Abstract
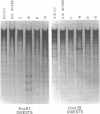
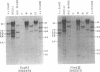
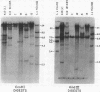
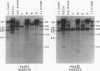
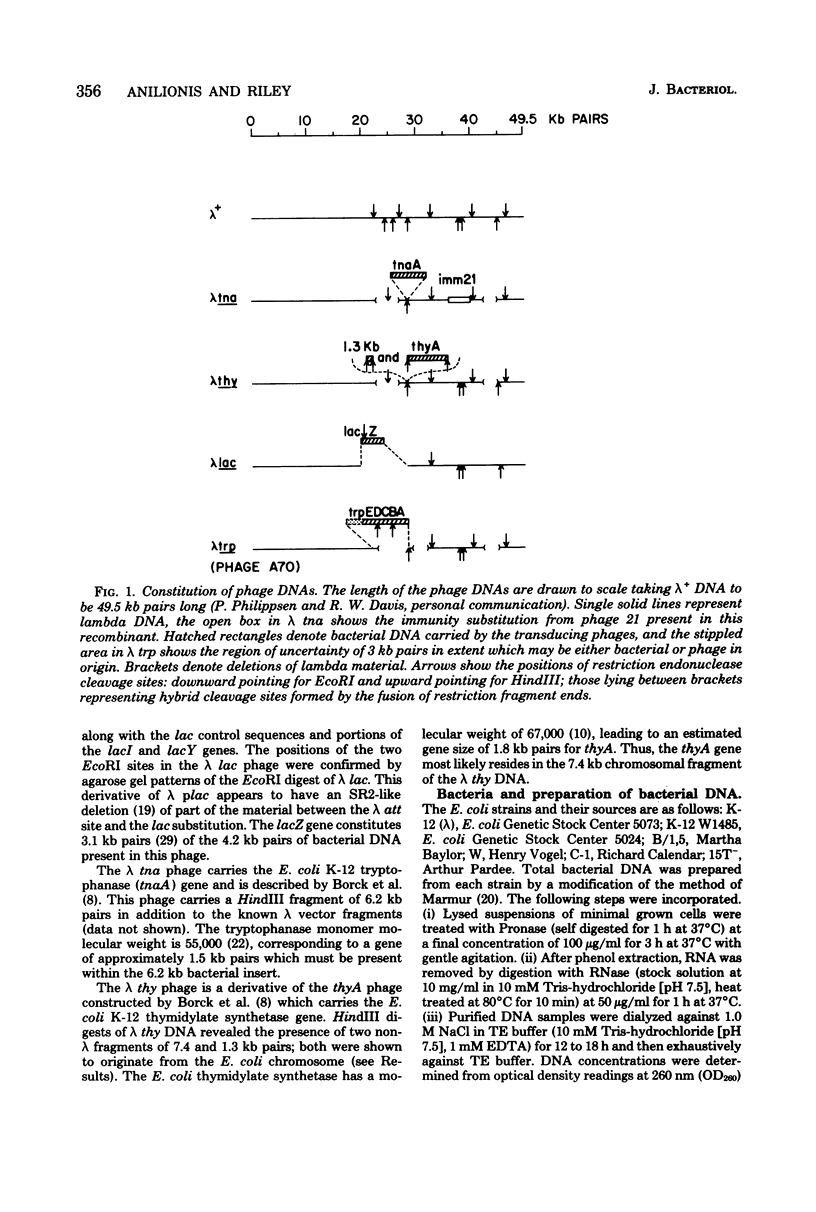
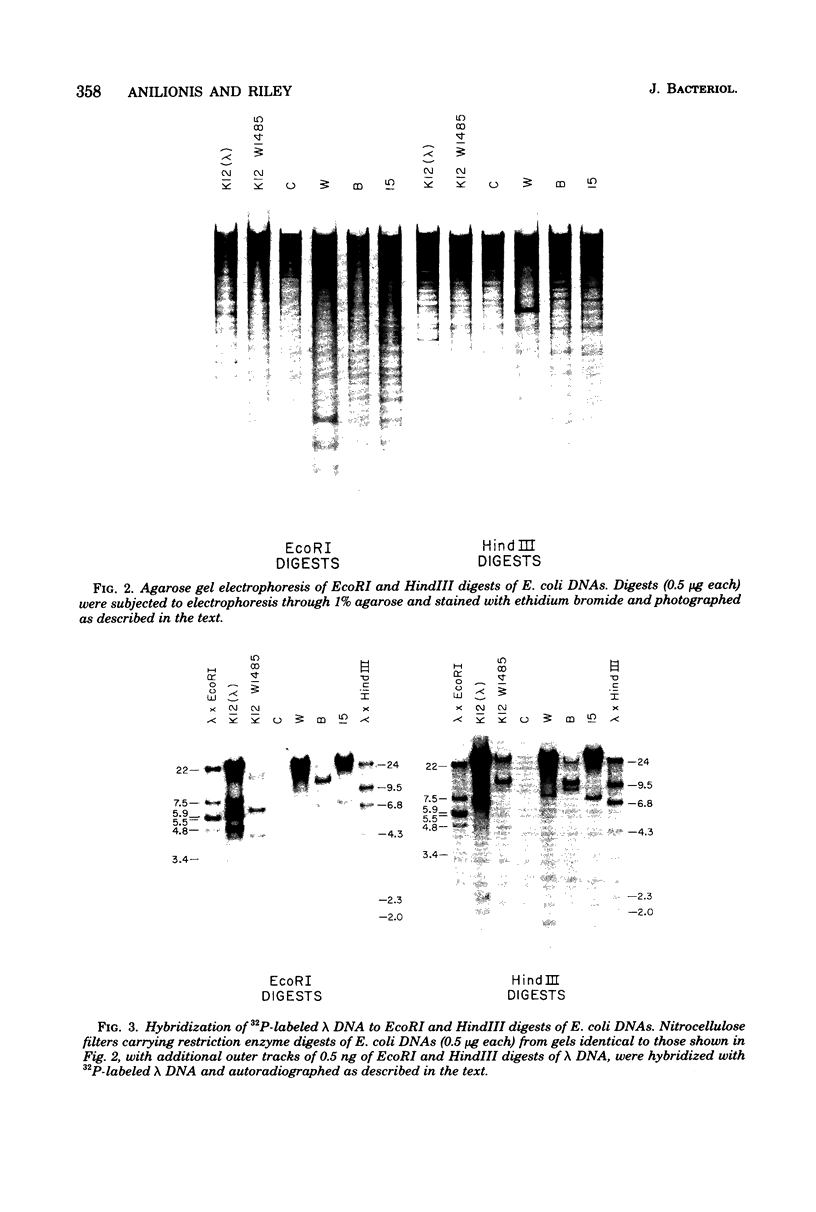
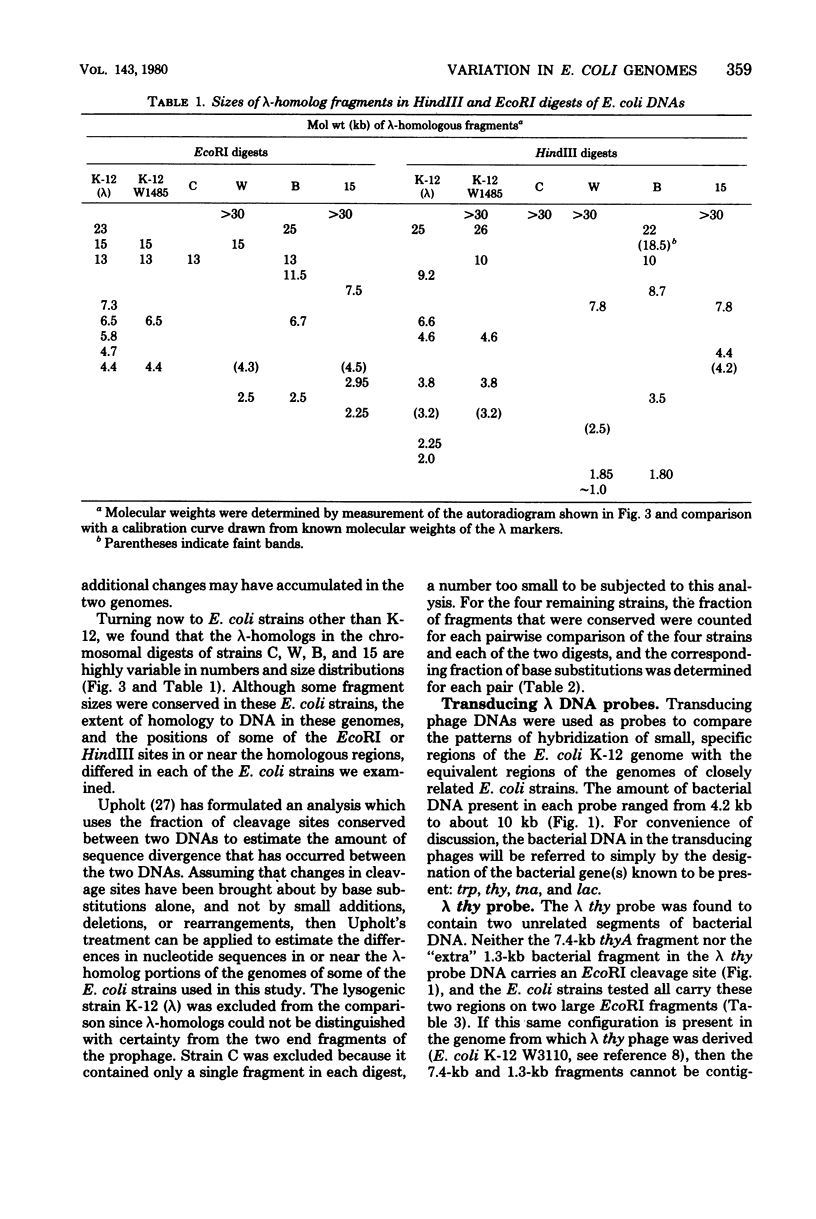
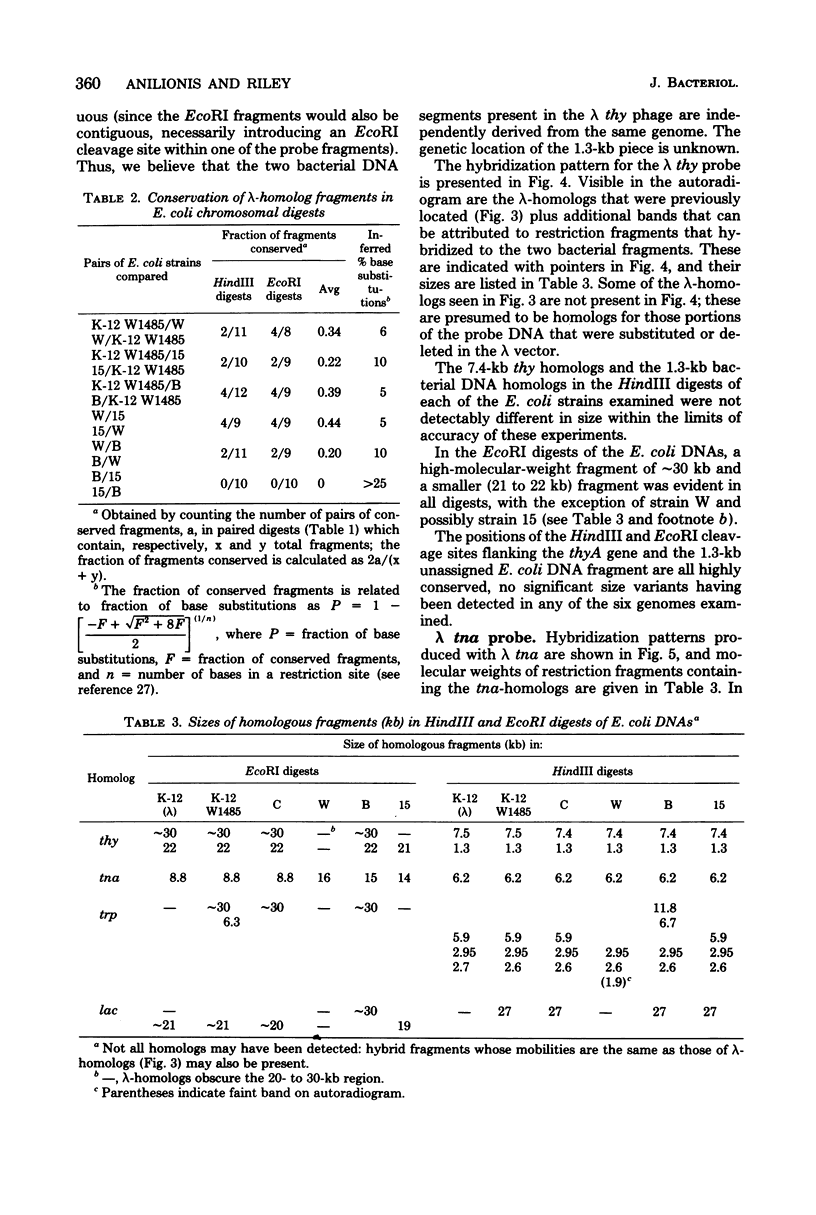
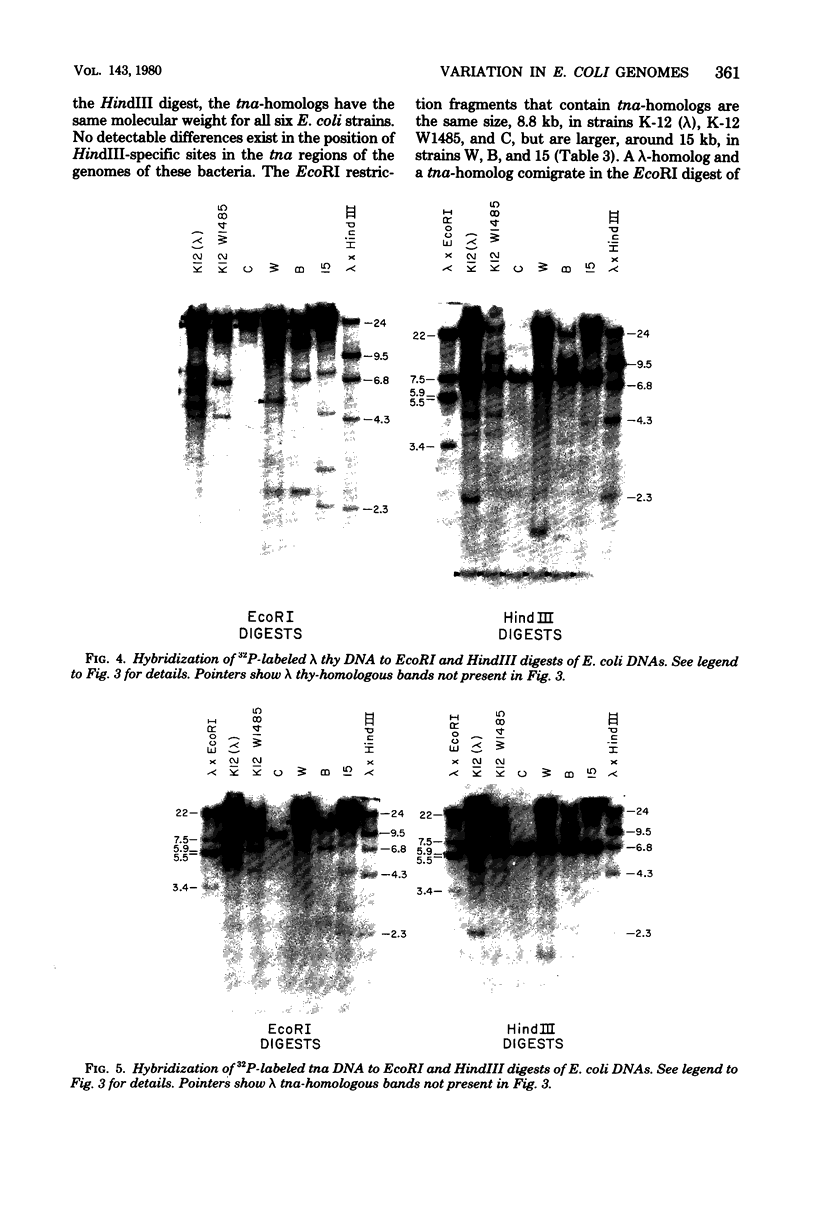
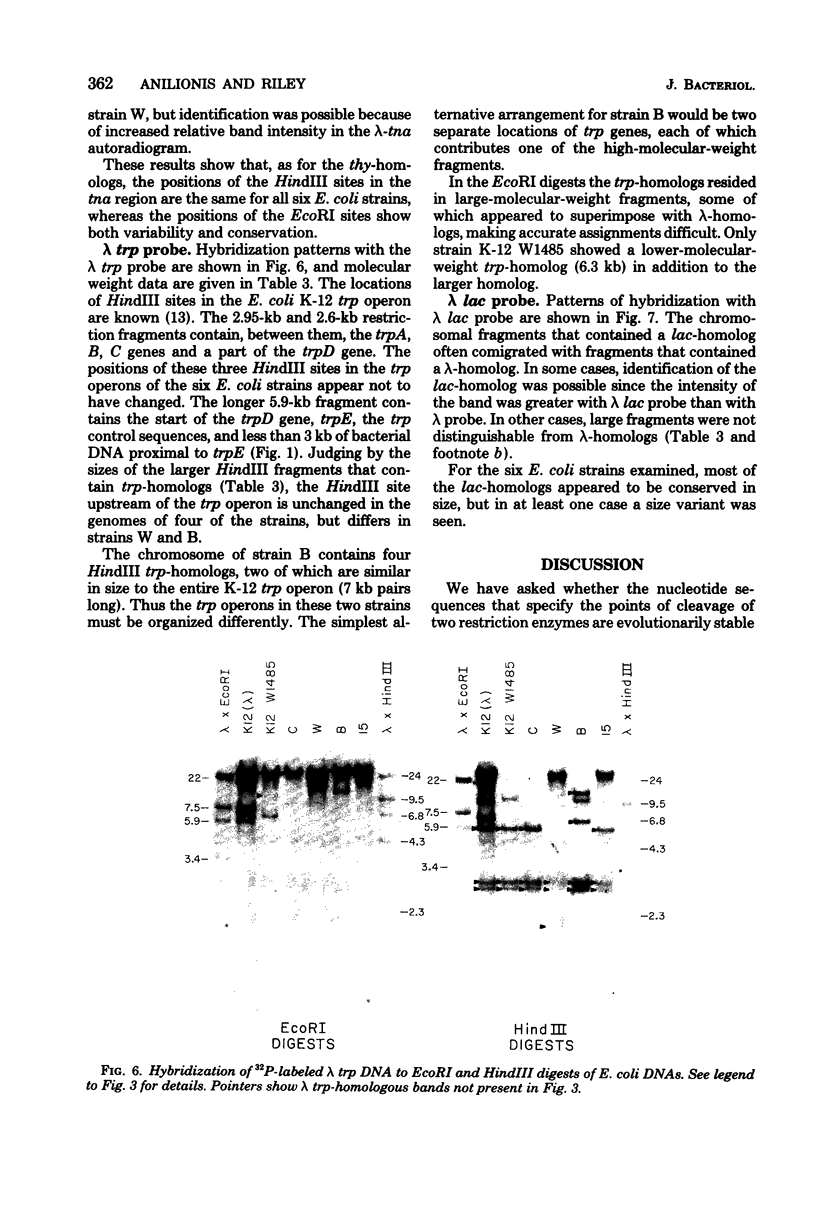
Changes in the patterns produced by annealing restriction endonuclease digests of bacterial genomes with probe deoxyribonucleic acids (DNAs) containing small portions of a bacterial genome provide sensitive indicator of the degree of nucleotide sequence relatedness that exists in localized regions of the genomes of closely related bacteria. We have used five probe DNAs to explore the relatedness of parts of the genomes of six laboratory Escherichi coli strains. A range in in the amount of variability in the positions of restriction enzyme cleavage sites in the selected portions of the genomes was found. Portions of the genome that are believed to be inacative were more variable than portions that