Abstract
Archaea make glutaminyl-tRNA (Gln-tRNAGln) in a two-step process; a non-discriminating glutamyl-tRNA synthetase (ND-GluRS) forms Glu-tRNAGln, while the heterodimeric amidotransferase GatDE converts this mischarged tRNA to Gln-tRNAGln. Many prokaryotes synthesize asparaginyl-tRNA (Asn-tRNAAsn) in a similar manner using a non-discriminating aspartyl-tRNA synthetase (ND-AspRS) and the heterotrimeric amidotransferase GatCAB. The transamidosome, a complex of tRNA synthetase, amidotransferase and tRNA, was first described for the latter system in Thermus thermophilus [Bailly, M., Blaise, M., Lorber, B., Becker, H.D. and Kern, D. (2007) The transamidosome: a dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagine biosynthesis. Mol. Cell, 28, 228–239.]. Here, we show a similar complex for Gln-tRNAGln formation in Methanothermobacter thermautotrophicus that allows the mischarged Glu-tRNAGln made by the tRNA synthetase to be channeled to the amidotransferase. The association of archaeal ND-GluRS with GatDE (KD = 100 ± 22 nM) sequesters the tRNA synthetase for Gln-tRNAGln formation, with GatDE reducing the affinity of ND-GluRS for tRNAGlu by at least 13-fold. Unlike the T. thermophilus transamidosome, the archaeal complex does not require tRNA for its formation, is not stable through product (Gln-tRNAGln) formation, and has no major effect on the kinetics of tRNAGln glutamylation nor transamidation. The differences between the two transamidosomes may be a consequence of the fact that ND-GluRS is a class I aminoacyl-tRNA synthetase, while ND-AspRS belongs to the class II family.
INTRODUCTION
Attaching the correct amino acid to its cognate tRNA is an essential step in maintaining the fidelity of protein synthesis. A group of enzymes, the aminoacyl-tRNA synthetases (aaRSs), pair amino acids with their cognate tRNA; each aaRS is specific for one amino acid:tRNA pair (1). However, glutaminyl-tRNA synthetase (GlnRS) is absent in all known archaea and most bacteria, while asparaginyl-tRNA synthetase (AsnRS) is absent in most prokaryotes (2). In these organisms, Gln-tRNAGln and/or Asn-tRNAAsn are formed by a tRNA-dependent amino acid transformation process catalyzed by amidotransferase (AdT) enzymes (2).
For Gln-tRNA synthesis ND-GluRS forms Glu-tRNAGln (3) which is then converted to Gln-tRNAGln by a glutamyl-tRNAGln amidotransferase (Glu-AdT) (4). In a similar manner, Asn-tRNAAsn is formed by the sequential action of ND-AspRS (5) and aspartyl-tRNAAsn amidotransferase (Asp-AdT) (6,7). In bacteria, the heterotrimeric AdT GatCAB can function for tRNA-dependent synthesis of Gln and Asn (2). In archaea, however, GatCAB is used solely for Asn-tRNAAsn formation (8), while the archaeal-specific heterodimeric enzyme, GatDE, serves as the Glu-AdT (9).
In the 80s, the existence of complexes of AdTs and ND-aaRSs was proposed (10); these complexes would allow substrate channeling (11) of the misacylated tRNA from the aaRS to the AdT. While a number of complexes between aaRSs and other proteins have been reported (12), it was only recently shown that a complex exists between ND-AspRS and GatCAB, the transamidosome (13). The interaction of these two proteins requires the presence of tRNAAsn and the complex is stable over the course of Asn-tRNA biosynthesis (13), protecting Asn-tRNAAsn from deacylation (13,14) and Asp-tRNAAsn from being recognized by elongation factor EF-Tu (13).
Similar complexes have been proposed for ND-GluRSs and AdTs (10,13,15,16). We report on such a complex between GatDE and ND-GluRS from the archaeon Methanothermobacter thermautotrophicus. This archaeal-specific transamidosome (ND-GluRS:GatDE) does not require tRNA to assemble, and specifically synthesizes Gln-tRNAGln. Rather than protecting Gln-tRNAGln from deacylation, the binding of GatDE with ND-GluRS sequesters the aaRS for Gln-tRNAGln formation. The differences between the two transamidosomes may be a consequence of ND-GluRS being a class I aaRS and ND-AspRS belonging to the class II family (17).
MATERIALS AND METHODS
Purification of M. thermautotrophicus GatDE, ND-GluRS, tRNAGln, tRNAGlu
Methanothermobacter thermautotrophicus tRNAGln2 and tRNAGlu transcripts were prepared in vitro as described previously (9). The tRNA isoacceptors were 32P-labeled on their 3′-terminus as described (18). GatDE, ND-GluRS and ND-AspRS were over-produced and purified as previously described (9,19,20).
Gel filtration chromatography
Size-exclusion chromatography was performed by FPLC using a Sephacryl S300 16/60 column (GE Healthcare) at 4°C equilibrated in 20 mM HEPES pH 7.2, 10 mM MgCl2, 50 mM KCl, 1 mM DTT and developed in the same buffer. For preparative assays, samples (2 ml) were prepared in the same buffer and pre-incubated for 30 min at room temperature with 20 µM of GatDE, GluRS, tRNAGln, Glu, ATP and/or Asn. The samples were loaded on a 120 ml column at 4°C at a flow rate of 0.5 ml/min and 1.5 ml fractions were collected. The optical density profile was monitored at 260 or 280 nm. The fractions corresponding to the elution of GluRS:GatDE complex were subsequently analyzed for aminoacylation and amidotransferase activities as described (8). Unbound tRNAGln occasionally eluted from the column with a second peak, possibly corresponding to dimerization of the tRNA.
Fluorescence anisotropy
Alexa fluor (AF) 488 tetrafluorophenyl ester (Molecular Probes, Invitrogen) was prepared in dimethyl sulfoxide, according to the manufacturer’s protocol. ND-GluRS or GatDE was incubated with AF for an hour at room temperature. Excess unreacted dye was immediately removed by passage through a 1 ml Sephadex G25 column (Amersham Biosciences). Remaining traces of dye were then removed by an overnight dialysis in a buffer containing 20 mM HEPES pH 7.2, 10 mM MgCl2, 50 mM KCl and 1 mM DTT. Fluorescently labeled protein was visualized on a 12% SDS polyacrylamide gel and subjected to ultraviolet illumination to confirm that the sample contained no free fluorophore. Prior to use in fluorescence anisotropy experiments, the activity of labeled protein was verified using the assays described below. Equilibrium dissociation constants were determined by measuring the fluorescence anisotropy of GluRS (20 nM) or GatDE (20 nM) as a function of increasing concentrations of an unlabeled protein (up to 2 µM) or tRNA (5 nM to 1 µM) (Table 1 and Supplementary Figure S1). Fluorescence anisotropy measurements were performed using a two-channel spectrofluorimeter (Proton Technology International) with linear polarizers. Samples were excited at 495 nm wavelength with linearly polarized light, and fluorescence emission was detected at 519 nm at two polarized orientations, parallel and perpendicular to the polarization of the excitation channel, slit widths of 5 nm, and the time- based function for 30 s. All of the measurements were carried out at least three times and the titration curves were fitted to a 1 : 1 binding stoichiometry.
