from supercoiled template. M, marker lane.
Supercoiling is known to facilitate G-quadruplex formation in DNA. In order to address whether template supercoiling affects G-quadruplex formation and transcription termination at CSB II, we compared the extent of premature termination on equimolar amounts of supercoiled and linearized DNA templates. Transcription termination occurred to a similar extent on linear and supercoiled template, as would be expected if the G-quadruplexes involved in termination form on the RNA transcript (Fig. 2B)
Native Gel Analysis of RNA Reveals G-Quadruplex Formation in the CSB II Sequence.
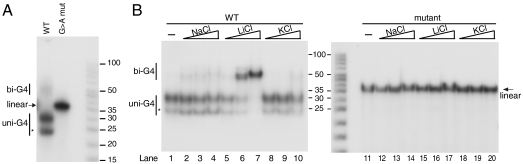
To directly monitor G-quadruplex formation and to prove that the CSB II sequence is sufficient for their formation, we designed RNA oligonucleotides containing either the wild-type CSB II sequence or a mutant version, with all Gs in CSB II replaced with As (Table 1). The oligonucleotides were 30 nucleotides in length and the 3′ ends correspond to the site previously shown to be the major 3′ end of transcripts terminated at CSB II (5). The oligonucleotides were 5′ end-labeled, incubated overnight to allow G-quadruplex formation, and separated on nondenaturing polyacrylamide gels. The mutant oligonucleotide migrated as a linear molecule of the expected molecular weight. In contrast, the WT CSB II oligonucleotide was separated into two major bands, both migrating faster than expected for a linear product (Fig. 3A). Faster migration on a native gel is characteristic of a unimolecular (intramolecular) G-quadruplex (11). Therefore, these two faster-running species (denoted uni-G4 and uni-G4*) may represent two different conformations of unimolecular G-quadruplexes involving different guanines where one conformation generates a more compact structure (uni-G4*). A small fraction of the Wt oligonucleotide exhibited lower electrophoretic mobility and was found at the level expected for a linear 50-mer. This species is indicative of a bimolecular G-quadruplex (11), because no dimer formation is expected to occur through standard Watson–Crick basepairing within the oligonucleotide.
Table 1.
Oligonucleotides used in the native gel analysis
| RNA oligo | Sequence |
| CSB II WT | GAAGCGGGGGAGGGGGGGUUUGGUGGAAAU |
| G → A mut | GAAGCAAAAAAAAAAAAAUUUAAUGGAAAU |
| 295–296 mut | GAAGCGGGGGAGGGGGGGUUUGGUAAAAAU |
| 305–306 mut | GAAGCGGGGGAGGGAAGGUUUGGUGGAAAU |
| 312–313 mut | GAAGCGGAAGAGGGGGGGUUUGGUGGAAAU |
Sequences with guanines in CSB II that are predicted to have G4-forming potential are underlined and mutated nucleotides are indicated in bold font. The numbering refers to the Cambridge reference sequence (29).
Fig. 3.
Native gel analysis of oligonucleotides containing wild-type or mutated CSB II. (A) Wt or G → A mutant RNA oligonucleotides (5 pmol total, of which 1.4 pmol labeled) were incubated overnight at 37 °C and separated on a 12% native gel. DNA size marker in the right-hand lane. bi-G4, bimolecular quadruplex; uni-G4, unimolecular quadruplex; asterisk denotes the more compact uni-G4* species. (B) RNA oligos (WT and mutant) were incubated in increasing concentrations of NaCl, LiCl, or KCl (0, 50, 150, or 1,000 mM) overnight at 37 °C. Reactions were separated on a 12% native gel. bi-G4, bimolecular quadruplex; uni-G4, unimolecular quadruplex; asterisk denotes a more compact G-quadruplex conformation. The arrow indicates the linear form of oligo, which is seen only with the mutant lacking the Gs required for quadruplex formation.
Next, we investigated effects of monovalent cations on the conformation of the RNA oligonucleotides by separating the species formed in increasing concentrations of Na+, K+, or Li+. In agreement with our transcription termination experiments, G-quadruplex formation was very efficient and regardless of the ionic conditions used, a clear majority of the WT oligonucleotide formed the unimolecular G-quadruplex that migrated faster than the linear oligo exemplified by the mutant. Only in the presence of ≥150 mM Li+ did the balance between the different conformations shift to favor the bimolecular G-quadruplex (Fig. 3B). In general, lithium ions do not favor the formation of G-quadruplexes; the bimolecular G4 RNA observed here in the presence of Li+ supports the finding that the G-quadruplex forming at CSB II is relatively stable regardless of the ions present, as exemplified above by the termination efficiency in Na+, Li+, and K+ (Fig. 2A). The mutant RNA oligo was not affected by the salt conditions and migrated as a linear molecule (Fig. 3B).
