Abstract
Objective
Podosomes, actin-rich structures, contribute to cell motility, matrix remodeling and tissue remodeling. We have shown that G-protein coupled receptor kinase 2-interacting protein 1 (GIT1) co-localizes with podosomes, and is important in podosome formation in endothelial cells. Src stimulates GIT1 tyrosine phosphorylation, which is critical for phospholipase C-γ (PLCγ) activation. Here we identified specific GIT1 tyrosines required for PLCγ activation and podosome formation in vascular smooth muscle cells (VSMC).
Methods and Results
We used phorbol 12, 13-dibutyrate (PDBU) to induce podosomes in A7r5 VSMC. GIT1 co-localized with podosomes and GIT1 knockdown using siRNA significantly reduced podosome formation. PDBU stimulated GIT1 tyrosine phosphorylation. GIT1 tyrosine-phosphorylation was dramatically decreased in SYF−/− cells, and also was reduced by pretreatment with the PKC and Src inhibitors, suggesting that GIT1 phosphorylation was dependent on PKC and Src. By mutation analysis of multiple tyrosines, we found that PDBU specifically increased GIT1-Y392 phosphorylation. Overexpression of GIT1 (Y392F), but not GIT1 (Y321F), decreased PDBU-mediated PLCγ activation and podosome formation without effect on ERK1/2 activation. Additionally, we provide evidence that GIT1 KO VSMC have markedly fewer podosomes upon PDBU treatment compared to WT VSMC. These data show that GIT1 is a key regulator of podosome formation in VSMC.
Conclusions
In conclusion, our data suggest that GIT1-Y392 phosphorylation is critical for PDBU-induced podosome formation by regulating PLCγ activation. We propose that specific signaling modules are assembled in a GIT1 phosphotyrosine-dependent manner as exemplified by PLCγ activation versus ERK1/2 activation.
Introduction
Podosomes are highly dynamic, actin-rich adhesion structures, shown to have an important function in tissue invasion, matrix remodeling and cell migration.1, 2 Podosomes were initially identified in monocyte-like cells, such as macrophages, osteoclasts and dendritic cells.2 Recently, podosomes were characterized in A7r5 smooth muscle cells.3-6 Many molecules have been detected in podosomes, including cytoskeletal components and their regulators, such as tyrosine kinases, serine-threonine kinases, integrins and RhoGTPases.7 Most of these molecules are localized to either the core or ring structure of podosomes. Although podosome architecture seems to be conserved, cell type-dependent differences in composition and regulation exist. A developing concept regarding podosomes and migration is that the formation of podosomes adjacent to membrane protrusions may induce ECM degradation and functionally promote cell migration.7 Phorbol esters such as phorbol dibutyrate (PDBU) are potent activators of PKC and Src signaling, and have been shown to increase podosome formation in A7r5 smooth muscle cells.3-5, 8
G protein coupled receptor kinase 2 interacting protein 1 (GIT1) was originally identified by its binding to GRK2 and effects on β-adrenergic receptor endocytosis.9 GIT1 has 5 functional domains, including a zinc finger domain responsible for ARF-GAP activity, three ankyrin repeats, a Spa2 homology domain (SHD), a synaptic localization domain (SLD), and a conserved carboxyl-terminal region that interacts with paxillin (PBS).10 Through these domains, GIT1 interacts with diverse proteins including ARF6, MEK1, phospholipase C-γ (PLCγ), p21-activated kinase (PAK)-interacting exchange factor (PIX) and paxillin. A major function of GIT1 is to regulate cytoskeletal dynamics during cell spreading and migration by interacting with specific binding partners and targeting them spatially.11, 12 Most importantly, our previous studies demonstrated the important role of GIT1 tyrosine phosphorylation in signal transduction, especially in PLCγ, MEK1-ERK1/2 and FAK activation.11, 13 Specifically, we showed that GIT1 was a substrate for c-Src that was tyrosine phosphorylated in response to angiotensin II (AngII) and epidermal growth factor (EGF).11 Phosphorylated GIT1 is required for the activation and localization of extracellular signal regulated kinases (ERK1/2) in focal adhesions and enhanced cell migration in response to EGF.11 Moreover, GIT1 associated with PLCγ and this interaction is required for PLCγ activation.13 PLCγ has been reported to associate with GIT1-βPIX complex, and tyrosine phosphorylation of the βPIX-GIT1 complex is essential for the interaction with PLCγ, the subsequent activation of PLCγ, and the progression to an elongated cell morphology.14 These data suggest a role for tyrosine phosphorylated GIT1 and PLCγ in cell protrusion and cell motility.
We previously showed that genetic deletion of GIT1 in mice leads to 60 % postnathal lethality due to impaired lung vasculature.15 Mechanistic studies suggested deficiencies in vascular formation specifically in the lung are due to endothelial cell defects in migration and apoptosis. The GIT1 knockout (KO) mouse phenocopies the PLCγ KO mouse suggesting a key role for PLCγ in GIT1 signaling. Recently, we showed that GIT1 and PLCγ co-localized with podosomes and were essential for podosome formation in endothelial cells. 16 In this study, we propose that GIT1 tyrosine (Y) phosphorylation specifically at Y392 plays an important role in podosome formation by regulating PLCγ activation.
Materials and Methods
An expanded Methods section is available in the Online Data Supplement.
Immunofluorescence staining and podosome quantification
A7r5 cells were treated with PDBU, fixed with 4% formaldehyde for 10 min, permeabilized with 0.05% Triton for 5 min, followed by blocking with 10% normal goat serum for 1 hour. Podosome formation was induced in primary VSMC as described 17. Briefly, GIT1 wild-type (WT) and KO VSMC (passages 2-5) were serum starved in 1% FBS overnight, stimulated with PDBU for 4 hrs, permeabilized with 0.1% triton and blocked with 5% BSA for 1 hr. Cells were then incubated with 2.5ug/ml TRITC-labeled phalloidin to stain for F-actin. In A7r5 cells, cells were incubated with GIT1 antibody (Invitrogen, 1:100) overnight followed by Alexa Fluor 488 anti-rabbit IgG (Molecular Probe, 1:1000). The numbers of cells containing podosomes were counted and analyzed by fluorescence microscopy. The podosomes in primary VSMC and A7r5 cells appear as multiple dot-like structures.2, 5 The cells containing five or more dot-like podosome were considered as podosome-positive cells. 100 cells per dish are examined and three dishes were included for each experiments. At least three independent experiments were repeated. The data are expressed as percentage of podosome positive cells.