Table 1.
Binding affinities between components of the archaeal-specific transamidosome
| AF-labeled proteina | Ligandb | KD (nM) |
|---|---|---|
| AF-GatDE alone | GluRS | 40 ± 5 |
| AF-GluRS alone | GatDE | 100 ± 22 |
| AF-GatDE alone | AspRS | ND |
| AF-AspRS alone | GatDE | ND |
| AF-GluRS alone | tRNAGln | 37 ± 4 |
| AF-GatDE alone | tRNAGln | ND |
| AF-GluRS alone | tRNAGlu | 80 ± 3 |
| AF-GatDE alone | tRNAGlu | ND |
| AF-GatDE + excess GluRSc | tRNAGln | 80 ± 14 |
| AF-GluRS + excess GatDEc | tRNAGln | 110 ± 9 |
| AF-GatDE + excess GluRSc | tRNAGlu | ND |
| AF-GluRS + excess GatDEc | tRNAGlu | ND |
Measurements were from three separate fluorescence anisotropy experiments as described in the ‘Materials and Methods’ section. Standard deviations are reported. ND stands for no detectable change in anisotropy under conditions studied.
aAlexa fluor 488 (AF)-labeled protein (20 nM) incubated with bIncreasing concentrations [protein (up to 2 µM) or tRNA (up to 1 µM)] indicated.
cEnzyme indicated added in excess (2 µM).
Gel shift assay
To examine ternary complex formation, except as otherwise indicated, [32P] tRNAGln (100 nM) or [32P] tRNAGlu (100 nM) was incubated at 37°C with or without ND-GluRS (1 µM) and/or GatDE (1–6 µM) in 50 mM HEPES–KOH buffer (pH 7.2), 50 mM KCl, 10 mM MgCl2 and 1 mM DTT, 10% glycerol, before adding 20 µl 5× loading buffer (100 mM Tris pH 7.5, 160 mM KCl, 0.5 mM EDTA pH 8, 50% glycerol, 12.5 mM DTT). The total volume of each sample was 60 µl. Samples were then loaded on a 6% polyacrylamide gel (330 × 430 × 0.5 mm) in 1× TBE buffer. After electrophoresis, the gel was exposed to an imaging plate (FujiFilms) and scanned on a Molecular Dynamics Storm 860 Phosphoimager. Visualization of GluRS:GatDE complex by native PAGE was performed by incubating GatDE (1 µM) with increasing concentrations of ND-GluRS (1–10 µM) in 20 mM HEPES pH 7.2, 10 mM MgCl2, 50 mM KCl, 1 mM DTT in a total volume of 20 µl at 37°C for 30 min. After incubation, 5 µl of 5× loading buffer (100 mM Tris pH 7.5, 160 mM KCl, 0.5 mM EDTA pH 8, 50% glycerol, 12.5 mM DTT) was added to the samples prior to loading on a 10% native polyacrylamide gel and run at 100 V for 8 h at 4°C in native running buffer (25 mM Tris, 0.2 M glycine, pH 8.0). The gels were stained with Coomassie Blue. The presence of recombinant GatDE in the upper band was established by immunoblot for the His6-tag on the AdT, and the presence of ND-GluRS in the band was established using AF-labeled ND-GluRS, which was visualized by UV illumination.
Aminoacylation assays
Were carried out as described (8) using the 32P-labeled tRNA/nuclease P1 assay (21,22). The experiments were carried out at 37°C. At this temperature, we were unable to detect deacylation of the relevant Glx-tRNA species during the time-course of these studies. For steady-state kinetic parameters, 50 nM of M. thermautotrophicus ND-GluRS was added in the presence or absence of either 2 µM GatDE or bovine serum album (BSA). The concentration of tRNAGln varied from 0.3125 µM up to 15 µM. For Glu-tRNAGlu activity, 80 nM 32P-labeled tRNAGlu was used with 50 nM ND-GluRS with or without GatDE (concentration varied from 7.1 nM to 1.8 µM) or BSA (1.8 µM) added.
Amidotransferase assays
The reactions were carried out and processed as described (23). Steady-state kinetic parameters were as described (8) with either 2 µM ND-GluRS or BSA added to the reaction mix. As noted previously (8), the experiments were carried out at 37°C. At this temperature, we were unable to detect deacylation of the relevant Glx-tRNAGln species during the time-course of these studies.
Pull-down assay
For the pull down assay, recombinant AF-labeled GluRS (0.8 mg) was mixed with an S100 fraction (40 ml) of E. coli extract from cells over-producing M. thermautotrophicus His-tagged GatDE and incubated in 50 mM HEPES–KOH buffer (pH 7.2), 50 mM KCl, 10 mM MgCl2 for 2 h at room temperature. Nickel-nitrilotriacetic acid (Ni-NTA) agarose beads (Qiagen) (500 µl), pre-equilibrated with the same buffer, were then incubated with the extract for an hour at room temperature. Following incubation, the beads were washed three times [50 mM HEPES–KOH buffer (pH 7.2), 50 mM KCl, 10 mM MgCl2], re-suspended in 100 µl SDS–PAGE loading buffer and incubated at 100°C for 5 min. Supernatants were resolved by electrophoresis on a 4–20% gradient polyacrylamide Tris–HCl gel and then visualized by UV illumination.
Nitrocellulose filter-binding assay
The ability of GatDE to compete with tRNAGlu for binding with ND-GluRS was measured using a nitrocellulose filter-binding assay (24). [32P] tRNAGlu (5 nM) was incubated with ND-GluRS (60 nM) in binding buffer [50 mM HEPES–KOH buffer (pH 7.2), 50 mM KCl, 10 mM MgCl2, and 1 mM DTT] with or without increasing concentrations of GatDE (7.8 nM to 2 µM) for 25 min at room temperature. The nitrocellulose membrane (MF-Millipore; Millipore Corp., Billerica, MA) was pre-soaked in binding buffer for 30 min at 4°C lightly shaking. A 96-well vacuum manifold (Hybri-dot 96; Whatman Biometra, Germany) was used to spot aliquots (6 µl) of the binding reaction onto the membrane. Each aliquot was then washed with binding buffer (200 µl). The experiments were repeated four times. The level of bound [32P] tRNAGlu was quantified by phosphorimaging. The fraction remaining bound was plotted as a function of GatDE concentration and fitted in Kaleidagraph to determine an apparent KI by non-linear regression.
RESULTS
Direct association of M. thermautotrophicus ND-GluRS with GatDE
The association of Thermus thermophilus ND-AspRS and GatCAB was shown to be tRNAAsn-dependent (13). The T. thermophilus ND-AspRS:tRNAAsn:GatCAB transamidosome structural model predicts that the two enzymes make extensive contact with the tRNA but only limited contact with one another due to the fact that AdT binds to the minor groove of the tRNA while ND-AspRS binds to the major groove (13). However, the modeling of the archaeal-specific transamidosome (GatDE:ND-GluRS:tRNAGln) proposes a greater interaction between the AdT and the aaRS, as both enzymes bind to the minor groove side of the tRNA and the insertion domain specific to GatE (25) forms a concave pocket in which ND-GluRS can fit (15), raising the possibility that GatDE and ND-GluRS could bind together even in the absence of tRNAGln.
