Abstract
MicroRNAs (miRs) regulate gene expression at the posttranscriptional level. To obtain some insights into the origins and evolutionary patterns of miR genes, we have identified miR genes in the genomes of 12 Drosophila species by bioinformatics approaches and examined their evolutionary changes. The results showed that the extant and ancestral Drosophila species had more than 100 miR genes and frequent gains and losses of miR genes have occurred during evolution. Although many miR genes appear to have originated from random hairpin structures in intronic or intergenic regions, duplication of miR genes has also contributed to the generation of new miR genes. Estimating the rate of nucleotide substitution of miR genes, we have found that newly arisen miR genes have a substitution rate similar to that of synonymous nucleotide sites in protein-coding genes and evolve almost neutrally. This suggests that most new miR genes have not acquired any important function and would become inactive. By contrast, old miR genes show a substitution rate much lower than the synonymous rate. Moreover, paired and unpaired nucleotide sites of miR genes tend to remain unchanged during evolution. Therefore, once miR genes acquired their functions, they appear to have evolved very slowly, maintaining essentially the same structures for a long time.
Keywords: birth-and-death evolution, gene duplication, gene regulation, multigene family, noncoding RNA, substitution rate
Introduction
MicroRNAs (miRs) constitute one of the major classes of noncoding RNAs that regulate gene expression at the posttranscriptional level (Ambros 2004; Bartel 2004; Lewis et al. 2005; Bartel 2009). They are first transcribed as primary miRs, which form hairpin (stem-loop) structures and undergo several processing steps to produce mature miRs with ∼22 nucleotides (nt) (fig. 1). These mature miRs interact with the transcripts of target genes and suppress their expression. After their first discovery in Caenorhabditis elegans (Lee et al. 1993), extensive studies have been conducted to understand their functional roles in gene regulatory systems (e.g., Reinhart et al. 2000; Li et al. 2006; Flynt and Lai 2008; Yekta et al. 2008).
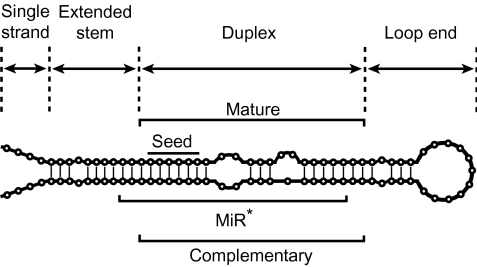
FIG. 1.—
Typical structure of miRs. Each miR consists of single-strand, extended-stem, duplex, and loop-end regions. The duplex structure that has ∼2 nt 3′ overhang is the final product of miRs, but only one of the two arms generally becomes mature miRs and the other arm called miR* is degraded. The seed sequence (second to seventh nucleotide in the mature region) is particularly important for target recognition in animals. Vertical rods indicate the paired nucleotide sites.
MiR genes are widely distributed in animals and land plants (Axtell and Bowman 2008) and several hypotheses have been proposed to explain their origins (Shabalina and Koonin 2008). The first hypothesis is that new miR genes originate from duplication of genetic elements such as miR and protein-coding genes. If a miR gene is duplicated, a resultant duplicate can become a new miR gene through some nucleotide substitutions (Tanzer and Stadler 2004). If a protein-coding gene (or any other genetic element) is duplicated in an inverted way, the resultant inverted duplicates form a hairpin structure and may also become a new miR gene. The second hypothesis is that terminal inverted repeats of transposable elements (TEs) become miR genes (Voinnet 2009). Indeed, it has been reported that several miR genes in animals and plants have originated from miniature inverted-repeat TEs (Piriyapongsa and Jordan 2007; Piriyapongsa and Jordan 2008). The third hypothesis is that random hairpin structures in intronic or intergenic regions become miR genes. Because there are hundreds of thousands of hairpin structures in the genomes of higher organisms (e.g., Bentwich et al. 2005; Felippes et al. 2008), some of them may become miR genes.
In Drosophila species, it has been suggested that most miR genes have originated from random hairpin structures and the contribution of gene duplication is negligibly small (Lu et al. 2008b). This conclusion was primarily obtained without examining miR genes generated by gene duplication. In practice, however, duplicated (paralogous) genes are known to exist. Therefore, we must consider the increase of miR genes due to gene duplication as well as the generation from random hairpin structures.
The purpose of this study is to examine the possible origins of miR genes and their evolutionary patterns in long-term Drosophila evolution. We used 12 Drosophila species whose genome sequences have been published (Clark et al. 2007). These species diverged at various evolutionary times from less than 1 MYA to over 60 MYA (Tamura et al. 2004). In addition, we used 152 miR genes, which have been experimentally identified and well confirmed in Drosophila melanogaster (e.g., Ruby et al. 2007a, 2007b; Stark et al. 2007; Griffiths-Jones et al. 2008). Using these data, we have identified miR genes in the 12 species with bioinformatics techniques and examined their evolutionary changes.
Materials and Methods
The names of 12 Drosophila species used in this study and their approximate divergence times are presented in figure 2. The genome sequence of D. melanogaster (release 5) was downloaded from Berkeley Drosophila Genome Project (http://fruitfly.org/). The genome sequences (CAF1) of other 11 species were downloaded from AAA database (http://rana.lbl.gov/drosophila/). We also downloaded 152 miR sequences in D. melanogaster from miRBase (release 13.0, http://microrna.sanger.ac.uk/; Griffiths-Jones et al. 2008). Using these sequences as queries, we conducted a BlastN search (Altschul et al. 1997) against each genome sequence with E-value ≤ 10−4.