Guanines at Nucleotide Positions 315–303 Are Required for Transcription Termination.
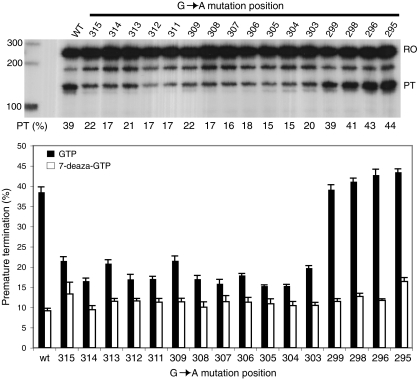
Generally, intramolecular G-quadruplexes can form in sequences containing four guanine runs separated by short spacers or loops. Furthermore, most of the guanines in the CSB II region are predicted to have the potential to contribute to a G-quadruplex (see Fig. 1A). In order to pinpoint the guanines in CSB II that are involved in the formation of the G-quadruplex that preterminates transcription, we created a series of constructs with each of the Gs in and around the CSB II element mutated to an A. Surprisingly, when these constructs were used as templates for in vitro transcription, we found that the mutation of any single G within the two G runs of CSB II clearly reduced termination (Fig. 4, mutations at nucleotide positions 315–303). In contrast, mutation of G299 at the end of CSB II or of the Gs immediately downstream of CSB II did not affect termination. It therefore seems that all 12 guanines in the stretches of 5 and 7 contiguous Gs in CSB II are important for the stability of the G-quadruplex structure, whereas downstream Gs do not contribute to the structure. The use of 7-deaza-GTP with these mutant templates resulted in only a small further decrease in transcription termination (Fig. 4 Lower), indicating that most of the G-quadruplex RNA is already destabilized due to the mutation of the single G nucleotides.
Fig. 4.
Mutations in the G stretches of CSB II reduce pretermination. (Upper) In vitro transcription using a series of mutant templates with a G-to-A mutation at the indicated position. Transcription reactions contained GTP and 50 mM KCl. RO, full-length runoff transcript; PT, preterminated transcript; PT(%), average percentage of pretermination in three independent experiments. (Lower) The percentage of prematurely terminated transcript in transcription assays carried out in 50 mM KCl with either GTP (black bars) or 7-deaza-GTP (white bars) on mutant or wild-type template. The nucleotide position of the G → A mutation is indicated. N = 3 and error bars represent standard error of the mean.
In accordance with these results, an RNA oligonucleotide with two guanines mutated to adenines just downstream of CSB II (positions 296–295) behaved essentially as wild type in native gel analysis (Fig. 5A). However, the 296–295 mutant was unable to form the unimolecular G4* variant, indicating that these guanines are involved in G-G hydrogen bonding within this particular quadruplex conformation but not the uni-G4 variant. In agreement with this observation, the 296–295 double mutation did not affect transcription termination in vitro (Fig. 5B). We can thus conclude from this observation and data in Fig. 4 that the uni-G4* variant is not essential for transcription pretermination.
Fig. 5.
CSBII mutants exhibiting altered G-quadruplex formation affect transcription pretermination accordingly. (A) Wild-type or mutant CSBII RNA oligonucleotides (5 pmol per reaction, of which 0,4 pmol labeled) with GG → AA alterations at nucleotide positions 296–295, 306–305, and 313–312 were incubated overnight at 37 °C and separated on a 12 % native gel. M, DNA marker. (B) DNA templates with GG → AA mutations corresponding to the ones in A were used in the transcription assay in parallel with wild-type and mutant 318–218 templates in the presence of 50 mM KCl and either GTP or 7-deaza-GTP. (C) Templates with mutations encompassing the poly-T (nt positions 291–286), poly-A (294–292), or both (294–286) runs downstream of CSB II were used in transcription assays in 50 mM KCl and either GTP or 7-deaza-GTP. RO, runoff transcript; PT, preterminated transcript; PT(%), average percentage of pretermination in three independent experiments; M, marker lane.
