Abstract
The Nrf2 (nuclear factor E2 p45-related factor 2) transcription factor responds to diverse oxidative and electrophilic environmental stresses by circumventing repression by Keap1, translocating to the nucleus, and activating cytoprotective genes. Nrf2 responses provide protection against chemical carcinogenesis, chronic inflammation, neurodegeneration, emphysema, asthma and sepsis in murine models. Nrf2 regulates the expression of a plethora of genes that detoxify oxidants and electrophiles and repair or remove damaged macromolecules, such as through proteasomal processing. However, many direct targets of Nrf2 remain undefined. Here, mouse embryonic fibroblasts (MEF) with either constitutive nuclear accumulation (Keap1−/−) or depletion (Nrf2−/−) of Nrf2 were utilized to perform chromatin-immunoprecipitation with parallel sequencing (ChIP-Seq) and global transcription profiling. This unique Nrf2 ChIP-Seq dataset is highly enriched for Nrf2-binding motifs. Integrating ChIP-Seq and microarray analyses, we identified 645 basal and 654 inducible direct targets of Nrf2, with 244 genes at the intersection. Modulated pathways in stress response and cell proliferation distinguish the inducible and basal programs. Results were confirmed in an in vivo stress model of cigarette smoke-exposed mice. This study reveals global circuitry of the Nrf2 stress response emphasizing Nrf2 as a central node in cell survival response.
INTRODUCTION
Nuclear factor E2 p45-related factor 2 (symbol Nfe2l2; commonly called Nrf2), a basic-region leucine zipper transcription factor (TF), binds to the cis-regulatory, antioxidant response element (ARE) and transcriptionally upregulates an environmental stress response gene battery (1). The Nrf2 response is critical for protection against pulmonary inflammation and emphysema, asthma, acute lung injury, hyperoxia, pulmonary fibrosis, innate immune response, hepatotoxicity, carcinogenesis, neurodegeneration, cardiovascular disease and aging (2–11). A decline in the Nrf2 pathway is associated with severe chronic obstructive pulmonary disease (COPD) (12–15). In addition, several classes of small molecules such as 1,2-dithiolethiones (e.g. oltipraz), isothiocyanates (e.g. sulforaphane) and triterpenoids (e.g. CDDO-Im) induce expression of Nrf2 target genes and protect against chemical carcinogenesis, acute hepatotoxicity and cigarette smoke (CS)-induced emphysema (16–19). On the other hand, studies show that many cancers such as non small cell (NSCLC) and squamous cell lung tumors develop inactivating mutations in the Keap1 and Nrf2 genes, respectively, thereby constitutively enhancing the Nrf2 pathway and promoting tumorigenecity and resistance to an array of chemotherapeutic compounds (20,21). Hence, due to the dual role of Nrf2 in carcinogenesis and degenerative chronic diseases and the diversity of target genes under a variety of stresses, understanding the pathway components and regulators is critical for effectively targeting the pathway for prophylactic and therapeutic purposes.
Gene-expression profiling studies using different tissue and cell-culture systems exposed to diverse conditions (chemical or environmental) reveal the pleiotropic properties of Nrf2 in stress response and cell survival. Keap1 (Kelch ECH associating protein 1), a cytosolic repressor of the Nrf2 pathway, plays a central role in regulation of the Nrf2 response. Under normal conditions, Nrf2 is targeted by Keap1, which promotes Nrf2 proteasomal degradation via interactions with an ubiquitin ligase (22). Keap1 further functions as a sensor of stress signals, through stress-induced oxidation of key cysteine residues that lead to conformational changes and the inability to bind Nrf2 (23). Nrf2 then accumulates in the nucleus, where it binds to AREs in a heterodimeric complex with one of a subset of the small Maf-family of TFs (24,25).
Using Nrf2-knockout (Nrf2−/−) murine models in different organs and with various stressors, Nrf2 was found to coordinately regulate a suite of targets genes such as antioxidant genes, xenobiotic-metabolizing enzymes, many of which have been traditionally classified as part of the phase II detoxification system, glutathione homeostasis, solute channels, proteome maintenance and innate immune response (8,26–32). Disruption of Keap1 (Keap1−/−) leads to enhanced nuclear accumulation of Nrf2 and elevated expression of Nrf2-regulated genes (33). Largely, the Nrf2 regulated genes are associated with pathways related to cell survival—batteries of proteins that provide protection against a variety of oxidative and electrophilic stressors.
