Abstract
NADH cytochrome b5 oxidoreductase (Ncb5or) is found in animals and contains three domains similar to cytochrome b5 (b5), CHORD-SGT1 (CS), and cytochrome b5 reductase (b5R). Ncb5or has an important function, as suggested by the diabetes and lipoatrophy phenotypes in Ncb5or null mice. To elucidate the structural and functional properties of human Ncb5or, we generated its individual b5 and b5R domains (Ncb5or-b5 and Ncb5or-b5R, respectively) and compared them with human microsomal b5 (Cyb5A) and b5R (Cyb5R3). A 1.25 Å x-ray crystal structure of Ncb5or-b5 reveals nearly orthogonal planes of the imidazolyl rings of heme-ligating residues His89 and His112, consistent with a highly anisotropic low spin EPR spectrum. Ncb5or is the first member of the cytochrome b5 family shown to have such a heme environment. Like other b5 family members, Ncb5or-b5 has two helix-loop-helix motifs surrounding heme. However, Ncb5or-b5 differs from Cyb5A with respect to location of the second heme ligand (His112) and of polypeptide conformation in its vicinity. Electron transfer from Ncb5or-b5R to Ncb5or-b5 is much less efficient than from Cyb5R3 to Cyb5A, possibly as a consequence of weaker electrostatic interactions. The CS linkage probably obviates the need for strong interactions between b5 and b5R domains in Ncb5or. Studies with a construct combining the Ncb5or CS and b5R domains suggest that the CS domain facilitates docking of the b5 and b5R domains. Trp114 is an invariant surface residue in all known Ncb5or orthologs but appears not to contribute to electron transfer from the b5R domain to the b5 domain.
Keywords: Cytochrome b, Electron Paramagnetic Resonance (EPR), Electron Transfer, FAD, Heme, Kinetics, NADH, Protein Structure, Reductase, X-ray Crystallography
Introduction
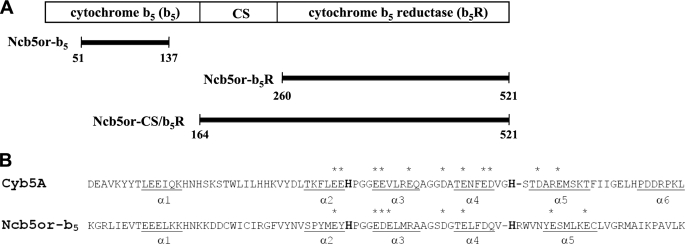
NADH cytochrome b5 oxidoreductase (Ncb5or; also named Cyb5R4, b5/b5R, or b5 + b5R) was cloned from humans as a natural fusion protein containing structural homolog of both cytochrome b5 (b5)3 and cytochrome b5 reductase (b5R) (1). Ncb5or is found only in animals and is expressed in a wide range of tissues and cells. Human Ncb5or contains 521 amino acid residues and three structural domains. The N-terminal b5 domain and the C-terminal b5R domain are linked by a CS (CHORD-SGT1) domain comprising ∼90 residues (Fig. 1A). Several natural fusion proteins are known that contain a cytochrome b5 domain and a second redox-active domain. Examples include sulfite oxidase (2–4) and Δ5 and Δ6 fatty acid desaturases (5, 6) in animals, nitrate reductase in algae (7) and plants (8, 9), and flavocytochrome b2 or lactate dehydrogenase (10) and Δ9 fatty acid desaturase (11) in bakers' yeast. However, Ncb5or is the only member of the cytochrome b5 superfamily known to contain three distinct domains. The function of the non-redox-active CS domain is presently unknown, although its primary sequence is distantly homologous to those in human heat shock protein 20 (HSP20, a co-chaperone of HSP90) and other CS family members (12).
FIGURE 1.
A, schematic diagram of individual domains in human Ncb5or. Cytochrome b5 and cytochrome b5 reductase domains are at the N and C terminus, respectively, with the CS domain in between Ncb5or-b5, Ncb5or-b5R, and Ncb5or-CS/b5R are three constructs used in this study. B, sequence alignment of human Ncb5or (bottom) and human Cyb5A (top) showing all helical segments (i.e. α1–α6 in Cyb5A and α1–α5 in Ncb5or-b5) as well as negatively charged surface residues in core 1 that are conserved in all vertebrate orthologs of each protein (asterisks). In boldface type are heme-ligating residues, His44 and His68 in Cyb5A and His89 and His112 in Ncb5or-b5.
The cytochrome b5 family includes two isoforms in vertebrates, one anchored to the membrane of the endoplasmic reticulum (Cyb5A) and the other anchored to the outer mitochondrial membrane (Cyb5B) (13). Membrane anchoring in Cyb5A and Cyb5B is accomplished by a hydrophobic C-terminal domain. The polar heme-binding domains of Cyb5A and Cyb5B have virtually identical folds with secondary structure elements occurring in the order β1-α1-β4-β3-α2-α3-β5-α4-α5-β2-α6 (14, 15). The structure has been described as comprising two hydrophobic cores separated by a five-stranded β-sheet (16). All cytochrome b5 proteins contain a redox active heme that is ligated by the imidazolyl side chains of two histidine residues. Heme binds in hydrophobic core 1 (α2–α5) and is sandwiched between two helix-loop-helix motifs. The heme ligands reside in the loops of these motifs, one between helices α2 and α3 and the other between helices α4 and α5 (Fig. 1B). Hydrophobic core 2 contains helix α1, which is located near the polypeptide N terminus as well as C-terminal helix α6. Rather substantial variations on this classic fold have been observed in a variety of b5 superfamily members, including the b5 domains of sulfite oxidase (3) and cytochrome b2 (10). The amino acid sequence homology of the b5 cores is very high among vertebrate orthologs of Cyb5A (supplemental Fig. 1), among Cyb5B family members (data not shown), and among Ncb5or orthologs (supplemental Fig. 2). In contrast, the b5 cores of human Cyb5A and human Ncb5or share only 31% identity and 52% similarity. In addition, heme ligand His112 in Ncb5or is displaced by one residue relative to the corresponding histidine residue in Cyb5A (Fig. 1B). This suggests substantial divergence of structural and functional properties of these two members of the cytochrome b5 superfamily.
Four b5R isoforms have been identified in humans, Cyb5R1-Cyb5R4 (1, 17). These and all other b5R family members belong to the ferredoxin-NADP+ reductase superfamily (18), and contain conserved amino acid residues responsible for FAD and NAD(P)H binding. Microsomal b5R (Cyb5R3) associates with both endoplasmic reticulum (ER) and outer mitochondrial membranes via a myristoyl group (19) and is the cognate reductase for both Cyb5A and Cyb5B (20). Ncb5or (Cyb5R4), the only b5R isoform in animals that contains more than one domain, is localized to the ER (21). Previous studies have shown that recombinant Ncb5or is soluble and that its heme is reduced instantaneously when excess NADH (or NADPH) is present (21). Kinetic measurements have revealed that human and mouse Ncb5or can reduce a number of artificial substrates in vitro, such as cytochrome c, methemoglobin, ferricyanide, and even molecular oxygen (1, 21).
Biochemical studies with in vitro reconstitution show that the Cyb5R3/Cyb5A pair serves as an electron source for stearoyl-CoA desaturase (SCD) in fatty acid desaturation (22). However, recent studies comparing normal and liver-specific Cyb5A knock-out mice have revealed no difference in the SCD index of liver microsomal lipids (23), and global Cyb5A knock-out mice do not display major phenotype in lipid metabolism (24). Notably, mice lacking the Ncb5or gene exhibit impaired SCD activity and develop early onset diabetes and lipoatrophy (25–27), suggesting that Ncb5or, not Cyb5A, functions in vivo as an electron donor in the SCD reaction. These discoveries have motivated us to further characterize the structural and functional properties of Ncb5or. Herein, we describe the results of studies with recombinant proteins representing the individual b5 and b5R domains of human Ncb5or (designated as Ncb5or-b5 and Ncb5or-b5R, respectively) as well as one containing both the CS and b5R domains (Ncb5or-CS/b5R). We report a 1.25 Å x-ray crystal structure of Ncb5or-b5, which reveals a heme environment that is unique among known cytochrome b5 superfamily members. We also report kinetic data showing that Ncb5or-b5 can be reduced by Ncb5or-b5R in the presence of excess NADH, albeit much less efficiently than the corresponding reaction in full-length Ncb5or or in Cyb5A/Cyb5R3. Finally, we provide evidence that the CS domain plays a role in facilitating interactions between the b5 and b5R domains.