We also investigated effects of mutations in the G runs of CSB II by replacing two guanines at positions 306–305 or 313–312 with adenines. Both these RNA oligos exhibited altered migration (Fig. 5A). The 296–295 mutation did not cause a major shift in the migration pattern when compared with the WT. In contrast, we observed a distinct change in the migration of the 306–305 mutant that was shifted toward the level of the linear G → A mutant oligo, supportive of the G-quadruplex being less stable or less likely to form in this mutant. Interestingly, the 313–312 mutant actually migrated even faster than the WT. Although we do not understand the reason for this unexpected behavior, one can speculate that it is due to a more compact folding of the oligonucleotide upon mutation of these two Gs near the end of the molecule. Although it is entirely possible that the 306–305 and 313–312 mutant migration patterns represent bona fide G-quadruplex structures (though involving different sets of guanines to the WT oligo), we speculate that they may be unstable or form too slowly to be functionally relevant in transcription termination. Given that there are many guanines within CSB II and flanking regions that are predicted to give rise to several alternative G-quadruplexes (as predicted by QGRS mapper), it is perhaps not surprising that mutating one pair of guanines does not completely abolish G-quartet formation, but rather allows the formation of alternative and less predisposed conformations that would not normally form in the wild-type oligo. These alternative G-quadruplexes are, however, not able to effectively support transcription termination because both the 306–305 and 313–312 double mutants caused a dramatic decrease in transcription termination in vitro (Fig. 5B). Taken together, we conclude that the most important quadruplex species to mediate transcription pretermination is the predominant unimolecular G-quadruplex (“uni-G4”) whose formation requires guanines within CSB II. The uni-G4* species, which was absent in all the mutant oligos including the 296–295 mutant, does not seem to be required for pretermination because the mutation of nucleotide positions 295 and 296 did not affect the quantity of the preterminated product in the transcription assay.
A Downstream Poly-(dA-dT) Stretch Modulates Transcription Termination at CSB II.
The mtDNA sequence just downstream of CSB II contains three contiguous As (nucleotide positions 294–292) followed by six Ts (positions 291–286). Furthermore, the transcription termination observed in the CSB II region has previously been mapped to between nucleotides 300–282 and thus overlaps with this region (5). In order to address whether these non-G mononucleotide runs contribute to the transcription termination caused by the G-quadruplex structure at CSB II, we constructed mutant templates with either the poly-dA stretch, the poly-dT stretch, or both these sequence elements mutated. As shown in Fig. 5C, the mutation of the poly-dT run (the 291–286 mutant) reduced the frequency of premature termination, whereas mutation of the three consecutive As (294–292 mutant) did not have the same effect. Instead, this mutation strengthened another terminated product only slightly shorter in size. The 294–286 mutation, covering the poly-dA and poly-dT runs, had a combined effect and resulted in a decrease in the major preterminated product as well as the appearance of the shorter preterminated transcript. As expected, these effects were independent of G-quadruplexes because the same changes were observed in the presence of 7-deaza-GTP. Our interpretation of these results is that the downstream mononucleotide stretches modulate transcription termination to some extent. The transcription termination observed at CSB II depends mainly on the Gs between positions 315–303, but downstream poly-dT stretch has a modest stimulatory effect on termination efficiency.
Discussion
We have here investigated the molecular mechanisms of the transcription termination and primer formation that takes place at nucleotide positions 300–282 immediately 3′ of CSB II (5). Our studies reveal a mechanism for transcription termination, not previously described in any other biological system. We demonstrate that the CSB II sequence has the potential to form G-quadruplex structures in the native transcript and that transcription termination is reduced in the presence of 7-deaza-GTP, which is known to reduce G-quadruplex formation. Conversely, the termination frequency increased in the presence of K+ ions, which stabilize G-quadruplexes through charge coordination (13). Mutation analysis confirmed the importance of each of the 12 guanines in the two G tracts of CSB II for termination and indicated the G-quadruplex as the main determinant of premature transcription termination, whereas the poly-dT run just downstream of CSB II was found to have a more modest stimulatory effect.