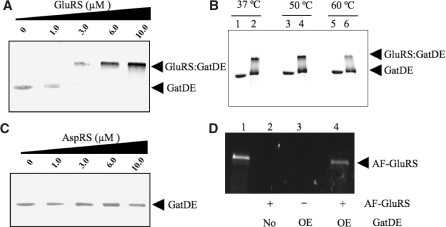
We thus investigated a direct association of M. thermautotrophicus ND-GluRS and GatDE through gel shift and pull-down assays (Figure 1), and gel filtration studies (Figure 2). In contrast to the T. thermophilus transamidosome, GatDE and ND-GluRS formed a complex in the absence of tRNA (Figures 1A and 2C, and Table 1) even at elevated incubation temperatures (Figure 1B). ND-GluRS had a strong tendency to aggregate in the wells of the gel and precipitate its binding partners (tRNA and GatDE), although bound ND-GluRS was less likely to precipitate than unbound aaRS. This aggregation by ND-GluRS prevented the use of gel-shift assays to quantitatively measure the affinity of the aaRS for its binding partners.
Figure 1.
Formation of the M. thermautotrophicus GluRS:GatDE binary complex. Electrophoretic mobility shift assays were performed between purified GatDE and ND-aaRS indicated as described in the ‘Materials and Methods’ section. Native PAGE (10%) separation of samples containing: (A) a stable concentration of GatDE (1 µM) and increasing concentrations of ND-GluRS (0–10 µM), (B) GatDE (3 µM) alone (lanes 1, 3 and 5) or both GatDE and GluRS (3 µM each) (lanes 2, 4 and 6) incubated at the temperatures indicated, and (C) a stable concentration of GatDE (1 µM) and increasing concentrations of ND-AspRS (0–10 µM), stained with Coomassie blue. (D) Ni-NTA pull-down with His6-tagged GatDE. Proteins were separated by SDS–PAGE and AF-GluRS was visualized by UV illumination. Escherichia coli extracts from cells either over-expressing His6-tagged GatDE (OE; lanes 3 and 4) or not (No; lane 2), were incubated with (+) or without (−) AF-GluRS (0.8 mg), and Ni-NTA agarose beads followed by three washes as described in the ‘Materials and Methods’ section. Purified AF-GluRS prior to Ni-NTA pull-down run in lane 1.
Figure 2.
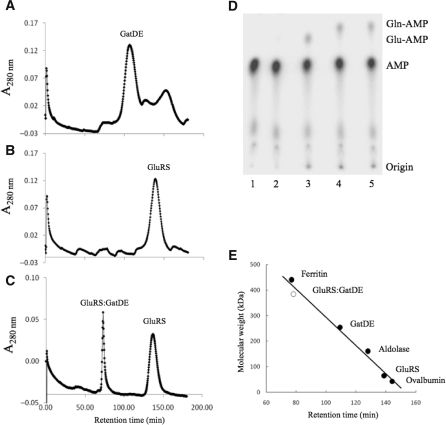
Gel filtration analysis of the archaeal GatDE:GluRS binary complex. Gel filtrations over a sephacryl S300 16/60 column were carried out as described in the ‘Materials and Methods’ section with GatDE (10 µM) and/or ND-GluRS (20 µM) added to the mix. Gel filtration analysis of (A) GatDE alone, (B) ND-GluRS alone and (C) GatDE and ND-GluRS pre-incubated together. (D) Combined aminoacylation/amidotransferase assay with 32P-labeled tRNAGln (100 nM), ATP (4 mM), l-Glu (2 mM) and Asn (2 mM) incubated with (lane 1) no enzyme, (lane 2) purified GatDE (250 nM), (Lane 3) purified ND-GluRS (250 nM), (Lane 4) purified ND-GluRS and GatDE (250 nM each) or (lane 5) the ND-GluRS:GatDE complex (400 nM) eluted from the gel filtration column (Figure 2C). (E) Calibration curve with known molecular weight standards (filled circle) and the ND-GluRS:GatDE complex (open circle).
Therefore, to measure the affinity of ND-GluRS and GatDE for one another we used fluorescence anisotropy (Table 1 and Supplementary Figure S1). ND-GluRS or GatDE were AF-labeled as described in the ‘Materials and Methods’ section. The label did not affect the activities other either enzyme. AF-labeled ND-GluRS had a KD of 100 ± 22 nM for GatDE and labeled GatDE had a KD for ND-GluRS of 40 ± 5 nM (Table 1). The insertion domain found in GatE but not GatB is predicted to preclude GatDE binding ND-AspRS (15). In agreement, we did not detect association of GatDE with ND-AspRS (Figure 1C and Table 1).
We next determined whether the complex could form in a crude extract. We incubated AF-labeled ND-GluRS in an extract from Escherichia coli overproducing His6-tagged GatDE before passing the sample over a Ni-NTA column. E. coli extract enabled us to test the achaeal tRNAGln-independence of the GatDE:ND-GluRS association as E. coli tRNA isoacceptors are not recognized by the archaeal ND-GluRS (26) and bacterial Glu-tRNAGln is a poor substrate for GatDE (8). Pull-down of the labeled ND-GluRS by a Ni-NTA column was observed only when His6-tagged GatDE was produced in E. coli (Figure 1D), consistent with our in vitro data indicating that ND-GluRS associates with GatDE even in the absence of M. thermautotrophicus tRNAGln.
GatDE forms a homodimer with the binding interface between the D-subunits (Figure 2A) (15,25), while ND-GluRS is a monomer (Figure 2B). The structural model of the archaeal-specific transamidosome (15) predicts that ND-GluRS makes contact with the E-subunit of GatDE and thus allows the AdT dimer to bind two ND-GluRS monomers. To investigate this, we incubated GatDE with a molar excess of ND-GluRS before running the mixture over a sephacryl S300 gel filtration column. Two peaks were seen (Figure 2C), one corresponding to monomeric ND-GluRS. The other peak corresponded to a protein complex with a molecular weight of 380 kDa, consistent with association of a GatDE homodimer with two ND-GluRS monomers. SDS-PAGE analysis confirmed the presence of GatDE and ND-GluRS in the fraction, and the presence of GatDE was further established by immunoblot for the His6-tag on the E-subunit (data not shown). The same fraction was able to form Gln-tRNAGln upon addition of tRNAGln, Glu, ATP and Asn (Figure 2D), confirming the presence of active ND-GluRS and GatDE in the complex.
