Abstract
Background/Aims
Some studies suggest that polymorphisms in angiotensin-converting enzyme (ACE), angiotensinogen (AGT), angiotensin II type I receptor (AGTR1) and angiotensin II type II receptor (AGTR2) genes may contribute to renal function variation.
Methods
Genotyping for single nucleotide polymorphisms (SNPs) in these candidate genes was performed in 2,847 participants from four racial/ethnic groups (African American, Chinese, White and Hispanic) without known cardiovascular disease in the Multi-Ethnic Study of Atherosclerosis. SNP and haplotype analyses were performed to determine associations between genotypes and cross-sectional renal function measurements, including urine albumin excretion (UAE) and estimated glomerular filtration rate (eGFR) using serum creatinine and cystatin C.
Results
Twenty-four ACE SNPs, 10 AGT SNPs, 15 AGTR1 SNPs and 6 AGTR2 SNPs were typed successfully. After adjusting for ancestry, age and gender, 3 SNPs (AGT M235T, AGT rs2148582 and AGTR1 rs2131127) showed associations with an empiric p value <0.05 with the same phenotype in multiple racial/ethnic groups, suggesting replication. The AGT M235T SNP has been shown previously to be associated with diabetic and hypertensive nephropathy. Conclusions: These data suggest that genetic polymorphisms in the renin-angiotensin system are associated with renal phenotypes in the general population, but that many associations differ across racial/ethnic groups.
Key Words: Renin-angiotensin, ACE, AGT, AGTR1, AGTR2, Albuminuria, Creatinine clearance epidemiology, Genetics
Introduction
The renin-angiotensin system plays an important role in sodium homeostasis, fluid balance and blood pressure [1, 2]. Sequencing of the angiotensin-converting enzyme (ACE), angiotensinogen (AGT), angiotensin II type I receptor (AGTR1) and angiotensin II type II receptor (AGTR2) genes has identified several polymorphisms that may contribute to variability in renal function [3,4,5,6,7,8,9,10]. An insertion/deletion polymorphism in ACE as well as the M235T polymorphism in AGT have been found to be associated with kidney disease in multiple studies [3, 5, 7,8,9,10]. However, with the exception of a recent study in Mexican American families [10], previous studies have focused on patients with diabetes or hypertension. To our knowledge, associations between single nucleotide polymorphisms (SNPs) in these genes and renal dysfunction have not been studied extensively in a multiracial general population, such as the Multi-Ethnic Study of Atherosclerosis (MESA).
Methods
Participants
MESA is an observational cohort study designed to investigate subclinical cardiovascular disease in people free of clinical cardiovascular disease. The study design has been described previously [11]. The current analysis includes 2,847 participants who were selected for genotyping from the MESA cohort to provide approximately equal numbers within each of the four racial/ethnic groups. Study participants in this subgroup consented to the use of their DNA. SNP selection and genotyping in MESA has been described previously [12].
Kidney Function Assessment
The measurement of cystatin C, creatinine and urine albumin excretion (UAE) has been described previously [13, 14]. Cystatin C was not available for 16 samples (0.6%), creatinine was not available for 5 samples (0.2%) and UAE was not available for 14 samples (0.5%). Estimated glomerular filtration rate (eGFR) was calculated using the Modification of Diet in Renal Disease (MDRD) equation: eGFR (ml/min/1.73 m2) = 175 × (Scr) − 1.154 × (Age) − 0.203 × (0.742 if female) × (1.210 if African American). Cystatin C eGFR was calculated using the following equation: eGFR = 127.7 × (Cystatin C–1.17) × (Age–0.13) × (0.91 if female) × (1.06 if African American) [15]. Albuminuria was defined as a ratio of ≥17 mg albumin per gram creatinine (mg/g) in men and ≥25 mg/g in women in a spot urine sample [16].