MATERIALS AND METHODS
Molecular Cloning and Site-directed Mutagenesis
On the basis of structure predictions using the online I-TASSER server (28), we assigned Lys51–Lys137 for Ncb5or-b5, Lys260–Ala521 for Ncb5or-b5R, and Gly164–Ala521 for Ncb5or-CS/b5R (schematic diagram in Fig. 1A). The cDNA fragment of wild-type Ncb5or-b5 (no polyhistidine tag) was synthesized and cloned into pET22b vector by Genscript Inc. (Piscataway, NJ). For comparison purposes, we required soluble proteins representing human Cyb5A and Cyb5R3. An expression plasmid for human erythrocyte cytochrome b5, which is identical to the soluble heme-binding domain of human Cyb5A with the exception of the C-terminal residue, was kindly provided to us by Dr. Grant Mauk (29) and is herein referred to as Cyb5A. Its cDNA was subcloned into pET19b vector. An expression construct of human Cyb5R3 (residues Ile34–Phe301) was generated in our laboratory on the basis of previously published reports (30). A 6-His tag was added to the NH2 terminus of Ncb5or-b5R, Ncb5or-CS/b5R, and human Cyb5R3 through the respective PCR primers and cloned into pET19b vector. Site-directed mutagenesis was performed to generate Ncb5or-b5 mutants, R113A and W114A, with the Stratagene QuikChange mutagenesis kit (La Jolla, CA) and the full-length wild-type Ncb5or cDNA as the PCR template. The mutated DNA fragment was then subcloned into the pET22b. Primers with the desired codon change were designed with Stratagene's software available on the World Wide Web. All oligonucleotides were synthesized by Integrated DNA Technology (Coralville, IA) and the sequences are available upon request.
Protein Preparation
Soluble forms of each recombinant protein were generated in E. coli BL21(DE3) cells that were transformed with the respective expression construct. For Ncb5or-b5 and Cyb5A, cells were grown at 37 °C in LB medium to an A600 of ∼0.7 before isopropyl 1-thio-β-d-galactopyranoside induction (1 mm) for 6 h at 25 °C. Cells were collected by centrifugation at 5000 × g (4 °C) for 30 min and used immediately, or the pellet was kept at −80 °C until ready for use. Human Cyb5A and Ncb5or-b5 were initially obtained as mixtures of holo (heme-bound) and apo (heme-free) forms. Hemin was added to the crude cell lysates in order to convert the apo forms to the holo forms (31). The holoprotein was purified to homogeneity by ion exchange, hydrophobic interaction, and size exclusion column chromatography consecutively using HiTrap Q HP, HiTrap Phenyl HP, and Superdex 200 columns at a flow rate of 1.0, 1.0, and 0.5 ml/min, respectively. For HiTrap Q HP, a linear gradient between 0 and 1 m NaCl in 20 mm Tris-HCl (pH 7.2) was used to elute all proteins in 20 column volumes. The fractions with red color were collected, pooled, and loaded directly onto HiTrap Phenyl HP to collect flow-through. For Superdex 200, the running buffer was 20 mm Tris-HCl (pH 7.2). All purifications were conducted using an ÄKTAxpress purification system (GE Healthcare) at 4 °C. SDS-PAGE was used to determine protein purity, and native PAGE was utilized to confirm the absence of residual apoprotein upon the completion of holo-Ncb5or-b5 and -Cyb5A purification (32). Small aliquots of concentrated Ncb5or-b5 and Cyb5A samples were flash frozen in liquid nitrogen and stored at −80 °C until use. For Ncb5or-b5R, Ncb5or-CS/b5R, and Cyb5R3, the same conditions were used, except cells were grown in TB medium and supplemented with 0.1 mm riboflavin (33). Expression was induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside overnight at 15 °C. Both Ncb5or-b5R and Cyb5R3 proteins were purified to homogeneity with Ni2+-NTA chelation and size exclusion column chromatography using HisTrap HP and Size Exclusion 100 columns at a flow rate of 1.0 and 0.5 ml/min, respectively. For HisTrap HP, the sample was loaded in 20 mm Tris-HCl, 500 mm NaCl, 10 mm imidazole (pH 8); washed with 20 mm imidazole for 10 column volumes; and then eluted with a linear gradient of 20–500 mm imidazole in 10 column volumes. The fractions with yellow color were collected and pooled. For Size Exclusion 100, the running buffer was 20 mm Tris-HCl, 500 mm NaCl, 0.1 mm EDTA (pH 7.2), and the yellow fractions were collected. The Ncb5or-CS/b5R was purified in one step by using affinity chromatography with Ni2+-NTA beads (Qiagen). The sample was loaded in 50 mm Tris-HCl, 300 mm NaCl (pH 8), washed with 10 mm imidazole (10 column volumes), and then eluted in 200 mm imidazole. The yellow fractions were collected and dialyzed exhaustively against 20 mm Tris-HCl, 500 mm NaCl, 0.1 mm EDTA (pH 7.2). Final yields of purified protein were 5–10 mg/liter for Ncb5or-b5, ≥20 mg/liter for Cyb5A, 2 mg/liter for Ncb5or-b5R, 1 mg/liter for Ncb5or-CS/b5R, and 10 mg/liter for Cyb5R3. All polypeptide products have the expected molecular weights by electrospray ionization mass spectrometry (University of Kansas Mass Spectrometry Laboratory). UV-visible spectra showed A413/A280 ratios of 4.1 and 6.4 for holo-Ncb5or-b5 and holo-Cyb5A, respectively. The FAD contents of Ncb5or-b5R, Ncb5or-CS/b5R, and Cyb5R3 were determined by A461 and used to represent enzyme concentrations.
Spectroscopy
UV-visible spectra were obtained using a Varian Cary 50 Bio spectrophotometer or a Varian 100 Bio equipped with a Peltier-thermostated multiple cell holder and a dedicated temperature probe accessory (±0.1 °C). The concentrations of heme and FAD were determined by the following ϵ values (mm−1 cm−1): 130 (413 nm) of oxidized heme in Ncb5or-b5 and Cyb5A (21), 10.5 (461 nm) of FAD in Ncb5or-b5R, Ncb5or-CS/b5R, and Cyb5R3 (33). Electron paramagnetic resonance (EPR) spectroscopy was performed on a Bruker Elexys E500 spectrometer operating at X-band frequencies. Temperature control was maintained by an Oxford ESR 900 continuous flow liquid helium cryostat interfaced with an Oxford ITC 503 temperature controller. Typical EPR parameters were as follows: sample temperature, 7 K; microwave frequency, 9.382 GHz; microwave power, 1 milliwatt; modulation frequency, 100 kHz; modulation amplitude, 5G. These conditions provided clean spectra without saturation of the signals. EPR data acquisition and background subtraction were performed using XeprView software (Bruker).