Results
GIT1 co-localizes with podosomes and mediates PDBU-induced podosome formation
PDBU stimulates podosome formation and increases cell motility by activating Src in A7r5 cells.7, 18, 19. Our lab previously demonstrated that GIT1, a Src substrate, mediated podosome formation in vascular endothelial cells.16 Therefore, we hypothesized that GIT1 co-localized with podosomes and mediated PDBU-induced podosome formation in A7r5 cells. To visualize podosomes, A7r5 cells were stimulated with 1 μM PDBU for 60 minutes and double stained with F-actin (Phalloidin) and GIT1 (Fig 1A) or cortactin (Supplemental Fig 1). In A7r5 cells, podosomes exist as dot-like structures.2, 5 As shown in Fig.1A, under control conditions, there were almost no podosomes in A7r5 cells as assayed by F-actin staining, and GIT1 was localized diffusely in the nucleus and cytosol. In response to PDBU, GIT1 translocated (Fig.1A, panel e) and co-localized with podosomes (Fig.1A, panel f). To provide further evidence that the PDBU induced structures were podosomes, A7r5 cells were double stained with F-actin and cortactin. As shown in Supplemental Fig.1 under control conditions cortactin did not show any podosome like structures (panels a-c). In response to PDBU, podosome like structures appeared that colocalized with both cortactin and F-actin in A7r5 cells (panels d-f). These results indicate that GIT1 localized in podosomes after PDBU stimulation in A7r5 cells.
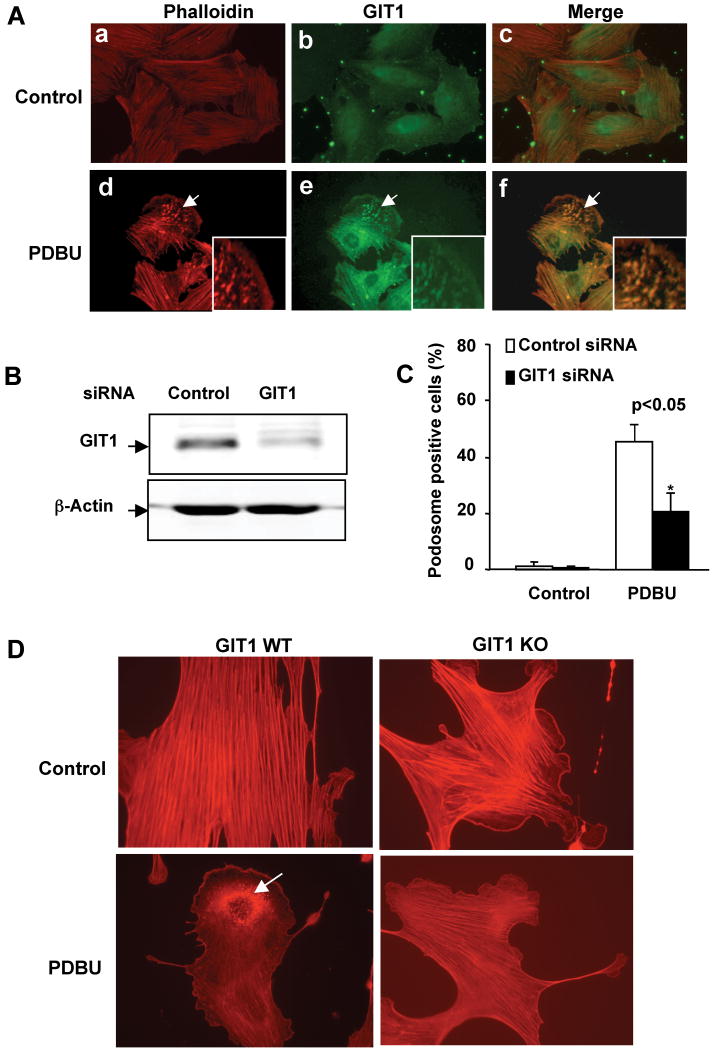
Figure 1. Role of GIT1 in PDBU-induced podosome formation.
(A) Representative immunofluorescent images showing F-actin (Phalloidin) and GIT1 localization in podosomes. A7r5 cells were starved for 6 hours and then treated with or without 1μM PDBU for 60 min. Cells were fixed with 4% formaldehyde and double-stained for either F-actin with TRITC-Phalloidin and GIT1. F-actin is in red and GIT1 is in green. Panels c and f are the merged images. Arrows show podosomes. Insets are enlarged images of podosome-enriched areas. (B) Effects of GIT1 siRNA on GIT1 expression. A7r5 cells were transiently transfected with either 100nM control siRNA or GIT1 siRNA per dish. After 48 hours, cell lysates were immunoblotted with anti-GIT1 antibody. Equal loading was confirmed by β-actin. (C) Effect of GIT1 siRNA on podosome formation. Podosomes were characterized and quantified as described in Methods. The data are expressed as percentage (%) of podosome positive cells (n=100 cells per dish). All values are expressed as mean ± SD of three independent experiments performed in triplicate. A p value < 0.05 was considered to be significant (*). (D) GIT1 WT and KO VSMC were cultured for 48 hrs, serum starved overnight and treated vehicle (DMSO) or PDBU for 4hrs. Cells were fixed and stained with TRITC-Phalloidin to identify podosomes (arrows).