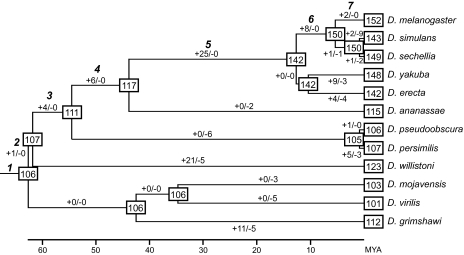
FIG. 2.—
Estimates of the numbers of miR genes in ancestral species and gains and losses of miR genes during Drosophila evolution. Numbers in squares show the numbers of miR genes in ancestral or extant species. Numbers along each branch indicate the numbers of gains (+) and losses (−) of miR genes, respectively. The time scale shown below the tree is from Tamura et al. (2004).
All hit sequences were classified into 152 groups of the D. melanogaster genes based on the E-values of the BlastN search. The sequences of each group were aligned using MUSCLE (Edgar 2004). We then used the following four criteria with different stringencies to eliminate non-miR genes. 1) Mature sequences (fig. 1) contain ≤2 nt indels compared with that of the D. melanogaster miR genes. This is a basic criterion and applied for all other criteria. 2) Free energy (FE) of the predicted hairpin structure is ≤−15 kcal/mol or P value in randomization test by RANDFOLD (Bonnet et al. 2004) is ≤0.2. 3) FE is ≤−15 kcal/mol and the P value is ≤0.2. 4) FE is ≤−15 kcal/mol and the P value is ≤0.05. The nucleotide sequences of all miR genes and their genomic locations are shown in miR_seqs.txt and supplementary tables S1–S12 (Supplementary Material online), respectively.
Based on the predicted hairpin structures by RNAFOLD (Mathews et al. 1999), we also extracted the complementary sequence, which is the opposite arm of the mature sequence in the duplex structure (fig. 1).
Results
Numbers of miR Genes in Drosophila Species
Our homology search identified more than 100 miR genes in each of the 12 species when search criterion 1 was used (table 1). However, the number of miR genes in a species decreased as the genetic divergence from D. melanogaster increased (roughly from top to bottom in table 1). This does not necessarily mean that D. melanogaster and Drosophila virilis have the largest and the smallest numbers of miR genes, respectively. We used only D. melanogaster miR sequences as queries for homology search, and therefore it is possible that we failed to identify miR genes, which exist in other species but not in D. melanogaster. In other words, the numbers of miR genes shown in table 1 are the minimum estimates, particularly in species distantly related to D. melanogaster. This is inevitable because the experimental identification of miR genes is still quite limited for the other 11 species. Nevertheless, Drosophila willistoni that diverged from D. melanogaster ∼62 MYA shows a larger number of miR genes than some other species (Drosophila ananassae, Drosophila pseudoobscura, and Drosophila persimilis) that diverged more recently from D. melanogaster. This suggests that expansion of miR genes has occurred in D. willistoni. It should be noted that several miR genes were regarded as non-miR genes when we used more stringent search criteria (table 1). This ambiguity is unavoidable because our computational approach can identify only potential candidates of miR genes.
Table 1.
Numbers of MiR Genes and Gene Families Identified in 12 Drosophila Species
| Number of Genes Under Different Criteria |
No. of Gene Familiesf | Average No. of Genes/Gene Familyf | ||||
| Species (MYAa) | Criterion 1 (Standard)b | Criterion 2c | Criterion 3d | Criterion 4e | ||
| Subgenus Sophophora | ||||||
| D. melanogaster (−) | 152 | 152 | 150 | 145 | 131 | 1.16 |
| D. simulans (5.4) | 143 | 141 | 137 | 132 | 124 | 1.15 |
| D. sechellia (5.4) | 149 | 147 | 145 | 139 | 127 | 1.17 |
| D. yakuba (12.8) | 148 | 148 | 144 | 137 | 120 | 1.23 |
| D. erecta (12.8) | 142 | 142 | 139 | 134 | 119 | 1.19 |
| D. ananassae (44.2) | 115 | 115 | 113 | 110 | 98 | 1.17 |
| D. pseudoobscura (54.9) | 106 | 106 | 103 | 100 | 87 | 1.22 |
| D. persimilis (54.9) | 107 | 107 | 106 | 103 | 84 | 1.27 |
| D. willistoni (62.2) | 123 | 123 | 121 | 120 | 86 | 1.43 |
| Subgenus Drosophila | ||||||
| D. mojavensis (62.9) | 103 | 103 | 101 | 97 | 88 | 1.17 |
| D. virilis (62.9) | 101 | 101 | 100 | 98 | 85 | 1.19 |
| D. grimshawi (62.9) | 112 | 112 | 111 | 109 | 85 | 1.32 |
Divergence time from D. melanogaster obtained by Tamura et al. (2004).
BlastN search with E-value ≤ 10−4 and ≤2 nt gaps in the mature region. This criterion was also applied for all other criteria.
FE of the predicted hairpin structure ≤−15 kcal/mol or P ≤ 0.2 in randomization test by RANDFOLD (Bonnet et al. 2004).
FE ≤ −15 kcal/mol and P ≤ 0.2.
FE ≤ −15 kcal/mol and P ≤ 0.05.