Although, many genes are regulated in an Nrf2-dependent manner, no large-scale studies have distinguished between direct and indirect targets of Nrf2, the latter perhaps emerging from Nrf2 cross-talk with other signaling pathways (34–36). In this report, we evaluated the concordance between global mRNA expression profiles at the transcriptomic levels (i.e., oligonucleotide microarray analyses) and genome-wide Nrf2-DNA-binding analysis using high-throughput methodologies (ChIP-Seq). This approach is important for establishing a systems level view of Nrf2-dependent processes occurring within cells. In the ChIP-Seq analysis, over 1200 Nrf2-bound regions were observed from Keap1−/− MEFs. These regions were strongly enriched for Nrf2-binding site patterns, with the motifs clustered near the maxima of the ChIP-Seq peaks. The gene-expression analysis identified 7155 genes significantly downregulated in Nrf2−/− compared with wild-type (WT) MEFs (basal targets) and 7832 significantly upregulated genes in Keap1−/− compared with WT MEFs (inducible targets), with 2489 modulated genes intersecting between the two datasets (common targets). Integrating the results from ChIP-Seq and expression profiling data, signatures were observed for gene groups that include detoxification response, cell proliferation, cell cycle and survival regulation networks. Strikingly, the proliferation and stress responses segregate between the direct Nrf2 target expression classes, with proliferation linked to basal expression and detoxification linked to inducibleactivation. The high-throughput results were validated using quantitative real-time polymerase chain reaction (qRT-PCR) and site-specific ChIP assays in MEFs and CS-exposed mouse lungs to demonstrate the Nrf2 response in vivo. The integrated analysis of the experimental data highlights the broad influence of Nrf2, providing a blueprint for the regulation of cytoprotective genes and genes involved in critical homeostatic function such as cell proliferation.
MATERIALS AND METHODS
Growth and viability of mouse embryonic fibroblasts
WT, Keap1−/−, and Nrf2−/− cell lines, were generated as described previously (33). Cells were grown to 90% confluency in 100-mm plates in Iscove’s modified Dulbecco’s medium (Invitrogen, CA) containing 10% FBS. Cells were harvested and processed for microarray and chromatin immunoprecipitation (ChIP) analyses as described below. Cell viability was assessed using the MTT assay as described previously (33).
CS exposure
WT and Nrf2−/− male mice (CD-1 strain, 8–10 weeks) were exposed to CS for 5 h/day using a TE-10 smoking machine (Teague Enterprises, Davis, CA) and 3R4F reference cigarettes (University of Kentucky, Tobacco Research Institute, Lexington, KY) as described previously (8). The study protocols were approved by the Johns Hopkins Institutional Review Board for animal studies.
ChIP
The ChIP was performed as described previously (37). MEFs were fixed ∼1% formaldehyde in cell growth media for 10 min followed by glycine fixation. Cells were then sonicated in ChIP–lysis buffer containing protease inhibitor cocktail after incubation on ice for 30 min. Upon sonication, the samples were spun at maximum speed for 12 min in a cold microcentrifuge. The supernatants were then addressed to as Input samples. A part of the input sample was precleared using 50% slurry of Protein A/G beads prepared in immunoprecipitation (IP) buffer for 2 h in cold, following which the beads were centrifuged out and supernatant was used for IP. Then the precleared samples were incubated with 2 µg of either of two distinct anti-Nrf2 antibodies that recognize different portions of Nrf2 protein. The rabbit-anti-Nrf2 monoclonal antibody from Epitomics Inc., CA, recognizes the C-terminal domain in all three cell lines, while the rabbit-anti-Nrf2 polyclonal antibody from Santa Cruz Biotech., CA, recognizes domains spanning the N-terminus in all three cell lines as well. For control, we used anti-rabbit IgG antibody for 1 h in cold. These antibodies have been used extensively in our laboratory for western and EMSA analysis [Figure 1A(i) and data not shown]. Protein A/G bead slurry was added to each reaction and was rotated overnight. Next day, beads were washed extensively, DNA was reverse crosslinked and eluted in elution buffer and stored at −80°C until used for sequencing or PCR validation assays. The eluted DNA was checked for 100- to 300-bp fragment enrichments using 8% polyacrylamide gel electrophoresis (data not shown).
Figure 1.