Crystallization and Structure Solution
Concentrated human Ncb5or-b5 (20 mg/ml in 20 mm Tris-HCl, pH 7.0) was screened for crystallization in Compact Jr. (Emerald Biosystems) sitting drop plates using 0.5 μl of protein and 0.5 μl of crystallization solution equilibrated against 100 μl of the latter. Red plate-shaped crystals were obtained in ∼3 days from the Wizard 2 screen (Emerald Biosystems) condition 45 (2 m (NH4)2SO4, 100 mm Tris-HCl, pH 7.0, 200 mm Li2SO4) at 4 °C. Crystal growth conditions were optimized using the pH buffer screen (Emerald Biosystems). Large single plate-shaped crystals were obtained from 2 m (NH4)2SO4, 100 mm sodium/potassium phosphate, pH 6.2, 200 mm Li2SO4 after ∼1 week at 4 °C. Crystals were equilibrated for 30 s in the same solution plus 25% glycerol and frozen for data collection at the Advanced Photon Source. Initial diffraction data were collected in house at 93 K using a Rigaku RU-H3 rotating anode generator (Cu-Kα) equipped with Osmic Blue focusing mirrors and a Rigaku Raxis IV++ image plate detector (University of Kansas Protein Structure Laboratory). Crystals obtained from the initial crystallization screen were used for data collection. The Matthews coefficient (34) (Vm = 2.2, 44.4% solvent) suggested that there were two molecules in the asymmetric unit. Additionally, the self-rotation function yielded a peak on the κ = 180° section at ω = 55.1°, φ = 180°, indicating the presence of a non-crystallographic 2-fold axis. Structure solution was carried out by molecular replacement with BALBES (35) in the space group P21, which produced a homology model for the rotation and translation searches from a high resolution structure of rat Cyb5B (Protein Data Bank entry 1EUE) (36). No solution was found in the space group P2. Initial refinement of the model following molecular replacement converged at r = 38.8%. The model was improved by automated building with BUCANNEER (37), which converged at r = 31.4%. A final model was obtained from subsequent rounds of structure refinement and manual model building. Atomic resolution diffraction data were collected at 100 K at the Advanced Photon Source Industrial Macromolecular Crystallography Association Collaborative Access Team beamline 17ID using an ADSC Quantum 210r CCD detector. Crystals obtained from the optimized growth conditions described above were used for synchrotron data collection. Intensities were integrated and scaled with the XDS (38) software package, and the models obtained from in-house diffraction data were used for molecular replacement with MOLREP (39). Refinement and model building were carried out with REFMAC (40) and COOT (41), respectively, and the final model was refined with anisotropic displacement parameters. Structure validation was conducted with Molprobity (42), and figures were prepared with the RIBBONS and CCP4MG packages (43, 44). There were two molecules in the asymmetric unit related by a non-crystallographic 2-fold axis. The final model was refined to 1.25 Å resolution and contained, in addition to the two polypeptide chains (denoted A and B), two heme molecules, two sulfate ions, and 73 water molecules. Both heme molecules in subunit A and B of Ncb5or-b5 adopt two orientations, as shown by a 2Fo − Fc electron density map (supplemental Fig. 3). The coordinates of human Ncb5or-b5 have been deposited in the Protein Data Bank (entry 3LF5). Crystallographic data are summarized in supplemental Table 1.
Electrostatic Map
The electrostatic maps of human Ncb5or-b5 and bovine Cyb5A were calculated from their x-ray structures using the Adaptive Poisson- Boltzmann Solver software plugin (45) in PyMOL (DeLano Scientific LLC). The necessary PQR files were generated from the Protein Data Bank files using the PDB 2PQR server (available on the World Wide Web) (46), and the pKa values were assigned using the PROPKA software (47). The parameters set for a typical calculation are as follows: internal dielectric constant = 2.0, external dielectric constant = 80.0, solvent probe radius = 1.4 Å, and temperature = 298 K. Visualization of electrostatic maps was performed with PyMOL.
Interdomain Electron Transfer
Interdomain electron transfer was measured under oxygen-free conditions in a regulated gas flow device that has been described previously (21). The substrate and reductant mixture (final volume 1.5 ml) was equilibrated with a slow stream of moisturized nitrogen (purity 99.999%, from Linweld (Kansas City, MO)) for 30 min in a reaction vessel. Upon the injection of a small aliquot of reductase (<5 μl), the complete mixture was pushed by N2 flow into a sealed quartz cuvette in a Varian Cary 50 spectrophotometer. Data collection was initiated after a dead time of <20 s due to mixing and reaction transfer. Reduction of heme was monitored by the increase of absorbance at the Soret band (ϵ424 = 120 mm−1 cm−1 (Ncb5or-b5) or 134 mm−1 cm−1 (Cyb5A)). Data were fit to a single exponential function and the resulting pseudo-first order rate constant was dissolved by the appropriate enzyme concentration to obtain the reported observed rate constants (min−1 μm−1). Two buffer conditions were examined: (i) 5 mm sodium phosphate and (ii) 50 mm sodium phosphate, pH 7.0, with ionic strength (μ) of 0.009 and 0.108 m, respectively.
Measurement of Michaelis-Menten Parameters
Ncb5or-b5 (0.7–100 μm) or Cyb5A (1.4–50 μm) was reduced by Ncb5or-b5R (140 nm) or Cyb5R3 (0.56 nm), respectively, in the presence of excess NADH (100 μm) in 5 mm sodium phosphate (pH 7.0, no air). All reactions were monitored by the absorbance increase of the Soret band (A424) when the concentration of substrate [S] was ≤10 μm or the β-peak (ϵ558 = 17.5 mm−1 cm−1 (Ncb5or-b5) or ϵ556 = 18.1 mm−1 cm−1 (Cyb5A)) when [S] was ≥10 μm. The Michaelis-Menten equation was used to fit V0 (initial rate) and [S] (substrate concentration) with SigmaPlot 10.0 software to generate Km and Vmax. Both V0 and the corresponding [S] are the actual values at the beginning of data collection.
All data points of the Cyb5A/Cyb5R3 pair fit well to the non-linear function, and the same results were obtained for the Cyb5A/Cyb5R3 pair whether or not a V0 = 0, [S] = 0 point was included. However, this was not the case with the Ncb5or pair, and consequently the (0, 0) point was omitted for data fitting. For reasons that have not been determined, a y intercept of 0.0061 ± 0.0011 was observed for the Ncb5or pair. The substrate is either Cyb5A or Ncb5or-b5, and the enzyme is either Cyb5R3 or Ncb5or-b5R.
Cytochrome c Reduction
Reduction of ferric horse cytochrome c (1.4 μm; Sigma) by the protein constructs described herein was performed in the presence of excess NADH (50 μm) in 5 mm phosphate (pH 7.0, no air). Reduction of heme was monitored by the increase of absorbance at 416 nm (Soret band). Data were fit to a single exponential function, and the resulting pseudo-first order rate constant was divided by the appropriate enzyme concentration (28 nm was used in all cases) to obtain the reported observed rate constants (min−1 μm−1).
RESULTS
Generation and Initial Characterization of Individual Redox Domains of Human Ncb5or
Recombinant proteins representing the individual redox domains of human Ncb5or (Fig. 1A), designated herein as Ncb5or-b5 and Ncb5or-b5R, were generated in order to (i) characterize the structure of the Ncb5or-b5 core and (ii) compare the nature of Ncb5or-b5/Ncb5or-b5R interactions with those exhibited by the well known Cyb5A/Cyb5R3 pair. We also generated a construct comprising the CS and b5R domains of Ncb5or (Ncb5or-CS/b5R) in order to explore possible roles played by the CS domain in interactions between the b5 and b5R domains. Human Ncb5or-b5 and human Cyb5A are virtually identical in stability, with thermal denaturation midpoints (Tm values) of 72 and 73.5 °C, respectively (supplemental Fig. 4). In contrast, Ncb5or-b5R is considerably less stable than Cyb5R3, as evidenced by its much lower expression yield and its much greater tendency toward loss of FAD and polypeptide aggregation during concentration following purification unless high salt levels were maintained (20 mm Tris-HCl, 500 mm NaCl, 0.1 mm EDTA, pH 7.0). Despite being obtained in a lower yield, Ncb5or-CS/b5R is more stable than Ncb5or-b5R. This suggests the possibility of favorable interactions between the CS and b5R domains, perhaps like those in full-length Ncb5or.
EPR Spectroscopy Indicates Different Heme Environments in Cyb5A and Ncb5or-b5
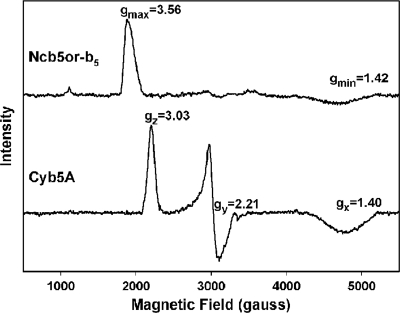
UV-visible spectra of Ncb5or-b5 (1) and Cyb5A (29) are quite similar and characteristic of low spin, bis-histidine heme ligation. However, a previously reported study comparing rat Cyb5A and Ncb5or revealed distinctly different EPR spectra (33). In the present study, we observed similar differences in EPR spectra for human Cyb5A and Ncb5or (Fig. 2). The EPR signals of ferric Cyb5A with g values of 3.03, 2.21, and 1.40 (Fig. 2, bottom) are typical of cytochrome b5 superfamily members and of most low spin, bis-histidine-ligated ferric heme proteins (supplemental Table 2). Such a spectrum reflects a rhombically distorted heme iron environment that arises when the dihedral angle between the planes of the His imidazolyl groups is <57° (48). In contrast, the EPR spectrum of ferric human Ncb5or-b5 is dominated by a signal at g = 3.56 (Fig. 2, top). This kind of spectrum, referred to as a highly anisotropic low spin (HALS) or “large gmax” spectrum (49, 50), is relatively rare for low spin bis-histidine-ligated heme proteins and indicative of iron in a tetragonally distorted environment (51, 52). HALS EPR spectra are defined as those in which the value of gz (gmax) is ≥3.2 and arise when the dihedral angle between the imidazolyl planes is >57° (48). The value of gmax for Ncb5or-b5 is close to the theoretical maximum of 3.8 (53).