Based on criterion 1.
We also counted the number of gene families in each species by using information given in miRBase (table 1). Here, a miR gene family is defined as a group of miR genes, of which the mature sequences are homologous to one another. The results show that each gene family consists of one or a few genes (on average 1.22 genes for all 12 species) except for the mir-2 family that contains seven to nine genes within species (see supplementary tables S1–S12, Supplementary Material online). This small number of miR genes per gene family has also been reported in other animals such as humans and mice (Li and Mao 2007). Yet, the average number (1.43) for D. willistoni was considerably greater than that for other Drosophila species, again suggesting species-specific duplication of miR genes in this species (particularly genes belonging to mir-959 and mir-964 gene families, see supplementary table S9, Supplementary Material online). In the following analyses, we used all miR genes identified by standard homology search (criterion 1 in table 1), but the results were essentially the same even when more stringent criteria were used.
Chromosomal Locations of miR Genes
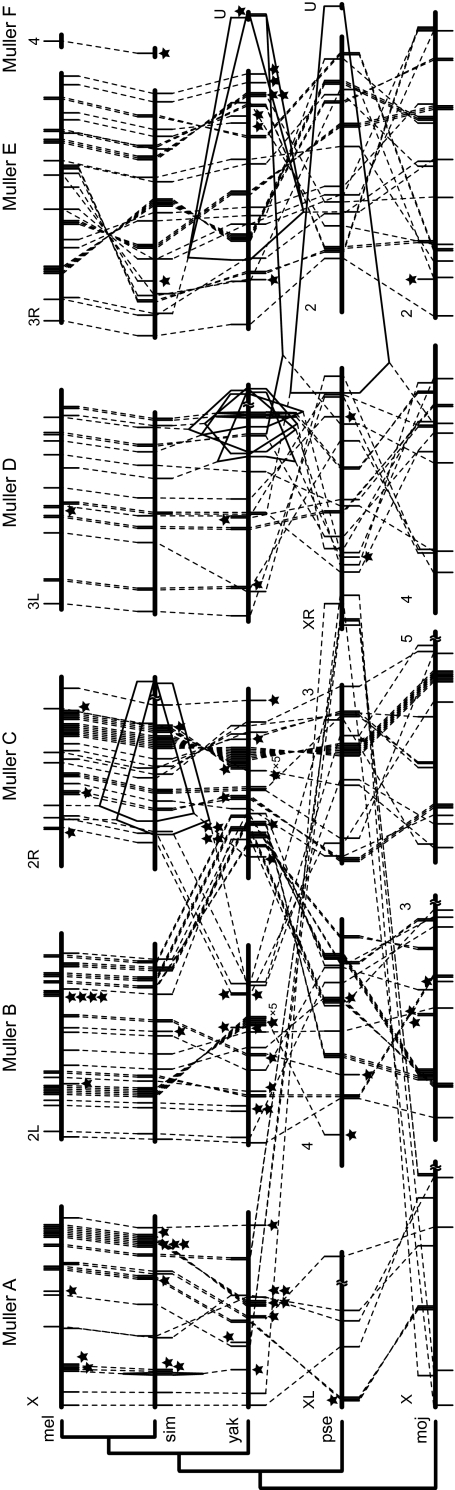
To examine the genomic locations of miR genes and their rearrangements during evolution, we mapped miR genes of D. melanogaster, Drosophila simulans, Drosophila yakuba, D. pseudoobscura, and Drosophila mojavensis on their chromosomes (fig. 3). In these species miR genes were widely distributed throughout the genome. In the comparison of D. melanogaster and D. simulans, which diverged ∼5.4 MYA, the order of orthologous miR genes (shown by broken lines) was perfectly conserved except for a change due to a chromosomal inversion in the chromosomal arm 3R of D. melanogaster (Schaeffer et al. 2008). Nevertheless, some miR genes were duplicated (solid lines) or missing (stars in the D. melanogaster genome) in D. simulans. Comparison of D. simulans and D. yakuba also shows gains and losses of miR genes. In addition, the order of orthologous genes has changed because of several chromosomal inversions and translocations. Yet, the changes were confined within the same chromosomal arm except for a change that was caused by a pericentric inversion between 2L and 2R in D. yakuba (Schaeffer et al. 2008). However, comparison of D. yakuba with D. pseudoobscura and D. pseudoobscura with D. mojavensis shows that the orders of orthologous genes have been shuffled even between different chromosomal arms and many gains and losses of miR genes have occurred. These results indicate that miR genes have been subject to birth-and-death evolution and their locations have changed considerably during Drosophila evolution.
FIG. 3.—
Chromosomal locations of miR genes and their orthologous and paralogous relationships in Drosophila melanogaster (mel), D. simulans (sim), D. yakuba (yak), D. pseudoobscura (pse), and D. mojavensis (moj). Rods above and below the chromosomes show the miR genes located on opposite strands. Broken and solid lines stand for orthologous and paralogous relationships of miR genes, respectively. Stars above and below the chromosomes represent the genes whose orthologous genes are absent in the upper and lower species, respectively. L and R indicate the left and right arms of chromosomes, respectively, whereas U indicates that the chromosomal location of the sequences remains undetermined. We also showed the Muller elements (A–F) above the chromosomes because the conservation of the gene contents within Muller elements are well supported (Schaeffer et al. 2008). We only showed the chromosomes where miR genes were located. The information about chromosomal assemblies is from Schaeffer et al. (2008). Scale is approximate.