contained functional genes: the sites in and near regions of homology to phage lambda DNA in the genome showed the greatest variability. These regions probably represent remnants of cryptic prophages. Variability was assessed pairwise among four of the E. coli strains and ranged from 5 to > 25% base pair substitutions in the lambda-related regions. In contrast, the endonuclease cleavage sites in the trp, tna, lac, thy regions, and one other as-yet-unidentified segment of the genome were more highly conserved. It seems likely that these sites lie in genetic locations that are subject to functional constraints.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendich A. J., McCarthy B. J. Ribosomal RNA homologies among distantly related organisms. Proc Natl Acad Sci U S A. 1970 Feb;65(2):349–356. doi: 10.1073/pnas.65.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D. E., Drummond M. Absence of DNA sequences homologous to transposable element Tn5 (Kan) in the chromosome of Escherichia coli K-12. J Bacteriol. 1978 Oct;136(1):419–422. doi: 10.1128/jb.136.1.419-422.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borck K., Beggs J. D., Brammar W. J., Hopkins A. S., Murray N. E. The construction in vitro of transducing derivatives of phage lambda. Mol Gen Genet. 1976 Jul 23;146(2):199–207. doi: 10.1007/BF00268089. [DOI] [PubMed] [Google Scholar]
- Brenner D. J., Falkow S. Genetics of the Enterobacteriaceae. C. Molecular relationships among members of the Enterobacteriaceae. Adv Genet. 1971;16:81–118. doi: 10.1016/s0065-2660(08)60355-7. [DOI] [PubMed] [Google Scholar]
- Brenner D. J., Fanning G. R., Skerman F. J., Falkow S. Polynucleotide sequence divergence among strains of Escherichia coli and closely related organisms. J Bacteriol. 1972 Mar;109(3):953–965. doi: 10.1128/jb.109.3.953-965.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner D. J., Fanning G. R., Steigerwalt A. G., Sodd M. A., Doctor B. P. Conservation of transfer ribonucleic acid and 5S ribonucleic acid cistrons in Enterobacteriaceae. J Bacteriol. 1977 Mar;129(3):1435–1439. doi: 10.1128/jb.129.3.1435-1439.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crusberg T. C., Leary R., Kisliuk R. L. Properties of thymidylate synthetase from dichloromethotrexate-resistant Lactobacillus casei. J Biol Chem. 1970 Oct 25;245(20):5292–5296. [PubMed] [Google Scholar]