Statistical Analysis
Means, medians and frequencies of demographic characteristics were calculated by racial/ethnic group. Minor allele frequencies (MAF) of each polymorphism were calculated and Hardy-Weinberg equilibrium (HWE) tests were performed. For each gene, a p value of 0.05 divided by the number of SNPs analyzed within the gene was used as the cutoff for HWE.
Haplotype analyses were performed on the four candidate genes. Haploview was used to evaluate linkage disequilibrium (LD), and haplotype blocks were defined according to the solid spine model. For SNPs in the same haplotypic block, haplotypes were reconstructed using PHASE [17].
Renal phenotypes were analyzed as continuous and dichotomous outcomes. Outcomes included eGFR using the MDRD equation and cystatin C eGFR as continuous variables and as dichotomous variables with a cutoff of <60 ml/min/m2 (the cutoff for stage 3 chronic kidney disease) and UAE as a log-transformed continuous variable and dichotomous variable. UAE was evaluated as a log-transformed variable because of a skewed distribution.
Cross-sectional associations between each SNP and haplotype and measures of renal function were determined using multiple linear and logistic regression, adjusting for age, gender and ancestry, and stratified by race/ethnicity. The additive model was used if the minor allele or haplotype frequency was >10%; the dominant model was used if the minor allele or haplotype frequency was ≤10%. For AGTR2, separate analyses were performed for men and women as the gene is located on the X chromosome.
Five principal components were included as covariates to adjust for ancestry. Principal components were obtained based on 199 ancestry informative markers and computed by EIGENSTRAT [18]. This method applies principal components analysis [19] to the SNP data to infer continuous axes of genetic variation. These principal components are then used as covariates in the regression model in the association analysis to account for ancestry differences among individuals.
Primary Analysis
Associations with a nominal p value of <0.05 were evaluated further via permutation testing. 1,000 permutation data sets were generated by shuffling phenotypic data, and association analysis was performed for the permutated datasets. Typically, the estimate of the empirical p value is obtained as r/n, where nis the number of replicate samples simulated, and ris the number of replicates that produce a test statistic greater than or equal to that calculated for the actual (unshuffled) data. A corrected formula, (r + 1)/(n + 1), is more accurate [20, 21]. For our primary analysis to identify significant SNP/phenotype associations, evidence of internal replication was demonstrated when an association was seen in the same direction with an empiric p value <0.05 after permutation testing in more than one racial/ethnic group.
For SNP/phenotype associations that were significant (p < 0.05) in the initial model (adjusting for age, gender and ancestry), we then used an additional model to adjust for hypertension and a separate model to adjust for diabetes. Hypertension was defined as a systolic blood pressure ≥140 mm Hg or diastolic blood pressure ≥90 mm Hg, or current use of antihypertensive medication. Diabetes was defined as fasting glucose ≥126 or use of diabetes medications.
Interaction tests for gender, hypertension and diabetes were performed on associations with an empiric p value <0.05 for the same phenotype in multiple racial/ethnic groups. For associations that showed an interaction with gender, hypertension or diabetes (p < 0.05), stratified analyses were performed according to the variable of interest.
Secondary Analysis
To improve study power and combine the evidence for association within each racial/ethnic group, we performed a meta-analysis by combining p values from each of the four racial/ethnic groups using the METAL program, which accounts for both strength and direction of association in the four racial/ethnic groups (http://www.sph.umich.edu/csg/abecasis/Metal/). Hispanics were excluded from the meta-analysis for the five SNPs that were not in HWE.
To determine the effective number of independent tests in the meta-analysis, a composite LD correlation was calculated directly from SNP genotypes [22]. The effective number of independent SNPs was 10 for ACE, 4 for AGT, 8 for AGTR1 and 4 for AGTR2, and the effective number of independent phenotypes was 4. The adjusted Bonferroni correction was calculated by dividing 0.05 by the effective number of independent tests [0.05/effective number of independent SNPs/effective number of independent phenotypes/two genders (AGTR2 only)]. Thus, for our secondary meta-analysis, cutoffs for a significant p value were 0.001 for ACE, 0.003 for AGT, 0.002 for AGTR1 and 0.002 for AGTR2.