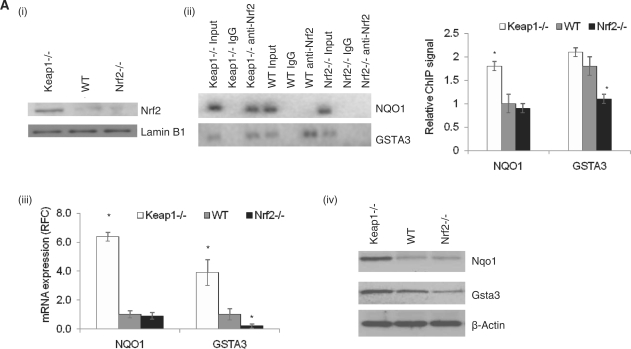
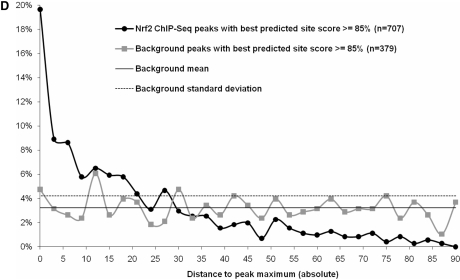
Nrf2 genome-wide ChIP-Seq dataset. (A) Keap1−/− MEFs have constitutive transcriptional activation of Nrf2 target genes (i) Nuclear Nrf2 protein levels in Keap1−/−, WT and Nrf2−/− MEFs using immunoblot analysis, (ii) ChIP–PCR and densitometry quantification analysis for known Nrf2-binding site in promoter of Nqo1 and Gsta3, (iii) qRT-PCR analysis of known Nrf2 targets, Nqo1 and Gsta3 and (iv) Immunoblot analysis for Nqo1 and Gsta3 in the MEFs. β-Actin was used as the loading control. (B) The distribution of Nrf2 ChIP-Seq peaks relative to the closest gene transcription start sites is similar to background. (C) Nrf2 ChIP-Seq peaks are enriched for high-scoring Nrf2 predicted motifs compared with background. (D) High-scoring predicted sites are preferentially located near the peak maximum height in the ChIP-Seq peaks whereas they are uniformly distributed in the background sequences. *P-value < 0.01, as analyzed by ANOVA analysis.
Massive parallel sequencing
We generated two DNA libraries using two Nrf2-specific antibodies [one monoclonal, rabbit-anti-Nrf2 (Epitomics Inc., CA), and one polyclonal, rabbit-anti-Nrf2 (Santa Cruz Biotech., CA)] incubated with crosslinked and sonicated DNA from Keap1−/− MEFs. A third control library contains the input DNA from Keap1−/− MEFs. The sequencing and reads alignment to the mouse genome (NCBI build 37) was performed at the Genome Science Centre (Vancouver, Canada) as described previously (38).
ChIP-Seq library peak finding
To identify significant Nrf2-bound regions in the data we used the FindPeaks software (39) with the following parameters: Triangle dist. = 100 low/200 median/300 high; Sub-peaks = 0.8; Trim-peaks = 0.2; Filter Duplicates = On; Filter out reads quality lower than 10. The false discovery rate estimation (FDR) was based on a Lander-Waterman calculation integrated in the FindPeaks software. An artifactual cluster of peaks was found at the beginning of chromosome 11 in all three libraries and was discarded from the analysis.
Peak overlap determination between the three ChIP-Seq libraries
The peak height is defined by the number of tags overlapping at a specific genomic location. All peaks above a height of 5 from each of the three libraries were considered in this analysis. Peaks were considered as overlapping when their maximum heights were located within 100 bp of each other.
Peak sequence extraction and analysis
One hundred nucleotides on each side of the peak maximum height location were extracted from the EnsEMBL database using Perl scripts. These sequences were analyzed for A, C, G, T and dinucleotide content. For the background, the sequences were extracted similarly for all peaks with a height ≥5 and only the ones with a composition similar to the Nrf2 ChIP-Seq peak sequences were retained.
To plot the peak distribution relative to transcription start sites (TSS), the closest TSS from each peak was extracted from the EnsEMBL database using Perl scripts and the distance computed.
The Nrf2-binding site prediction was performed by scanning each sequence with the Nrf2-binding profile based on known binding sites (Figure 2A) using the TFBS Perl modules (40). For the peaks containing a predicted site with a score at least equal to 0.85, the absolute distance from the closest end of the site to the peak maximum height position was computed.
Figure 2.