If tandem duplication is important for generating new miR genes, many miR genes should be clustered in the genome. Our data show that 43% of miR genes are clustered on average with at least one other miR gene (≤3 kb apart; supplementary table S13, Supplementary Material online). Particularly, D. willistoni shows a higher proportion of clustered genes (54%) than other species, largely because a cluster containing mir-959 and mir-964 orthologs has been duplicated several times (supplementary table S9, Supplementary Material online). (Of course, this could be due to assembly errors.) In all species examined, however, the genes included in a cluster were largely nonhomologous and belonged to different gene families (supplementary table S13, Supplementary Material online). Many clustered genes are therefore likely to have originated from non-miR sequences. Nevertheless, the proportion of gene increase by tandem duplication within clusters was on average estimated to be as large as 31% (see supplementary table S13, Supplementary Material online for details). This would be the minimum estimate because some gene clusters generated by tandem duplication may have been dispersed by chromosomal rearrangements. Therefore, tandem duplication has also been important for increasing the number of miR genes within clusters.
Gains and Losses of miR Genes
Estimates of the numbers of miR genes in ancestral species and gains and losses of miR genes during Drosophila evolution are presented in figure 2. These estimates were obtained by the parsimony method. For example, if a miR gene was present in D. melanogaster and D. simulans but absent in other species, we assumed that the gene was generated in the ancestor of these two species (branch 6 in fig. 2) and lost in Drosophila sechellia.
The number of miR genes in the most recent common ancestor of 12 Drosophila species was estimated to be 106, although this number should be a minimum estimate as mentioned above. We found that D. willistoni gained 21 genes during the evolution, and D. melanogaster acquired 46 miR genes after the 12 Drosophila species split into the two subgenera (branches 2–7 in fig. 2). In addition, many gene gains were observed in other lineages. We also found many losses of miR genes in various lineages. Note that there was no gene loss in the lineage to D. melanogaster and no gene gain in some other lineages. This has occurred because of the limitation of our homology search. We would find losses and gains in these lineages if experimental data on miR genes become available for all 12 Drosophila species and are used for homology search as queries.
We classified the 46 gene gains observed in the lineage to D. melanogaster into gene gains within a gene family and gene gains generating new gene families. If a new gene belongs to one of the preexisting miR gene families, the gene is most likely to be generated by gene duplication. However, our results show that 89% (41/46) of gene gains have generated new gene families (table 2). These gains have occurred in intronic and intergenic regions in almost equal frequencies. Essentially the same results were obtained even when we considered all 152 miR genes in D. melanogaster, although three genes were located in untranslated regions of protein-coding genes (supplementary table S14, Supplementary Material online). These results suggest that many miR genes have originated from random hairpin structures in intronic or intergenic regions. Yet, note that 11% of the miR genes have clearly been derived from duplication of miR genes. Also, all gene gains observed in the lineages of non-melanogaster species have obviously originated by gene duplication (see also solid lines in fig. 3) because homology search was used for detecting these gene gains. Therefore, the duplication of miR genes has apparently contributed to produce new miR genes as well.
Table 2.
Genomic Locations and Possible Origins of 46 MiR Genes that Have Been Gained During Evolution of Drosophila melanogaster
MiR genes generated by duplication of miR genes. (Newly arisen genes showed sequence similarity to preexisting miR genes.)
MiR genes derived from non-miR sequences. (Newly arisen genes showed no significant sequence similarity to preexisting miR genes.)
Similarity of miR Genes to Protein-Coding Genes and TEs
We also examined the possibilities that miR genes have originated from protein-coding genes and TEs. If this is the case, miR genes are likely to show sequence similarity to them. We therefore examined the similarity of miR genes to every protein-coding gene using a BlastN search with E-value ≤ 10−4. In this analysis, we used only the protein-coding genes in D. melanogaster (dmel-all-gene-r5.16.fasta in FlyBase, http://flybase.org/) because gene annotations appeared to be incomplete in other species. The results showed that none of the miR genes in D. melanogaster has significant sequence similarity to protein-coding genes. This suggests that Drosophila miR genes have not originated from inverted duplicates of protein-coding genes.
Similarly, we examined the sequence similarity between miR genes and TEs by using RepeatMasker (open-3.2.8, http://www.repeatmasker.org/) with default settings. The results were negative except for two miR genes in D. yakuba. Both of them (numbers 146 and 148 in our annotation, which were orthologs of mir-10) showed sequence similarity to jockey, a retrotransposon (Mizrokhi et al. 1988). However, only parts of the miR genes were alignable with the jockey element, and mir-10 orthologs in other 11 species and one ortholog (number 118) in D. yakuba did not show significant sequence similarity to jockey. In addition, these two genes were regarded as non-miR genes when we used more stringent search criteria (see supplementary table S4, Supplementary Material online). It is therefore unlikely that the mir-10 gene originated from jockey. In any case, the contribution of TEs to miR genes appears to be negligible in Drosophila species.