Results
Online supplementary table 1 describes demographic characteristics (for all online supplementary material see www.karger.com/doi/10.1159/000315866). The mean age was 62 years, and about half of the participants were female. Hypertension was present in 44%, with a higher prevalence of hypertension (56%) in African Americans. Diabetes was present in 24%.
Twenty-four of 26 ACE SNPs, 10 of 11 AGT SNPs, 15 of 16 AGTR1 SNPs and 6 of 8 AGTR2 SNPs were typed successfully. Online supplementary figure 1 shows the location of SNPs in the genes and online supplementary figures 2–5 show LD among SNPs and haplotype block structure. Two of 24 ACE SNPs were not polymorphic. Online supplementary table 2 summarizes SNPs and MAF. Five SNPs in Hispanics were not in HWE.
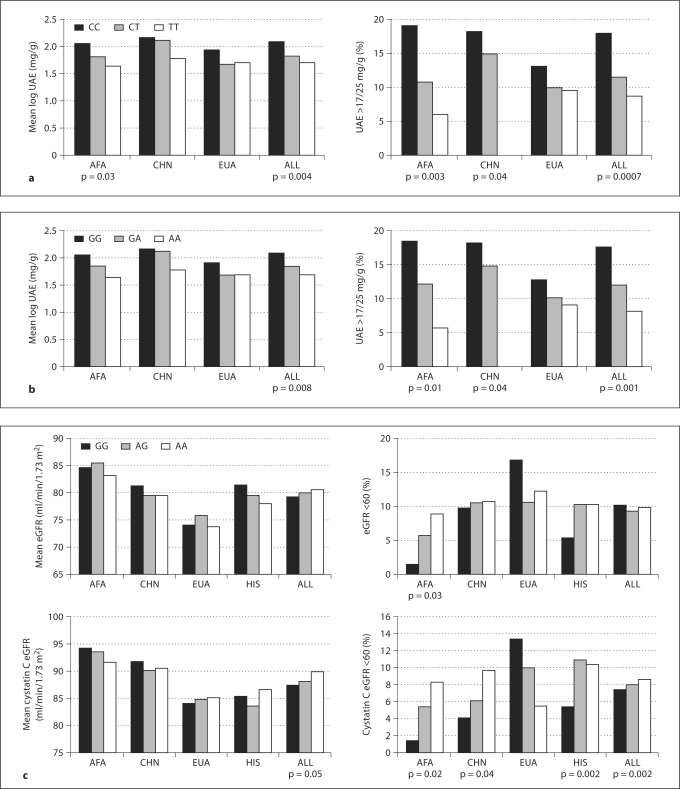
Online supplementary table 3 summarizes associations between SNPs and renal phenotypes, adjusted for age, gender and ancestry, as well as stratified by racial/ethnic group and combined. While there were numerous associations at a p value <0.05 in all 4 genes, only 3 SNPs (AGT M235T, AGT rs2148582 and AGTR1 rs2131127) showed associations in the same direction with an empiric p value <0.05 within the same phenotype in multiple racial/ethnic groups (table 1, fig. 1). In AGT, SNPs M235T and rs2148582 were associated with UAE ≥17/25 mg/g in both African Americans and Chinese (fig. 1a, b, table 1). These SNPs are located in the same haplotype block in both African Americans and Chinese (D’ = 0.91 for African Americans, 0.99 for Chinese). Furthermore, in African Americans, M235T was associated not only with UAE ≥17/25 mg/g but also with mean log UAE, and similar trends were seen in Whites. Both SNPs were not in HWE in Hispanics. Haplotype analysis showed that in African Americans, the third most common haplotype for the haplotype block which harbors both M235T and rs2148582 was associated with both UAE and eGFR (online supplementary table 4).
Table 1.