To evaluate the specific role of GIT1 in PDBU-induced podosome formation, we depleted GIT1 with GIT1 specific siRNA in A7r5 cells. After transfection with GIT1 siRNA for 48 hours, GIT1 expression was reduced by 70%, whereas control siRNA had no effect (Fig.1B). To induce podosomes, A7r5 cells were treated with PDBU for 60 minutes. As shown in Fig.1C, the number of podosome positive cells was significantly increased by PDBU from 2±1% to 45±6% in the control siRNA group. In contrast, podosome positive cells were significantly reduced to 21±6% in the GIT1 siRNA treatment group. Furthermore, to confirm that genetic deletion of GIT1 affects podosome formation, we treated VSMC isolated from GIT1 WT and KO mice with PDBU to induce podosomes (Fig 1D). We observed that 8 ±2% of GIT1 WT VSMC formed punctate F-actin rich podosomes. In contrast, only 1% of cells from GIT1 KO VSMC formed podosomes. These results show that GIT1 is an important mediator for PDBU-induced podosome formation.
PDBU-induced GIT1 tyrosine phosphorylation is PKC- and Src-dependent
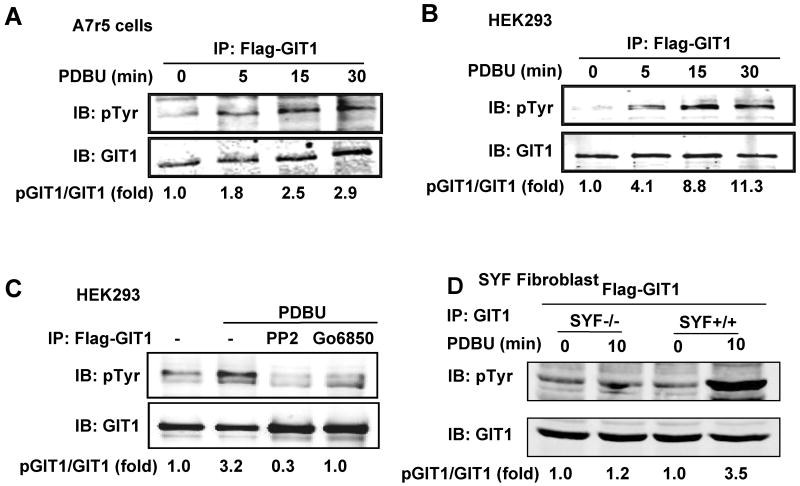
We previously showed that c-Src activation of MEK1 was dependent upon GIT1 tyrosine phosphorylation.11 Therefore we determined whether PDBU–induced podosome formation required GIT1 tyrosine phosphorylation by c-Src. We transfected A7r5 cells with Flag-GIT1 and then stimulated the cells with 1 μM PDBU for various times. As shown in Fig.2A, PDBU increased tyrosine phosphorylation of GIT1 at 5 min that was maintained for 30 min. A similar result was observed in HEK293 cells, which have low endogenous GIT1 (Fig.2B). Pretreatment with PKC inhibitor Go6850 or Src inhibitor PP2 significantly decreased tyrosine phosphorylation of GIT1 (Fig.2C), suggesting that the phosphorylation of GIT1 induced by PDBU is dependent on both PKC and Src. To confirm the role of Src in PDBU-induced GIT1 tyrosine phosphorylation, we used SYF+/+ and SYF-/- fibroblast cell lines. As shown in Fig.2D, PDBU significantly stimulated tyrosine phosphorylation of GIT1 in SYF+/+ cells. In contrast, there was a dramatic decrease in SYF-/- cells in response to PDBU, demonstrating that GIT1 tyrosine phosphorylation is dependent on Src.
Figure 2. Role of PKC and Src in PDBU-induced GIT1 phosphorylation.
PDBU-induced GIT1 tyrosine phosphorylation in A7r5 (A) and in HEK293 (B) cells. Flag-GIT1 (WT) was transfected into A7r5 or HEK293 cells for 24 hours. The cells were starved for 6 hours prior to being treated with or without 1μM PDBU for various times. (C) Effect of PKC and Src inhibition on PDBU-induced GIT1 tyrosine phosphorylation. After transfection with Flag-GIT1 (WT), HEK293 cells were pretreated with 10 μM PP2 (Src inhibitor) or 5 μM Go6850 (PKC inhibitor) for 30 min and then treated with or without 1μM PDBU for 10 min. (D) Role of Src in PDBU-induced GIT1 tyrosine phosphorylation. SYF+/+ and SYF-/- fibroblasts were infected with Flag-GIT1 (WT) adenovirus for 24 hours and then starved for 6 hours followed by treatment with or without 1μM PDBU for 10 min. Lysates were immunoprecipitated with GIT1 antibody and then immunoblotted with 4G10 antibody to detect pGIT1 (top panels). To confirm equal protein immunoprecipitation, the blot was reprobed with GIT1 antibody (lower panels). The blots were analyzed by densitometry using LiCor software. Fold changes normalized to 0 min (A-B) or to the first lane 29 as shown below each blot (n=2-3).
GIT1 Y392 phosphorylation is involved in PDBU-induced podosome formation
A recent study identified eight possible tyrosine phosphorylation sites in GIT1, which are conserved among different species, suggesting the key functions of these phosphorylation sites.20 Among these tyrosines, Y293 and Y392 were suggested to be potential Src phosphorylation sites. Of importance, we showed previously that GIT1 aa 250-420 (encompassing the SHD and part of the SLD domain) was required for GIT1 interaction with PLCγ and PLCγ activation.13 In addition, our previous data demonstrated that Y321, which is also in the SHD domain, was required for EGF and AngII-stimulated Src-dependent ERK1/2 activation.11 Therefore, these three tyrosines (Y293, Y321 and Y392) of GIT1 are likely Src phosphorylation sites and may play important roles in podosome formation.