FIGURE 2.
EPR spectra of human Ncb5or-b5 (top) and human Cyb5A (bottom). Ncb5or-b5 exhibits a HALS signal with gmax = 3.56 and gmin = 1.42 (1866 and 4683 gauss, respectively). Cyb5A shows a classic low spin axial heme spectrum with g values of 3.03 (z), 2.21 (y), and 1.40 (x), at 2183, 3016, and 4800 gauss, respectively.
Atomic Structure of Human Ncb5or-b5
A 1.25 Å x-ray crystal structure was obtained for Ncb5or-b5 with two protein molecules in the asymmetric unit. Although a solution structure of human Cyb5A determined by NMR has been deposited in the Protein Data Bank (entry 2I96), its x-ray crystal structure is not known. We will therefore use the 1.5 Å crystal structure of the bovine Cyb5A lipase fragment (Protein Data Bank entry 1CYO) (14) for comparison herein with our structure of Ncb5or-b5. The heme-binding domains of human and bovine Cyb5A differ at only four of the 87 residues that are involved in specific packing interactions in the bovine Cyb5A crystal structure (residues 6–92, supplemental Fig. 1). Two of those differences involve residues in the heme-binding pocket (core 1 or α2–α5), which is the focus of our current structural comparison. Neither of these two residues has been proposed to be involved in interactions with Cyb5R3, and furthermore, both are conservative mutations and result in no significant change in chemical properties (His27 and Met75 in human Cyb5A are replaced by Tyr and Leu, respectively, in bovine Cyb5A). The residue numbers used for Cyb5A in this report reflect those in the native protein sequences rather than the commonly employed numbering system that was devised by Mathews for the bovine Cyb5A lipase fragment in which Ser6 is designated Ser1 (54, 55).
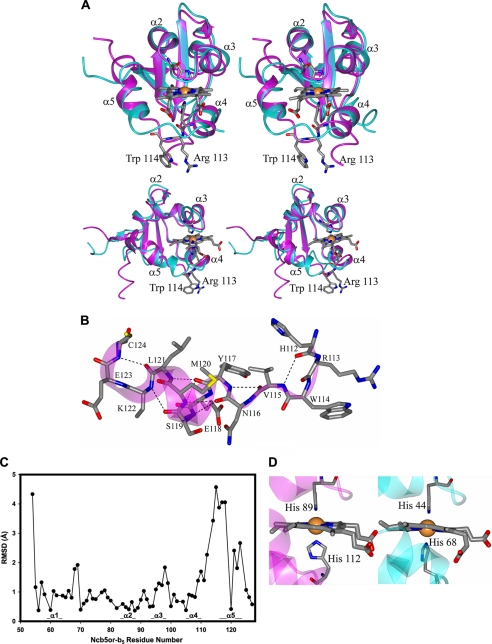
The structural overlay in Fig. 3A reveals that Ncb5or-b5 exhibits the same general fold as Cyb5A, with two hydrophobic cores separated by a five-stranded β-sheet. Ncb5or-b5 has three Cys residues, whereas Cyb5A does not have any, but none of the Cys residues in Ncb5or-b5 are involved in disulfide bridges or covalent linkages to heme. The heme in Ncb5or-b5 is ligated by the imidazolyl side chains of His89 and His112, confirming previously reported mutagenesis studies on rat Ncb5or (33). In addition, Fig. 3A shows that the backbone conformation in helices α2 and α3 and the intervening loop containing His89 in Ncb5or-b5 is similar to that in the corresponding regions of Cyb5A. The loop containing His89 in Ncb5or and His44 in Cyb5A is part of the characteristic “HPGG” motif that is conserved in all known eukaryotic members of the cytochrome b5 superfamily. Helix α4 is also similar in length and orientation in Ncb5or-b5 and in Cyb5A.
FIGURE 3.
A, stereo views of the overlay of human Ncb5or-b5 (magenta) and bovine Cyb5A (Protein Data Bank entry 1CYO; cyan). The helices are labeled using the standard notation (α2–α5) of Cyb5A. The top panel is viewed perpendicular to the face of the heme binding pocket with the heme propionates clearly visible, and the bottom panel is rotated ∼45°. B, loop and α5 region of Ncb5or-b5. Hydrogen bonds are indicated by dashed lines. Electron density for Lys122 was not observed; therefore, the side chain was truncated. Secondary structure is represented as a magenta ribbon. C, root mean square deviations (RMSD) (Å) between Cα atoms of Ncb5or-b5 (chain A) reported here and bovine Cyb5A (Protein Data Bank entry 1CYO). The helices in the heme binding core of Ncb5or (α1–α5) are marked. For this comparison, residues Val4 to Gly77 of 1CYO were superimposed onto Leu54–Gly127 of Ncb5or-b5. D, zoomed in view of the heme binding pockets for Ncb5or-b5 (left) and bovine Cyb5A (right). The imidazole rings of the histidine residues that ligate the heme iron atom in Ncb5or-b5 are nearly orthogonal to each other.
Two distinct differences in polypeptide conformation between Ncb5or-b5 and Cyb5A are observed. First, a C-terminal helix in Ncb5or-b5 corresponding to α6 in Cyb5A (core 2) is absent. The last segment of secondary structure in Ncb5or-b5 is β-sheet strand β2, which terminates at residue Met129, and the last residue in the polypeptide involved in packing interactions with the b5 core is Ala130. The remaining six C-terminal residues in the Ncb5or-b5 structure extend into solvent. Second, a striking difference in polypeptide conformation is observed within core 1 of Ncb5or-b5 in the vicinity of the second His ligand (His112 in Ncb5or-b5; His68 in Cyb5A) and the ensuing polypeptide segment that corresponds to helix α5 in Cyb5A. In mammalian Cyb5A orthologs, helix α4 is followed by a highly conserved V66GHS69 loop containing ligand His68, and this in turn is followed by the 9-residue helix α5 that merges with the central β-sheet. The corresponding region in all known mammalian Ncb5or orthologs contains the invariant sequence H112RWVNYESMLKEC124 (supplemental Fig. 2). DSSP analysis (56) and visual inspection of the structural model indicate that, in this sequence, heme ligand His112 is the C-terminal residue in helix α4, and that α4 is followed by a turn/loop segment comprising residues Arg113–Asn116, which features a hydrogen bond between the amide NH of Val115 and the carbonyl group of His112. The next element of secondary structure indicated by the DSSP analysis is an α-helix of approximately one turn comprising residues Tyr117–Met120 but which involves no i/i + 4 hydrogen bond among those residues. Rather, the α-CO group of Asn116, which does not have a helical backbone conformation, forms a hydrogen bond with the α-NH of Met120. This short α-helix is followed by a kink at Leu121 and then a one-turn 310 helix comprising Lys122–Cys124. Leu121 forms a hydrogen bond with the backbone NH of Cys124 in the 310 helix. We consider the eight residues from Tyr117–Cys124 to comprise a kinked helix, which we designate α5. The single turn of α-helix in this kinked helix is canted at an angle of ∼45° relative to the nine-residue helix α5 in Cyb5A (Fig. 3B). The major conformational difference between Ncb5or-b5 and Cyb5A in the α5 region can clearly be seen from a plot of root mean square deviation of the Cα atoms in the two proteins (Fig. 3C). Whereas the average root mean square deviation value between Cα atoms is 1.62 Å, the largest difference in polypeptide backbone conformation between the two structures occurs between residues Arg113 and Ser119 (root mean square deviation 2.5–4.1 Å).