β-Coefficients and p values for three SNPs with significant associations for the same phenotype in multiple racial/ethnic groups, adjusted for age, gender and ancestry
| Continuous |
Dichotomous |
|||||
|---|---|---|---|---|---|---|
| P | β-coefficient | P | β-coefficient | |||
| ACT M235T | eGFR | AFA | 0.06 | 2.27 | 0.06 | −0.67 |
| eGFR | CHN | 0.64 | 0.50 | 0.76 | 0.08 | |
| eGFR | EUA | 0.9 | −0.14 | 0.54 | −0.10 | |
| eGFR | HIS | HWE | HWE | HWE | HWE | |
| eGFR | combined | 0.50 | 0.31 | 0.14 | −0.15 | |
| Cys C eGFR | AFA | 0.27 | 1.66 | 0.43 | −0.26 | |
| Cys C eGFR | CHN | 0.38 | 1.13 | 0.24 | −0.45 | |
| Cys C eGFR | EUA | 0.41 | −0.76 | 0.72 | 0.07 | |
| Cys C eGFR | HIS | HWE | HWE | HWE | HWE | |
| Cys C eGFR | combined | 0.49 | 0.38 | 0.29 | −0.10 | |
| UAE | AFA | 0.03a,c,e,f | −0.18 | 0.003a | −0.66 | |
| UAE | CHN | 0.17 | −0.11 | 0.04a,d | −0.45 | |
| UAE | EUA | 0.16 | −0.07 | 0.35 | −0.17 | |
| UAE | HIS | HWE | HWE | HWE | HWE | |
| UAE | combined | 0.004 | −0.06 | 0.007b | −0.15 | |
| ACT rs2148582 | eGFR | AFA | 0.24 | 1.43 | 0.07 | −0.63 |
| eGFR | CHN | 0.52 | 0.68 | 0.99 | 0.00 | |
| eGFR | EUA | 0.99 | 0.01 | 0.51 | −0.11 | |
| eGFR | HIS | HWE | HWE | HWE | HWE | |
| eGFR | combined | 0.54 | 0.25 | 0.07 | −0.18 | |
| Cys C eGFR | AFA | 0.27 | 1.64 | 0.12 | −0.56 | |
| Cys C eGFR | CHN | 0.27 | 1.44 | 0.23 | −0.47 | |
| Cys C eGFR | EUA | 0.55 | −0.55 | 0.55 | 0.11 | |
| Cys C eGFR | HIS | HWE | HWE | HWE | HWE | |
| Cys C eGFR | combined | 0.36 | 0.49 | 0.20 | −0.11 | |
| UAE | AFA | 0.06 | −0.15 | 0.01a | −0.57 | |
| UAE | CHN | 0.19 | −0.11 | 0.04a,c,d | −0.46 | |
| UAE | EUA | 0.15 | −0.07 | 0.37 | −0.17 | |
| UAE | HIS | HWE | HWE | HWE | HWE | |
| UAE | combined | 0.008 | −0.05 | 0.001b | −0.14 | |
| AGTRl rs2131127 | eGFR | AFA | 0.22 | −1.28 | 0.03a | 0.59 |
| eGFR | CHN | 0.3 | −0.81 | 0.87 | 0.03 | |
| eGFR | EUA | 0.91 | 0.12 | 0.18 | −0.24 | |
| eGFR | HIS | 0.3 | −0.98 | 0.06 | 0.41 | |
| eGFR | combined | 0.11 | −0.85 | 0.15 | 0.13 | |
| Cys C eGFR | AFA | 0.06 | −2.43 | 0.02a | 0.64 | |
| Cys C eGFR | CHN | 0.68 | −0.39 | 0.04a | 0.52 | |
| Cys C eGFR | EUA | 0.48 | −0.68 | 0.2 | −0.27 | |
| Cys C eGFR | HIS | 0.31 | −0.99 | 0.002a | 0.72 | |
| Cys C eGFR | combined | 0.05 | −1.05 | 0.002 | 0.31 | |
| UAE | AFA | 0.56 | −0.04 | 0.72 | 0.06 | |
| UAE | CHN | 0.96 | 0.00 | 0.66 | −0.07 | |
| UAE | EUA | 0.17 | −0.07 | 0.22 | −0.26 | |
| UAE | HIS | 0.09 | 0.11 | 0.83 | 0.03 | |
| UAE | combined | 0.90 | 0.00 | 0.58 | −0.05 | |
Additional symbols indicate whether p values remained significant after permutation testing and after adjusting for diabetes or hypertension, and whether interactions were present with diabetes or gender. No interactions were seen with hypertension. β-Coefficients are in the same direction for dichotomous traits with a positive β-coefficient indicating that the minor allele is associated with worse renal function (a higher percentage with low eGFR or proteinuria). However, for continuous traits, a negative eGFR β-coefficient indicates that the minor alíele is associated with worse renal function (i.e. lower eGFR), which generally corresponds with a positive UAE β-coefficient (i.e. more proteinuria). Values in italics represent significant (p < 0.05) associations for the same phenotype in multiple racial/ethnic groups.