Evolutionary Rates of miR Genes
To examine the extent of conservation of miR genes after their origination, we next studied the rates of nucleotide substitution for the mature, complementary, and other (loop end, extended stem, and single strand, hereafter LES) regions (fig. 1). (We analyzed the complementary region instead of the miR* region because the miR* sequences for several miR genes have not been determined even in D. melanogaster.) We also considered the seed sequence (positions 2–7) separately from other parts of the mature sequence because the seed sequence is known to be most critical for target recognition (see Bartel 2009 for review). Moreover, to examine the relationships between substitution rate and the time after birth of a miR gene, we estimated the substitution rate for a group of miR genes, which were generated in each branch of the lineage to D. melanogaster (1–6 in fig. 2). In this analysis, we used 110 orthologous groups of miR genes, which contained no paralogs and computed the substitution rate for each of them. For example, suppose that a miR gene was generated on branch 6, and only D. melanogaster, D. simulans, and D. sechellia have the gene (fig. 2). If the numbers of nucleotide substitutions per site (Jukes and Cantor 1969) for the mature region between D. melanogaster and D. simulans and between D. melanogaster and D. sechellia are 0.04 and 0.06, respectively, the average becomes (0.04 + 0.06)/2 = 0.05. As the divergence time between D. melanogaster and D. simulans (or D. sechellia) has been estimated to be 5.4 MYA, the substitution rate for the mature region of the miR gene can be estimated by 0.05/(5.4 × 106 × 2) = 4.6 × 10−9/site/year. For comparison, we also estimated the substitution rates at synonymous and nonsynonymous sites of 12,285 orthologous protein-coding genes (release FB2009_03 in FlyBase). The modified Nei-Gojobori method (Zhang et al. 1998) with transition/transversion ratio of 2 was used for computing the numbers of nucleotide substitutions per synonymous and nonsynonymous sites.
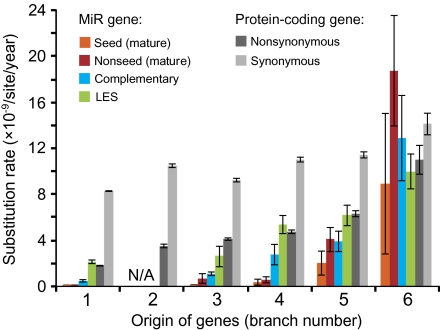
The results show that there is a negative correlation between the substitution rate and the time after the birth of miR genes (P < 0.001 by t-test; fig. 4). In other words, old miR genes have evolved much slower than new miR genes, suggesting that old miR genes have more important functions than new ones. For miR genes generated in branches 1–5, the evolutionary rate was lowest in the mature region (orange and red bars in fig. 4), intermediate in the complementary region (blue bars), and highest in the LES region (green bars), which is consistent with the previous studies (Ehrenreich and Purugganan 2008; Lu et al. 2008a). The seed sequence (orange bars) showed an even lower rate compared with the other mature region (red bars). For example, the rate for seed sequences was as small as 2.0 × 10−11/site/year for the miR genes generated in branch 1. Note that even the LES region showed a substitution rate comparable with the nonsynonymous substitution rate in protein-coding genes (dark gray bars). This indicates that there are some functional constraints even in the LES region. By contrast, for miR genes that originated in branch 6, the rates of mature, complementary, and LES regions were nearly the same (∼12 × 10−9) and were similar to the synonymous substitution rate (14.1 × 10−9) of protein-coding genes (light gray bars).
FIG. 4.—
Substitution rates of miR and protein-coding genes that originated in each branch (1–6 in fig. 2). We analyzed 110 and 12,285 orthologous groups of miR and protein-coding genes, respectively. Error bars indicate the standard errors. “N/A” indicates that there was no such orthologous group of miR genes in the data set. The numbers of orthologous miR genes analyzed for each branch are as follows: 74 for branch 1, 0 for branch 2, 4 for branch 3, 6 for branch 4, 19 for branch 5, and 7 for branch 6.
To show the extent of natural selection for each miR region in more detail, we computed the average r/rS (= w) ratio for each miR region, where r is the substitution rate for each miR region and rS is the synonymous substitution rate of protein-coding genes (table 3). If we assume that synonymous substitutions are neutral, the w values of >1, 1, and <1 suggest positive, neutral, and purifying selection, respectively. The results show that w is much lower than 1 for old miR genes, suggesting strong purifying selection for these genes (table 3 for the statistical significance). For example, 99% (1 − w = 1 − 0.01) of mutations in the mature region of miR genes generated in branch 1 are likely to have been deleterious and eliminated by purifying selection. Even for the LES region, 74% (1 − 0.26) of mutations appear to have been deleterious. The same trend was observed for miR genes that originated in branches 1–5. By contrast, miR genes that originated in branch 6 show that w is close to 1. Therefore, new miR genes appear to have evolved in a more or less neutral fashion.
Table 3.
w (= r/rS) Values for Each Region of MiR Genes
| Brancha | Matureb | Complementary | LESc |
| 1 | 0.01* | 0.06* | 0.26* |
| 2d | — | — | — |
| 3 | 0.06* | 0.12* | 0.29 |
| 4 | 0.05* | 0.26* | 0.49* |
| 5 | 0.30* | 0.35* | 0.55* |
| 6 | 1.11 | 0.91 | 0.71 |
P (r = rS) < 0.05 by t-test after Bonferroni correction for multiple testing.
Branch numbers correspond to those in figure 2.
Entire mature sequence was considered without separating seed and other parts of the mature sequence.
Loop-end, extended-stem, and single-strand regions.
There was no such orthologous gene group in the data set.