Formation of the ND-GluRS:GatDE:tRNAGln ternary complex
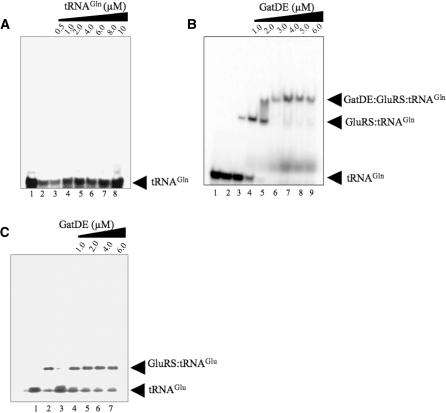
To be functional, the ND-GluRS:GatDE binary complex must bind tRNAGln. Therefore we investigated whether a ND-GluRS:GatDE:tRNAGln ternary complex could form. ND-GluRS readily binds tRNAGln with a KD of 37 ± 4 nM (Table 1). However, M. thermautotrophicus GatDE by itself has a poor affinity for unaminoacylated tRNAGln (Figures 3A and 4B, and Table 1). No change in anisotropy was observed for AF labeled-GatDE with increasing concentrations of tRNAGln (Table 1). However, in the presence of excess ND-GluRS, the apparent affinity of GatDE for tRNAGln significantly increases (KD of 80 ± 14 nM), suggesting the ND-GluRS:GatDE binary complex is able to efficiently bind tRNAGln.
Figure 3.
Formation of the ND-GluRS:GatDE:tRNAGln complex. Electrophoretic mobility shift assays were performed between purified ND-GluRS and GatDE with 32P-labeled tRNAGln or tRNAGlu as described in the ‘Materials and Methods’ section. (A) 32P-labeled tRNAGln (8 µM) incubated with no enzyme (lane 1) or increasing concentrations of 32P-labeled tRNAGln incubated with GatDE (1 µM; lanes 2–8). (B) 32P-labeled tRNAGln (100 nM) incubated with (lane 1) no enzyme, (lane 2) GatDE (6.0 µM), (lane 3) ND-GluRS (1 µM) or (lanes 4–9) ND-GluRS (1 µM) and GatDE (1.0–6.0 µM). (C) 32P-labeled tRNAGlu (100 nM) incubated with (lane 1) no enzyme, (lane 2) ND-GluRS (1 µM), (lane 3) GatDE (6 µM) or (lanes 4–7) ND-GluRS (1 µM) and GatDE (1.0–6.0 µM).
Figure 4.
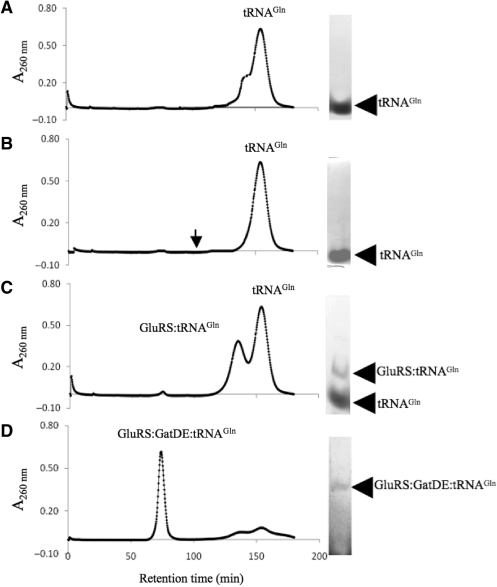
Gel filtration analysis of the archaeal GatDE:GluRS:tRNAGln complex. Gel filtrations over a sephacryl S300 16/60 column were carried out as described in the ‘Materials and Methods’ section with tRNAGln (20 µM) and protein(s) indicated (GatDE and/or ND-GluRS) (20 µM). (A) Gel filtration and native PAGE analysis of free tRNAGln. (B) Gel filtration and native PAGE analysis of a sample containing GatDE with tRNAGln. The arrow indicates how long approximately GatDE is retained on the column (Figure 2A). (C) Gel filtration and native PAGE analysis of a sample containing GluRS and tRNAGln. (D) Gel filtration and native PAGE analysis of a sample containing GluRS, GatDE and tRNAGln.
Consistent with this interpretation, a gel shift assay with stable concentrations of ND-GluRS and tRNAGln, but with increasing concentrations of GatDE, yielded a ternary complex of ND-GluRS:GatDE:tRNAGln (Figure 3B). In further agreement, a complex of ND-GluRS, GatDE, and tRNAGln eluted from a sephacryl S300 gel filtration column (Figure 4C) after pre-incubating the three compenents together. Pre-incubation of GatDE with ND-GluRS prior to addition of tRNAGln to the reaction mix did not appear to alter ternary complex formation (data not shown).
Stability of the ternary complex with aminoacylated tRNA
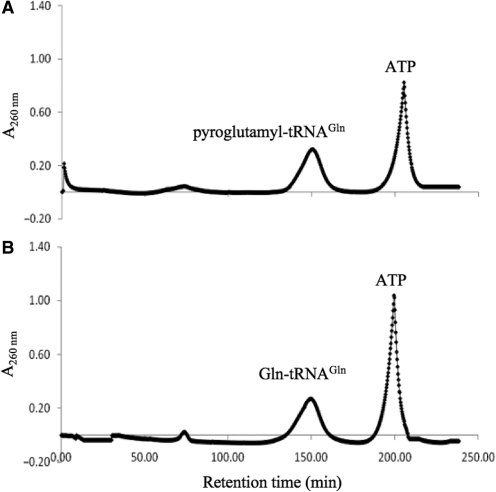
The T. thermophilus ND-AspRS:tRNAAsn:GatCAB transamidosome is stable after aminoacylation even in the presence of ATP (13). However, when we glutamylated tRNAGln, the ternary complex appeared to mostly dissociate (Figure 5A). Whether this is due to Glu-tRNAGln formation is unclear as residual ATP was still present. Under those conditions, GatDE readily converts Glu-tRNAGln to pyroglutamyl-tRNAGln (pGlu-tRNAGln) (19), a breakdown product of activated intermediate in the transamidation pathway. In the case of tRNA-dependent Asn synthesis, the activated intermediate readily converts back to Asp-tRNAAsn (19) without an amide donor and thus might not have been observed in the earlier T. thermophilus study (13). Formation of pGlu-tRNAGln would explain why no GatDE:Glu-tRNAGln binary complex eluted from the gel filtration column (Figure 5A). Following transamidation, the T. thermophilus transamidosome also remains intact (13), however under similar conditions the archaeal-specific transamidosome releases Gln-tRNAGln (Figure 5B). In both experiments, a minor peak was observed (at approximately 75 min) that might correspond to ND-GluRS:GatDE binding residual unacylated tRNAGln. Under the conditions of the gel filtration chromatography (see ‘Materials and Methods’ section), deacylation of the relevant aa-tRNA species was minimal (<5%) over the time-range used in the studies.
Figure 5.
Gel filtration analysis after aminoacylation and transamidation. Gel filtrations over a sephacryl S300 16/60 column were carried out as described in the ‘Materials and Methods’ section with tRNAGln, GatDE and ND-GluRS, (20 µM each) and either (A) ATP (4 mM) and l-Glu (3.5 mM), or (B) ATP (4 mM), l-Glu (3.5 mM) and Asn (4 mM) added to the incubation mixture.