Association confirmed by permutation testing using empiric p value <0.05
significant in meta-analysis using adjusted Bonferroni correction
no longer significant (using p < 0.05) after adjusting for hypertension
no longer significant (using p < 0.05) after adjusting for diabetes
interaction (p < 0.05) with diabetes
interaction (p < 0.05) with gender. HWE = Not in Hardy-Weinberg equilibrium; AFA = African American; CHN = Chinese; EUA = White; HIS = Hispanic.
Fig. 1.
AGT M235T (a), AGT rs2148582 (b), AGTR1 rs2131127 (c) and renal phenotypes. Hispanics were not in HWE for the AGT M235T and AGT rs2148582 SNPs. Therefore, the p values and means for the combined (ALL) group for these SNPs exclude Hispanics. AFA = African American; CHN = Chinese; EUA = White; HIS = Hispanic.
In AGTR1, SNP rs2131127 was associated with cystatin C eGFR <60 in African Americans, Chinese and Hispanics and with creatinine eGFR <60 in African Americans (fig. 1c, table 1).
Three SNPs in the meta-analysis which combined the 4 racial/ethnic groups were significant using the adjusted Bonferroni correction: AGT M235T for UAE >17/25 mg/g (p = 0.0007), AGT rs2148582 for UAE >17/25 mg/g (p = 0.001) and AGTR2 rs5950584 in males for mean log UAE (p = 0.0002). The AGT M235T and rs2148582 SNPs also showed associations with UAE >17/25 mg/g in both African Americans and Chinese, as described above. AGTR2 rs5950584 is not polymorphic in the Chinese and has a low MAF in Whites (0.004) and Hispanics (0.09), which limits the ability to detect significant associations. The p value for the association between this SNP and log UAE was 0.0008 in white males.
Online supplementary table 4 summarizes the associations between haplotypes and renal phenotypes, adjusted for age and gender. In general, the haplotype analysis supports the SNP analysis results. Most haplotypes that had significant (p < 0.05) associations with renal phenotypes included SNPs that were associated with renal phenotypes in the SNP analysis.
Discussion
We found associations between four renin-angiotensin system polymorphisms and measures of renal function in the general population. We observed associations with AGT M235T, AGT rs2148582 and UAE >17/25 mg/g in both our primary analysis (African Americans and Chinese) and our secondary meta-analysis. We observed associations with AGTR1 rs2131127 and cystatin C eGFR<60 in our primary analysis (African Americans, Chinese and Hispanics). In addition, we observed an association with AGTR2 rs5950584 and mean log UAE in males in our secondary meta-analysis.
Substantial overlap existed among polymorphisms associated with eGFR using creatinine and the MDRD equation and polymorphisms associated with cystatin C eGFR. However, the polymorphisms associated with eGFR and albuminuria were different, suggesting different genetic associations for impairment of the nephron's filtration versus barrier mechanisms. Furthermore, associations with continuous and dichotomous traits were different. All associations which replicated involved dichotomous traits. Discrepancies in the continuous versus dichotomous associations may be due to a threshold effect, whereby SNPs may be associated with pathology (as reflected by dichotomous traits), but may not be associated with normal variation within a nonpathological range (as reflected by continuous traits).