The Bis-histidine Ligands in Ncb5or Are Nearly Orthogonal to Each Other
As noted above, Cyb5A and Ncb5or-b5 exhibit quite similar backbone conformations in helices α2 and α3 and the intervening loop containing the HPGG motif. As a result, the imidazolyl side chains of His44 in Cyb5A and His112 in Ncb5or-b5 adopt similar orientations with respect to the heme (Fig. 3D). However, the imidazolyl side chain of His112 in Ncb5or-b5 is substantially rotated relative to that of His68 in Cyb5A, which is probably a consequence of the different locations of these His residues in their respective polypeptides. The dihedral angle between the planes of the imidazolyl rings of His44 and His68 in bovine Cyb5A (Fig. 3D, right) is 21.2°, similar to values in other structurally characterized Cyb5A orthologs and consistent with observation of a rhombic EPR spectrum (supplemental Table 2). In contrast, the imidazolyl rings of His89 and His112 in Ncb5or-b5 are nearly orthogonal (dihedral angles of 83.2 and 81.3° in molecules A and B of the asymmetric unit, respectively) (supplemental Table 2), consistent with observation of a HALS EPR spectrum having a gmax value near the theoretical limit of 3.8.
Differences in Solvent-exposed Residues in Core 1 of Cyb5A and Ncb5or-b5
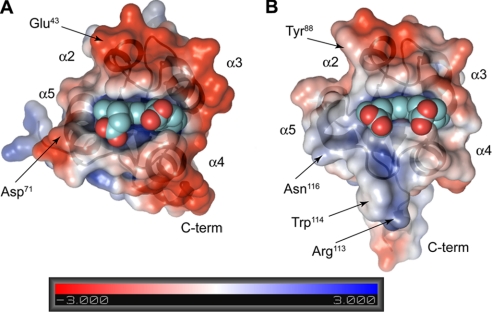
A number of studies have shown that docking between Cyb5A and Cyb5R3 involves electrostatic interactions between the side chains of negatively charged residues in Cyb5A and positively charged residues in Cyb5R3 (57–59). The negatively charged residues in Cyb5A suggested and demonstrated to mediate its docking with Cyb5R3 are located in core 1 of the protein, comprising helices α2–α5 and the associated loops containing the heme axial ligands. Comparison of mammalian Cyb5A amino acid sequences reveals 11 invariant negatively charged residues (Glu and Asp; Fig. 1B) in this region. The resulting high density of negative charge in core 1 can clearly be seen in an electrostatic map of bovine Cyb5A calculated from its crystal structure (Fig. 4A). The corresponding region of Ncb5or-b5 contains eight conserved Glu and Asp residues (Fig. 1B), which correlates to a comparatively smaller negative charge density in α2–α5 of core 1 (Fig. 4B).
FIGURE 4.
Electrostatic surface maps of bovine Cyb5A (A) and human Ncb5or-b5 (B) show the two proteins in the same orientation as that in Fig. 3A (top). The negatively charged surface in Cyb5A interacting with Cyb5R3 is shown along with a much more weakly charged corresponding surface in Ncb5or-b5. Both proteins are oriented identically with the heme propionates clearly visible.
It is worth noting two additional solvent-exposed residues near the front edge of the Ncb5or-b5 heme-binding pocket, Arg113 and Trp114. Both of these residues are invariant among known examples of mammalian Ncb5or, and Trp114 is invariant among known Ncb5or orthologs from all species (supplemental Fig. 2). There are no amino acid side chains directed toward solvent in the segment of Cyb5A corresponding most closely to that containing Arg113 and Trp114 in Ncb5or. Moreover, there is no positively charged amino acid residue in the region of Cyb5A thought to be extensively involved in docking with Cyb5R3. We therefore generated the R113A and W114A mutants for comparison with the wild-type protein in interdomain electron transfer studies, described below.
Interdomain Electron Transfer
Although previous steady state kinetics studies with Cyb5A and Cyb5R3 have typically been performed in the presence of air (30, 59), we observed that reduction of Ncb5or-b5 by Ncb5or-b5R only occurred at a detectable rate when air was excluded from the system. One likely contributing factor is that the reduction potential of Ncb5or-b5 (−108 mV versus standard hydrogen electrode or SHE) (21) is much more negative than that of Cyb5A (−9 mV versus SHE) (29), making its reduced form much more susceptible to autoxidation. Consequently, all studies reported herein were performed under an inert atmosphere (see “Materials and Methods”). For reasons that will become clear below, we also needed to perform comparative kinetics studies at low ionic strength (5 mm sodium phosphate; μ = 0.009 m).
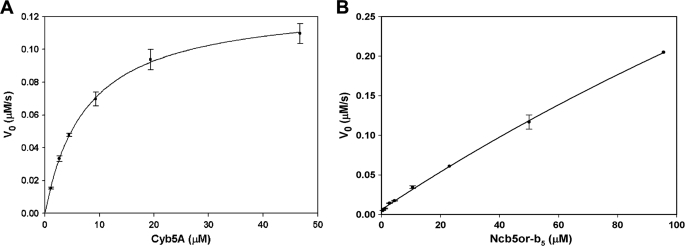
Consistent with previous studies (30), reduction of Cyb5A by Cyb5R3 obeyed Michaelis-Menten kinetics (Fig. 5A and Table 1). In contrast, saturation of Ncb5or-b5R by Ncb5or-b5 was not approached even when [Ncb5or-b5] reached 100 μm, the highest concentration we could use in our system (Fig. 5B). As a consequence of the nearly linear V0 versus [S] plot, the kcat and Km values obtained for the Ncb5or-b5/Ncb5or-b5R pair have relatively high S.E. values and should therefore be considered as rough estimates (Table 1). Our results reveal a much higher Km value for the Ncb5or-b5/Ncb5or-b5R pair than for the Cyb5R3/Cyb5A pair (Table 1), suggesting a considerably weaker interaction in the former. It should be noted that the b5 domain in intact Ncb5or is reduced instantaneously following the addition of excess NADH (1).
FIGURE 5.
Kinetics of heme reduction of Cyb5A (A) and Ncb5or-b5 (B) under low ionic strength (0.009 m). The initial rate (V0) is plotted against substrate concentration [S]. Cyb5A (1.4–50 μm) was reduced by Cyb5R3 (0.56 nm), and Ncb5or-b5 (0.7–100 μm) was reduced by Ncb5or-b5R (140 nm). Each data point with error bar (S.E.) is averaged from 2–4 independent reactions. The substrate is either Cyb5A or Ncb5or-b5, and the enzyme is either Cyb5R3 or Ncb5or-b5R.
TABLE 1.
Kinetic properties of Cyb5A/Cyb5R3 and Ncb5or-b5/Ncb5or-b5R reduction
The raw data of V0 versus [S] were fit to the Michaelis-Menten equation to generate Km and kcat (Vmax/[E]). The substrate is either Cyb5A or Ncb5or-b5, and the enzyme is either Cyb5R3 or Ncb5or-b5R.
| Substrate/Enzyme | [E] | Km | kcat | kcat/Km |
|---|---|---|---|---|
| nm | μm | s−1 | μm−1s−1 | |
| Cyb5A/Cyb5R3 | 0.56 | 7.6 ± 0.3 | 230 ± 3 | 30.1 |
| Ncb5or-b5/ Ncb5or-b5R | 140 | 498 ± 129 | 8.8 ± 1.9 | 0.018 |
The efficiency of Cyb5A reduction by Cyb5R3 decreased 4-fold when the solution ionic strength was increased from 0.009 m (buffer i) to 0.108 m (buffer ii), as measured by kinetic studies performed at a single substrate concentration (Table 2). This is indicative of a strong electrostatic component to docking between the molecules, consistent with previous reports (60, 61). A similar decrease in catalytic efficiency was observed in analogous experiments performed with Ncb5or-b5 and Ncb5or-b5R (Table 2), suggesting that electrostatic interactions contribute to docking between this redox pair as well.
TABLE 2.
Observed rate constants (Kobs, min−1 μm−1) of interdomain electron transfer as a function of environmental ionic strength
Ncb5or-b5 or Cyb5A (1.4 μm each) was reduced by Ncb5or-b5R or Cyb5R3, respectively, in the presence of excess NADH (50 μm) in buffer with various ionic strength (pH 7.0, no air).
| Buffer | Ionic strength, μ | Cyb5A/Cyb5R3 | Ncb5or-b5/Ncb5or-b5R |
|---|---|---|---|
| m | |||
| i | 0.009 | 1400 ± 90a | 4.20 ± 0.17b |
| ii | 0.108 | 331 ± 16a | 1.13 ± 0.06b |
a 0.56 nm enzyme used.
b 140 nm enzyme used..