The ND-AspRS:tRNAAsn:GatCAB complex remaining intact after aminoacylation and transamidation allows the transamidosome to protect Asp-tRNAAsn and especially Asn-tRNAAsn from deacylation (13,14). Given that the archaeal-specific ternary complex may not remain intact over the course of Gln-tRNAGln formation, we tested whether the presence of both GatDE and ND-GluRS protected Glu-tRNAGln and Gln-tRNAGln from deacylation (Supplementary Table S1). In contrast to the complexes with ND-AspRS and GatCAB, the presence of both ND-GluRS and GatDE did not protect the Glx-tRNAGln species from deacylation consistent with the observation that the ternary complex dissociated after Gln-tRNAGln formation (Figure 5B). In fact, in the case of Gln-tRNAGln, when both enzymes were present t1/2 decreased to 1.6 ± 0.1 min from 3.1 ± 0.1 min in the absence of protein (Supplementary Table 1). However, GatDE was able to provide some protection of Glu-tRNAGln. In the presence of only ND-GluRS, the t1/2 of Glu-tRNAGln was 1.7 ± <0.1 min, which increased to 3.5 ± 0.2 min in the presence of GatDE with the aaRS. The best protection was obtained with GatDE alone (t1/2 of 4.6 ± 0.5 min versus 3.3 ± 0.2 min without either enzyme present). The results suggest that GatDE, but not the binary complex of ND-GluRS:GatDE, binds Glu-tRNAGln and protects it from deacylation.
The ND-AspRS:tRNAAsn:GatCAB complex remaining intact over the course of RNA-dependent Asn biosynthesis also has an effect on the kinetics of the aaRS and AdT (13,14). Thermus thermophilus ND-AspRS aminoacylates tRNAAsn 8-fold faster when in complex with GatCAB. (13). Also, the rate-limiting step becomes product release (13), indicative of a stable complex during both aminoacylation and transamidation. Helicobacter pylori GatCAB has a 2-fold lower KM for Asp-tRNAAsn in the presence of ND-AspRS (14). In the case of the M. thermautotrophicus ND-GluRS:GatDE complex, we did not detect any significant change in kinetic behavior (Supplementary Figure S2, Tables 2 and 3). ND-GluRS was as active in the presence of GatDE as BSA (Table 2) and in the presence of GatDE product release did not appear to be rate-limiting (Supplementary Figure S2). Like in the T. thermophilus transamidosome (13), the rate of transamidation was faster than that of aminoacylation (Tables 2 and 3). For GatDE, we detected a slight increase in kcat when ND-GluRS was present (1.3-fold), but no change in KM as compared to when BSA was added (2.3 ± 0.6 µM and 2.4 ± 1.0 µM, respectively; Table 3).
Table 2.
Kinetic data for glutamylation activity of M. thermautotrophicus ND-GluRS for tRNAGln
| Enzyme | KM (µM) | kcat (min−1) | kcat/KM (min−1/µM) |
|---|---|---|---|
| GluRS | 2.1 ± 0.4 | 0.7 ± 0.1 | 0.3 ± 0.1 |
| GluRS + GatDEa | 2.4 ± 0.6 | 0.6 ± 0.1 | 0.3 ± 0.1 |
| GluRS + BSAb | 2.4 ± 0.5 | 0.6 ± <0.1 | 0.3 ± 0.1 |
Measurements were from three to four separate experiments. Standard deviations are reported. Reactions with M. thermautotrophicus ND-GluRS (50 nM) were carried out as described in the ‘Materials and Methods’ section at 37°C in the presence of excess ATP (4 mM), Glu (1 mM) and Asn (2 mM). The concentration of tRNAGln varied from 0.31 µM up to 15 µM.
aGatDE (2.0 µM) or bBSA (2.0 µM) was added to the reaction mix.
Table 3.
Kinetic data for transamidase activity of M. thermautotrophicus GatDE for Glu-tRNAGln
| Enzyme | KM (µM) | kcat (s−1) | kcat/KM (s−1/µM) |
|---|---|---|---|
| GatDEa | 1.7 ± 0.3 | 0.9 ± 0.1 | 0.5 ± 0.1 |
| GatDE + GluRSb | 2.3 ± 0.6 | 1.2 ± 0.1 | 0.6 ± 0.1 |
| GatDE + BSAc | 2.4 ± 1.0 | 0.9 ± 0.1 | 0.4 ± 0.2 |
Measurements were from three to four separate experiments. Standard deviations are reported. Reactions with M. thermautotrophicus GatDE (10 nM) were carried out as described in the ‘Materials and Methods’ section at 37°C in the presence of excess ATP (4 mM) and Asn (2 mM). Concentration of Glu-tRNAGln varied from 0.41 µM up to 13 µM.
aFrom reference 8.
bND-GluRS (2.0 µM) or cBSA (2.0 µM) was added to the reaction mix.
Presence of GatDE decreases ND-GluRS affinity for tRNAGlu
ND-GluRS in addition to recognizing tRNAGln (KD = 37 ± 4 nM) also uses tRNAGlu (KD = 80 ± 3 nM) as a substrate (Table 1). Methanothermobacter thermautotrophicus GatDE, however specifically amidates Glu-tRNAGln and thus has a greater affinity for tRNAGln than tRNAGlu, rejecting the latter tRNA based on its enlarged D-loop (15). As expected, GatDE was unable to form a ternary complex with ND-GluRS and tRNAGlu (Figure 3D). In the presence of excess GatDE, we were unable to saturate fluorescently labeled-ND-GluRS with increasing concentrations of tRNAGlu (Table 1). This is in contrast to tRNAGln, which was bound by the labeled ND-GluRS with similar affinities in the presence and absence of GatDE (KD = 110 ± 9 nM and 37 ± 4 nM, respectively). These results suggest that the association of GatDE with ND-GluRS enables the aaRS to be specific for tRNAGln over tRNAGlu. Consistent with that interpretation, increasing concentrations of GatDE decreased tRNAGlu binding to ND-GluRS in a nitrocellulose filter-binding assay (Supplementary Figure S3) with an apparent KI of 33 ± 5 nM. Also, in the presence of GatDE (1.8 µM), the Glu-tRNAGlu activity of ND-GluRS decreased almost in half (Supplementary Figure S4). In contrast, the rate of Glu-tRNAGln formation by ND-GluRS was unaffected by the presence of GatDE (Supplementary Figure S2, Table 2). The apparent KI of GatDE for ND-GluRS in the Glu-tRNAGlu reaction was 31 ± 15 nM, similar to the KD of GatDE for ND-GluRS (40 ± 5 nM, Table 1).
DISCUSSION
The archaeal transamidosome: a dynamic complex for RNA-dependent Gln biosynthesis
The T. thermophilus transamidosome (ND-AspRS:tRNAAsn:GatCAB) is a ribonucleoprotein (RNP) due to the fact the association of the two proteins is tRNA-dependent (13). The archaeal-specific transamidosome (ND-GluRS:GatDE), however, is not an RNP as tRNA is not essential for the two enzymes to bind one another, making the ND-GluRS:GatDE complex similar in this respect to the O-phosphoseryl-tRNA synthetase (SepRS):SepCysS complex for Cys-tRNACys formation in methanogenic archaea (27). Given the differences between the ND-GluRS:GatDE complex and the T. thermophilus complex, we suggest that a transamidosome is a complex between a ND-aaRS and an AdT in the process of amide aa-tRNA formation.