Moreover, associations varied markedly across racial/ethnic groups. These differences may be due to differences in LD patterns between the racial/ethnic groups, differences in MAF which impact the power of the analyses, and differences in unmeasured interactions with environmental factors. This variation in associations highlights the importance of studying these polymorphisms in diverse populations such as MESA.
AGT M235T was associated with renal phenotypes in both African American and Chinese populations. Associations between this SNP and renal function have been identified in previous studies in patients with hypertension or diabetes. Our study was population-based and included participants with hypertension, participants with diabetes, as well as participants without hypertension or diabetes. The findings of this study are consistent with results of previous studies in patients with hypertension or diabetes, highlighting the potential influence of this SNP on renal function [3, 5, 7, 8, 10, 23].
Our study did not identify any associations within ACE that were significant for the same phenotype in multiple racial/ethnic groups. This is surprising given the extensive literature suggesting an association between the ACE insertion/deletion polymorphism and renal disease. However, these prior studies have been limited to patients with diabetes; ACE insertion/deletion has not been convincingly linked to progression of nondiabetic nephropathy [14]. In our study, only one quarter of the participants had diabetes. We did find numerous associations within ACE that were present in individual racial/ethnic groups. While these associations did not replicate across multiple racial/ethnic groups, they may still be true positives, due to differences in LD, MAF or environmental interactions among the racial/ethnic groups, and will require further confirmation.
We identified one AGTR1 SNP (rs2131127) with associations that were significant (p < 0.05) for the same phenotype in multiple racial/ethnic groups. The associations between this SNP and renal phenotypes will need to be confirmed in further studies.
We did not identify any associations in AGTR2 for the same phenotype in multiple racial/ethnic groups. Using the Bonferroni correction in the meta-analysis, there were no significant associations in females, but there was a significant association in males for AGTR2 rs5950584 and mean log UAE. The association between this SNP and UAE will need to be confirmed in further studies. The power to detect significant associations was limited in AGTR2 because the analyses were conducted separately in men and women, and many of the SNPs had low MAF.
Five SNPs were not in HWE in Hispanics. This lack of HWE may be due to the diverse origins of the Hispanic population. MESA recruited participants from six different field centers across the United States, and the origins of the Hispanic population are unique in these different sites.
We used eGFR and UAE to assess kidney function. Although these are standard measures of kidney function, they do have limitations. While eGFR has good accuracy <60 ml/min/m2, the accuracy of eGFR is decreased >60 ml/min/m2[24]. Furthermore, UAE can be transiently increased by factors such as urinary tract infection, hemodynamic stress and metabolic abnormalities [24, 25].
This investigation has many strengths. It is a large multiracial population-based study that evaluates the influence of renin-angiotensin system polymorphisms on renal function. Multiple polymorphisms throughout each gene were evaluated individually and through haplotype analysis. Numerous associations with renal phenotypes with a p value <0.05 were identified. Because we adjusted for ancestry, the SNP/phenotype associations we identified as significant are more likely to be truly associated with kidney function than spurious associations resulting from population stratification. Many associations seen were in noncoding regions of the gene and are likely in LD with causal variants or may regulate gene expression. Some associations may be false positives since multiple statistical tests were performed. However, three SNPs were associated with renal phenotypes in multiple racial/ethnic groups in the stratified analyses. These SNPs are more likely to be true positives since they have been replicated. M235T has been identified in previous studies that were not population based. Associations seen with other SNPs will need to be confirmed in future studies.
In conclusion, numerous associations between SNPs and haplotypes in ACE, AGT, AGTR1 and AGTR2 and renal function have been identified. Future studies are needed to elucidate which polymorphisms are causal variants and mechanisms through which these effects are mediated. Enhanced understanding of these genetic polymorphisms may enable screening of asymptomatic patients to determine their genetic risk for renal dysfunction. Additional studies will be needed to assess whether these at-risk patients may benefit from aggressive blood pressure control and/or treatment with ACE inhibitors or angiotensin-receptor blockers, which have been demonstrated to slow progression of kidney disease [26].
