Abstract
High density tiling arrays are an effective strategy for genome-wide identification of transcription factor binding regions. Sliding window methods that calculate moving averages of log ratios or t-statistics have been useful for the analysis of tiling array data. Here, we present a method that generalizes the moving average approach to evaluate sliding windows of p-values by using combined p-value statistics. In particular, the combined p-value framework can be useful in situations when taking averages of the corresponding test-statistic for the hypothesis may not be appropriate or when it is difficult to assess the significance of these averages. We exhibit the strengths of the combined p-values methods on Drosophila tiling array data and assess their ability to predict genomic regions enriched for transcription factor binding. The predictions are evaluated based on their proximity to target genes and their enrichment of known transcription factor binding sites. We also present an application for the generalization of the moving average based on integrating two different tiling array experiments.
Keywords: transcription factor, binding sequence, tiling array, combined p-value
1. Introduction
Genomic tiling arrays are an adaptation of microarray technology where probes on the array are not only representative of genes but are designed to span the entire genome at regular intervals (Royce et al., 2006). Applications include the mapping of transcribed sequences and DNA methylation sites (Yazaki et al., 2007). Tiling arrays can also be used to map regions containing sequences that are bound by transcription factors, proteins important for regulating transcription, the first stage of gene expression. Transcription factors interact with their binding sequences in the genome and can either activate or repress the transcription of target genes. The identification of these binding sequences is important for understanding gene expression regulation and tiling arrays offer a high-throughput approach for determining their locations in the genome.
Tiling arrays are often used to probe sequences obtained from chromatin immunoprecipitation (ChIP). In ChIP, DNA fragments binding to the transcription factor of interest are enriched using antibodies specific to that transcription factor. The use of ChIP followed by hybridization to an array is called ChIP-chip. An alternative to the initial ChIP step is the DNA adenine methyltransferase ID (DamID) method (van Steensel et al., 2001). DamID is based on a fusion protein of the transcription factor of interest to the Dam enzyme, which methylates adenines in the sequence GATC along the genome. When the fused transcription factor-Dam protein binds to the respective binding sequence for the transcription factor, the fused Dam enzyme will deposit methylation tags close to the binding sequence. The methylated sequences can then be isolated using methyl sensitive restriction enzymes. DamID is useful when an antibody for the protein of interest is not available to perform ChIP.
For either of the initial procedures, DamID or ChIP, the result of the tiling array experiment is a series of intensity measurements along the genome, typically evenly spaced (see example in Figure 1). Computational methods are available for analyzing this type of tiling array data to predict regions of transcription factor binding (see review by Liu, 2007). Many methods are based on sliding window averages of the intensity values across the genome (Buck et al., 2005, Keleş et al., 2006).
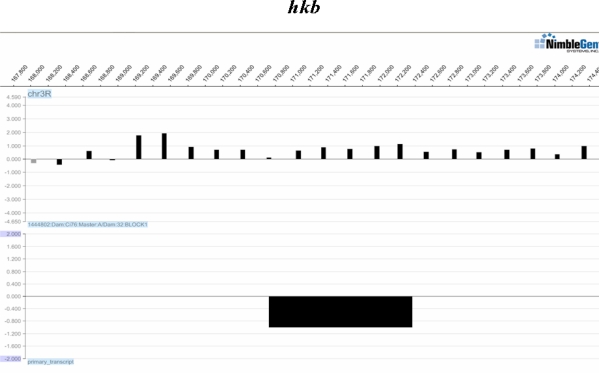
Figure 1:
Example of Data. The chromosome arm and positions are indicated on top. The first row shows log ratio intensity values (y-axis) in a region of the chromosome for one replicate. The second row indicates the location of the hkb gene (y-axis is for illustrative purposes). Plots were produced using the SignalMap software (NimbleGen).
The strengths of the sliding window approaches are that they are simple and fast. However, when initially introduced they did not account for dependencies among neighboring probes, which occur because genomic fragments bound by the transcription factor, obtained by DamID or ChIP, may span multiple probes. The average fragment length is 500–1000 base pairs (bp) for ChIP and 2000–5000 bp for DamID (Buck and Lieb, 2004, van Steensel et al., 2001), while probes may be 25–75 bp long separated by 100–300 bp depending on the platform. Using ChIP-chip data, Bourgon (2006) illustrated that probes more than 1000 bp apart are positively correlated if the average fragment length is 500 bp. An alternative method for tiling array analysis that takes first order dependencies into account is the use of hidden Markov models (Ji and Wong, 2005, Munch et al., 2006). However, computation may become costly if longer range dependencies are taken into account. More recently, incorporating correlations among probes into sliding window averages has been shown to improve performance in the work of Kuan et al. (2008) (see also Bourgon, 2006).
In a typical analysis of tiling arrays, each probe is assessed for positive intensity (or log ratio intensity) to test whether intensity values significantly differ from zero at a location. However, with more complicated experimental designs, other hypotheses may be evaluated using different types of statistics, e.g., an F-statistic when there are multiple factors, or non-parametric test statistics. In these cases, a moving average of the statistic may not be appropriate. Furthermore, the length of the sliding windows that are typically used may not have a large enough sample size for the average to be approximately normally distributed. Therefore, in this work, we develop a generalization of the sliding window average method that allows for the “averaging” of results from a hypothesis test at each probe. Our method is based on “averaging” of p-values instead of test statistics and thus expands the capabilities of current moving average approaches.
Once a p-value resulting from a hypothesis test of interest is determined at each probe, we present a sliding window “average” of the p-values based on the application of combined p-value statistics (Loughin, 2004) that also incorporates correlation adjustments (Bourgon, 2006, Kuan et al., 2008). We refer to our method as a “generalized” moving average and make evaluations on DamID tiling array data for transcription factors where there are target genes and binding site information to confirm predictions of transcription factor binding regions.
The paper is organized as follows. In Section 2, we describe the three benchmark data sets. In Section 3 we describe the Methods, including the method for generating p-values at each probe, the two combined p-value statistics that are applied for the generalized moving average across probes and the three metrics for evaluating predictions. In Section 4, we show two sets of results. The first set evaluates the performance of the generalized moving average compared with other methods, including what we define as the “standard” moving average approach for taking averages of signal intensities, log ratios or t-statistics. To compare the different methods in this application, the results are based on t-statistics or t-test p-values for the standard and generalized moving average methods respectively. The second set of results illustrates an application of the generalized moving average to an analysis question that may not be straightforward for the standard moving average methods. In Section 5, we end with a conclusion and discussion of the methods.
2. Data
2.1. Ci Data Sets
Two data sets are based on a study of DNA binding activity of Cubitus interruptus (Ci), the mediating transcription factor of the Hedgehog (Hh) signaling pathway in Drosophila melanogaster (fruit fly). Two distinct forms of the Ci protein have repressor and activator activities with respect to transcription. In cells which receive the Hh ligand, the full length form of Ci is converted into a transcriptional activator, directly mediating the up-regulation of known Hh targets. This process is required for a variety of developmental events, both in the embryo and larva. In cells that do not receive the Hh ligand, full-length Ci is processed into a repressor form. Since both forms of Ci retain the same DNA binding motif and recognize the same DNA sequence, it is thought that both forms bind the same DNA targets. However, the extent to which both Ci forms bind the same sequences is unknown.