GatDE and ND-GluRS associating in an RNA-independent manner may be due to an insertion domain found in GatE but not in its paralog GatB (9,25,28). The insertion and catalytic domains of GatE form a concave pocket (25) that structural modeling predicts could accommodate ND-GluRS (15), increasing the surface for the AdT to bind the aaRS with. The tRNA-independent association maybe further aided by the fact that ND-GluRS is a class I aaRS whereas ND-AspRS is a class II aaRS. Class I aaRSs like AdTs bind to the minor groove of the tRNA acceptor stem whereas class II aaRSs bind to the major groove (13,15,17). Consequently, GatDE and ND-GluRS both bind to the same side of tRNAGln (15) while GatCAB and ND-AspRS likely recognize opposite sides of tRNAAsn (13). Thus, while GatDE and ND-GluRS can associate together, possibly enhanced by the GatE specific insertion domain, and recognize tRNAGln, ND-AspRS and GatCAB can only make minimal contact with one another and still specifically bind tRNAAsn preventing them from associating without tRNA (13).
Besides not requiring tRNA to form, the archaeal-specific ND-GluRS:GatDE transamidosome also differs from the ND-AspRS:tRNAAsn:GatCAB complex following aminoacylation and transamidation. The transamidosome for Asn-tRNAAsn synthesis is stable through product formation (13), leading to (i) an increase in ND-AspRS activity (13), (ii) product release becoming rate-limiting (13), (iii) protection of Asp-tRNAAsn and especially Asn-tRNAAsn from deacylation (13,14) and (iv) an enhancement of the KM of GatCAB for Asp-tRNAAsn (14). Similar results were found with the SepRS:SepCysS:tRNACys complex which is also stable after aminoacylation (27). However the archaeal-specific transamidosome, did not protect Glu-tRNAGln nor Gln-tRNAGln from deacylation and does not affect the kinetics of either ND-GluRS or GatDE. Taken all together, this suggests that the ND-GluRS:GatDE complex is not stable through Gln-tRNAGln formation. In agreement we did not detect ND-GluRS:GatDE bound to Gln-tRNAGln in our gel filtration studies.
The kinetic and deacylation data also suggest the archaeal-specific complex is not stable after aminoacylation. GatDE protects Glu-tRNAGln from deacylation whereas the addition of ND-GluRS actually increases the deacylation rate of the aa-tRNA species, possibly due to the aaRS favoring tRNAGln over Glx-tRNAGln species. This deacylation is probably not significant in vivo due to the presence of GatDE and EF-1α. Also, unlike ND-AspRS and GatCAB (14), the presence of ND-GluRS does not decrease the KM of GatDE for its mischarged substrate (Glu-tRNAGln). Thus, GatDE may remain bound to Glu-tRNAGln while ND-GluRS dissociates after aminoacylation.
Such a scenario would fit the structural modeling of the transamidosomes (10,13,15,16). While ND-GluRS and GatDE both binding to the tRNAGln minor groove may allow the enzymes to associate without the tRNA present, such an orientation of the proteins would hinder the movement of 3′ aminoacylated end of the tRNA acceptor stem from the aminoacylation active site of the ND-aaRS to the catalytic pocket of the AdT, as the direct route would be blocked by ND-GluRS. In the case of ND-AspRS and GatCAB, the fact they bind to opposite sides of the tRNA enables the 3′-end of acceptor stem to easily flip from the aminoacylation site in the aaRS to the transamidation site of the AdT (13). ND-GluRS dissociating from the complex after aminoacylation would eliminate the steric hindrance and allow the 3′ glutamylated end of tRNAGln to bind in the catalytic pocket of GatE for transamidation. The putative bacterial ND-GluRS:GatCAB complex may behave in a similar manner as it is expected that GatCAB binds tRNA like GatDE (16). However, given GatB lacks the insertion domain found in GatE, the association of ND-GluRS with GatCAB may require tRNAGln.
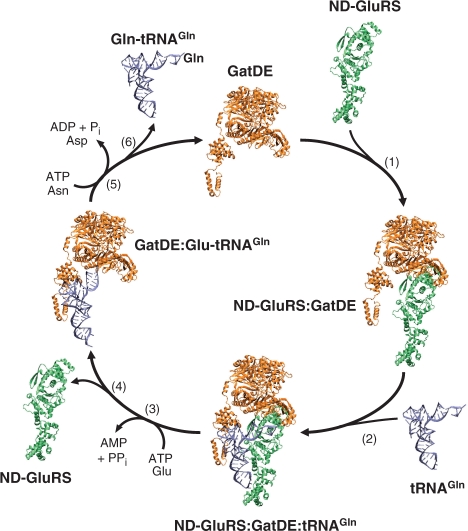
Given the above, we propose the following model for RNA-dependent biosynthesis of Gln in archaea (Figure 6). First, GatDE and ND-GluRS associate with the aaRS binding in the concave pocket formed by the GatE-specific insertion domain. Next, the ND-GluRS:GatDE complex recognizes tRNAGln with the 3′-end of the acceptor stem in the active site of the aaRS, while GatDE makes contact with the D-loop of the tRNA to distinguish tRNAGln from tRNAGlu (15). Alternatively, ND-GluRS could first find tRNAGln followed by GatDE binding. Following aminoacylation, ND-GluRS dissociates from the complex allowing the 3′ glutamylated end of tRNAGln to flip into the GatE catalytic pocket. GatDE recognition of the A1-U72 (15) may properly position the glutamyl-moiety for transamidation as well as serve as a final proofreading step to ensure the enzyme does not transamidate Glu-tRNAGlu. Binding of Glu-tRNAGln is expected to trigger conformational changes in GatDE to enable the AdT to liberate ammonia from free Asn or Gln to use in amidating the mischarged tRNA species (19,25). After transamidation, GatDE releases Gln-tRNAGln to be used in protein synthesis and the AdT is free to bind ND-GluRS and start the cycle again. Alternatively, GatDE could also remain unbound to transamidate any free Glu-tRNAGln in the cell.
Figure 6.
Cycle of RNA-dependent biosynthesis of Gln in Archaea. (1) GatDE (orange) associates with ND-GluRS (lime). (2) ND-GluRS:GatDE binary complex binds tRNAGln (silver-blue) to form the ternary complex. (3) In the transamidosome (ND-GluRS:GatDE:tRNAGln), ND-GluRS glutamylates tRNAGln. (4) ND-GluRS dissociates from the complex allowing the 3′ CCA-end of the tRNA to flip into the GatDE kinase active site. (5) GatDE transamidates the tRNA-bound Glu to Gln. (6) Gln-tRNAGln is released from GatDE. For clarity only one GatDE monomer is shown.