In initial rate studies, we observed that Cyb5R3 reduces Cyb5A ∼70-fold more rapidly than Ncb5or-b5 (Table 3). In contrast, Ncb5or-b5R did not exhibit specificity toward its cognate redox partner, instead reducing Cyb5A ∼4-fold more rapidly than Ncb5or-b5. When Ncb5or-b5R was replaced by the Ncb5or-CS/b5R construct, we observed a 95-fold decrease in the initial rate of Cyb5A reduction while reduction of Ncb5or-b5 was diminished only 8-fold (Table 3). Thus, Ncb5or-CS/b5R reduces Ncb5or-b5 nearly 3-fold more rapidly than it reduces the non-cognate electron acceptor Cyb5A. Ncb5or-CS/b5R, Ncb5or-b5R, and Cyb5R3 all reduce the nonspecific substrate cytochrome c with essentially identical rate constants (Table 4). The fact that Ncb5or-CS/b5R is less efficient than Ncb5or-b5R at reducing Ncb5or-b5 therefore cannot be ascribed to deactivation of the b5R domain by the presence of the CS domain. Indeed, as noted above, Ncb5or-CS/b5R is more stable than Ncb5or-b5R.
TABLE 3.
Initial rate (μm/min/μm) of interdomain electron transfer
Ncb5or-b5 (wild type and W114A and R113A mutants) or Cyb5A (1.4 μm each) was reduced by Ncb5or-b5R, Ncb5or-CS/b5R, or Cyb5R3, in the presence of excess NADH (50 μm) in 5 mm sodium phosphate (pH 7.0, no air). All of the values in the table were normalized by reductase concentrations.
| Cyb5A | Ncb5or-b5 | Ncb5or-b5W114A | Ncb5or-b5R113A | |
|---|---|---|---|---|
| Cyb5R3 | 1730 ± 63a | 24.60 ± 1.73b | 11.06 ± 1.23b | 24.52 ± 2.27b |
| Ncb5or-b5R | 22.97 ± 1.67b | 5.69 ± 0.29c | 2.80 ± 0.22c | 4.63 ± 0.10c |
| Ncb5or-CS/b5R | 0.24 ± 0.01d | 0.69 ± 0.06d | 0.17 ± 0.02d | 0.42 ± 0.04d |
a 0.56 nm reductase used.
b 28 nm reductase used.
c 140 nm reductase used.
d 500 nm reductase used.
TABLE 4.
Observed rate constants (min−1 μm−1) of cytochrome c reduction
Single enzyme or mixture was used to reduce cytochrome c (1.4 μm) in the presence of excess NADH (50 μm) and 5 mm phosphate (pH 7.0, no air). An enzyme concentration of 28 nm (each protein) was used in all cases. NA, not available.
| Cyb5R3 | Ncb5or-b5R | Ncb5or-CS/b5R | Ncb5or | |
|---|---|---|---|---|
| Cytochrome c alone | 10.39 ± 0.50 | 9.32 ± 0.36 | 12.41 ± 0.57 | 34.70 ± 1.07 |
| Cytochrome c + Ncb5or-b5 | 14.09 ± 0.64 | 9.69 ± 0.49 | 13.27 ± 0.39 | NA |
As noted above, there are no amino acid side chains directed toward solvent in the segment of Cyb5A corresponding most closely to that containing Arg113 and Trp114 in Ncb5or. To investigate possible roles of these residues in interactions between the b5 and b5R domains of Ncb5or, we generated and characterized the R113A and W114A mutants of our Ncb5or-b5 construct. The R113A mutant was reduced by Ncb5or-b5R with nearly identical catalytic efficiency (Table 3), whereas the W114A mutation decreased the rate of Ncb5or-b5 reduction by ∼3-fold. A similar -fold change was observed for both mutants when they were reduced by Ncb5or-CS/b5R (Table 3). Neither mutation had a measurable effect on protein stability as determined in thermal denaturation studies (supplemental Fig. 4). Small differences in far-UV and Soret region CD spectra of the WT, R113A, and W114A proteins suggest that the mutations may cause subtle alterations in local structure, however, with the W114A mutant appearing to exert a larger effect (supplemental Fig. 5).
DISCUSSION
The cytochrome b5 fold was first revealed when Mathews et al. (54, 55) determined the x-ray crystal structure of the lipase fragment of bovine Cyb5A. On the basis of that structure, the x-ray crystal structure of the two-domain protein yeast flavocytochrome b2, a three-dimensional model of tobacco nitrate reductase, and amino acid sequences of multiple other b5 superfamily members, Lederer (62) aptly described the cytochrome b5 fold as an adaptable module. In the present work, this adaptability is highlighted by (i) the low sequence identity between the b5 cores of human Ncb5or and human Cyb5A (Fig. 1B), which results from substantial differences in residues with buried and with solvent-exposed (Fig. 4) side chains; (ii) the absence of a helix in Ncb5or corresponding to C-terminal α6 in Cyb5A; (iii) a distinct difference in polypeptide secondary structure in the vicinity of this second His ligand and the subsequent stretch of polypeptide leading into the central β-sheet in the two proteins (Fig. 3A); and (iv) a substantially larger dihedral angle between the planes of the two axial His ligands in Ncb5or than in Cyb5A (Fig. 3D). It can reasonably be argued that iii and iv in the preceding list are both consequences of the conserved difference in location of the second heme axial ligand in the amino acid sequences of Ncb5or (His112) and Cyb5A (His68) (Fig. 1B and supplemental Figs. 1 and 2).
Some of the structural differences described above are almost certainly linked to the divergent functional roles of Cyb5A and Ncb5or. Both proteins have been implicated to function in fatty acid desaturation (22, 26), but more studies are needed to delineate their specific roles. We turn our attention to the differences in His ligand orientations. The dihedral angle between the planes of the His ligands in Ncb5or-b5 is close to the maximum value of 90°, accounting for its very high gmax value in EPR spectra, and is by far the largest among structurally characterized members of the cytochrome b5 superfamily (supplemental Table 2). In fact, it is similar to the largest interplanar angles previously reported for natural heme proteins: heme bL in the cytochrome bc1 complex of the mitochondrial respiratory chain (50, 52, 63) and proximal heme b in quinol:fumarate reductase (64, 65). These and all other previously reported bis-histidine ligated b-hemes exhibiting HALS spectra are known or expected to be located in a membrane-embedded region of its protein, with the heme ensconced in a four-helix bundle exhibiting a left-handed twist (51). The two His ligands reside in diametrically opposed helices of the bundle, which cross over the top and bottom faces of the heme. To the best of our knowledge, Ncb5or is the only member of the cytochrome b5 superfamily demonstrated to exhibit a HALS EPR spectrum and/or orthogonal His ligands. It differs from other b-type heme proteins in this category in two key ways. First, although available evidence suggests that Ncb5or may be loosely associated with the ER membrane (21), its b5 domain has a highly polar surface and therefore probably extends into solvent. Indeed, a solvent-exposed location would seem to be essential for this domain's role in shuttling electrons from the b5R domain to its downstream partner or partners. Second, although heme is surrounded by a four-helix bundle in Ncb5or-b5, the helices do not cross the heme faces and do not exhibit a left-handed twist, and only one of the two His ligands is formally in a helix (albeit the C-terminal residue).
Perpendicular histidine ligands are clearly not essential for interdomain electron transfer in Ncb5or, as evidenced by the fact that Ncb5or-b5R reduces its cognate partner Ncb5or-b5 less efficiently than it reduces Cyb5A. The difference in dihedral angle between the His ligand planes in Ncb5or and Cyb5A may instead be important for tailoring the biophysical properties of the two proteins for optimal physiological function. One possibility is that the perpendicular His ligands in Ncb5or-b5 contribute to its substantially more negative redox potential (−108 mV versus SHE) (21) in comparison with Cyb5A (−9 mV versus SHE) (29). This is intriguing in light of molecular orbital arguments suggesting that changing from parallel to perpendicular His ligands should be accompanied by a significant positive shift in redox potential (51, 66). Ncb5or-b5 will provide the opportunity to firmly establish the relationship between ligand orientation and redox potential in bis-histidine-ligated heme proteins.
An alternative possibility is that the difference in location of the second His ligands in the Ncb5or and Cyb5A polypeptide sequences plays a more important functional role than the difference in His ligand orientations, due to its apparent effect on nearby polypeptide secondary structure. In this context, it is worth noting that the region encompassing ligand His112 through the C terminus of helix α5 in Ncb5or orthologs from vertebrates has a higher degree of conservation than do the other helices in the heme binding pocket (α2–α4; supplemental Fig. 2). This is in contrast to a lower degree of conservation in helix α5 than α2–α4 among Cyb5A orthologs from vertebrates (supplemental Fig. 1). This secondary structure difference, coupled with divergence in primary structure, results in decidedly different protein surfaces in Cyb5A and Ncb5or-b5 in the vicinity of the second His ligand and the adjacent polypeptide, including α5 (Fig. 4).