The involvement of a G-rich sequence and a T run in termination resembles the mechanism of intrinsic (Rho-independent) termination, where termination is determined by the presence of two elements: a GC-rich palindromic sequence followed by an oligo-dT sequence (24). In the nascent RNA, the GC-rich sequence folds into a stable hairpin, weakening RNA-to-polymerase contacts. At the same time, the oligo(rU) stretch of the nascent RNA is positioned in the RNA-DNA hybrid-binding site of the polymerase, resulting in a weak rU-dA hybrid. Together, these two factors cause destabilization of the elongation complex and termination of transcription (reviewed in ref. 25). Our data suggest that the pretermination of mitochondrial transcription near CSB II occurs in an analogous way. RNA forms a G-quadruplex during transcription, which in turn weakens interactions with the POLRMT, similar to the role assigned to the hairpin structure required in intrinsic termination.
We do not yet understand why transcription termination in mitochondria relies on a G-quadruplex structure instead of the canonical hairpin loop, found in so many other systems, but it is possible that the G-quadruplex structure may have role also after transcription is terminated. Normally, nascent RNA transcripts do not remain hybridized to the DNA template, but are pried off as the DNA strands reanneal. The mitochondrial primer RNA appears to be an exception to this rule because many different studies have demonstrated that the primer remains stably associated with the DNA template at yeast, mouse, and human CSB II (26–28). The molecular basis for this extraordinary stability is not known, but a hybrid G-quadruplex between RNA and DNA may explain why the transcribed RNA primer remains associated with the DNA template.
It is worth noting that also nucleotides 387–370 just upstream of CSB III have the potential to form G-quadruplexes (as predicted by QGRS mapper). Because CSB III only has a minor effect on transcription termination in vitro (5), we have focused our efforts on studies of the functionally more important CSB II element. There are also naturally occurring polymorphisms in CSBII, including a 7-bp-long deletion of the G-rich region starting at position 303 (www.mitomap.org). These polymorphisms are not homoplasmic, and how they affect initiation of DNA synthesis has not been analyzed. Transcription termination and primer formation may be directed from other regions in the absence of a functional CSBII sequence. We can, e.g., not exclude the possibility that CSB III may contribute to the formation of higher order G-quadruplex structures and transcription termination in vivo.
Materials and Methods
Bioinformatic Approaches.
For prediction of potential G-quadruplex-forming sequences, the human mitochondrial genome was analyzed by QGRS Mapper (22) using standard settings.
Preparation of Templates for Transcription.
Base pairs 1–477 of human mtDNA were cloned into pUC18 and linearized with SspI (New England Biolabs) prior to use in the transcription reaction. Mutant templates were prepared by PCR site-directed mutagenesis using Pfu polymerase (Stratagene) and standard cloning techniques. The 7-deaza-dGTP-containing templates were produced by amplifying nucleotides 1–407 of human mtDNA in a PCR reaction that contained 0.06 mM 7-deaza-dGTP (New England Biolabs) instead of dGTP.
In Vitro Transcription.
Recombinant POLRMT, TFAM, and TFB2M were expressed in insect cells and purified as described previously (23). Transcription reactions were carried out in the presence of [α-32P]-UTP as in Falkenberg et al. (23), but with addition of NaCl, KCl, or LiCl where indicated in the figure legends. Transcripts were purified and run on 6% acrylamide/urea gels in 1× Tris/Borate/EDTA buffer (45 mM Tris-borate, 1 mM EDTA), in parallel with 100-bp DNA marker (NEB). Gels were visualized on X-ray film and radioactive signal was quantified using the FLA-7000 analyzer (Fujifilm). Pretermination efficiency was calculated by dividing the signal of the preterminated transcript by the sum of the preterminated and runoff signals.
Gel Analysis of G-Quadruplex RNA Structures.
RNA oligonucleotides were purchased from Dharmacon (Table 1) and purified by polyacrylamide gels by UV shadowing. Oligo sequences are listed in Table 1. Oligonucleotides were 5′-end-labeled with T4 polynucleotide kinase and [γ-32P]-ATP. Unincorporated label was removed on a Sephadex G-25 column (GE Healthcare). For the formation of G4 RNA we essentially followed Sen and Gilbert (11) with some modifications. Per reaction, a total of 5 pmol of oligonucleotides (of which 0.1–1.4 pmol was labeled) were incubated in TE (10 mM Tris-Cl pH 7.5, 1 mM EDTA) at 95 °C for 5 min, then rapidly cooled on ice. Oligonucleotides were then incubated at 1 μM final concentration in TE overnight at 37 °C. To study the effect of salts, NaCl, LiCl, or KCl were included at 50, 150, or 1,000 mM concentration. All reactions were resolved under nondenaturing conditions on 12% polyacrylamide gels in Tris-Borate-EDTA buffer containing 10 mM KCl. Low Molecular Weight DNA size marker was purchased from USB Corporation.