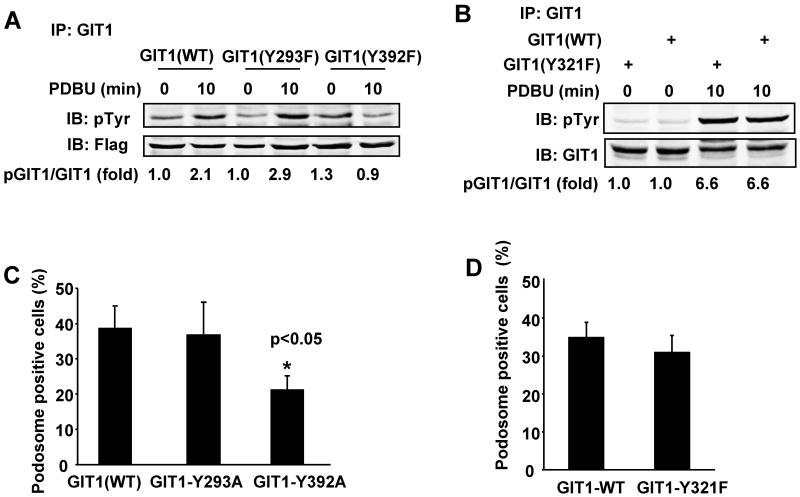
To define the specific roles of Y293, Y392 and Y321, we mutated them individually to phenylalanine, transfected the mutants into HEK293 cells, and stimulated the cells with PDBU. As shown in Fig.3A-B, PDBU stimulated tyrosine phosphorylation of GIT1 (WT), GIT1 (Y293F) and GIT1 (Y321F). However, there was no significant increase in phosphorylation of GIT1 (Y392F).of GIT1 (Y393F). We We next investigated whether mutation of GIT1-Y392 specifically impaired podosome formation. We overexpressed GFP-GIT1 mutants in A7r5 cells and then stimulated the cells with PDBU. After staining with F-actin (red fluorescence), the percentage of podosome positive cells was determined only in cells expressing GFP (green fluorescence). As shown in Supplemental Fig.2, GIT1 (WT), GIT1 (Y293A) and GIT1(Y321F) translocated and co-localized with podosomes. In contrast, GIT1 (Y392A) expressing cells had no podosomes. Quantification of these results showed that only GIT1 (Y392A) significantly reduced PDBU-induced podosome positive cells (from 38±4% to 22±3%; Fig.3C and 3D). These data suggest that tyrosine phosphorylation of GIT1 at Y392, but not at Y293 and Y321, is important for podosome formation.
Figure 3. Role of GIT1-Y293, -Y321, and –Y392 in PDBU-induced GIT1 tyrosine phosphorylation and podosome formation.
Effects of GIT1 tyrosine mutants on PDBU-induced GIT1 tyrosine phosphorylation. HEK293 cells were transfected with Flag-GIT1 (WT), Flag-GIT1 (Y293F), and Flag-GIT1 (Y392F) (A) or Xpress-GIT1 (WT) or Xpress-GIT1 (Y321F) (B) for 24 hours. Cells were starved for 6 hours and then treated with or without 1μM PDBU for 10 min. Cell lysates were immunoprecipitated with Flag antibody or Xpress antibody, then immunoblotted using 4G10 antibody to detect pGIT1 (top panel), and reprobed to detect GIT1 (lower panel). The blots were analyzed by densitometry using LiCor software. Fold changes normalized to the first lane are shown below the blots (n=2-3). Effects of GIT1 tyrosine mutants on PDBU-induced podosome formation. A7r5 cells were transfected with different GIT1 mutants for 24 hours followed by treatment with or without 1μM PDBU for 60 min. Cells were stained for GIT1 and podosomes. Percentage of podosome positive cells (%) were counted and analyzed only in cells expressing GIT1 mutant (with green fluorescence). Quantified data are presented as percentages of podosome positive cells in GIT1 mutant positive cells (n=100 cells per dish). All values are expressed as mean ± SD of three independent experiments performed in triplicate. A p value < 0.05 was considered to be statistically significant (*)
GIT1 Y392 phosphorylation mediates podosome formation by regulating PLCγ activation
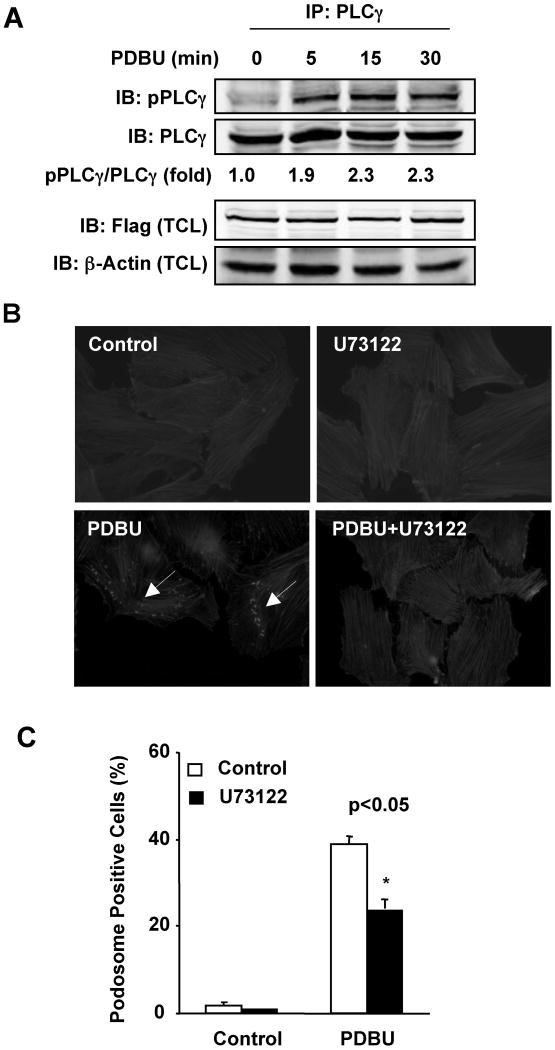
Our previous data showed that GIT1 interacted with PLCγ via the SHD domain and part of the SLD domain (aa 250-420). Both Src-dependent GIT1 phosphorylation and GIT1-PLCγ interaction are required for PLCγ activation.13 To determine the role of GIT1 in PLCγ activation, we transfected GIT1 and PLCγ into HEK293 cells and then stimulated cells with PDBU. As shown in Fig.4A, PDBU significantly increased phosphorylation of PLCγ at 5 min that was maintained for 30 min. To provide further evidence for an essential role of PLCγ in podosome formation in A7r5 cells, we pretreated cells with the PLCγ inhibitor U73122. PDBU-stimulated podosome formation was significantly reduced from 39±1% to 24±2% (Fig.4B-C).
Figure 4. Role of PLCγ in PDBU-induced podosome formation.