Studies have shown that Cyb5A/Cyb5R3 recognition has a strong electrostatic component involving interactions between negatively charged residues on Cyb5A and positively charged residues on Cyb5R3 (57–59). Our ionic strength studies have shown that recognition between the b5 and b5R domains of Ncb5or also involves a significant electrostatic component. In this context, it is noteworthy that there are 11 negatively charged residues in the four-helix bundle surrounding heme in Cyb5A but only eight in the corresponding region of Ncb5or-b5 (Fig. 1B). Mutagenesis studies have been performed by others to replace 10 of the 11 Glu and Asp residues in core 1 of Cyb5A by Ala (supplemental Table 3). Results suggest that only Glu43 and Asp71 are involved in strong interactions with Cyb5R3 (58, 59). As indicated in Fig. 4, both of these residues are near the front of the heme binding pocket, in close proximity to and pointing in the same general direction as the heme propionate group that is thought to be important in Cyb5R3 recognition as well. Notably, the residues in human Ncb5or-b5 corresponding to Glu43 and Asp71 in Cyb5A are uncharged (Tyr88 and Asn116, respectively). Asn116 is an invariant residue among all known Ncb5or orthologs, whereas residue 88 is either Tyr or Phe (supplemental Fig. 2). As a consequence, there is much lower negative charge density near the front edge of heme in Ncb5or-b5 (Fig. 4B) than in Cyb5A (Fig. 4A), the region of each protein likely to undergo the most extensive interactions with the cognate reductase proteins for electron transfer (59, 67). The much larger Km value obtained in our studies for the Ncb5or-b5/Ncb5or-b5R pair than for the Cyb5A/Cyb5R3 pair may therefore have a major contribution from weaker electrostatic interactions. Such a large difference in Km values for these pairs is perhaps not surprising, given that Cyb5A and Cyb5R3 must diffuse through the ER membrane prior to docking, whereas the b5 and b5R domains in intact Ncb5or are held in proximity by the intervening CS domain (1). A low Km value for the b5 and b5R domains of Ncb5or might reasonably be expected to impair the ability of Ncb5or to deliver electrons to its downstream partner or partners.
On the basis of studies with Cyb5A and Cyb5R3, it can be inferred that the residues most likely to be involved in docking of the Ncb5or b5 domain to its b5R domain are located at the front of the four-helix bundle that surrounds heme. A major difference in this region of Ncb5or-b5 in comparison with Cyb5A is the presence of Trp114, which is invariant among all known Ncb5or orthologs in animals, and the adjacent positively charged residue Arg113, which is invariant among known mammalian Ncb5or orthologs. Both residues have fully solvent-exposed side chains, whereas the corresponding region of Cyb5A is devoid of exposed side chains. Solvent-exposed residues in proteins, if not constrained by structural or functional demand, are more subject to random mutation than buried residues. Our CD and thermal denaturation data indicate that Arg113 and Trp114 do not play essential structural or stabilizing roles in human Ncb5or. It can therefore reasonably be assumed that they are necessary for function. Our mutagenesis studies strongly suggest that Arg113 is not involved in the b5-b5R interaction in Ncb5or. Trp114 appears to play little if any role in this interaction either because the 3-fold decrease in electron transfer rate constant observed for the W114A mutant seems insufficiently large to explain the invariance of Trp114 among known Ncb5or proteins. These conclusions ultimately need to be verified in rapid kinetics studies of full-length Ncb5or and its R113A and W114A mutants. Full-length proteins will also be necessary to examine possible roles of Arg113, Trp114, and other highly conserved Ncb5or-b5 residues in interactions between Ncb5or and its likely downstream partner SCD. This represents a significant challenge, given that SCD is an integral membrane protein. Efforts are under way to develop an in vivo reconstitution system with primary hepatocyes from Ncb5or knock-out mice for this purpose. We prefer this experimental system over the classical in vitro reconstitution for two reasons: (i) the in vivo system reflects more accurately the native desaturation pathway, considering two recent papers that show no major phenotype in lipid metabolism of mice lacking Cyb5A in the liver (23) or in the whole body (24), in contrast to the in vitro data (22); (ii) The SCD enzyme is an integral membrane protein that is hard to purify in an active form for biochemical assays.
The physical interaction between the b5 and b5R domains of Ncb5or is clearly different from that in the Cyb5A and Cyb5R3 complex. The CS domain has been shown to be functionally important to allow the electron flow from the b5R to the b5 domain in full-length Ncb5or (1). When the individual domains are separated and mixed in vitro, Ncb5or-b5R is able to reduce Ncb5or-b5 at a slow rate. The electron transfer in the Ncb5or-b5/Ncb5or-b5R pair requires complex formation involving electrostatic interaction, which may be weaker than those in the Cyb5A/Cyb5R3 pair. Nc5or-CS/b5R selectively reduces Ncb5or-b5 over Cyb5A, in contrast to Ncb5or-b5R, but the presence of the CS domain also retards reduction of Ncb5or-b5. Coupled with the finding that the CS domain does not affect the rate of cytochrome c reduction, this observation leads us to conclude that the role of the CS domain in Ncb5or is more complex than simply keeping the b5 and b5R domains in close proximity. At the very least, the CS domain appears to play a role in mediating docking of the b5 and b5R domains. We are currently working to determine the three-dimensional structure of full-length Ncb5or in order to address this and other questions related to its functions.
Supplementary Material
Acknowledgments
We thank Drs. H. Franklin Bunn (Brigham and Women's Hospital) for generous support, Richard L. Schowen (University of Kansas) for helpful suggestions on calculation, and Grant Mauk (University of British Columbia) for providing the plasmid construct of human erythrocyte cytochrome b5.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 DK067355 (to H. Franklin Bunn and H. Z.) and by National Institutes of Health Centers of Biomedical Research Excellence–Protein Structure and Function award 5P20 RR17708 (to the University of Kansas, R. P. Hanzlik, P. I.). This work was also supported by American Heart Association Grant-in-Aid 0755879T (to B. R. G.). Use of the Advanced Photon Source was supported by the United States Department of Energy under Contract W-31-109-Eng-38, and use of the Industrial Macromolecular Crystallography Association Collaborative Access Team beamline 17-ID was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with the Center for Advanced Radiation Sources at the University of Chicago.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5 and Tables 1–3.
The atomic coordinates and structure factors (code 3LF5) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- b5
- cytochrome b5
- b5R
- cytochrome b5 reductase
- ER
- endoplasmic reticulum
- SCD
- stearoyl-CoA desaturase
- HALS
- highly anisotropic low spin.