Function of the archaeal-specific ND-GluRS:GatDE transamidosome
Both the archaeal-specific and T. thermophilus transamidosomes provide a means for mischarged tRNA generated by ND-aaRSs to be channeled to the AdT (10,13,15), keeping the mischarged tRNA from being recognized by elongation factors and compromising the fidelity of protein synthesis. In addition, the T. thermophilus ND-AspRS:tRNAAsn:GatCAB complex is thought to protect Asn-tRNAAsn from deacylation until it can be used in translation (13). The archaeal ND-GluRS:GatDE complex does not appear to carry out this function as the presence of both enzymes actually accelerated the deacylation rate of Gln-tRNAGln. Instead, the binding of GatDE to ND-GluRS appears to sequester the aaRS for Gln-tRNAGln formation. In the presence of GatDE, ND-GluRS has a much higher affinity for tRNAGln than tRNAGlu (>10-fold), while in the absence of the AdT, ND-GluRS has similar affinities for both tRNAs. By increasing the concentration of GatDE relative to ND-GluRS, a cell can potentially favor Gln-tRNAGln formation over Glu-tRNAGlu.
Sequestering GluRS in a complex for a particular role in a cell is not unusual. For example, in Saccharomyces cerevisiae Arc1p sequesters GluRS in the cytoplasm for Glu-tRNAGlu synthesis as otherwise the aaRS is targeted to the mitochondrion to be used with an AdT to form Gln-tRNAGln (29). In chordates, GluRS is fused with the aaRS for Pro (GluProRS) and is part of the multi-synthetase complex (MSC) with seven other aaRSs for protein synthesis (12). In humans upon phosphorylation, GluProRS is released from the MSC and associates with NS1-associated protein 1, ribosomal protein L13a, and glyceraldehyde-3-phosphate dehydrogenase to silence translation of certain mRNAs related to the inflammatory response (30). In Chlamydomonas reinhardtii and Mycobacterium tuberculosis glutamyl-tRNA reductase (GluTR) binds GluRS likely to divert Glu-tRNAGlu synthesis from use in translation to tetrapyrrole biosynthesis (31,32). Given that many archaea posses GluTR (33), there may be competition for binding with ND-GluRS between GatDE and GluTR.
Evolutionary perspective
In archaea, two AdTs are encoded to form the amide aa-tRNA species needed for translation (8); GatDE serves as the Glu-AdT (9) while GatCAB is the Asp-AdT (8). It is still unclear what enabled such segregation of function, but phylogenetics suggests early archaea emerged from the last universal communal ancestor with both AdTs (28). Consistent with our biochemical data, structural modeling indicates the insertion domain specific to GatE prevents GatDE from binding ND-AspRS while still being able to associate with ND-GluRS (15). It is thus possible that the acquisition of the insertion domain by GatE may have favored GatDE serving as the Glu-AdT in early archaea as the AdT could complex with ND-GluRS but not ND-AspRS. Ancestral GatCAB however could still associate with ND-AspRS for tRNA-dependent Asn biosynthesis. Under such a scenario, the selective pressure to have a separate Glu-AdT and Asp-AdT would then have favored GatCAB as the Asp-AdT. Over time, GatDE would co-evolve with tRNAGln and ND-GluRS, and GatCAB with tRNAAsn and ND-AspRS, leading to the modern differences in tRNA recognition by the two archaeal AdTs (15,34,35).
Caution must be taken with regard to whether the functions of the transamidosomes mentioned above were initially selected for. For example, GatDE sequestering ND-GluRS and ND-AspRS:GatCAB protecting Asn-tRNAAsn from deacylation maybe evolutionary spandrels (36), a consequence perhaps of GluRS being a class I aaRS and AspRS being a class II enzyme as described above, with the actual selection to enable substrate channeling. Preventing mischarged tRNA from being used in protein synthesis may have also not been initially selected for. It is speculated that aaRS:AdT complexes were the mechanisms by which Gln and Asn were added to the genetic code with the codons for Gln and Asn initially coding for Glu and Asp respectively (13,15,37). Thus, the initial selection for an aaRS:AdT complex may have been for RNA-dependent synthesis of Gln and Asn for translation, and preventing mischarged tRNA from being used for translation as a consequence of the association of the two enzymes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Institute of General Medical Sciences and the United States Department of Energy (to D.S.). Funding for open access charge: National Institute of General Medical Sciences American Recovery and Reinvestment Act grant GM022854-34S1.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to A. Nakamura (Hokkaido University), H. Oshikane and O. Nureki (University of Tokyo), and L. Randau, I. Heinemann, P. O’Donoghue and J. Ling (Yale University) for many stimulating discussions. We thank A. Miranker, D. Blaho and A. Corajarena (Yale University) for assistance with the two-channel spectrofluorimeter.
REFERENCES
- 1.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Sheppard K, Yuan J, Hohn MJ, Jester B, Devine KM, Söll D. From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res. 2008;36:1813–1825. doi: 10.1093/nar/gkn015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lapointe J, Duplain L, Proulx M. A single glutamyl-tRNA synthetase aminoacylates tRNAGlu and tRNAGln in Bacillus subtilis and efficiently misacylates Escherichia coli tRNAGln1in vitro. J. Bacteriol. 1986;165:88–93. doi: 10.1128/jb.165.1.88-93.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilcox M, Nirenberg M. Transfer RNA as a cofactor coupling amino acid synthesis with that of protein. Proc. Natl Acad. Sci. USA. 1968;61:229–236. doi: 10.1073/pnas.61.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker HD, Reinbolt J, Kreutzer R, Giegé R, Kern D. Existence of two distinct aspartyl-tRNA synthetases in Thermus thermophilus. Structural and biochemical properties of the two enzymes. Biochemistry. 1997;36:8785–8797. doi: 10.1021/bi970392v. [DOI] [PubMed] [Google Scholar]
- 6.Curnow AW, Ibba M, Söll D. tRNA-dependent asparagine formation. Nature. 1996;382:589–590. doi: 10.1038/382589b0. [DOI] [PubMed] [Google Scholar]
- 7.Becker HD, Kern D. Thermus thermophilus: a link in evolution of the tRNA-dependent amino acid amidation pathways. Proc. Natl Acad. Sci. USA. 1998;95:12832–12837. doi: 10.1073/pnas.95.22.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheppard K, Sherrer RL, Söll D. Methanothermobacter thermautotrophicus tRNAGln confines the amidotransferase GatCAB to asparaginyl-tRNAAsn formation. J. Mol. Biol. 2008;377:845–853. doi: 10.1016/j.jmb.2008.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tumbula DL, Becker HD, Chang WZ, Söll D. Domain-specific recruitment of amide amino acids for protein synthesis. Nature. 2000;407:106–110. doi: 10.1038/35024120. [DOI] [PubMed] [Google Scholar]