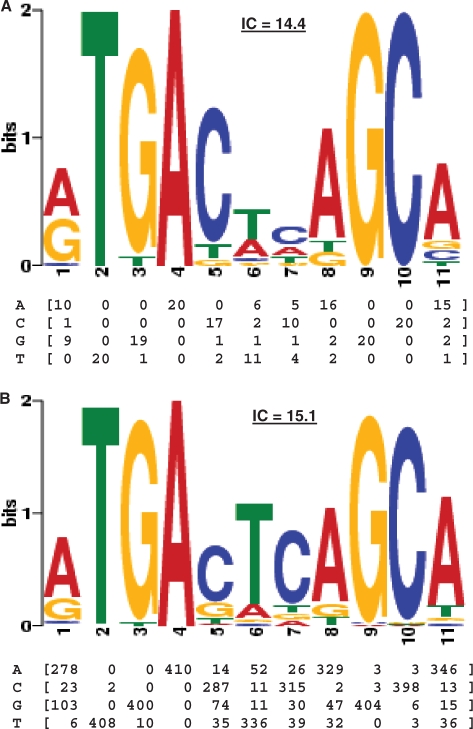
Nrf2-binding profiles. (A) The original Nrf2-binding profile was defined by the alignment of the 20 known binding sites described in Table 1. (B) The motif generated by the MEME motif discovery algorithm (50) on the Nrf2 ChIP-Seq dataset is highly similar to the original profile with a higher information content (IC; in bits). The position Frequency Matrix (PFM) captures the count of each nucleotide found at each position in the motif and the logo provides a visualization of the data. The information content is the sum of the information content of each position and is computed as described previously (61).
Statistical analysis
The statistical significance of the differences observed between the Nrf2 ChIP-Seq dataset and the background were computed using a Wilcoxon rank sum test with continuity correction using the R package. The Fisher’s exact test demonstrating greater overlaps than expected by chance between various datasets were performed using the fisher. test() function in the R package with the option alternative = ‘greater’.
Transcription factor-binding site overrepresentation
We used a modified version of the oPOSSUM system (41) in which the overrepresentation is calculated directly on the ChIP-Seq sequences rather than on conserved regions located around user-specified genes. The overrepresentation was calculated through the comparison with the background sequences from the control library. The binding profiles were extracted from the JASPAR 2008 vertebrate core collection (42) using a score threshold of 80%.
MEME motif discovery analyses
The MEME web service (http://meme.sdsc.edu) was ran with the following options: -dna -mod zoops -nmotifs 1 -minw 6 -maxw 50 -time 7200 -maxsize 60000 –revcomp. The obtained motifs were compared with the original Nrf2-binding profile using the online STAMP service (http://www.benoslab.pitt.edu/stamp/). Motifs were aligned using the Smith–Waterman algorithm and compared with the Pearson Correlation Coefficient similarity measure.
Oligonucleotide microarray
Total RNA was purified using the RNeasy Mini kit (Qiagen, Valencia, CA) after isolation using TRIzol reagent (Life Technologies, Inc., Grand Island, NY). Murine Genome MOE 430A GeneChip arrays (Affymetrix, Santa Clara, CA), which contain probes for detecting ∼14 500 well-characterized genes and 4371 expressed sequence tags (ESTs) were used to probe the RNA obtained from the cells using the procedure described previously (10).
Microarray data analysis
Data analysis was performed using open source ‘R’ Statistical software (version 2.6.1) through the Bioconductor project (43). The raw data files were loaded using affy package while probe intensities and normalizations were done using Robust Multichip Averaging (gcRMA) method. The first approach was to test the internal consistency and to explore the relationship among samples and underlying features of gene expression comparing 21 536 unique gene probe sets across three replicates for control and treatment. The corresponding gene names and annotation information was extracted from ‘mouse4302’ annotation package provided by Bioconductor. Differential expression analysis was conducted using Significance Analysis of Microarray (SAM) using the same R statistical package (version 1.25). SAM calculates a score for each gene on the basis of the change in expression relative to the standard deviation of all measurements by computing a t-statistic. SAM then performs a set of permutations to determine the false discovery rate (FDR) with adjustment method for multiple testing. The reported list of ranked genes have a ‘delta value’ which defines the threshold of false positive in the validated dataset, which was adjusted to a stringent FDR <1 (44). Seed Clustering was performed with set seeds (determined from differential expression analysis and known active AREs). The seed clustering calculates a distance matrix between genes and based on indicated seeds (clusters), places the gene with the minimum distance to a seed into the seed’s cluster.