Acknowledgments.
This work was supported by the Swedish Research Council, the Swedish Cancer Society, the European Commission (fp6 EUMITOCOMBAT), the Swedish Strategic Foundation, the Göran Gustafsson Foundation, the Knut and Alice Wallenberg Foundation, and the Medical Research Council.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
References
- 1.Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 2.Clayton DA. Replication and transcription of vertebrate mitochondrial DNA. Annu Rev Cell Biol. 1991;7:453–478. doi: 10.1146/annurev.cb.07.110191.002321. [DOI] [PubMed] [Google Scholar]
- 3.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 4.Chang DD, Clayton DA. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc Natl Acad Sci USA. 1985;82:351–355. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pham XH, et al. Conserved sequence box II directs transcription termination and primer formation in mitochondria. J Biol Chem. 2006;281:24647–24652. doi: 10.1074/jbc.M602429200. [DOI] [PubMed] [Google Scholar]
- 6.Wanrooij S, et al. Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc Natl Acad Sci USA. 2008;105:11122–11127. doi: 10.1073/pnas.0805399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuste JM, et al. Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol Cell. 2010;37:67–78. doi: 10.1016/j.molcel.2009.12.021. [DOI] [PubMed] [Google Scholar]
- 8.Lee DY, Clayton DA. RNase mitochondrial RNA processing correctly cleaves a novel R loop at the mitochondrial DNA leading-strand origin of replication. Gene Dev. 1997;11:582–592. doi: 10.1101/gad.11.5.582. [DOI] [PubMed] [Google Scholar]
- 9.Lee DY, Clayton DA. Initiation of mitochondrial DNA replication by transcription and R-loop processing. J Biol Chem. 1998;273:30614–30621. doi: 10.1074/jbc.273.46.30614. [DOI] [PubMed] [Google Scholar]
- 10.Kiss T, Filipowicz W. Evidence against a mitochondrial location of the 7-2/MRP RNA in mammalian cells. Cell. 1992;70:11–16. doi: 10.1016/0092-8674(92)90528-k. [DOI] [PubMed] [Google Scholar]
- 11.Sen D, Gilbert W. Guanine quartet structures. Method Enzymol. 1992;211:191–199. doi: 10.1016/0076-6879(92)11012-8. [DOI] [PubMed] [Google Scholar]
- 12.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc Natl Acad Sci USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 14.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 15.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eddy J, Maizels N. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006;34:3887–3896. doi: 10.1093/nar/gkl529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tornaletti S, Park-Snyder S, Hanawalt PC. G4-forming sequences in the non-transcribed DNA strand pose blocks to T7 RNA polymerase and mammalian RNA polymerase II. J Biol Chem. 2008;283:12756–12762. doi: 10.1074/jbc.M705003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: The beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuzmine I, Gottlieb PA, Martin CT. Structure in nascent RNA leads to termination of slippage transcription by T7 RNA polymerase. Nucleic Acids Res. 2001;29:2601–2606. doi: 10.1093/nar/29.12.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kikin O, D’Antonio L, Bagga PS. QGRS Mapper: A web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falkenberg M, et al. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 24.d’Aubenton Carafa Y, Brody E, Thermes C. Prediction of rho-independent Escherichia coli transcription terminators: A statistical analysis of their RNA stem-loop structures. J Mol Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 25.Nudler E, Gottesman ME. Transcription termination and anti-termination in E. coli. Genes Cells. 2002;7:755–768. doi: 10.1046/j.1365-2443.2002.00563.x. [DOI] [PubMed] [Google Scholar]
- 26.Lee DY, Clayton DA. Properties of a primer RNA-DNA hybrid at the mouse mitochondrial DNA leading-strand origin of replication. J Biol Chem. 1996;271:24262–24269. doi: 10.1074/jbc.271.39.24262. [DOI] [PubMed] [Google Scholar]
- 27.Xu B, Clayton DA. A persistent RNA-DNA hybrid is formed during transcription at a phylogenetically conserved mitochondrial DNA sequence. Mol Cellular Biol. 1995;15:580–589. doi: 10.1128/mcb.15.1.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu B, Clayton DA. RNA-DNA hybrid formation at the human mitochondrial heavy-strand origin ceases at replication start sites: An implication for RNA-DNA hybrids serving as primers. EMBO J. 1996;15:3135–3143. [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]