(A) PDBU induced PLCγ phosphorylation. Flag-GIT1 and PLCγ were co-transfected into HEK293 cells for 24 hours. Cells were starved for 6 hours and treated with or without 1μM PDBU for the indicated times. Cell lysates were immunoprecipitated with PLCγ antibody and then immunoblotted with pPLCγ (Y783) antibody and reprobed with PLCγ antibody. The total cell lysates (TCL) were detected with Flag antibody to show the Flag-GIT1 expression. Equal loading was confirmed by immunoblotting for β-actin. The blots were analyzed by densitometry using LiCor software. Fold changes normalized to the first lane are shown below the blots (n=2-3). (B and C) Effects of PLCγ inhibitor (U73122) on podosome formation. A7r5 cells were pretreated with 5μM U73122 for 30 min prior to treatment with or without 1μM PDBU for 60 min. Cells were then stained for podosomes (B). Quantified data are presented as percentages of podosome positive cells (n=100 cells per dish) (C). All values are expressed as mean ± SD of three independent experiments performed in triplicate. A p value < 0.05 was considered to be significant (*)
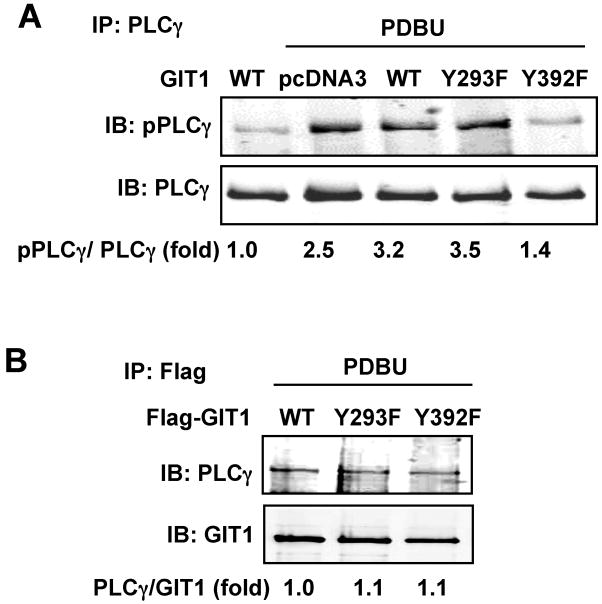
To further investigate the specific role for GIT1 Y392 phosphorylation in PLCγ activation, we transfected HEK293 cells with GIT1 mutants and stimulated the cells with PDBU. As shown in Fig.5A, PDBU significantly stimulated PLCγ activation in cells expressing GIT1 (WT) and GIT1 (Y293F). However, there was no increase in PLCγ phosphorylation in GIT1 (Y392F) overexpressing cells. These data show that only GIT1-Y392 was critical for PLCγ activation in response to PDBU. To investigate whether GIT1-Y392 phosphorylation was required for interaction between GIT1 and PLCγ, we transfected HEK293 cells with GIT1 mutants and PLCγ. As shown in Fig. 5B, GIT1 Y293F and Y392F had no effect on the interaction between GIT1 and PLCγ, suggesting that phosphorylation of GIT1 at Y392 regulates PLCγ activation, but not GIT1-PLCγ interaction. GIT1 Y321F had no effect on either PLCγ activation or on GIT1- PLCγ interaction (Supplemental Fig 3). These data together indicate that PLCγ activation is critical for PDBU-induced podosome formation, and that GIT1-Y392 phosphorylation contributes to podosome formation via PLCγ activation.
Figure 5. Role of GIT1 tyrosine phosphorylation in PLCγ activation and GIT1-PLCγ interaction.
(A) Effects of GIT1 tyrosine mutants on PLCγ activation. HEK293 cells were co-transfected with PLCγ together with pCDNA3 vector, Flag-GIT1 (WT), Flag-GIT1 (Y293F), or Flag-GIT1 (Y392F) (B) for 24 hours and then starved for 6 hours. Cells were treated with or without 1μM PDBU for 10 min and cell lysates were immunoprecipitated with PLCγ antibody and immunoblotted with pPLCγ antibody (top panel). To confirm equal protein immunoprecipitation, the blot was reprobed with PLCγ antibody (lower panel). (B) Effects of GIT1 tyrosine mutants on GIT1-PLCγ interaction. HEK293 cells were co-transfected with PLCγ together with Flag-GIT1 (WT), Flag-GIT1 (Y293F), or Flag-GIT1 (Y392F) for 24 hours and starved for 6 hours. After treatment with 1μM PDBU for 10 min, cell lysates were immunoprecipitated with Flag antibody for GIT1 and then immunoblotted with PLCγ antibody. To confirm equal protein immunoprecipitation, the blot was reprobed with GIT1 antibody. The blots were analyzed by densitometry using LiCor software. Fold changes normalized to the first lane are shown below the blots (n=2-3).