Validation of ChIP-Seq data by ChIP–PCR
Selected peaks from promoters of genes from different functional groups were validated by ChIP–PCR for selected regions. Primers were selected on both strands a 100 bp from the Nrf2 ChIP-Seq peaks. The same immunoprecipitated DNA as analyzed in the ChIP-Seq assay was used for PCR amplification using the specific gene promoter PCR assay. PCR primers for each of the PCR reaction are shown in Supplementary Table S1. PCR amplifications were carried out with a 95°C denaturation step for 5 min followed by 35 cycles with a cycle consisting of 95°C denaturation for 30 s, 58°C annealing step for 45 s and 72°C extension step for 45 s each, followed by a final elongation step at 72°C for 7 min. The PCR products were then run on a 1.2% Agarose gel and visualized with ethidium bromide staining. The data are represented as the relative ChIP signal in a cell line. They are calculated by taking the ratio of fold difference in (PCR amplification in specific antibody ChIP samples as compared with PCR amplification with genomic DNA input sample in each cell line) versus (PCR amplification in IgG samples for each cell line as compared with PCR amplification with genomic DNA input in that respective cell line). The significance of ChIP signal in Keap1−/− or Nrf2−/− cell lines are calculated as compared with WT cells using Student’s t-test and represented at P-value < 0.01.
Validation of microarray data by qRT-PCR
Selected genes from different functional groups were validated by qRT-PCR. Total RNA from cells used for microarray was purified using the Qiagen RNeasy kit (Qiagen, Valencia, CA) for mRNA isolation. qRT-PCR was then performed using Assay-on-Demand primers and probe sets from Applied Biosystems (Foster City, CA) as described previously (10). We used the ABI 7000 Taqman system (Applied Biosystems) to perform these assays. β-actin was used as a normalization control. The mRNA data are represented as relative fold change (RFC). RFC are calculated as the ratio of target gene expression (fold change of particular gene mRNA expression in experimental sample/control sample) versus (fold change of β-actin mRNA expression in experimental sample/control sample). Control sample for MEF gene-expression data are WT MEFs and WTAir for mice mRNA expression data.
Cell proliferation assay
Cell proliferation was measured by QuantosTM cell proliferation assay (Stratagene, La Jolla, CA) and cell proliferation ELISA, BrDU chemiluminescence assay (Roche Applied science, Indianapolis, IN) as per manufacturer’s instructions.
Caspase activity assay as a marker of cell death
Caspase activity for caspase-3 and -7 was measured using Apo-ONE homogeneous caspase-3 and -7 assay kit (Promega, WI).
Cell senescence assay
Cell senescence was measured using a fluorimetric assay to examine senescence associated beta-galactosidase activity from Cell Biolabs Inc. CA.
Immunoblot assay
Immunoblots were performed using antibodies for Nrf2, Srxn1, Cdkn1a, Cdkn2a (Santa Cruz Biotechnology, CA), Nqo1, Gsta3, Txnrd1, Gsta4, Gstm1, Als2, Bmpr1a, Igf1, Npnt, Pdgfc, Vegfc (Abcam Inc., MA). β-actin and Lamin B1 (Santa Cruz Biotechnology, CA) were used as loading controls. These immunoblots were performed using protocols as described previously with Nrf2, Srxn1, Cdkn1a, Cdkn2a and β-actin primary antibodies used at 1:500 dilution; Nqo1, Gsta3, Txnrd1, Gsta4, Gstm1, Als2, Bmpr1a, Igf1, Npnt, Pdgfc, Vegfc used at 1:200 dilution while Lamin B1 (β-Lamin) was used at 1:1000 dilutions (13).
RESULTS
Genetic model of Nrf2 activation
MEFs derived from Keap1−/−, Nrf2−/−, as well as WT mice were used as models in our studies. Nrf2 is constitutively elevated in nuclei of Keap1−/− MEFs as compared with WT or Nrf2−/− MEFs [Figure 1A(i)]. Consequently, Nrf2 target site (ARE) binding, as observed by ChIP [Figure 1A(ii)] and Nrf2-regulated transcriptional activity, measured as target gene mRNA expression by qRT-PCR [Figure 1A(iii)] and protein expression by immunoblot [Figure 1A(iv)], are significantly higher in Keap1−/− MEFs when compared with WT and Nrf2−/− MEFs. There is basal nuclear translocation of Nrf2 in WT MEFs at low levels that leads to expression of a subset of Nrf2 targets which will be referred to as basal Nrf2 targets. This is the case for Gsta3, which displays Nrf2 binding to its promoter in WT MEFs and differential expression between WT and Nrf2−/− MEFs [Figure 1A(ii–iv)]. In contrast, Nqo1 is an inducible target of Nrf2, as Nrf2 binds to its promoter and selectively induces its expression only in Keap1−/− MEFs [Figure 1A (ii–iv)].