To identify the genomic regions that interact with either the activator or repressor form of Ci, transgenic fly strains were constructed that express a constitutively active form of Ci fused to the DAM enzyme. The DamID technique results in the local methylation of DNA regions that are specific to the activator form of Ci. The methylated genomic fragments were isolated from fly embryos, PCR amplified, and labeled with a fluorescent dye. As a control comparison, methylated DNA from embryos expressing DAM alone were treated in the same manner. For Ci Activator (CiA) and Ci Repressor (CiR) separately, three independent replicates of labeled DamCi/Dam alone samples, which includes one dye-swap Dam/DamCI to account for dye bias, were hybridized to Roche NimbleGen 385K Whole-Genome Tiling Arrays for Drosophila (UCSC DM2 version, FlyBase v4.0). The Nimble-Gen 385,000 feature arrays consist of 60mer oligos spaced approximately 300 bp apart spanning the whole genome. Hybridized arrays were scanned and fluorescent intensity ratios were calculated for the basis of this study (Biehs et al. submitted manuscript).
Note that the Ci transcription factor in general, regardless of form, will be referred to as Ci and that the collection of data for the two forms, CiA and CiR, will be referred to as the Ci data sets. The individual data from one or the other Ci form will be referred by its abbreviation respectively, CiA or CiR.
2.2. Prospero Data Set
A third data set is based on a study of DNA binding activity of Prospero (Pros), a transcription factor that helps mediate the choice between stem cell self-renewal or differentiation in Drosophila melanogaster (Choksi et al., 2006). The DamID technique was also used in this study to identify genomic regions bound by Pros and the tiling array has a similar design to the NimbleGen array used in the Ci study, with details provided in Choksi et al. (2006). The Pros data set was provided by the authors and consists of four replicates, two of which are dye-swaps.
3. Methods
3.1. Pre-processing
All analyses were performed in R 2.7.1 (R Development Core Team, 2005) and Bioconductor 2.2 (Gentleman et al., 2004). R code developed for the methods described in 3.3 is available upon request. The data were normalized, applying the “loess” option for within-array normalization and “Aquantile” option for between-array normalization based on the methods used in Wormald et al. (2006). Normalization results were also inspected visually. At each probe, accounting for the dye swap replicate(s), the log ratios between the target and control sample were calculated using the limma package (Smyth, 2005).
3.2. P-values across Replicates
This work presents an application of existing combined p-value methods for the analysis of tiling array data. The purpose of introducing these methods to tiling array analysis, where moving averages are commonly calculated to identify enrichment regions in the genome, is that they can be used to determine a “moving” average of p-values from any hypothesis test performed at each probe. The combined p-value methods applied and described below are general for any test performed at a probe. In Sections 4.1 and 4.2, the results are based on t-tests. The use of t-tests facilitates comparisons with other methods that can be applied using the t-statistic, the test statistic of the t-test. Then, in Section 4.3, we show results from a more general application.
3.2.1. T-test Application
For the results in Sections 4.1 and 4.2, we are interested in testing the null hypothesis that the mean log ratio intensity is equal to 0, H0i: μi = 0 for each probe i. Because only positive intensity signals are indicative of enriched binding sequences, we only consider the one-sided alternative HAi: μi > 0 and use a one-sided one-sample t-test. However, tiling array experiments may have small sample sizes (e.g., n = 3 in the Ci data set) and estimates of the standard error in the t-statistic denominator can be unstable. Several modifications of the t-statistic have been suggested (Smyth, 2005, Tusher et al., 2001) that pool information from all genes to obtain more stables variance estimates. We applied an empirical Bayes procedure (Smyth, 2005), which is available in the limma package in R. First, a linear model for the replicates was fit for each probe i with the lmFit function and then moderated t-statistics t̃i were returned for each probe using the eBayes function.
Under the null hypothesis, assuming a linear model fit, independence between probes, specified prior distributions and approximate normality of the estimators, the moderated t-statistic follows a t-distribution with augmented degrees of freedom (Smyth, 2005). These assumptions are often violated in microarray and tiling array experiments, especially with small sample size. We found evidence of this as well, since the calculated moderated t-statistic values are more extreme than expected (see example in Figure S1). Therefore, the moderated t-statistics t̃ were further normalized using , where t̃* is the mode of the distribution of t̃ and sd(t̃neg) is the standard deviation of a symmetrical distribution based on the negative values, t̃neg, of t̃. The mode and sd were both calculated after removing the most extreme 5% probes. The mode is used because the distribution of t̃ tends to be asymmetrical around zero and the standard deviation is calculated based on the negative values because they are considered to be background (Buck and Lieb, 2004). This procedure follows techniques used in other tiling array analysis methods (Buck and Lieb, 2004, Kuan et al., 2008) and the effect is illustrated in Figure S1, where now the t-statistics more closely follow the expected distribution, but there are still more extreme positive values indicative of binding regions. One-sided test p-values, pi, were then determined by the moderated t-statistic and augmented degrees of freedom returned by the eBayes function.
3.2.2. New Application
In Section 4.3, we show an example where a t-statistic or corresponding p-value of a t-test is not applicable for the analysis. To predict binding enrichment regions for CiA-CiR, we can perform a Union-Intersection Test (Casella and Berger, 2002), and at each probe i (where k = 1, 2). A p-value for this test is determined by the Stouffer-Lipták Test (without adjustments) for each probe (see Section 3.3 below). This test was selected over the Fisher’s Combined Probability Test, which puts relatively more emphasis on small p-values (Loughin, 2004) and may be problematic here since the enrichment patterns between the data sets vary so widely. Now at each probe, a p-value has been determined for the test of interest, and we can apply the generalized moving average methods described below in Section 3.3.
3.3. P-values across Probes - Combined P-value Approach
Because transcription factor bound fragments are typically longer than array probes, it is of interest to find a genomic region spanning multiple probes with high signal intensity called an enrichment region (ER). Several authors have used a sliding window procedure to find ERs, where an average intensity value is calculated for a series of consecutive probes. We have adapted the sliding window procedure so that consecutive p-values across the region are combined rather than working with a test statistic such as the average intensity across the region. In particular, for each probe, a window size of w adjacent probes is considered on each side of the probe. A combined p-value is then based on the l = 2 * w + 1 p-values in the w-neighborhood for that probe. We discuss the selection of w below.
There are l tests in the w-neighborhood and the combined null hypothesis H0 is that each of the i = 1,…, l independent null hypotheses H0i is true (Loughin, 2004). The combined alternative HA is that at least one of the null hypotheses H0i is false. A combined p-value test statistic is then calculated for each probe i based on the l p-values. This test statistic can be used to test H0 vs HA (i.e., the mean intensity is equal to zero for all probes in the window versus at least one probe in the window has mean intensity greater than zero). Significant probes according to the combined p-values are predicted to be in ERs. Following a review of other applications of combined p-value methods, we detail several different combined p-value test-statistics CP and variations of these test statistics that account for dependent hypotheses.
3.3.1. Use of Combined P-values
Combined p-value methods are commonly used for meta-analysis and have a long history (see reviews in Folks, 1984, Hedges and Olkin, 1985 and Loughin, 2004), with Fisher’s Combined Probability Test dating back to 1932. In genomics, combined p-values are often used to integrate data in a meta-analysis of different studies (e.g., in Rhodes et al., 2002). But there have also been applications of the combined p-value methods within a study. For example, Zaykin et al. (2002) combine p-values for neighboring genomic markers in a genetic association study using the dependence correction described in 3.3.3 and a truncated p-value product method. In the area of microarray analysis, Affymetrix probe level p-values were combined to obtain probe set level p-values (Hess and Iyer, 2007). For tiling arrays, Ghosh et al. (2006) calculate Wilcoxon signed-rank test p-values across probes and then combine the p-values across multiple replicates. In this work, we perform tests using the replicates and then combine the p-values across probes. Below, we describe the combined p-value methods that are applied in this work.