GIT1 tyrosine phosphorylation is not required for PDBU-stimulated ERK1/2 activation
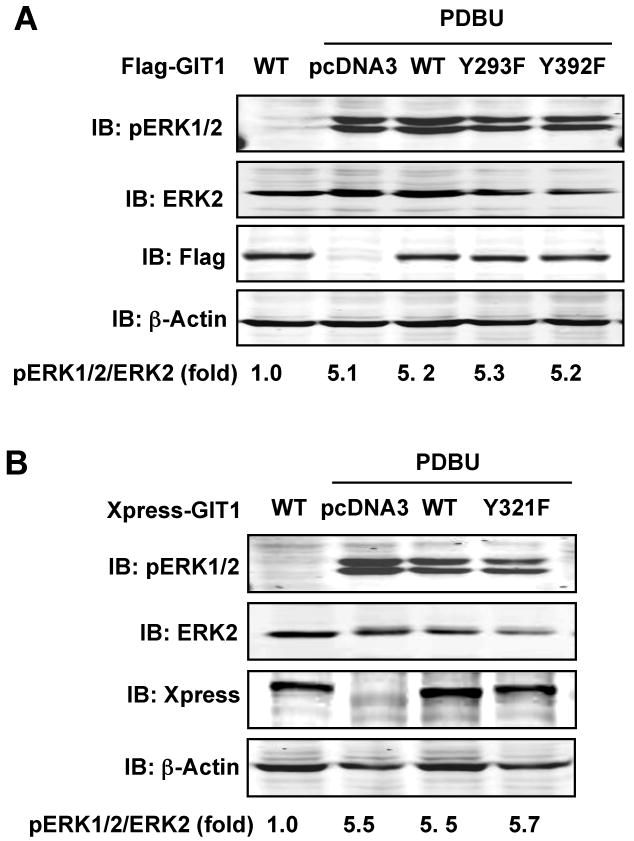
Our previous studies have demonstrated that GIT1 associates with MEK1 and ERK1/2 via the SHD domain, and that Src-induced GIT1-Y321 phosphorylation is required for ERK1/2 activation upon EGF stimulation.11 Therefore, we examined the role of these tyrosine phosphorylation sites in PDBU-induced ERK1/2 activation. The effect of GIT1 mutants on ERK1/2 activation induced by PDBU was studied in HEK293 cells. As shown in Fig.6A and B, PDBU significantly stimulated ERK1/2 phosphorylation. However, there was no significant difference in ERK1/2 activation between GIT1(WT) and pcDNA3 expressing cells after treatment with PDBU, indicating that expression of wild type GIT1 does not enhance PDBU-induced ERK1/2 activation. Consistent with this result, all three GIT1 mutants did not alter ERK1/2 activation induced by PDBU (Fig.6A-B). Together these data suggest that GIT1 does not regulate PDBU-mediated ERK1/2 activation.
Figure 6. Role of GIT1 tyrosine phosphorylation in PDBU-induced ERK1/2 activation.
HEK 293 cells were transfected with (A) pcDNA3, Flag-GIT1 (WT), Flag-GIT1 (Y293F), or Flag-GIT1 (Y392F) or (B) pcDNA3, Xpress-GIT1 (WT), or Xpress-GIT1 (Y321F) for 24 hours and starved for 6 hours. After treatment with or without 1μM PDBU for 10 min, cell lysates were immunoblotted with specific pERK1/2 antibody (upper panels) and reprobed with ERK2 antibody (lower panels). Flag (or Xpress) antibody blot shows the expression of various GIT1 plasmids, and β-actin antibody blot shows equal loading. The blots were analyzed by densitometry using LiCor software. Fold changes normalized to the first lane are shown below the blots (n=2-3).
Discussion
The major finding of this study is that Src-mediated tyrosine phosphorylation of GIT1-Y392 may be critical for podosome formation by stimulating PLCγ activation. Evidence to support this mechanism includes: 1) GIT1 co-localized with podosomes induced by PDBU (Fig.1A). 2) Knockdown of GIT1 with siRNA inhibited PDBU-induced A7r5 smooth muscle cell podosome formation (Fig.1C). 3) PDBU increased GIT1 tyrosine phosphorylation, which was PKC- and Src-dependent (Fig.2). 4) GIT1-Y392 was critical in PDBU-induced podosome formation and PLCγ activation (Figs 3-5). 5) PLCγ was an important mediator for podosome formation induced by PDBU (Fig.4). GIT1-Y392 appears to be the major tyrosine phosphorylation site in response to PDBU stimulation. This is because PDBU-stimulated GIT1 tyrosine phosphorylation was almost completely blocked in the cells overexpressing GIT1-Y392F mutant (Fig.3). Since PDBU-induced GIT1 tyrosine phosphorylation was prevented by Src inhibitor PP2 (Fig.2C), the data suggest that GIT1-Y392 phosphorylation is mediated by Src. The role of GIT1-Y392 in podosome formation was further demonstrated by the findings that (1) PLCγ function was essential for podosome formation; and (2) PDBU-induced PLCγ phosphorylation (activation) and podosome formation were significantly reduced by overexpressing GIT1-Y392 mutants. The mechanism by which GIT1-Y392 mutants are able to inhibit PLCγ activation and podosome formation is very likely due to that GIT1-Y392 phosphorylation is critical for mediating PLCγ activation/podosome formation. GIT1-Y392 mutants cannot be phosphorylated and thus function as dominant negative molecules that inhibit endogenous GIT1 function. However, it should be noted that GIT1-Y392 mutation changes the confirmation of GIT1, which is independent of Y392 phosphorylation. Nevertheless, our findings in this study demonstrate an important role of GIT1 in PLCγ activation and podosome formation and suggest that GIT1-Y392 is a critical site.
We propose that phosphorylated GIT1 at Y392 specifically mediates the assembly of a signaling complex in podosomes. In addition to PLCγ, PIX may a candidate protein in the complex. It has been shown that on a basement membrane-type matrix, PLCγ mediates cell spreading by activating Cdc42 and Rac1 via regulating assembly of a PLCγ-GIT1-PIX complex.14 Since podosome formation is known to be dependent on GIT1, PLCγ, and small GTPases, PLCγ could be an important component in the PLCγ-GIT1-PIX complex and activation of small GTPases. Another likely candidate is calmodulin kinase kinase (CaMKK). For example, it has been shown that spinogenesis is dependent on CaMKK-CaMKI-PIX-Rac1 signaling pathway in a Ca2+-dependent manner.21 In hippocampal neurons, activation of CaMKI phosphorylated PIX at Ser576 and enhanced its GEF activity, resulting in the activation of Rac1, and promoting spines and synapse formation.21 Our lab recently also demonstrated that GIT1 mediated AngII induced PLCγ-Ca2+-CamKII-HDAC5 activation by recruiting these proteins in a complex.22 Thus, it is possible that the GIT1-PLCγ complex activates CaMKK and recruits CaMKI to podosomes, where CaMKI phosphorylates and activates PIX consequently enhancing Rac1 activity and podosome formation. Finally, it was also shown that constitutively active PAK can rescue neuronal spine formation in the presence of CaMKI inhibition or DN-PIX, suggesting that PAK is another candidate mediator in GIT1-PLCγ-CaMKK signaling.22 More importantly, constitutively active PAK is able to induce podosome formation.20, 23. Therefore, these observations collectively suggest that GIT1 may mediate podosome formation via PLCγ-CaMKK-PIX-Rac1-PAK signaling pathway.