Genome-wide identification of Nrf2 target sites
We performed a ChIP-Seq experiment with Keap1−/− MEFs, in which Nrf2 is protected from ubiquitination and degradation, constitutively activating the expression of Nrf2 target genes. We generated two DNA libraries using two Nrf2-specific antibodies (one monoclonal and one polyclonal). A third control library contains the input DNA. We obtained 13 998 222, 14 068 662, and 16 375 760 mapped reads for the polyclonal, monoclonal and control libraries, respectively. Clusters of overlapping tags were identified as peaks using the FindPeaks software (39). Peak height thresholds were identified for each library corresponding to a stringent FDR ≤0.001. The monoclonal antibody selected library was of superior quality. A peak-height threshold of nine specified the required stringency with 1128 peaks meeting the criteria (Supplementary Table S2). In contrast, the polyclonal antibody selected library required a threshold of 11 to achieve the stringency with only 271 peaks meeting the criteria (Supplementary Table S2). Only 40 of the 271 peaks were unique to the polyclonal library. We focused on the monoclonal collection. A final set of 886 monoclonal peaks not overlapping the control library peaks were selected for further analysis. As the polyclonal library is complementary to the monoclonal library, we added those peaks that overlapped between the two libraries (peak height ≥5) but were absent from the control library (370 peaks). This resulted in a total of 1256 ChIP-Seq peaks representing a high-confidence set of Nrf2-binding sites. In addition to these nuclear genome-wide Nrf2-binding sites, we identified nine significant Nrf2 ChIP-Seq peaks in the mitochondrial genome. These peaks represent the first report of Nrf2 transcriptional activity in the mitochondria. They were not included in the bioinformatics sequence analyses below because of the different properties of the nuclear and mitochondrial genomes.
To confirm the quality of the results, we assessed the recovery of bona fide Nrf2-binding sites in the human, mouse or rat genomes compiled from the literature into the PAZAR regulatory annotation repository (18). Ten of the corresponding mouse regions overlap one of the ChIP-Seq peaks and while four do not overlap a peak, a peak can be found in proximity (Table 1, Supplementary Figure S1). This finding of 50% sensitivity (considering only the peaks that strictly overlap a site) is highly significant (Fisher’s exact test; P < 2.2e–16) and is a strong indication of the quality of the results, all the more so as the literature derived studies were performed in a variety of species and cell types.
Table 1.
Overlap of Nrf2 ChIP-Seq peaks with known Nrf2-binding sites annotated in the PAZAR database (http://www.pazar.info)
| Gene symbol | PAZAR annotation | ChIP-Seq peak |
|---|---|---|
| Fth1 | RS0144992 | Overlaps known BS |
| Ftl1 | RS0144993 | Overlaps known BS |
| Gsta3 | RS0145004 | Overlaps known BS |
| Gstp1/Gstp2 | RS0145008 | Overlaps known BS |
| Mafg | RS0145006 | Overlaps known BS |
| Nqo1 | RS0145007 | Overlaps known BS |
| Txn1 | RS0144999 | Overlaps known BS |
| Gclc | RS0144994 | Overlaps known BS + Additional peak |
| Hmox1 | RS0145005 | Overlaps known BS + Additional peak |
| Txnrd1 | RS0145000 | Overlaps known BS + Additional peak |
| Abcc1 | RS0144989 | Nearby known BS |
| Abcc2 | RS0145001 | Nearby known BS |
| Ets1 | RS0144991 | Nearby known BS |
| Gclm | RS0144995 | Nearby known BS |
| Akr1b3 | RS0145002 | No peak |
| Gnai2 | RS0144996 | No peak |
| Gpx2 | RS0144997 | No peak |
| Gsta1 | RS0145003 | No peak |
| Nqo2 | RS0144998 | No peak |
| Akr1c1 | RS0144990 | Mouse ortholog difficult to assign |
Focusing on 201-bp sequences (100-bp flanking each side of the maximum height position), we determined that these sequences exhibit a nucleotide composition bias with 57% (A or T), 43% (G or C) and a ratio of [CG]/[C or G] of 0.024. Thus, we selected the background sequences from the control library with similar composition and obtained a control sample of 5225 sequences with an overall composition of 56% (A or T), 44% (G or C), and a ratio of [CG]/[C or G] of 0.027.
Comparing the positional distribution of the 1256 Nrf2-binding sites with the 5225 background sequences relative to the closest gene transcription start site (TSS) showed no significant difference (Figure 1B). The concentration bias in favor of proximal TSS observed in both the Nrf2 and control sequence sets is likely a reflection of the open chromatin state of these regions making the DNA more susceptible to breakage in the shearing step of the procedure. Similar enrichment bias for transcription start regions has been reported as systematic in ChIP-Seq experiments (45). Similarly, the chromosomal distribution of the peaks showed no significant difference with the background (data not shown).