Supplementary Material
Supplementary Figures
Supplementary Tables
Acknowledgements
This research was supported by contracts N01-HC-95159 through N01-HC-95169 (MESA) and RO1 HL071205 (MESA Family) from the National Heart, Lung, and Blood Institute. The authors thank the other investigators, staff and participants of the MESA study for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org.
References
- 1.Hall JE, Mizelle HL, Woods LL. The renin-angiotensin system and long-term regulation of arterial pressure. J Hypertens. 1986;4:387–397. doi: 10.1097/00004872-198608000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Sealy JE, Laragh JH. The renin-angiotensin-aldosterone system for normal regulation of blood pressure and sodium and potassium homeostasis. In: Laragh JH, Brenner BM, editors. Hypertension: Pathophysiology, Diagnosis, and Management. New York: Raven Press; 1995. [Google Scholar]
- 3.Fabris B, Bortoletto M, Candido R, Barbone F, Cattin MR, Calci M, Scanferla F, Tizzoni L, Giacca M, Carretta R. Genetic polymorphisms of the renin-angiotensin-aldosterone system and renal insufficiency in essential hypertension. J Hypertens. 2005;23:309–316. doi: 10.1097/00004872-200502000-00013. [DOI] [PubMed] [Google Scholar]
- 4.Hsu CC, Bray MS, Kao WH, Pankow JS, Boerwinkle E, Coresh J. Genetic variation of the renin-angiotensin system and chronic kidney disease progression in black individuals in the atherosclerosis risk in communities study. J Am Soc Nephrol. 2006;17:504–512. doi: 10.1681/ASN.2005050468. [DOI] [PubMed] [Google Scholar]
- 5.Osawa N, Koya D, Araki S, Uzu T, Tsunoda T, Kashiwagi A, Nakamura Y, Maeda S. Combinational effect of genes for the renin-angiotensin system in conferring susceptibility to diabetic nephropathy. J Hum Genet. 2007;52:143–151. doi: 10.1007/s10038-006-0090-5. [DOI] [PubMed] [Google Scholar]
- 6.Pettersson-Fernholm K, Frojdo S, Fagerudd J, Thomas MC, Forsblom C, Wessman M, Groop PH. The AT2 gene may have a gender-specific effect on kidney function and pulse pressure in type I diabetic patients. Kidney Int. 2006;69:1880–1884. doi: 10.1038/sj.ki.5000348. [DOI] [PubMed] [Google Scholar]
- 7.Prasad P, Tiwari AK, Kumar KM, Ammini AC, Gupta A, Gupta R, Sharma AK, Rao AR, Nagendra R, Chandra TS, Tiwari SC, Rastogi P, Gupta BL, Thelma BK. Chronic renal insufficiency among Asian Indians with type 2 diabetes: I. Role of RAAS gene polymorphisms. BMC Med Genet. 2006;7:42. doi: 10.1186/1471-2350-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogus JJ, Moczulski D, Freire MB, Yang Y, Warram JH, Krolewski AS. Diabetic nephropathy is associated with AGT polymorphism T235: results of a family-based study. Hypertension. 1998;31:627–631. doi: 10.1161/01.hyp.31.2.627. [DOI] [PubMed] [Google Scholar]
- 9.Ruggenenti P, Bettinaglio P, Pinares F, Remuzzi G. Angiotensin converting enzyme insertion/deletion polymorphism and renoprotection in diabetic and nondiabetic nephropathies. Clin J Am Soc Nephrol. 2008;3:1511–1525. doi: 10.2215/CJN.04140907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thameem F, Voruganti VS, He X, Nath SD, Blangero J, MacCluer JW, Comuzzie AG, Abboud HE, Arar NH. Genetic variants in the renin-angiotensin system genes are associated with cardiovascular-renal-related risk factors in Mexican Americans. Hum Genet. 