- Evans R., Seeley N. R., Kuempel P. L. Loss of rac locus DNA in merozygotes of Escherichia coli K12. Mol Gen Genet. 1979 Oct 1;175(3):245–250. doi: 10.1007/BF00397223. [DOI] [PubMed] [Google Scholar]
- Goldberg A. R., Howe M. New mutations in the S cistron of bacteriophage lambda affecting host cell lysis. Virology. 1969 May;38(1):200–202. doi: 10.1016/0042-6822(69)90148-2. [DOI] [PubMed] [Google Scholar]
- Hopkins A. S., Murray N. E., Brammar W. J. Characterization of lambdatrp-transducing bacteriophages made in vitro. J Mol Biol. 1976 Nov 15;107(4):549–569. doi: 10.1016/s0022-2836(76)80082-4. [DOI] [PubMed] [Google Scholar]
- Hori H., Osawa S. Evolutionary change in 5S RNA secondary structure and a phylogenic tree of 54 5S RNA species. Proc Natl Acad Sci U S A. 1979 Jan;76(1):381–385. doi: 10.1073/pnas.76.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippen K., Shapiro J. A., Beckwith J. R. Transposition of the lac region to the gal region of the Escherichia coli chromosome: isolation of lambda-lac transducing bacteriophages. J Bacteriol. 