Nrf2-binding site motifs are enriched within the ChIP-Seq identified regions. A binding profile reflecting the properties of the literature-derived Nrf2-binding site collection was constructed (Figure 2A). Figure 1C displays the distribution of the top scoring matches to the Nrf2-binding profile in the Nrf2 peaks and the control peaks. The background shows a normal distribution centered around a score of 0.8 (Figure 1C). The score distribution for the Nrf2 bound peaks is skewed towards higher scoring sites with more than 55% of the sequences presenting Nrf2 sites with a score ≥0.85, a stringent threshold (Figure 1C), and is significantly different from the background (Wilcoxon rank sum test; P < 2.2e–16). Binding site predictions suffer from high rates of false positives (46), as evident in the 7% of control peaks containing an Nrf2 profile match with a score ≥0.85. A positional bias was observed in the positive collection (Figure 1D). In the 707 Nrf2 ChIP-Seq peaks that have a predicted Nrf2-binding site scoring at least 0.85, 65% contain the site within 22 bp of the peak maximum height, whereas in the 379 background sequences, the sites are distributed uniformly across the sequences. The difference proves statistically significant (Wilcoxon rank sum test; P < 2.2e–16). In conclusion, the results demonstrate that the Nrf2 ChIP-Seq dataset is highly enriched for Nrf2-binding sites as defined by the enriched presence and positional distribution of Nrf2-binding site motifs.
Binding site enrichment is a powerful tool to identify relationships between characterized TFs and sets of genes or sequences determined from genome-scale profiling experiments. In addition to the dramatic enrichment for Nrf2-binding sites, we observed enrichment for Fos and Mafb-binding sites within the Nrf2 collection (Table 2). This result is consistent with the fact that Nrf2 is known to heterodimerize with Maf proteins (47) and a subset of Nrf2-binding sites have been shown to overlap AP-1 response elements, AP-1 being a complex composed of Fos and Jun proteins (48).
Table 2.
TFBS overrepresentation in the Nrf2 ChIP-Seq peaks using the oPOSSUM system and the vertebrate JASPAR profiles
| TF | TF Class | Background gene hits | Target gene hits | Background TFBS rate | Target TFBS rate | Z-score | Fisher score |
|---|---|---|---|---|---|---|---|
| NRF2 | bZIP | 1222 | 849 | 0.0144 | 0.0546 | 169.7 | 5.94E–185 |
| Fos | bZIP | 3566 | 1063 | 0.0449 | 0.0736 | 69.97 | 1.87E–31 |
| Mafb | bZIP, MAF | 4125 | 1081 | 0.0604 | 0.0839 | 49.77 | 5.23E–08 |
| Lhx3 | HOMEO | 2066 | 610 | 0.0219 | 0.0331 | 38.67 | 1.28E–08 |
| MEF2A | MADS | 1192 | 344 | 0.0124 | 0.0165 | 18.4 | 6.47E–04 |
| Cebpa | bZIP | 2706 | 704 | 0.0421 | 0.0492 | 17.99 | 7.36E–03 |
| CREB1 | bZIP | 1121 | 313 | 0.0143 | 0.0178 | 14.98 | 6.74E–03 |
| HNF1A | HOMEO | 435 | 143 | 0.006 | 0.0083 | 14.91 | 6.79E–04 |
| IRF1 | TRP-CLUSTER | 1220 | 338 | 0.0157 | 0.0194 | 14.8 | 6.90E–03 |
| Myf | bHLH | 1765 | 472 | 0.0238 | 0.0283 | 14.79 | 9.90E–03 |
| ESR1 | NUCLEAR RECEPTOR | 76 | 33 | 0.0013 | 0.0023 | 14.43 | 4.39E–03 |
| NFIL3 | bZIP | 1034 | 296 | 0.012 | 0.0146 | 12.2 | 2.68E–03 |
Only significant hits are reported (Z-score >10 and Fisher score <0.01).
To investigate whether the current binding model for Nrf2 (Figure 2A)—the model used to predict the binding sites in the above analyses—reflects the data contained in the Nrf2 ChIP-Seq dataset, we ran a motif discovery analysis on the 1256 sequences. Motif discovery software is extremely sensitive to noise (49). Thus, we focused our analysis on the 45-bp region central to the peaks as we had determined that most true binding sites were located within ∼22 bp of the peak maximum (Figure 1D). We used the MEME motif discovery tool (50) to perform the analysis of these sequences and obtained a motif very similar to the original one (Figure 2B, E-value = 4.996e–15). The most remarkable difference lies in the central TC pattern that is stronger in the ChIP-Seq data and leads to an overall stronger profile (higher information content; Figure 2) that could lead to more specific predictions.