3.3.2. Independent Tests
Under the null hypothesis, the p-value pi for a test-statistic with a continuous null distribution is uniformly distributed in the interval [0,1]. A general framework for combining p-values based on this feature are quantile combination methods described in Loughin (2004). In this framework, a parametric cumulative distribution function F is chosen and the p-values are transformed into quantiles according to qi = F−1 (pi), for i = 1,…,l. The combined test statistic is a sum of independent and identically distributed random variables qi each of which follows the corresponding probability density function for F. This can be used to obtain the p-value for CP using the additivity property of independent and identically distributed random variables. Below, two common combined p-value test-statistics are described that fall under this framework.
-
Fisher’s Combined Probability Test
In Fisher’s Combined Probability Test (Fisher, 1932), F is selected as the cumulative χ2 distribution with 2 degrees of freedom. Therefore, and qi = F−1(pi) = −2ln(1 – pi) and each qi follows the probability density function for a χ2 distribution with 2 degrees of freedom. The combined p-value test statistic follows a χ2 distribution with 2l degrees of freedom due to the additivity property of independent χ2. The combined p-value is p*= F*(CP), where F* is the cumulative distribution function for the χ2 distribution with 2l degrees of freedom. These results also follow directly from the fact that -2 times the logarithm of a uniformly distributed random variable is χ2 with 2 degrees of freedom. Alternatively, note that if , then p* = 1 – F*(CP).
-
Stouffer-Lipták Test
In the Stouffer-Lipták test (Lipták, 1958, Stouffer et al., 1949), F is selected as the cumulative standard normal distribution N(0,1). Therefore, F(x) =Π(x), where Π is the cumulative distribution function of the standard normal, and qi = Π −1 (pi). Each qi follows the probability density function of a standard normal and using the additivity property of independent random variables, the combined p-value test statistic follows a standard normal distribution. The combined p-value is p* = Φ(CP). Note that if pi is based on a t-test, then for large sample sizes when the distribution of the t-statistic is similar to the normal distribution, the Stouffer-Lipták test will be equivalent to the “standard” moving average approach where t-statistics are averaged.
3.3.3. Dependent Tests
Due to the dependencies in intensity values for neighboring probes, we considered variations of the two combined p-value test statistics that account for correlations among probes as introduced first in Kuan et al. (2008). Since the correlations are positive, if adjustments are not made, the test statistics tend to be inflated and therefore statistical significance is exaggerated.
-
Dependence Adjustment to Fisher’s Combined Probability Test
When the pi’s are dependent, the distribution of Fisher’s Combined Probability test statistic CP can be approximated by a scaled χ2 distribution, , with scaling factor c and degrees of freedom f, such that (Brown, 1975). Equating E[CP] with and Var(CP) with , we can solve for c and f and obtain
These terms depend on the mean and variance for CP which are(1)
and(2)
since .(3) Exact computation of the covariance terms in Var(CP) requires numerical integration but Kost and McDermott (2002) provide approximations by fitting a polynomial regression to the true values using a grid approach ranging values for the degrees of freedom (9 ≤ v ≤ 125) and the correlations −0.98 ≤ ρ ≤ 0.98). Based on their approximations, when the p-values pi are determined from t-statistics with v degrees of freedom and where the correlations, ρjk between probes j and k are known, then(4) This approximation can be used to calculate Var(CP), which is then used to calculate c and f. The combined p-value for Cp is then determined by using the approximating distribution . As stated above, the approximations are based on the assumption that the p-values are determined from t-statistics with v degrees of freedom. In Sections 4.1 and 4.2 we present results based on applying this correction to p-values from moderated t-statistics for the three data sets where the degrees of freedom v vary from 7.53 for CiR, 8.69 for CiA to 9.39 for Pros, which are slightly below or within the grid values for the approximations.
-
Dependence Adjustment to Stouffer-Lipták Test
This dependence adjustment is based on transforming the correlated quantiles Q = {qi} into independent quantiles Q* as in Zaykin et al. (2002), and then applying the Stouffer-Lipták test on Q*. To perform this transformation, the pi’s are assumed to be correlated according to a non-degenerate correlation matrix ∑. Assuming that ∑ is positive definite, the Cholesky factor C exists such that ∑ = CC′. By applying the transformation Q* = C−1 Q, the ’s are now independent and follow a standard normal distribution (Zaykin et al., 2002). The Stouffer-Lipták test can then be applied to the ’s. This method is used for Section 4.3 since this adjustment, unlike the adjustment for the Fisher’s Combined Probability test, does not depend on the assumption that the p-values are determined from t-statistics.
3.3.4. Auto Correlation
Both dependence adjustments rely on correlations between probes. The correlation of the moderated t-statistic between neighboring probes j and k is determined using the entire data. However, since regions of interest are contained within the entire chromosome, to estimate the correlation between probes, we use an estimation procedure robust to outliers in R available in the robust–ts package (Spangl et al., 2009), which is based on the work in Spangl (2008). The auto correlation is calculated on each chromosome arm using the “ACF” function in robust-ts with the Spearman’s rank option, for each lag x = 1,2,…. Probes were removed from the calculation that neighbored gaps (defined as more than 700 base pairs between probes). For probes k > j, the correlation is estimated as ρjk = ACF(k – j). We use window sizes of 3–8 which would correspond to a maximum lag of 16. Since spacings between probes are roughly 300bp, these window sizes correspond to total lengths of ∼ 1800–4800 (2*w*300), which is the range of average fragment lengths in DamID experiments.
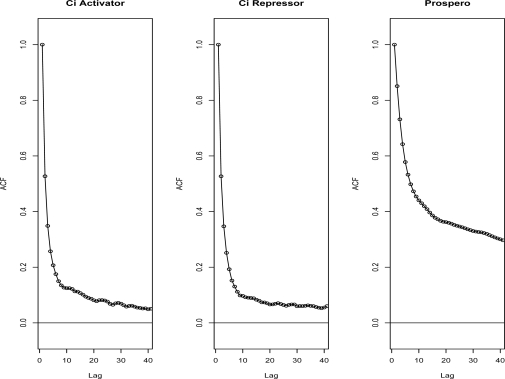
Figure 2 shows that the auto correlation drops under .2 after a lag of 5 probes for chromosome arm 2R for the Ci data sets and under .4 after a lag 13 for the Pros data set. Results are similar for other chromosomes arms (data not shown). Even with the robust alternative, all data sets had unusually high correlation (> 0) at far lags, especially for the Pros data set, suggestive of a long-term memory process. These may be a feature of DamID data sets where the fragment lengths tend to be longer. To use the auto correlation to estimate the correlation between probes, we are assuming that the probes are regularly spaced. Although there are exceptions, most of the probes are evenly spaced approximately 300 bp from each other: 81% of the probes are spaced within 350 bp of the neighboring probes and 99% are spaced within 600 bp. In the few cases where there are large gaps between probes (> 700 bp) a combined p-value was not calculated for the probes at the border of the gaps or probes within w probes of the gaps.