2008;124:557–559. doi: 10.1007/s00439-008-0581-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, Greenland P, Jacob DR, Jr., Kronmal R, Liu K, Nelson JC, O'Leary D, Saad MF, Shea S, Szklo M, Tracy RP. Multi-ethnic study of atherosclerosis: objectives and design. Am J Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 12.McGeachie M, Ramoni RL, Mychaleckyj JC, Furie KL, Dreyfuss JM, Liu Y, Herrington D, Guo X, Lima JA, Post W, Rotter JI, Rich S, Sale M, Ramoni MF. Integrative predictive model of coronary artery calcification in atherosclerosis. Circulation. 2009;120:2448–2454. doi: 10.1161/CIRCULATIONAHA.109.865501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kramer H, Jacobs DR, Jr, Bild D, Post W, Saad MF, Detrano R, Tracy R, Cooper R, Liu K. Urine albumin excretion and subclinical cardiovascular disease. The Multi-Ethnic Study of Atherosclerosis. Hypertension. 2005;46:38–43. doi: 10.1161/01.HYP.0000171189.48911.18. [DOI] [PubMed] [Google Scholar]
- 14.Moran A, Katz R, Jenny NS, Astor B, Bluemke DA, Lima JA, Siscovick D, Bertoni AG, Shlipak MG. Left ventricular hypertrophy in mild and moderate reduction in kidney function determined using cardiac magnetic resonance imaging and cystatin C: the multi-ethnic study of atherosclerosis (MESA) Am J Kidney Dis. 2008;52:839–848. doi: 10.1053/j.ajkd.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stevens LA, Coresh J, Schmid CH, Feldman HI, Froissart M, Kusek J, Rossert J, Van Lente F, Bruce RD, 3rd, Zhang YL, Greene T, Levey AS. Estimating GFR using serum cystatin C alone and in combination with serum creatinine: a pooled analysis of 3,418 individuals with CKD. Am J Kidney Dis. 2008;51:395–406. doi: 10.1053/j.ajkd.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattix HJ, Hsu CY, Shaykevich S, Curhan G. Use of the albumin/creatinine ratio to detect microalbuminuria: implications of sex and race. J Am Soc Nephrol. 2002;13:1034–1039. doi: 10.1681/ASN.V1341034. [DOI] [PubMed] [Google Scholar]
- 17.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 19.Jackson JE. A User's Guide to Principal Components. Hoboken: Wiley-Interscience; 2003. [Google Scholar]
- 20.Davison AC, Hinkley DV. Bootstrap Methods and Their Application. Cambridge: Cambridge University Press; 1997. [Google Scholar]
- 21.North BV, Curtis D, Sham PC. A note on the calculation of empirical p values from Monte Carlo procedures. Am J Hum Genet. 2002;71:439–441. doi: 10.1086/341527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32:361–369. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]
- 23.Boright AP, Paterson AD, Mirea L, Bull SB, Mowjoodi A, Scherer SW, Zinman B. Genetic variation at the ACE gene is associated with persistent microalbuminuria and severe nephropathy in type 1 diabetes: the DCCT/EDIC Genetics Study. Diabetes. 2005;54:1238–1244. doi: 10.2337/diabetes.54.4.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevens LA, Levey AS. Current status and future perspectives for CKD testing. Am J Kidney Dis. 2009;53:S17–S26. doi: 10.1053/j.ajkd.2008.07.047. [DOI] [PubMed] [Google Scholar]
- 25.Stevens LA, Coresh J, Greene T, Levey AS. Assessing kidney function – measured and estimated glomerular filtration rate. N Engl J Med. 2006;354:2473–2483. doi: 10.1056/NEJMra054415. [DOI] [PubMed] [Google Scholar]
- 26.K/DOQI clinical practice guidelines on hypertension and antihypertensive agents in chronic kidney disease. Am J Kidney Dis. 2004;43:S1–S290. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Tables