REFERENCES
- 1.Zhu H., Qiu H., Yoon H. W., Huang S., Bunn H. F. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14742–14747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kessler D. L., Rajagopalan K. V. (1972) J. Biol. Chem. 247, 6566–6573 [PubMed] [Google Scholar]
- 3.Kisker C., Schindelin H., Pacheco A., Wehbi W. A., Garrett R. M., Rajagopalan K. V., Enemark J. H., Rees D. C. (1997) Cell 91, 973–983 [DOI] [PubMed] [Google Scholar]
- 4.Rudolph M. J., Johnson J. L., Rajagopalan K. V., Kisker C. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 1183–1191 [DOI] [PubMed] [Google Scholar]
- 5.Cho H. P., Nakamura M., Clarke S. D. (1999) J. Biol. Chem. 274, 37335–37339 [DOI] [PubMed] [Google Scholar]
- 6.Cho H. P., Nakamura M. T., Clarke S. D. (1999) J. Biol. Chem. 274, 471–477 [DOI] [PubMed] [Google Scholar]
- 7.Cannons A. C., Barber M. J., Solomonson L. P. (1993) J. Biol. Chem. 268, 3268–3271 [PubMed] [Google Scholar]
- 8.Hyde G. E., Crawford N. M., Campbell W. H. (1991) J. Biol. Chem. 266, 23542–23547 [PubMed] [Google Scholar]
- 9.Barber M. J., Desai S. K., Marohnic C. C., Hernandez H. H., Pollock V. V. (2002) Arch. Biochem. Biophys. 402, 38–50 [DOI] [PubMed] [Google Scholar]
- 10.Xia Z. X., Shamala N., Bethge P. H., Lim L. W., Bellamy H. D., Xuong N. H., Lederer F., Mathews F. S. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 2629–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell A. G., Martin C. E. (1995) J. Biol. Chem. 270, 29766–29772 [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Ranea J. A., Mirey G., Camonis J., Valencia A. (2002) FEBS Lett. 529, 162–167 [DOI] [PubMed] [Google Scholar]
- 13.Lederer F., Ghrir R., Guiard B., Cortial S., Ito A. (1983) Eur. J. Biochem. 132, 95–102 [DOI] [PubMed] [Google Scholar]
- 14.Durley R. C., Mathews F. S. (1996) Acta Crystallogr. D Biol. Crystallogr. 52, 65–76 [DOI] [PubMed] [Google Scholar]
- 15.Rodríguez-Marañón M. J., Qiu F., Stark R. E., White S. P., Zhang X., Foundling S. I., Rodríguez V., Schilling C. L., 3rd, Bunce R. A., Rivera M. (1996) Biochemistry 35, 16378–16390 [DOI] [PubMed] [Google Scholar]
- 16.Mathews F. S., Gerwinsky E. W., Argos P. (1979) in The Porphyrins (Dolphin D. ed) Vol. 7, pp. 107–147, Academic Press, Inc., New York [Google Scholar]
- 17.Yubisui T., Naitoh Y., Zenno S., Tamura M., Takeshita M., Sakaki Y. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 3609–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karplus P. A., Daniels M. J., Herriott J. R. (1991) Science 251, 60–66 [PubMed] [Google Scholar]
- 19.Borgese N., Longhi R. (1990) Biochem. J. 266, 341–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pietrini G., Carrera P., Borgese N. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 7246–7250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu H., Larade K., Jackson T. A., Xie J., Ladoux A., Acker H., Berchner-Pfannschmidt U., Fandrey J., Cross A. R., Lukat-Rodgers G. S., Rodgers K. R., Bunn H. F. (2004) J. Biol. Chem. 279, 30316–30325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strittmatter P., Spatz L., Corcoran D., Rogers M. J., Setlow B., Redline R. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 4565–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finn R. D., McLaughlin L. A., Ronseaux S., Rosewell I., Houston J. B., Henderson C. J., Wolf C. R. (2008) J. Biol. Chem. 283, 31385–31393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLaughlin L. A., Ronseaux S., Finn R. D., Henderson C. J., Wolf C. R. (2010) Mol. Pharmacol. 78, 269–278 [DOI] [PubMed] [Google Scholar]
- 25.Xie J., Zhu H., Larade K., Ladoux A., Seguritan A., Chu M., Ito S., Bronson R. T., Leiter E. H., Zhang C. Y., Rosen E. D., Bunn H. F. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10750–10755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larade K., Jiang Z., Zhang Y., Wang W., Bonner-Weir S., Zhu H., Bunn H. F. (2008) J. Biol. Chem. 283, 29285–29291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Larade K., Jiang Z. G., Ito S., Wang W., Zhu H., Bunn H. F. (2010) J. Lipid Res. 51, 53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y. (2008) BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lloyd E., Ferrer J. C., Funk W. D., Mauk M. R., Mauk A. G. (1994) Biochemistry 33, 11432–11437 [DOI] [PubMed] [Google Scholar]
- 30.Roma G. W., Crowley L. J., Barber M. J. (2006) Arch. Biochem. Biophys. 452, 69–82 [DOI] [PubMed] [Google Scholar]
- 31.Falzone C. J., Mayer M. R., Whiteman E. L., Moore C. D., Lecomte J. T. (1996) Biochemistry 35, 6519–6526 [DOI] [PubMed] [Google Scholar]
- 32.Cowley A. B., Rivera M., Benson D. R. (2004) Protein Sci. 13, 2316–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis C. A., Dhawan I. K., Johnson M. K., Barber M. J. (2002) Arch. Biochem. Biophys. 400, 63–75 [DOI] [PubMed] [Google Scholar]
- 34.Matthews B. W. (1968) J. Mol. Biol. 33, 491–497 [DOI] [PubMed] [Google Scholar]
- 35.Long F., Vagin A. A., Young P., Murshudov G. N. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wirtz M., Oganesyan V., Zhang X., Studer J., Rivera M. (2000) Faraday Discuss. 116, 221–234; discussion 257–268 [DOI] [PubMed] [Google Scholar]
- 37.Cowtan K. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 38.Kabsch W. (1988) J. Appl. Crystallogr. 21, 67–72 [Google Scholar]
- 39.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 40.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 41.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 42.Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 43.Carson M. (1997) Methods Enzymol. 277, 493–505 [PubMed] [Google Scholar]
- 44.Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 45.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dolinsky T. J., Nielsen J. E., McCammon J. A., Baker N. A. (2004) Nucleic Acids Res. 32, W665–W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bas D. C., Rogers D. M., Jensen J. H. (2008) Proteins 73, 765–783 [DOI] [PubMed] [Google Scholar]
- 48.Yatsunyk L. A., Dawson A., Carducci M. D., Nichol G. S., Walker F. A. (2006) Inorg. Chem. 45, 5417–5428 [DOI] [PubMed] [Google Scholar]
- 49.Migita C. T., Iwaizumi M. (1981) J. Am. Chem. Soc. 103, 4378–4381 [Google Scholar]
- 50.Salerno J. C. (1984) J. Biol. Chem. 259, 2331–2336 [PubMed] [Google Scholar]
- 51.Berry E. A., Walker F. A. (2008) J. Biol. Inorg. Chem. 13, 481–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zoppellaro G., Bren K. L., Ensign A. A., Harbitz E., Kaur R., Hersleth H. P., Ryde U., Hederstedt L., Andersson K. K. (2009) Biopolymers 91, 1064–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salerno J. C., Leigh J. S. (1984) J. Am. Chem. Soc. 106, 2156–2159 [Google Scholar]
- 54.Mathews F. S., Levine M., Argos P. (1971) Nat. New Biol. 233, 15–16 [DOI] [PubMed] [Google Scholar]
- 55.Mathews F. S., Levine M., Argos P. (1972) J. Mol. Biol. 64, 449–464 [DOI] [PubMed] [Google Scholar]
- 56.Kabsch W., Sander C. (1983) Biopolymers 22, 2577–2637 [DOI] [PubMed] [Google Scholar]
- 57.Strittmatter P., Hackett C. S., Korza G., Ozols J. (1990) J. Biol. Chem. 265, 21709–21713 [PubMed] [Google Scholar]
- 58.Shirabe K., Nagai T., Yubisui T., Takeshita M. (1998) Biochim. Biophys. Acta 1384, 16–22 [DOI] [PubMed] [Google Scholar]
- 59.Kawano M., Shirabe K., Nagai T., Takeshita M. (1998) Biochem. Biophys. Res. Commun. 245, 666–669 [DOI] [PubMed] [Google Scholar]
- 60.Meyer T. E., Shirabe K., Yubisui T., Takeshita M., Bes M. T., Cusanovich M. A., Tollin G. (1995) Arch. Biochem. Biophys. 318, 457–464 [DOI] [PubMed] [Google Scholar]
- 61.Yantsevich A. V., Gilep A. A., Usanov S. A. (2008) Biochemistry 73, 1096–1107 [DOI] [PubMed] [Google Scholar]
- 62.Lederer F. (1994) Biochimie 76, 674–692 [DOI] [PubMed] [Google Scholar]
- 63.Iwata S., Lee J. W., Okada K., Lee J. K., Iwata M., Rasmussen B., Link T. A., Ramaswamy S., Jap B. K. (1998) Science 281, 64–71 [DOI] [PubMed] [Google Scholar]
- 64.Lancaster C. R., Kröger A., Auer M., Michel H. (1999) Nature 402, 377–385 [DOI] [PubMed] [Google Scholar]
- 65.Madej M. G., Nasiri H. R., Hilgendorff N. S., Schwalbe H., Lancaster C. R. (2006) EMBO J. 25, 4963–4970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walker F. A., Huynh B. H., Scheidt W. R., Osvath S. R. (1986) J. Am. Chem. Soc. 108, 5288–5297 [Google Scholar]
- 67.Nishida H., Miki K. (1996) Proteins 26, 32–41 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.