Figure 2:
Auto Correlation for Chromosome Arm 2R in the Three Data Sets. The y-axis is the robust estimate of the auto correlation described in Methods. The x-axis is the lag measured in number of probes
For chromosome arm 2L, which has overall length ∼ 22 megabases, there is a ∼ 1.5 megabase stretch of the chromosome arm with probes every ∼ 100 bp in addition to those that are every ∼ 300 bp. In this overlapping region, the lag-3 correlation of the 100 bp density probes is similar to the lag-1 correlation of the 300 bp density probes (data not shown). Since the locations of the 100 bp spaced probes fall within 1–2 bp of the 300 bp spaced probes, overlapping probes are averaged to obtain one set of probe p-values that are regularly spaced 100 bp apart in this region. The auto correlation is calculated separately for this region to determine the combined p-values. The rest of chromosome arm 2L, which has ∼ 300 bp spacing between probes, is analyzed in the same manner as the other chromosome arms. Finally, the combined p-value methods assume stationarity, i.e., the same auto correlation is used across the entire chromosome. We examined this assumption by dividing the chromosomes into blocks and calculating the auto correlation for each block. Except for the blocks at chromosome ends, the auto correlation appeared to be consistent (data not shown).
3.4. Determining Enrichment Regions
Each probe is associated with a combined p-value using one of the methods described above. P-values were corrected for false discovery rate (FDR) control using the Benjamini & Hochberg procedure (Benjamini and Hochberg, 1995). Then, ERs were defined by scanning probes in order of the genome. If a probe had adjusted p-values below some set cutoff a new ER was formed. The next probe passing the cutoff is then evaluated to see if it is within w probes and 700 bp of the last probe in an ER. If so, it is added to that ER, otherwise a new ER is designated. This procedure is continued in a step-wise fashion until the last probe on the chromosome arm is evaluated. Several different FDR cutoffs were selected to construct ERs in Section 4.1. Alternatively, results are also presented where a set number of top probes ranked by a specific procedure are used to construct ERs in Section 4.2. Note that the FDR cutoffs are used to indicate false discovery control at the probe-level and not at the level of ERs.
3.5. Comparison with Other Methods
We compare the generalized moving approach, based on combined p-values, with other existing methods for tiling array analysis. First, comparisons are made with a “standard” moving average approach based on taking averages of signal intensities, log ratios or t-statistics. For this purpose, we use the CMARRT (Correlation, Moving Average, Robust and Rapid method on Tiling array) software based on a method that also incorporates correlations among probes in the moving average. In CMARRT, a Gaussian approximation is used to determine a p-value for the moving average statistic where the variance includes covariance terms determined from the auto correlation function. The R package for CMARRT (Kuan et al., 2008) was run with the window.opt option set to fixed.probe and the frag.length option set to 2000, 2600, 3200, 3800, 4400 and 5000, which corresponds approximately to the window sizes described above of 3–8, respectively, assuming probes are spaced ∼ 300 bp apart. As with the combined p-value methods, the input for CMARRT was the moderated t-statistic described in Section 3.2, which was also suggested by the CMARRT manual for two-channel arrays such as NimbleGen. One line of the original code of the ma.stat function was edited to accommodate the larger ∼300 bp spacing between probes in the NimbleGen arrays. CMARRT was also run with the independent option to get results based on a simple moving average without correlation adjustment. The ER definition described above was applied to the correlation and independent based p-values for probes returned by CMARRT in two ways. First, only FDR adjusted probes that survived a specified FDR control cutoff were used for the results in Section 4.1. Second, only FDR adjusted probes that were in a ranked list of a set number of probes were used for the results in Section 4.2.
Another existing software for tiling array analysis is TileMap, where a first order hidden Markov model can be used to combine neighboring probes and identify regions that have hybridization patterns of interest. As with the other methods, input for TileMap was the moderated t-statistic for each data set. We ran the HMM option with Expected hybridization length set to 10 (for a typical hybridization region of 3000) and Maximal gap allowed set to 700 to correspond to settings used in the other methods or to settings that are appropriate for the DamID data. For each probe, the HMM option returns posterior probabilities that a probe is in a region of interest. We used the direct posterior probability approach in Newton et al. (2004) to control the FDR. The ER definition described above was applied to probes in two ways as with CMARRT. Note that the HMM option does not have a window option, so when TileMap results are plotted with the other methods for different window sizes, the same results are repeated. TileMap only takes first order dependencies into account and does not incorporate longer range correlations as do the various dependence adjusted procedures. In Figures 3 and 4, we also included the TileMap ER predictions directly based on using the HMM option. Since there is no FDR cutoff in this case, the x-axis locations of these results are for illustrative purposes only.
Figure 3:
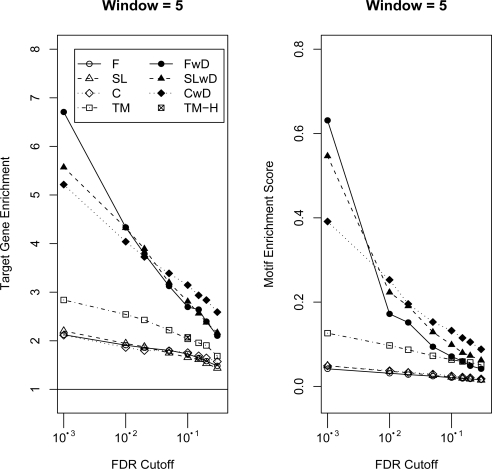
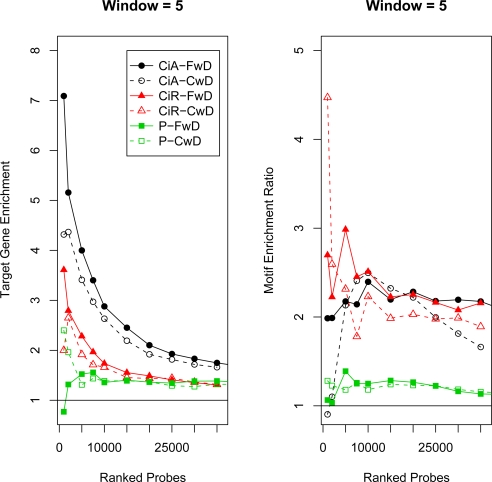
Target Gene and Motif Enrichment. The x-axis corresponds to probe-level FDR cutoffs for determining ERs. On the left, the y-axis corresponds to the “target gene enrichment ratio” (see Methods). Values greater than one, indicated by the line, correspond to relatively more target genes predicted than expected based on their frequency in the genome. On the right, the y-axis corresponds to the “motif enrichment score”, which is the motif enrichment ratio corrected for overall ER lengths (see Methods). Larger values correspond to relatively more motifs in the ERs than expected based on the frequency of the genome. The results are displayed for four combined p-value statistics, Fisher’s Combined Probability Test (F), Fisher’s Combined Probability Test with Dependence (FwD), Stouffer-Lipták Test (SL), Stouffer-Lipták Test with Dependence (SLwD), two moving average methods CMARRT (C), CMARRT with Dependence (CwD) and TileMap (TM) using FDR cutoffs or the HMM prediction method (TM-H). See Figures S2 and S3 for all window sizes.