Previously we showed that GIT1-Y321 is important for EGF-induced ERK1/2 phosphorylation 11 and localization in focal adhesions.12 In this study we found that GIT1-Y321 does not contribute to PDBU-induced ERK1/2 activation (Fig.6) nor to PDBU-induced podosome formation in A7r5 cells (Supplemental Fig.2). However, we found that GIT1-Y392 plays an important role in PDBU-induced PLCγ activation and podosome formation in A7r5 cells (Figs. 3-5). These results support an important concept that Src-mediated GIT1 phosphorylation at Y392 versus Y321 acts as a switch for activation of PLCγ versus ERK1/2 in smooth muscle cells, which may be mediated by the assembly of distinct phospho-tyrosine specific signaling modules. For example, proteins interacting with GIT1-phospho-Y321 are required for MEK1-ERK1/2 activation in focal adhesions while proteins associating with GIT1-phospho-Y392 are involved in PLCγ activation and regulation of podosome formation. The important role of GIT1-Y392 is further supported by a recent finding that phosphorylation of GIT1-Y392 was required for interaction between Grb4 and GIT1 in response to ephrin B in rat hippocampal neurons.24 Mutation of GIT1-Y392 or disruption of Grb4-GIT1 interaction impaired localization of GIT1 at synapses, impairing spine morphogenesis and synapse formation.24 These data suggest that tyrosine phosphorylation of GIT1, especially Y392, plays a critical role in regulation of neuronal function. The fact that there are multiple functionally different tyrosine phosphorylation sites in GIT1 is consistent with the evolving concept that GIT1 is a multidomain scaffold protein that plays an essential role in multiple cellular processes.25 Our recent publication characterizing the phenotype of the GIT1 global knockout (KO) mouse revealed a critical role for GIT1 in pulmonary vascular development, likely via effects on endothelial cell function.15 The GIT1 KO also showed important alterations in bone and brain function 26. Recently Quintavalle et al provided evidence for podosome formation in vivo and suggested a critical role for these actin rich structures in many physiological and pathophysiological processes 17. Podosomes are likely to be important for VSMC functions such as migration that occurs from the media to form an intima in pathologic conditions such as restenosis, atherosclerosis and carotid intima-media thickening 27. To place the biochemical findings of the present study in the context of tissue-specific alterations in the GIT1 KO mouse, we propose a model that incorporates GIT1 protein-protein interactions, GIT1 subcellular localization, and cell-specific GIT1 functions as shown in Supplemental Fig.4.
While our data suggest an important role for Src-GIT1 in PDBU-induced podosome assembly, there are likely other signaling pathways involved in podosome formation. Examples include the MEK1/ERK1/2/caldesmon signaling cascade shown to regulate PDBU-induced podosome dynamics.4 PDBU is able to activate PKC directly, which may in turn activate MEK1/ERK1/2 signaling pathways via Ras/Raf and promote increase podosome formation, in a Src-independent manner.4 In addition, it has been reported that ERK5 promoted Src-induced podosome formation by increasing the MEF2-dependent expression of RhoGAP and RhoGAP7/DLC-1 in NIH 3T3 cells.28 These data suggest that other proteins may also contribute to podosome formation by acting as Src downstream targets, which are independent of GIT1. These GIT1-independent mechanisms may provide an explanation why inhibiting Src-GIT1-PLCγ signaling is unable to completely block PDBU-induced podosome formation.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Keigi Fujiwara, Dr. Joe Miano, and Dr. Mike Zuscik for helpful suggestions and thoughtful discussion of data.
Sources of Funding: This work was supported by National Institutes of Health grant HL 63462 to Dr. Bradford C. Berk and HL77789 to Dr. Chen Yan.
Footnotes
Disclosures: None
References
- 1.McNiven MA, Baldassarre M, Buccione R. The role of dynamin in the assembly and function of podosomes and invadopodia. Front Biosci. 2004;9:1944–1953. doi: 10.2741/1348. [DOI] [PubMed] [Google Scholar]
- 2.Linder S, Aepfelbacher M. Podosomes: adhesion hot-spots of invasive cells. Trends Cell Biol. 2003;13:376–385. doi: 10.1016/s0962-8924(03)00128-4. [DOI] [PubMed] [Google Scholar]
- 3.Brown D, Dykes A, Black J, Thatcher S, Fultz ME, Wright GL. Differential actin isoform reorganization in the contracting A7r5 cell. Can J Physiol Pharmacol. 2006;84:867–875. doi: 10.1139/y06-027. [DOI] [PubMed] [Google Scholar]
- 4.Gu Z, Kordowska J, Williams GL, Wang CL, Hai CM. Erk1/2 MAPK and caldesmon differentially regulate podosome dynamics in A7r5 vascular smooth muscle cells. Exp Cell Res. 2007;313:849–866. doi: 10.1016/j.yexcr.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hai CM, Hahne P, Harrington EO, Gimona M. Conventional protein kinase C mediates phorbol-dibutyrate-induced cytoskeletal remodeling in a7r5 smooth muscle cells. Exp Cell Res. 2002;280:64–74. doi: 10.1006/excr.2002.5592. [DOI] [PubMed] [Google Scholar]
- 6.Kaverina I, Stradal TE, Gimona M. Podosome formation in cultured A7r5 vascular smooth muscle cells requires Arp2/3-dependent de-novo actin polymerization at discrete microdomains. J Cell Sci. 2003;116:4915–4924. doi: 10.1242/jcs.00818. [DOI] [PubMed] [Google Scholar]