Substitution Patterns of miR Genes
It is known that the duplex structure of mature and miR* regions is very important for several steps of miR maturation (see Lau and MacRae 2009 for review). For this reason, the proportion of paired sites (A–U, G–C, and G–U pairs) between mature and complementary regions was as high as ∼80% on average (supplementary fig. S1, Supplementary Material online). Also, the proportions of paired sites were essentially the same between old and new miR genes (supplementary fig. S1, Supplementary Material online). To clarify how miR genes have maintained a stable proportion of paired sites during evolution, we examined the patterns of nucleotide substitution in mature and complementary regions. In this analysis, we used 91 orthologous groups of miR genes, which contained no paralogs and no gaps in the mature and complementary regions. For each ortholog, we inferred the nucleotide sequences of ancestral species by using the likelihood method (Yang et al. 1995) and counted the numbers of substitutions at paired (A–U, G–C, and G–U pairs) and unpaired sites (all other pairs) separately.
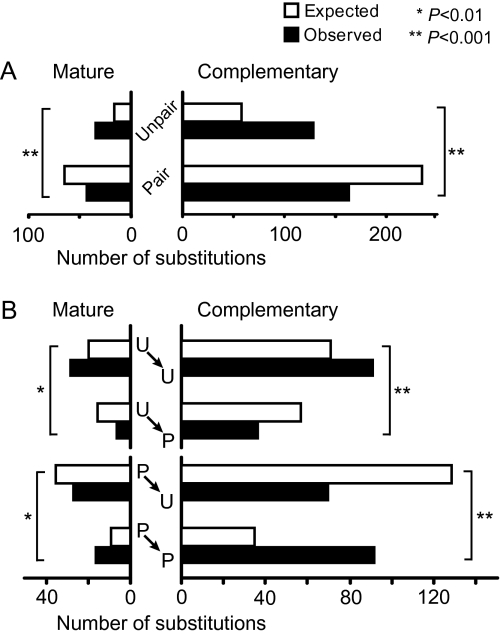
The results have shown that the numbers of substitutions at paired and unpaired sites are similar in both mature and complementary regions (fig. 5A), even though the number of paired sites is about four times larger than that of unpaired sites (supplementary fig. S1, Supplementary Material online). Therefore, nucleotide substitutions have occurred more frequently at unpaired sites than at paired sites in both mature and complementary regions (P < 0.001 by χ2 test). These results suggest that paired sites are under stronger functional constraints than unpaired sites.
FIG. 5.—
Substitution patterns in mature and complementary regions of Drosophila miR genes. (A) Numbers of substitutions at paired and unpaired sites. (B) Numbers of different types of substitutions: paired to paired (P→P), paired to unpaired (P→U), unpaired to paired (U→P), and unpaired to unpaired (U→U). We analyzed 91 orthologous groups of genes, which contained no paralogs and no gaps in the mature and complementary regions. Open and solid bars indicate the expected and observed numbers of substitutions, respectively. The expected numbers of substitutions were computed under the assumption of equal rate among different types of substitutions. Asterisks indicate the statistical significance (* for 1% and ** for 0.1% level) of the difference between the expected and observed numbers of substitutions by χ2 test.
We then examined whether the pairing status (paired or unpaired) changes when a nucleotide substitution occurs. The results showed a strong tendency that both paired and unpaired statuses remain unchanged after nucleotide substitutions more often than expected by chance (fig. 5B). These results suggest that the positions of paired and unpaired sites in the duplex structure have been more or less stable during evolution. We actually found that the proportion of paired sites is on average lower in the middle part of the duplex structure than in the upper (5′) and lower (3′) parts (supplementary fig. S2, Supplementary Material online). This is consistent with the idea that small internal loops at the middle portion of the duplex structure are important for the accurate miR biogenesis (Han et al. 2006). Therefore, it appears that miR genes have kept essentially the same structures for a long time.
Discussion
In this study, we have examined the evolutionary dynamics of miR genes in Drosophila species. Although available data are still limited, we found that at least 100 miR genes existed in the common ancestor of the 12 Drosophila species used and frequent gains and losses of miR genes have occurred during evolution. This birth-and-death evolution (Nei and Rooney 2005) of miR genes in Drosophila species is similar to the evolutionary mode of protein-coding genes such as olfactory receptor (Guo and Kim 2007; Nozawa and Nei 2007) and odorant-binding protein (Vieira et al. 2007) genes.
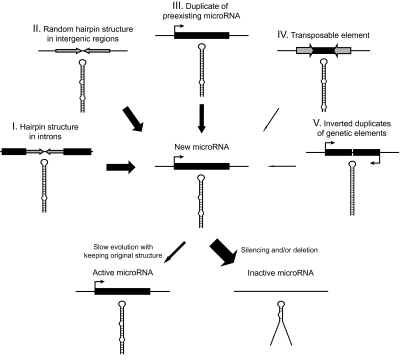
We have shown that many miR genes have been derived from non-miR sequences. Of the miR genes generated from non-miR sequences, about one-half have occurred in intronic regions of protein-coding genes (I in fig. 6). Intronic regions may be easy to generate miR genes because they can be co-transcribed with the protein-coding genes. Some miR genes in introns are actually known to be processed by the spliceosome and called mirtrons (Berezikov et al. 2007; Okamura et al. 2007; Ruby et al. 2007a; Zhu et al. 2008). Nevertheless, the advantage of miR genes existing in protein-coding genes is still unclear. Behura (2007) examined the physical association of miR genes and their overlapping protein-coding genes in the genomes of D. melanogaster, mosquito (Anopheles gambiae), and honey bee (Apis mellifera) and found that only two orthologous miR genes were located within the introns of the same protein-coding genes in all the three species. Therefore, further studies are necessary to understand the importance of miR genes located in introns.