Figure 4:
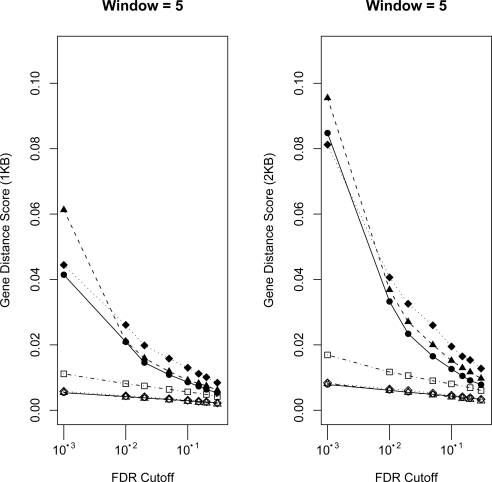
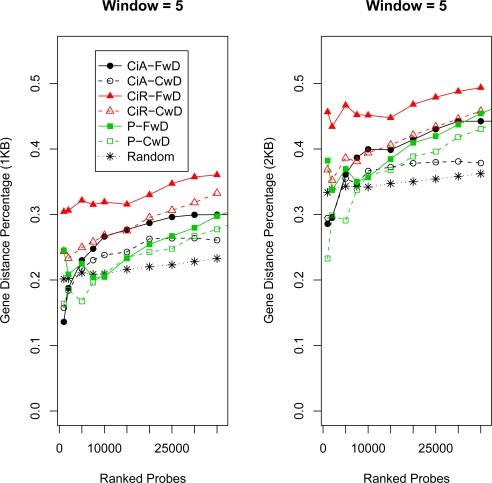
Gene Distance Score. The x-axis corresponds to probe-level FDR cutoffs for determining ERs. The y-axis corresponds to the “gene distance score” corrected for overall ER lengths (see Methods). Larger values correspond to relatively more ERs close to genes. Distances less than or equal to 1KB and 2KB are used. See Figure 3 for details on legend and Figure S4 and S5 for all window sizes.
3.6. Evaluation
All methods were applied to the CiA, CiR and Pros tiling array data described above. For both transcription factors a consensus motif has been identified and target genes have been predicted using gene expression data. Taking advantage of these two sources of external information, we have used three methods to evaluate the methods of ER prediction.
3.6.1. Target Gene Enrichment
Using gene locations from the v4.0 FlyBase collection (Wilson et al., 2008), a list of genes associated with an ER were determined by extracting the closest upstream and downstream gene to the ER and any genes that overlap an ER. This procedure resulted in at least two genes predicted for each ER. Genes appearing multiple times for different ERs were only counted once. The collection of all genes predicted to be near or at ER, called “ER genes”, were then compared to a set of genes determined by a series of gene expression experiments to be likely targets of Ci or Pros called “target genes”.
Ci is the mediating transcription factor of the Hedgehog signaling pathway. Target genes were identified by having to show a median gene expression fold induction of ∼ 1.4 in genetic backgrounds where Hedgehog signaling was retained compared to a situation where Hedgehog signaling was absent (Biehs et al., submitted manuscript). We only examined the 199 of 230 target genes that also had annotation in the v4.0 FlyBase collection. The Pros target genes were obtained from the list of the authors’ differentially expressed genes (Choksi et al., 2006) that had at least log fold 2 change (n=214). Of these, 201 could be mapped to annotations in the v4.0 FlyBase collection.
The overlap of ER genes with the predicted Ci and Pros target genes were evaluated using the “target gene enrichment ratio”, which is equal to the percentage of target genes that intersect the ER genes divided by percentage of all genes that are ER genes. For example using Ci, if one set of ERs has 2695 neighboring ER genes (∼ 20% of the 13472 total genes), which intersect with 60 target genes, (∼ 30% of the 199 target genes), then target gene enrichment is .
Gene Ontology (GO) analysis for Table S1 was performed using the GOstat software (Beissbarth and Speed, 2004) with default options and “False Discovery Rate correction (Benjamini)” for multiple testing correction.
3.6.2. Motif Enrichment
A consensus motif for Ci “tgggtggtc” has been identified both by its occurrence in known Ci enhancers (Alexandre et al., 1996, Kinsler and Vogelstein, 1990) and by transcription-factor binding affinity experiments (Hallikas et al., 2006). A consensus motif for Pros “taagacg” is reported in Choksi et al. (2006). We searched for occurrences of the motifs along with its reverse complement in the set of ER sequences. This observed number was compared to the expected number, which is the frequency of occurrences of the motif in the entire genome multiplied by the total length of the ER sequences,
| (5) |
The “motif enrichment ratio” is the ratio of observed versus expected counts.
Motif enrichment is sensitive to the lengths and number of ERs. Especially for the longer Ci consensus, there may be few or no occurrences if the total length of predicted ERs are short. In Figure 3, where the total ER lengths for each predicted method may be different and may bias the results, the motif enrichment value is divided by the total length of the ER regions for a set of predictions. This number is then multiplied by 105 for plotting purposes and is referred to as the “motif enrichment score.”
3.6.3. Distance to Gene
For each ER, the shortest distance to a gene was determined by comparing the ER locations with all transcription start positions from the v4.0 FlyBase collection of genes. Then, different distance cutoffs were used to find the “gene distance percentage”, which is the percentage of ERs whose closest gene, upstream or downstream, is less than or equal to a defined distance (e.g., 1KB and 2KB).
This metric is also sensitive to the lengths and number of ERs. For a large set of randomly selected ERs, this metric increases because the fly genome is relatively gene dense (median distance between transcription start sites is 3500–4500bp depending on the chromosome arm) and by chance ERs will be selected that are close to genes (see Results). Therefore, as with the motif enrichment score, the percentage is divided by the total length of the ER regions for a set of predictions. This number, the “gene distance score” is then multiplied by 105 for plotting purposes in Figure 4. To evaluate the locations of random ERs to genes, ERs were constructed from a set number of random probes selected over all chromosomes. This was repeated 10 times and for each set of 10 ERs, based on a set number of probes, the average percentage of ERs within a distance to a transcription start were tallied and displayed in Figures 6, S8 and S9.
Figure 6:
Gene Distance Percentage for Top Predictions. The x-axis corresponds to probe-level FDR cutoffs for determining ERs. The y-axis corresponds to the “gene distance percentage” (see Methods). Larger values correspond to relatively more ERs close to genes. Distances less than or equal to 1KB and 2KB are used. See Figure 5 for details and Figure S8 and S9 for all window sizes.
4. Results
We report the evaluation of different ER prediction methods based on independent sources of target gene and motif data (see Methods). Although in theory, the generalized moving average approach based on combined p-values is designed for any hypothesis test, to facilitate comparison with existing methods, in Sections 4.1 and 4.2, we apply the method using p-values based on t-tests. Then, we run other methods such as CMARRT and TileMap using t-statistics, the test statistic for the t-test. The purpose of this comparison is to observe whether any performance is lost by using a generalized moving average based on p-values instead of the test statistics themselves. Then, in Section 4.3, we present an example of an application that can be more difficult to assess using other methods, but fits into the generalized moving average framework. We use three metrics to evaluate predicted sets of ERs: by target gene enrichment, motif enrichment and proportion within a set distance to a gene. For all three metrics, larger values are more favorable.
4.1. Evaluation of Moving Average Methods and Effect of Dependence Adjustment
First, the generalized moving average method, based on combining p-values from t-tests, is compared to an existing or “standard” moving average approach, based on averaging t-statistics. In particular, two different options of the combined p-value method, Fisher’s Combined Probability Test (F) or Stouffer-Lipták Test (SL) are compared to the moving average procedure of CMARRT (C). Second, the effect of incorporating correlations among probes is evaluated by comparing the different moving average procedures with or without the dependence adjustments and by evaluating a third procedure, TileMap (see Methods). All results in this section are based on the Ci Activator data set.