- 10.Schön A, Kannangara CG, Gough S, Söll D. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature. 1988;331:187–190. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- 11.Srivastava DK, Bernhard SA. Metabolite transfer via enzyme-enzyme complexes. Science. 1986;234:1081–1086. doi: 10.1126/science.3775377. [DOI] [PubMed] [Google Scholar]
- 12.Hausmann CD, Ibba M. Aminoacyl-tRNA synthetase complexes: molecular multitasking revealed. FEMS Microbiol. Rev. 2008;32:705–721. doi: 10.1111/j.1574-6976.2008.00119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailly M, Blaise M, Lorber B, Becker HD, Kern D. The transamidosome: a dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagine biosynthesis. Mol. Cell. 2007;28:228–239. doi: 10.1016/j.molcel.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 14.Huot JL, Balg C, Jahn D, Moser J, Émond A, Blais SP, Chênevert R, Lapointe J. Mechanism of a GatCAB amidotransferase: aspartyl-tRNA synthetase increases its affinity for Asp-tRNAAsn and novel aminoacyl-tRNA analogues are competitive inhibitors. Biochemistry. 2007;46:13190–13198. doi: 10.1021/bi700602n. [DOI] [PubMed] [Google Scholar]
- 15.Oshikane H, Sheppard K, Fukai S, Nakamura Y, Ishitani R, Numata T, Sherrer RL, Feng L, Schmitt E, Panvert M, et al. Structural basis of RNA-dependent recruitment of glutamine to the genetic code. Science. 2006;312:1950–1954. doi: 10.1126/science.1128470. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura A, Sheppard K, Yamane J, Yao M, Söll D, Tanaka I. Two distinct regions in Staphylococcus aureus GatCAB guarantee accurate tRNA recognition. Nucleic Acids Res. 2010;38:672–682. doi: 10.1093/nar/gkp955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnez JG, Moras D. Structural and functional considerations of the aminoacylation reaction. Trends Biochem. Sci. 1997;22:211–216. doi: 10.1016/s0968-0004(97)01052-9. [DOI] [PubMed] [Google Scholar]
- 18.Sheppard K, Akochy PM, Salazar JC, Söll D. The Helicobacter pylori amidotransferase GatCAB is equally efficient in glutamine-dependent transamidation of Asp-tRNAAsn and Glu-tRNAGln. J. Biol. Chem. 2007;282:11866–11873. doi: 10.1074/jbc.M700398200. [DOI] [PubMed] [Google Scholar]
- 19.Feng L, Sheppard K, Tumbula-Hansen D, Söll D. Gln-tRNAGln formation from Glu-tRNAGln requires cooperation of an asparaginase and a Glu-tRNAGln kinase. J. Biol. Chem. 2005;280:8150–8155. doi: 10.1074/jbc.M411098200. [DOI] [PubMed] [Google Scholar]
- 20.Min B, Pelaschier JT, Graham DE, Tumbula-Hansen D, Söll D. Transfer RNA-dependent amino acid biosynthesis: an essential route to asparagine formation. Proc. Natl Acad. Sci. USA. 2002;99:2678–2683. doi: 10.1073/pnas.012027399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolfson AD, Pleiss JA, Uhlenbeck OC. A new assay for tRNA aminoacylation kinetics. RNA. 1998;4:1019–1023. doi: 10.1017/s1355838298980700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolfson AD, Uhlenbeck OC. Modulation of tRNAAla identity by inorganic pyrophosphatase. Proc. Natl Acad. Sci. USA. 2002;99:5965–5970. doi: 10.1073/pnas.092152799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheppard K, Akochy PM, Söll D. Assays for transfer RNA-dependent amino acid biosynthesis. Methods. 2008;44:139–145. doi: 10.1016/j.ymeth.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riggs AD, Suzuki H, Bourgeois S. Lac repressor-operator interaction. I. Equilibrium studies. J. Mol. Biol. 1970;48:67–83. doi: 10.1016/0022-2836(70)90219-6. [DOI] [PubMed] [Google Scholar]
- 25.Schmitt E, Panvert M, Blanquet S, Mechulam Y. Structural basis for tRNA-dependent amidotransferase function. Structure. 2005;13:1421–1433. doi: 10.1016/j.str.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 26.Santoro SW, Anderson JC, Lakshman V, Schultz PG. An archaebacteria-derived glutamyl-tRNA synthetase and tRNA pair for unnatural amino acid mutagenesis of proteins in Escherichia coli. Nucleic Acids Res. 2003;31:6700–6709. doi: 10.1093/nar/gkg903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang CM, Liu C, Slater S, Hou YM. Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNACys. Nat. Struct. Mol. Biol. 2008;15:507–514. doi: 10.1038/nsmb.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheppard K, Söll D. On the evolution of the tRNA-dependent amidotransferases, GatCAB and GatDE. J. Mol. Biol. 2008;377:831–844. doi: 10.1016/j.jmb.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frechin M, Senger B, Brayé M, Kern D, Martin RP, Becker HD. Yeast mitochondrial Gln-tRNAGln is generated by a GatFAB-mediated transamidation pathway involving Arc1p-controlled subcellular sorting of cytosolic GluRS. Genes Dev. 2009;23:1119–1130. doi: 10.1101/gad.518109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ray PS, Arif A, Fox PL. Macromolecular complexes as depots for releasable regulatory proteins. Trends Biochem. Sci. 2007;32:158–164. doi: 10.1016/j.tibs.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 31.Jahn D. Complex formation between glutamyl-tRNA synthetase and glutamyl-tRNA reductase during the tRNA-dependent synthesis of 5-aminolevulinic acid in Chlamydomonas reinhardtii. FEBS Lett. 1992;314:77–80. doi: 10.1016/0014-5793(92)81465-x. [DOI] [PubMed] [Google Scholar]
- 32.Paravisi S, Fumagalli G, Riva M, Morandi P, Morosi R, Konarev PV, Petoukhov MV, Bernier S, Chênevert R, Svergun DI, et al. Kinetic and mechanistic characterization of Mycobacterium tuberculosis glutamyl-tRNA synthetase and determination of its oligomeric structure in solution. FEBS J. 2009;276:1398–1417. doi: 10.1111/j.1742-4658.2009.06880.x. [DOI] [PubMed] [Google Scholar]
- 33.Moser J, Schubert WD, Beier V, Bringemeier I, Jahn D, Heinz DW. V-shaped structure of glutamyl-tRNA reductase, the first enzyme of tRNA-dependent tetrapyrrole biosynthesis. EMBO J. 2001;20:6583–6590. doi: 10.1093/emboj/20.23.6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailly M, Giannouli S, Blaise M, Stathopoulos C, Kern D, Becker HD. A single tRNA base pair mediates bacterial tRNA-dependent biosynthesis of asparagine. Nucleic Acids Res. 2006;34:6083–6094. doi: 10.1093/nar/gkl622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Namgoong S, Sheppard K, Sherrer RL, Söll D. Co-evolution of the archaeal tRNA-dependent amidotransferase GatCAB with tRNAAsn. FEBS Lett. 2007;581:309–314. doi: 10.1016/j.febslet.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gould SJ, Lewontin RC. The spandrels of San Marco and the Panglossian paradigm: a critique of the adaptationist programme. Proc. Roy. Soc. Lond. B. Biol. Sci. 1979;205:581–598. doi: 10.1098/rspb.1979.0086. [DOI] [PubMed] [Google Scholar]
- 37.Di Giulio M. The origin of the genetic code: theories and their relationships, a review. Biosystems. 2005;80:175–184. doi: 10.1016/j.biosystems.2004.11.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.