FIG. 6.—
Possible evolutionary scenario of Drosophila miR genes. Thickness of solid arrows roughly indicates the importance of the processes.
The remaining half of the new miR genes from non-miR sequences are likely to have been derived from random hairpin structures in intergenic regions (II in fig. 6). Obviously, if we consider a single hairpin structure, the possibility of the hairpin to become a miR gene would be negligibly small because the promoter region of a miR gene also must be generated. However, there are hundreds of thousands of hairpin structures in the D. melanogaster genome (Stark et al. 2007; Lu et al. 2008b). In addition, a substantial fraction of a genome appears to be transcribed in Drosophila species (Manak et al. 2006). Therefore, it is possible that some of these hairpins in intergenic regions have evolved into new miR genes.
However, we also found that duplication of miR genes has played significant roles in the origin of miR genes (III in fig. 6). The proportion of new miR genes derived by gene duplication was ∼10% in the D. melanogaster lineage for the last 60 Myr. Many gene gains by duplication were also observed in the lineages leading to other species. Moreover, ∼30% of gene gains within clusters can be explained by tandem duplication, although many clusters were generated before the divergence of Drosophila species. Note that these are minimum estimates because the new miR genes may have diverged considerably from the original miR genes so that their similarity is no longer detectable.
This finding about the role of gene duplication is different from that of Lu et al. (2008b), who studied the generation of miR genes in D. melanogaster, D. simulans, and D. pseudoobscura. They suggested that miR genes have originated almost exclusively from random hairpin structures and the contribution of gene duplication is very small. This happened primarily because they used their own experimental data for their analysis and did not really consider the possibility of miR genes being derived by duplication. By contrast, we considered both possibilities for origins of miR genes from random hairpin structures and by gene duplication. In addition, we used miR genes, which satisfied very stringent criteria and were listed in the miRBase (e.g., Ruby et al. 2007b; Stark et al. 2007), whereas Lu et al. (2008b) analyzed their own experimental data, in which miR genes were identified with more relaxed criteria. Therefore, it is possible that their data contained many non-miR sequences and gave a biased conclusion (see Berezikov et al. 2010). Furthermore, we studied the long-term evolution of miR genes, whereas Lu et al. (2008b) were primarily interested in the short-term evolution. If the short-term evolution is considered, the probability of generation of new genes by duplication would certainly be much lower than that from random hairpin structures. Indeed, when we reanalyzed their data, none of the species-specific miR genes they identified in D. melanogaster and D. simulans had paralogous genes. However, most miR genes derived from random hairpin structures seem to disappear quickly (Lu et al. 2008b). Therefore, if the long-term evolution is considered, gene duplication appears to become more important for the origin of miR genes than previously thought. It is possible that miR genes generated by gene duplication have survived longer than those derived from random hairpin structures.
After the birth of a miR gene, there seem to be two different modes of evolution. We have shown that new miR genes have evolved in a more or less neutral fashion. Therefore, a majority of these genes may not have acquired any function and may be transcribed at very low levels in an unregulated fashion. New miR genes were actually shown to be expressed at a lower level compared with old ones (Lu et al. 2008b). By contrast, the rate of nucleotide substitution of old miR genes is very low compared with that of protein-coding genes. We also found that once the structure of miR genes is established it tends to be kept for a long evolutionary time. Therefore, miR genes evolve almost neutrally at the initial stage of evolution and many of them appear to become inactive (fig. 6). Only a few of them acquire solid functions and evolve very slowly under strong purifying selection, keeping their original structures.
Using the McDonald–Kreitman (MK) test (McDonald and Kreitman 1991), Lu et al. (2008a) concluded that positive selection is the major force of evolution of miR genes, particularly at the initial stage. They estimated that ∼80% of nucleotide substitutions in the new genes, which were generated during Drosophila evolution, have occurred by positive selection. However, because the results of the MK test can be interpreted in many different ways without positive selection (e.g., Eyre-Walker 2002; Hughes 2008; Nei M, Suzuki Y, Nozawa M, unpublished data), the conclusion obtained by this test is generally unreliable. In fact, we did not find any signature of positive selection even in the new miR genes. Therefore, positive selection is unlikely to be a major force in evolution of Drosophila miR genes.
It should be noted that our study is based on a computational approach to identify the miR genes and therefore our results may contain a certain fraction of false positives. In addition, there must be other miR genes, which are unidentified in this study. To obtain a complete picture of miR gene evolution in Drosophila species, extensive experimental identification of miR genes is necessary for many different species. Nevertheless, this bioinformatics approach must be a good starting point for identifying potential miR genes in a genome. In fact, our estimates of ancestral gene numbers roughly agree with those obtained by a recent experimental study (Berezikov et al. 2010).
The study of evolution of miR genes has just begun. It is therefore important to collect more data from various groups of organisms and derive general conclusions. It is already known that miR gene families in plants contain more member genes than those in animals (Li and Mao 2007), suggesting that the contribution of gene duplication for the formation of new genes is greater in plants than in animals. Note also that the contribution of TEs for the formation of miR genes may be greater in mammals and land plants than in insects because the former genomes are known to harbor a larger number of TEs than the latter genomes. At this stage, it is important to consider various possibilities of miR gene evolution.