Figure 3 and Figure S2 show the target gene enrichment value for the different combinations of tests, window size and FDR cutoffs for determining ERs. Each FDR cutoff (at the probe-level) on the x-axis determines a set of probes that are used to construct ERs. For all tests, the target gene enrichment increases as the FDR cutoff becomes smaller, indicating that as the threshold for significant probes becomes more stringent, relatively more target genes are included in the ER gene set than expected by chance. As the FDR cutoff becomes more stringent, the dependent tests consistently have higher enrichment values than the independent tests for all window sizes and for both the combined p-value and moving average methods. By not accounting for the dependence, p-values are much smaller and inflate the significance of a probe. For example, using window size 4 a FDR cutoff of 10−12 for Fisher’s Combined Probability Test is necessary to achieve a similar size of predicted ERs as a FDR cutoff of 10−3 for Fisher’s Combined Probability Test with Dependence (FwD).
The maximum value of target gene enrichment, ∼ 7.2, occurs for window size 7 and FwD. In general, there is a slight advantage for FwD over Stouffer-Lipták Test with Dependence (SLwD) and both have higher target gene enrichment values than the t-statistic moving average method, CMARRT, with dependence (CwD) except for large FDR cutoffs. The three methods without dependence adjustments perform very similarly (F, SL and C) and achieve a maximum value of target gene enrichment of ∼ 2.0. They appear to be less sensitive to window size than the methods with dependence. For comparison, TileMap (TM) was also applied and performs slightly better than the unadjusted versions, but not as well as the adjusted methods.
Figure 3 and Figure S3 show the enrichment of the Ci transcription factor binding site motif (see Methods) for the different combinations of tests, window size and probe-level FDR cutoffs for determining ERs. Again, the unadjusted methods perform more poorly compared to the dependence adjusted methods. In general, the unadjusted methods identify many and long ERs; so although their motif enrichment ratio may be high, after adjusting for the overall length of the ERs, they have relatively worse motif enrichment score than the adjusted methods. Within the dependence adjusted methods, depending on the window size, one of the combined p-value methods (FwD or SLwD) performs slightly better than the moving average approach (CwD) for smaller window sizes and FDR cutoffs. For larger window sizes, all three methods perform similarly. TileMap again performs slightly better than the unadjusted methods, but not as well as the adjusted methods.
Figure 4 and Figures S4 and S5 show the enrichment of ERs close to transcription start sites. These results are consistent with the previous metrics in that the adjusted methods tend to predict ERs closer to genes, after correcting for overall ER length, compared to the unadjusted methods and TileMap and that there is also some advantage for SLwD for smaller windows and FDR cutoffs, but this diminishes for larger windows and FDR cutoffs.
4.2. Evaluation of Additional Data Sets
The generalized and existing moving average methods that incorporate correlations among probes appear to make the best ER predictions using the three evaluation metrics on the CiA data set. To explore their performance on two additional data sets, these methods were also applied to data sets based on Ci Repressor (CiR) and Prospero (Pros) (see Methods). However, because these are more challenging data sets, there were very few predictions. For example, at window size 3 and relatively relaxed probe-level FDR cutoff of 0.10, only 7 ERs were predicted for a total of 9113 bases using CwD and none using FwD. This may be due to relatively smaller enrichment signal in the genome for these data sets or other factors that are data set specific (e.g., Pros has relatively high auto correlation values compared to the Ci data sets, see Figure 2).
Therefore, for these two data sets, we took a set number of top probes, ranked by the respective method, and constructed ERs based on those probes. These ERs were compared using the previously described evaluation criteria. This was also repeated on the original Ci activator (CiA) data set. In addition to exploring the behavior of the methods on the noisier data sets, the advantage of this comparison is that because the same number of probes are being used, the total ER lengths should roughly be the same. For example, for window size 3, using the top 1000 probes, FwD has 240 ERs (251234bp) while CwD has 215 ERs (229699bp), which are roughly the same magnitude. Therefore, ER total length corrections are not necessary as in Figures 3 and 4. We compared one of the generalized moving average methods based on combined p-values with dependence (FwD) with the moving average method with dependence (CwD).
Figures 5 and S6 show the target gene enrichment for all three data sets and two different methods. FwD performs the best on the CiA and CiR data set for all window sizes with enrichment value of 7.0. CwD achieves the highest value of 2.5 for Pros. In general, the results on Pros are strikingly worse. Either the target gene list provided by the authors has too many false positives or negatives, the enrichment signal for Pros is relatively low or the high auto correlation makes prediction difficult.
Figure 5:
Target Gene and Motif Enrichment for Top Predictions. The x-axis corresponds to the number of top ranked probes used to construct ERs. On the left, the y-axis corresponds to the “target gene enrichment ratio” (see Methods). Values greater than one, indicated by the line, correspond to relatively more target genes predicted than expected based on their frequency in the genome. On the right, the y-axis corresponds to the “motif enrichment ratio” (see Methods). Values greater than one, indicated by the line, correspond to relatively more motifs in the ERs than expected by what is observed in the overall genome. The results are displayed for Fisher’s Combined Probability Test with Dependence (FwD) and the moving average method CMARRT with Dependence (CwD). See Figure 3 for details and Figures S6 and S7 for all window sizes.
For motif enrichment in the CiA data set (Figures 5 and S7), except for window size 3, where FwD achieves the maximum enrichment, FwD does better at the higher and lower ranked probes, but then CwD achieves larger values in the middle of the probe rankings. CwD also achieves the highest motif enrichment ratio for CiR, with some advantage for FwD in the middle ranked probes. For Pros the results are all poor, perhaps due to the specificity of the published consensus motif for Pros (the authors of the original Pros study also did not find evidence for enrichment of the motif, personal communication). However, FwD achieves the maximum value of ∼ 1.6 for window size 8.
For the gene distance metric in Figure 6 and Figures S8 and S9, all methods are very close, but FwD performs best for all three data sets for the highest ranking probes. The differences between the methods is smaller for the Pros data set and the differences also decrease as the window size becomes smaller. Based on the gene density of the Drosophila genome, with median distance 3500–4500bp between transcription start sites, even random ERs may be within a certain distance to a gene. Roughly ∼ 20% and ∼ 35% of random ERs are within 1KB and 2KB to a transcription start site respectively, with a slight increase as more probes are used to construct the ERs (see Methods and Figures S8 and S9). After a certain set of top probes are included, almost all methods for all data sets find ERs closer to genes than the random sets.
4.3. New Application
In the previous sections, the results of the generalized moving average methods showed either comparable or slightly enhanced performance to standard moving average methods. Therefore, for the t-test application, there does not appear to be a loss in performance by using results of the hypothesis test instead of the test-statistic themselves and in some cases there appears to be a gain. However, the generalized moving average approach may also be useful in situations where it is difficult to assess significance of the moving average of certain statistics and/or asymptotic normality does not hold. In these situations, if a problem can be expressed as a hypothesis test at each probe and a p-value can be derived, then the generalized moving approach can be applied as an alternative.
We present an example using the Ci data sets, where it is also of interest to identify enrichment regions that overlap between the two forms of the Ci transcription factor. Since both forms have the same DNA binding domain, it would be expected that they would bind to similar sequences. On the other hand, the overall shape of the two protein forms are vastly different and you might expect that one is capable of binding to low affinity sites better than the other. These differences may also explain why the two forms have very different levels of enrichment across the genome. To explore this problem in more detail, we are interested in finding ERs that overlap between the two forms. One strategy would be to take the two sets of ERs and examine the genomic regions where they overlap, or the overlap of genes nearby the ERs. However, as discussed above, using even relatively relaxed FDR cutoffs, very few or no ERs are predicted for CiR for most methods. Therefore, we have analyzed the two data sets to make integrated predictions of ERs for CiA and CiR.