1971 Oct;108(1):5–9. doi: 10.1128/jb.108.1.5-9.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAISER A. D. The production of phage chromosome fragments and their capacity for genetic transfer. J Mol Biol. 1962 Apr;4:275–287. doi: 10.1016/s0022-2836(62)80005-9. [DOI] [PubMed] [Google Scholar]
- Kaiser K., Murray N. E. Physical characterisation of the "Rac prophage" in E. coli K12. Mol Gen Genet. 1979 Sep;175(2):159–174. doi: 10.1007/BF00425532. [DOI] [PubMed] [Google Scholar]
- Lee F., Bertrand K., Bennett G., Yanofsky C. Comparison of the nucleotide sequences of the initial transcribed regions of the tryptophan operons of Escherichia coli and Salmonella typhimurium. J Mol Biol. 1978 May 15;121(2):193–217. doi: 10.1016/s0022-2836(78)80005-9. [DOI] [PubMed] [Google Scholar]
- Malamy M. H., Fiandt M., Szybalski W. Electron microscopy of polar insertions in the lac operon of Escherichia coli. Mol Gen Genet. 1972;119(3):207–222. doi: 10.1007/BF00333859. [DOI] [PubMed] [Google Scholar]
- McConaughy B. L., Laird C. D., McCarthy B. J. Nucleic acid reassociation in formamide. Biochemistry. 1969 Aug;8(8):3289–3295. doi: 10.1021/bi00836a024. [DOI] [PubMed] [Google Scholar]
- Morino Y., Snell E. E. The subunit structure of tryptophanase. I. The effect of pyridoxal phosphate on the subunit structure and physical properties of tryptophanase. J Biol Chem. 1967 Dec 10;242(23):5591–5601. [PubMed] [Google Scholar]
- Nichols B. P., Yanofsky C. Nucleotide sequences of trpA of Salmonella typhimurium and Escherichia coli: an evolutionary comparison. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5244–5248. doi: 10.1073/pnas.76.10.5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sato K., Campbell A. Specialized transduction of galactose by lambda phage from a deletion lysogen. Virology. 1970 Jul;41(3):474–487. doi: 10.1016/0042-6822(70)90169-8. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Upholt W. B. Estimation of DNA sequence divergence from comparison of restriction endonuclease digests. Nucleic Acids Res. 1977;4(5):1257–1265. doi: 10.1093/nar/4.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Fox G. E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]