- 7.Linder S. The matrix corroded: podosomes and invadopodia in extracellular matrix degradation. Trends Cell Biol. 2007;17:107–117. doi: 10.1016/j.tcb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Gimona M, Kaverina I, Resch GP, Vignal E, Burgstaller G. Calponin repeats regulate actin filament stability and formation of podosomes in smooth muscle cells. Mol Biol Cell. 2003;14:2482–2491. doi: 10.1091/mbc.E02-11-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Premont RT, Claing A, Vitale N, Freeman JL, Pitcher JA, Patton WA, Moss J, Vaughan M, Lefkowitz RJ. beta2-Adrenergic receptor regulation by GIT1, a G protein-coupled receptor kinase-associated ADP ribosylation factor GTPase-activating protein. Proc Natl Acad Sci U S A. 1998;95:14082–14087. doi: 10.1073/pnas.95.24.14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Natarajan K, Yin G, Berk BC. Scaffolds direct Src-specific signaling in response to angiotensin II: new roles for Cas and GIT1. Mol Pharmacol. 2004;65:822–825. doi: 10.1124/mol.65.4.822. [DOI] [PubMed] [Google Scholar]
- 11.Yin G, Haendeler J, Yan C, Berk BC. GIT1 functions as a scaffold for MEK1-extracellular signal-regulated kinase 1 and 2 activation by angiotensin II and epidermal growth factor. Mol Cell Biol. 2004;24:875–885. doi: 10.1128/MCB.24.2.875-885.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin G, Zheng Q, Yan C, Berk BC. GIT1 is a scaffold for ERK1/2 activation in focal adhesions. J Biol Chem. 2005;280:27705–27712. doi: 10.1074/jbc.M502271200. [DOI] [PubMed] [Google Scholar]
- 13.Haendeler J, Yin G, Hojo Y, Saito Y, Melaragno M, Yan C, Sharma VK, Heller M, Aebersold R, Berk BC. GIT1 mediates Src-dependent activation of phospholipase Cgamma by angiotensin II and epidermal growth factor. J Biol Chem. 2003;278:49936–49944. doi: 10.1074/jbc.M307317200. [DOI] [PubMed] [Google Scholar]
- 14.Jones NP, Katan M. Role of phospholipase Cgamma1 in cell spreading requires association with a beta-Pix/GIT1-containing complex, leading to activation of Cdc42 and Rac1. Mol Cell Biol. 2007;27:5790–5805. doi: 10.1128/MCB.00778-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pang J, Hoefen R, Pryhuber GS, Wang J, Yin G, White RJ, Xu X, O'Dell MR, Mohan A, Michaloski H, Massett MP, Yan C, Berk BC. G-protein-coupled receptor kinase interacting protein-1 is required for pulmonary vascular development. Circulation. 2009;119:1524–1532. doi: 10.1161/CIRCULATIONAHA.108.823997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Taba Y, Pang J, Yin G, Yan C, Berk BC. GIT1 mediates VEGF-induced podosome formation in endothelial cells: critical role for PLCgamma. Arterioscler Thromb Vasc Biol. 2009;29:202–208. doi: 10.1161/ATVBAHA.108.174391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quintavalle M, Elia L, Condorelli G, Courtneidge SA. MicroRNA control of podosome formation in vascular smooth muscle cells in vivo and in vitro. J Cell Biol. 2010;189:13–22. doi: 10.1083/jcb.200912096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreau V, Tatin F, Varon C, Genot E. Actin can reorganize into podosomes in aortic endothelial cells, a process controlled by Cdc42 and RhoA. Mol Cell Biol. 2003;23:6809–6822. doi: 10.1128/MCB.23.19.6809-6822.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreau V, Tatin F, Varon C, Anies G, Savona-Baron C, Genot E. Cdc42-driven podosome formation in endothelial cells. Eur J Cell Biol. 2006;85:319–325. doi: 10.1016/j.ejcb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Webb DJ, Kovalenko M, Whitmore L, Horwitz AF. Phosphorylation of serine 709 in GIT1 regulates protrusive activity in cells. Biochem Biophys Res Commun. 2006;346:1284–1288. doi: 10.1016/j.bbrc.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 21.Saneyoshi T, Wayman G, Fortin D, Davare M, Hoshi N, Nozaki N, Natsume T, Soderling TR. Activity-dependent synaptogenesis: regulation by a CaM-kinase kinase/CaM-kinase I/betaPIX signaling complex. Neuron. 2008;57:94–107. doi: 10.1016/j.neuron.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pang J, Yan C, Natarajan K, Cavet ME, Massett MP, Yin G, Berk BC. GIT1 mediates HDAC5 activation by angiotensin II in vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2008;28:892–898. doi: 10.1161/ATVBAHA.107.161349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nayal A, Webb DJ, Brown CM, Schaefer EM, Vicente-Manzanares M, Horwitz AR. Paxillin phosphorylation at Ser273 localizes a GIT1-PIX-PAK complex and regulates adhesion and protrusion dynamics. J Cell Biol. 2006;173:587–589. doi: 10.1083/jcb.200509075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segura I, Essmann CL, Weinges S, Acker-Palmer A. Grb4 and GIT1 transduce ephrinB reverse signals modulating spine morphogenesis and synapse formation. Nat Neurosci. 2007;10:301–310. doi: 10.1038/nn1858. [DOI] [PubMed] [Google Scholar]
- 25.Hoefen RJ, Berk BC. The multifunctional GIT family of proteins. J Cell Sci. 2006;119:1469–1475. doi: 10.1242/jcs.02925. [DOI] [PubMed] [Google Scholar]
- 26.Menon P, Deane R, Sagare A, Lane SM, Zarcone TJ, O'Dell MR, Yan C, Zlokovic BV, Berk BC. Impaired spine formation and learning in GPCR kinase 2 interacting protein-1 (GIT1) knockout mice. Brain Res. 2010;1317:218–226. doi: 10.1016/j.brainres.2009.11.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lener T, Burgstaller G, Crimaldi L, Lach S, Gimona M. Matrix-degrading podosomes in smooth muscle cells. Eur J Cell Biol. 2006;85:183–189. doi: 10.1016/j.ejcb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 28.Schramp M, Ying O, Kim TY, Martin GS. ERK5 promotes Src-induced podosome formation by limiting Rho activation. J Cell Biol. 2008;181:1195–1210. doi: 10.1083/jcb.200801078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blaukat A, Ivankovic-Dikic I, Gronroos E, Dolfi F, Tokiwa G, Vuori K, Dikic I. Adaptor proteins Grb2 and Crk couple Pyk2 with activation of specific mitogen-activated protein kinase cascades. J Biol Chem. 1999;274:14893–14901. doi: 10.1074/jbc.274.21.14893. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.