We express the problem as a hypothesis test at each probe and p-values are determined such that the generalized moving average approach can be applied to the resulting p-values (see Methods). We use the Stouffer-Lipták Test combined p-value test (see Methods) with dependence adjustment and predict 1657 ERs for CiA-CiR using window size 4 and FDR cutoff of .01, which resulted in 1029 associated ER genes. In Table S1, the GO terms associated with the ER genes range from general categories (e.g., multicellular organismal process) to specific (e.g., neurogenesis or wing disc development). However, the developmental programs that require Hh signaling in the fly are reflected in the ER genes grouped according to their GO terms. For example, Hh signaling has largely been characterized in the context of the fly wing imaginal disc and we identify a group of genes that fall into this category. In addition, Hh signaling has shown to be required for the development of specialized cells in the nervous system and we identify genes that are involved in that process (see Table S1, neurogenesis and generation of neurons). Although several conserved signaling pathways participate in the above mentioned processes, we attribute these findings to the identification of Hh target genes. This conclusion is based in part to the presence of known Hh target genes in these lists.
5. Conclusion and Discussion
We have described a new method for analyzing tiling arrays which generalizes the moving average approach to scenarios where a problem can be presented as a hypothesis test at each probe and a moving “average” can be applied to the p-values resulting from this test. Our method was applied to data for the Ci and Pros transcription factors, where we had information on both their target genes and binding sites.
The first set of results based on a t-test were used to compare the performance of the method to the standard moving average approaches where the moving average of a log-ratio or t-statistic can be evaluated (Sections 4.1–4.2). In general, we found that the results were comparable between the methods on the three metrics used. Since the t-statistic moving average approach is a special case that falls into the generalized moving average framework, it is reassuring that the results were consistent. Furthermore, for large sample sizes, the Stouffer-Lipták test should be equivalent to the standard moving average approach.
The generalized moving average generally performed at least as well as the existing moving average methods, with some variation in the best performer due to the data set and evaluation criteria. As in Kuan et al. (2008), an adjustment was made based on correlations between probes and our results (Figures 3–4) are consistent with previous work that showed improvement in predictions due to the dependence adjustments (Bourgon, 2006, Kuan et al., 2008). TileMap, which only accounts for first order dependencies, performs more similarly to the unadjusted methods than the dependence adjusted methods.
The ER predictions were compared using several evaluation metrics (target gene enrichment, motif enrichment and distance to gene). These types of information are useful for investigators to evaluate the quality of ER predictions. However, there are caveats regarding their use. First, the consensus is based on current knowledge and may not completely specify the binding specificity and second, the sets of target genes are likely to suffer from both false positives and false negatives. This may be especially true for the Pros results which are strikingly worse than the Ci results. However, the Pros results may also be due to to a relatively poorer enrichment signal or considerably higher auto correlation, which may make prediction difficult. Despite the inherent problems with the different benchmarks, using the combination of the three can still be useful for comparing methods.
The initial set of results can also be used to evaluate how different cutoffs and options affect the results. As expected, the more stringent FDR control cutoffs provided more accurate ER predictions based on the metrics. However, there are two caveats regarding these cutoffs. First, many moving averages methods including those applied here use Benjamini & Hochberg FDR control assuming tests are independent despite tests being locally correlated. The effect of correlation may make FDR estimation unstable and little is known about how to account for correlation (Efron, 2007, Schwartzman and Liny, 2009). Second, the FDR cutoffs are at the probe-level not the ER-level, which is also common for many tiling array analysis methods. It would be more ideal for investigators to have FDR cutoffs that correspond to the ER-level test.
We also evaluated the effect of the window size option. Windows sizes of 3–8 were selected to correspond to a typical DamID fragment length of 1800–4800. The combined p-value methods appear to be more sensitive to window length. Larger window sizes result in the prediction of longer ERs in general and better target gene enrichment, but worse motif enrichment and gene distance scores. Intermediate window sizes in the range 4–5 appear to balance these two effects.
Finally, two different versions of the combined p-value methods were introduced; Fisher’s Combined Probability Test and Stouffer-Lipták Test. For the most part, there appears to be an advantage of FwD compared to SLwD on smaller window sizes based on the evaluation metrics. In general, Fisher’s Combined Probability Test puts relatively more emphasis on small p-values than the Stouffer-Lipták Test (Loughin, 2004). This would be more of a problem when the p-values that are combined are very disparate in magnitude, as in the analysis of the two Ci data sets in Section 4.3.
Although t-statistics were used in the first set of results, the use of the generalized moving average need not depend on that specific type of test as long as the problem can be expressed in a hypothesis testing framework and p-values can be determined for each probe. In the second set of results (Section 4.3), we presented another application to illustrate this point and the generalized moving average with dependence adjustment could be applied across probes. Using a standard moving average in this scenario may not be appropriate, since the moderated t-statistics at each probe from the two different experiments (CiA and CiR) have different degrees of freedom and it is of interest to identify regions where there is enrichment of at least one or both signals. Alternatively, TileMap provides the flexibility to make comparisons under multiple experimental methods (e.g., mutant 1 < wild type < mutant 2), but in this application, it was not easily adapted to test for binding under the two conditions (personal communication). In this context, the generalized moving average method provided an alternative analysis method with predictions near genes that are consistent with annotation for known Hh targets.
Supplementary Materials
Footnotes
This work was supported in part by NIH K01 AA016922 (KK) and R01 GM077407 (TK). The support of facilities by NIH R24 AA013162 to Boris Tabakoff is gratefully acknowledged. We thank Tony Southall for providing the Prospero data set, Bernhard Spangl and Peter Ruckdeschel for their assistance with the “robust-ts” package and Gary Grunwald, Elizabeth Siewert and Nichole Carlson for helpful discussions.
References
- Alexandre C, Jacinto A, Ingham P. “Transcriptional Activation of hedgehog Target Genes in Drosophila is Mediated Directly by the Cubitus interruptus Protein, a Member of the GLI Family of Zinc Finger DNA-Binding Proteins,”. Genes and Development. 1996;10:2003–2013. doi: 10.1101/gad.10.16.2003. [DOI] [PubMed] [Google Scholar]
- Beissbarth T, Speed T. “GOstat: Find Statistically Overrepresented Gene Ontologies within a Group of Genes,”. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. “Controlling the False Discovery Rate - A Practical and Powerful Approach to Multiple Testing,”. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- Bourgon R. Chromatin-Immunoprecipitation and High-Density Tiling Microarrays: A Generative Model, Methods for Analysis, and Methodology Assessment in the Absence of a ”Gold Standard”. 2006. PhD thesis, University of California, Berkeley.