Supplementary Material
Supplementary tables S1–S14, figures S1 and S2, and miR_seqs.txt are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Jian Lu, Stephen Schaeffer, and Chung-I Wu for their valuable advices during our study. We also thank Michael Axtell, Hielim Kim, Chungoo Park, Yoshiyuki Suzuki, Naoko Takezaki, and Zhenguo Zhang for their comments on earlier versions of the manuscript. This work was supported by National Institutes of Health Grant [GM020293 to M.Nei] and Japan Society for the Promotion of Science [to M.Nozawa].
References
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Bowman JL. Evolution of plant microRNAs and their targets. Trends Plant Sci. 2008;13:343–349. doi: 10.1016/j.tplants.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behura SK. Insect microRNAs: structure, function and evolution. Insect Biochem Mol Biol. 2007;37:3–9. doi: 10.1016/j.ibmb.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Bentwich I, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, et al. Evolutionary flux of canonical microRNAs and mirtrons in Drosophila. Nat Genet. 2010;42:6–9. doi: 10.1038/ng0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet E, Wuyts J, Rouze P, Van de Peer Y. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics. 2004;20:2911–2917. doi: 10.1093/bioinformatics/bth374. [DOI] [PubMed] [Google Scholar]
- Clark AG, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich IM, Purugganan MD. Sequence variation of micro-RNAs and their binding sites in Arabidopsis. Plant Physiol. 2008;146:1974–1982. doi: 10.1104/pp.108.116582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A. Changing effective population size and the McDonald-Kreitman test. Genetics. 2002;162:2017–2024. doi: 10.1093/genetics/162.4.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felippes FF, Schneeberger K, Dezulian T, Huson DH, Weigel D. Evolution of Arabidopsis thaliana microRNAs from random sequences. RNA. 2008;14:2455–2459. doi: 10.1261/rna.1149408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Kim J. Molecular evolution of Drosophila odorant receptor genes. Mol Biol Evol. 2007;24:1198–1207. doi: 10.1093/molbev/msm038. [DOI] [PubMed] [Google Scholar]
- Han J, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Near neutrality leading edge of the neutral theory of molecular evolution. Ann N Y Acad Sci. 2008;1133:162–179. doi: 10.1196/annals.1438.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HM, editor. Mammalian protein metabolism. New York: Academic; 1969. p. 21–132. [Google Scholar]
- Lau PW, MacRae IJ. The molecular machines that mediate microRNA maturation. J Cell Mol Med. 2009;13:54–60. doi: 10.1111/j.1582-4934.2008.00520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li A, Mao L. Evolution of plant microRNA gene families. Cell Res. 2007;17:212–218. doi: 10.1038/sj.cr.7310113. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, et al. Adaptive evolution of newly emerged micro-RNA genes in Drosophila. Mol Biol Evol. 2008a;25:929–938. doi: 10.1093/molbev/msn040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, et al. The birth and death of microRNA genes in Drosophila. Nat Genet. 2008b;40:351–355. doi: 10.1038/ng.73. [DOI] [PubMed] [Google Scholar]
- Manak JR, et al. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Mizrokhi LJ, Georgieva SG, Ilyin YV. jockey, a mobile Drosophila element similar to mammalian LINEs, is transcribed from the internal promoter by RNA polymerase II. Cell. 1988;54:685–691. doi: 10.1016/s0092-8674(88)80013-8. [DOI] [PubMed] [Google Scholar]
- Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M, Nei M. Evolutionary dynamics of olfactory receptor genes in Drosophila species. Proc Natl Acad Sci USA. 2007;104:7122–7127. doi: 10.1073/pnas.0702133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piriyapongsa J, Jordan IK. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS One. 2007;2:e203. doi: 10.1371/journal.pone.0000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piriyapongsa J, Jordan IK. Dual coding of siRNAs and miRNAs by plant transposable elements. RNA. 2008;14:814–821. doi: 10.1261/rna.916708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007a;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, et al. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007b;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer SW, et al. Polytene chromosomal maps of 11 Drosophila species: the order of genomic scaffolds inferred from genetic and physical maps. Genetics. 2008;179:1601–1655. doi: 10.1534/genetics.107.086074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina SA, Koonin EV. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol. 2008;23:578–587. doi: 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, et al. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res. 2007;17:1865–1879. doi: 10.1101/gr.6593807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
- Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J Mol Biol. 2004;339:327–335. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- Vieira FG, Sanchez-Gracia A, Rozas J. Comparative genomic analysis of the odorant-binding protein family in 12 Drosophila genomes: purifying selection and birth-and-death evolution. Genome Biol. 2007;8:R235. doi: 10.1186/gb-2007-8-11-r235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Yang Z, Kumar S, Nei M. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics. 1995;141:1641–1650. doi: 10.1093/genetics/141.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Tabin CJ, Bartel DP. MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nat Rev Genet. 2008;9:789–796. doi: 10.1038/nrg2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rosenberg HF, Nei M. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc Natl Acad Sci USA. 1998;95:3708–3713. doi: 10.1073/pnas.95.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QH, et al. A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res. 2008;18:1456–1465. doi: 10.1101/gr.075572.107. [DOI] [PMC free article] [PubMed] [Google Scholar]