- Brown M. “A Method for Combining Non-Independent, One-Sided Tests of Significance,”. Biometrics. 1975;31:987–992. doi: 10.2307/2529826. [DOI] [Google Scholar]
- Buck M, Lieb J. “ChIP-chip: Considerations for the Design, Analysis, and Application of Genome-Wide Chromatin Immunoprecipitation Experiments,”. Genomics. 2004;83:349–360. doi: 10.1016/j.ygeno.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Buck M, Nobel A, Lieb J. “ChIPOTle: A User-Friendly Tool for the Analysis of ChIP-chip Data,”. Genome Biology. 2005;6 doi: 10.1186/gb-2005-6-11-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casella G, Berger R. Statistical Inference. second edition Belmont, CA: Duxbury Press; 2002. [Google Scholar]
- Choksi S, Southall T, Bossing T, Edoff K, de Wit E, Fischer B, van Steensel B, Micklem G, Brand A. “Prospero Acts as a Binary Switch between Self-Renewal and Differentiation in Drosophila Neural Stem Cells,”. Developmental Cell. 2006;6:775–789. doi: 10.1016/j.devcel.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Efron B. “Correlation and Large-Scale Simultaneous Hypothesis Testing,”. Journal of the American Statistical Association. 2007;102:93–103. doi: 10.1198/016214506000001211. [DOI] [Google Scholar]
- Fisher R. Statistical Methods for Research Workers. 4th Edition ed. London: Oliver and Boyd; 1932. [Google Scholar]
- Folks J. “Combination of independent tests,”. In: Krishnaiah P, editor. Handbook of Statistics 4: Nonparametric Methods. Amsterdam: Elsevier; 1984. pp. 113–121. [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Sawitzki AJRG, Smith C, Smyth G, Yang LTJYH, Zhang J. “Bioconductor: Open Software Development for Computational Biology and Bioinformatics,”. Genome Biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Hirsch H, Sekinger E, Struhl K, Gingeras T. “Rank-Statistics Based Enrichment-Site Prediction Algorithm Developed for Chromatin Immunoprecipitation on Chip Experiments,”. BMC Bioinformatcs. 2006;7 doi: 10.1186/1471-2105-7-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, Taipale J. “Genome-wide Prediction of Mammalian Enhancers Based on Analysis of Transcription-Factor Binding Affinity,”. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- Hedges L, Olkin I. Statistical Methods for Meta-Analysis. London: Academic Press; 1985. [Google Scholar]
- Hess A, Iyer H. “Fisher’s Combined P-value for Detecting Differentially Expressed Genes using Affymetrix Expression Arrays,”. BMC Genomics. 2007;8 doi: 10.1186/1471-2164-8-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Wong W. “TileMap: Create Chromosomal Map of Tiling Array Hybridizations,”. Bioinformatics. 2005;21:3629–3636. doi: 10.1093/bioinformatics/bti593. [DOI] [PubMed] [Google Scholar]
- Keleş S, van der Laan M, Dudoit S, Cawley S. “Multiple Testing Methods for ChIP-Chip High Density Oligonucleotide Array Data,”. Journal Computational Biology. 2006;13:579–613. doi: 10.1089/cmb.2006.13.579. [DOI] [PubMed] [Google Scholar]
- Kinsler K, Vogelstein B. “The GLI Gene Encodes a Nuclear Protein which Binds Specific Sequences in the Human genome,”. Molecular and Cellular Biology. 1990;10:634–642. doi: 10.1128/mcb.10.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost J, McDermott M. “Combining Dependent P-values,”. Statistics & Probability Letters. 2002;60:183–190. doi: 10.1016/S0167-7152(02)00310-3. [DOI] [Google Scholar]
- Kuan P, Chun H, Keleş S. “CMARRT: A Tool for the Analysis of ChIP-chip Data from Tiling Arrays by Incorporating the Correlation Structure,”. In: Altman R, Dunker A, Hunter L, Murray T, Klein T, editors. Pacific Symposium on Biocomputing. 2008. pp. 515–526. World Scientific. [PMC free article] [PubMed] [Google Scholar]
- Lipták T. “On the Combination of Independent Tests,”. Magyar Tudomnyos Akadmia Matematikai Kutat Intezetenek Kozlemenyei. 1958;3:1971–1977. [Google Scholar]
- Liu XS. “Getting Started in Tiling Microarray Analysis,”. PLoS Computational Biology. 2007;3 doi: 10.1371/journal.pcbi.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughin T. “A Systematic Comparison of Methods for Combining P-values from Independent Tests,”. Computational Statistics & Data Analysis. 2004;47:467–485. doi: 10.1016/j.csda.2003.11.020. [DOI] [Google Scholar]
- Munch K, Gardner P, Arctander P, Krogh A. “A Hidden Markov Model Approach for Determining Expression from Genomic Tiling Micro Arrays,”. BMC Bioinformatics. 2006;7 doi: 10.1186/1471-2105-7-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton M, Noueiry A, Sarkar D, Ahlquist P. “Detecting Differential Gene Expression with a Semiparametric Hierarchical Mixture Method,”. Biostatistics. 2004;5:155–176. doi: 10.1093/biostatistics/5.2.155. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2005. URL http://www.R-project.org. [Google Scholar]
- Rhodes DR, Barrette TR, Rubin MA, Ghosh D, Chinnaiyan AM. “Meta-Analysis of Microarrays: Interstudy Validation of Gene Expression Profiles Reveals Pathway Dysregulation in Prostate Cancer,”. Cancer Res. 2002;62:4427–4433. [PubMed] [Google Scholar]
- Royce T, Rozowsky J, Luscombe N, Emanuelsson O, Yu H, Zhu X, Snyder M, Gerstein M. “Extrapolating Traditional DNA Microarray Statistics to Tiling and Protein Microarray Technologies,”. Methods in Enzymology. 2006;411:282–311. doi: 10.1016/S0076-6879(06)11015-0. [DOI] [PubMed] [Google Scholar]
- Schwartzman A, Liny X. “The Effect of Correlation in False Discovery Rate Estimation,”. 2009. Harvard University Biostatistics Working Paper Series 106, Harvard University. [DOI] [PMC free article] [PubMed]
- Smyth G. “Limma: Linear Models for Microarray Data,”. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- Spangl B. On Robust Spectral Density Estimation. 2008. Ph.D. thesis, Vienna University of Technology.
- Spangl B, Boudt K, Ruckdeschel P, Fried R, Agostinelli C, Kohl M. “robust-ts: Robust Time Series project,”. 2009. http://r-forge.r-project.org/projects/robust-ts/.
- Stouffer S, Suchman E, DeVinnery L, Star S, Williams R, Jr, Lumsdaine A, Lumsdaine M, Smith M, Janis I, Cottrell L., Jr . The American Soldier, Volume I: Adjustment During Army Life. Princenton, NJ: Princeton University Press; 1949. [Google Scholar]
- Tusher V, Tibshirani R, Chu G. “Significance Analysis of Microarrays Applied to the Ionizing Radiation Response,”. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Delrow J, Henikoff S. “Chromatin Profiling Using Targeted DNA Adenine Methyltransferase,”. Nature Genetics. 2001;27:304–308. doi: 10.1038/85871. [DOI] [PubMed] [Google Scholar]
- Wilson R, Goodman J, Strelets V FlyBase Consortium. “FlyBase: Integration and Improvements to Query Tools,”. Nucleic Acids Research. 2008;36:D588–D593. doi: 10.1093/nar/gkm930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormald S, Hilton D, Smyth G, Speed T. “Proximal Genomic Localization of STAT1 Binding and Regulated Transcriptional Activity,”. BMC Genomics. 2006;7:254. doi: 10.1186/1471-2164-7-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki J, Gregory B, Ecker J. “Mapping the Genome Landscape Using Tiling Array Technology,”. Current Opinion in Plant Biology. 2007;10:534–542. doi: 10.1016/j.pbi.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaykin D, Zhivotovsky L, Westfall P, Weir B. “Truncated Product Method for Combining P-values,”. Genetic Epidemiology. 2002;22:170–185. doi: 10.1002/gepi.0042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.