Microarray based Nrf2 genome-wide target selection and characterization
Genome-wide microarray experiments were performed to identify both basal and inducible Nrf2 target genes. Basal Nrf2 target genes are transcribed in untreated normal cells and are thus expected to be downregulated in Nrf2−/− MEFs as compared with WT (Figure 3A). Inducible Nrf2 targets are transcribed in response to stress conditions (such as oxidative stress), when Nrf2 is released from Keap1 repression, and are thus expected to be upregulated in Keap1−/− MEFs compared with WT (Figure 3A). From differential expression experiments, we identified 7155 genes significantly downregulated in Nrf2−/− compared with WT MEFs (‘basal’) and 7832 significantly upregulated genes in Keap1−/− compared with WT MEFs (‘inducible’) (Figure 3B). Focusing on the intersection of the two sets, 2489 genes are both basally expressed and inducible, referred to as ‘common’ targets (Figure 3B).
Figure 3.
Global transcriptional profiling of MEF genetic models to decipher comprehensive Nrf2-dependent transcriptional targets. (A) Scheme depicts definitions for classification of Nrf2 target genes as inducible and/or basally regulated by Nrf2. Basal targets: differentially expressed genes between WT versus Nrf2−/− MEFs; Inducible targets: differentially expressed genes between Keap1−/− versus WT MEFs; and Common targets: genes satisfying both the former comparison criteria. (B) The panel represents the overlap of differentially expressed genes across experiments. The orange circle represents genes that were downregulated in Nrf2−/− MEFs compared with WT (basal targets) while the blue circle shows the genes upregulated in Keap1−/− MEFs compared with WT (inducible targets). (C) The panel represents the overlap between the microarray datasets and the ChIP-Seq dataset and intersections represent the genes common in the two datasets to identify Nrf2 direct (basal, inducible and common) transcriptional targets.
Integrated analysis of Nrf2 direct target genes
We hypothesized that genes directly modulated by Nrf2 would contain a ChIP-Seq peak in their vicinity and show altered regulation in the microarray studies. We extracted the flanking genes, from both sides, of the Nrf2 ChIP-Seq peaks totaling 1738 genes. We identified 654 unique genes overlapping between ChIP-Seq and basal Nrf2 target dataset (genes downregulated in Nrf2−/− versus WT comparison, 7155 genes) (Supplementary Table S3, Figure 3C) and 645 genes overlap between ChIP-Seq and inducible Nrf2 targets (genes upregulated in Keap1−/− versus WT comparison, 7832 genes) (Supplementary Table S4, Figure 3C). The intersection of the peak-flanking genes with the 2489 basal and inducible expression modulated genes is statistically significant (Fisher’s exact test; P = 3.469e–06) with an integrated set of 244 genes (Supplementary Table S5, Figure 3C).
The MEME motif discovery analysis described above was ran on the ChIP-Seq peak sequences located near each of the three Nrf2 target gene subsets (basal, inducible and common), but no significant difference was observed in the obtained profile.
Gene Ontology (GO) term over-representation analyses were performed for ‘basal’ and ‘inducible’ groups independently using the DAVID annotation analysis system (51) and identified term enrichment for biological processes. Of important note, the inducible and basal gene sets exhibited distinct maximally enriched processes, with ‘cell proliferation’ dominating the basal genes and ‘response to oxidative stress’ the most prominent and only statistically-significant for the inducible genes (see Supplementary Tables S6 and S7 for 654 basal and 645 inducible target genes, respectively). In addition to the regular GO over-representation analysis, DAVID provides a clustering function that forms sets of overlapping gene categories, again highlighting ‘response to stress’ and ‘cell proliferation’ for the two categories (Table 3).
Table 3.
Functional clustering of gene annotations using the DAVID resource reveals an enrichment for genes involved in cell proliferation in the basal target gene set and glutathione metabolism, involved in response to stress, in the inducible target gene set
| Category | Term | Count | Percentage | P-value | FDR |
|---|---|---|---|---|---|
| Top cluster for the set of 654 basal target genes (Geometric mean: 5.23E–6) | |||||
| GOTERM_BP_4 | GO:0042127∼regulation of cell proliferation | 33 | 5.05 | 7.47E–07 | 1.27E–03 |
| GOTERM_BP_4 | GO:0008284∼positive regulation of cell proliferation | 21 | 3.21 | 9.00E–06 | 1.53E–02 |
| GOTERM_BP_4 | GO:0048522∼positive regulation of cellular process | 54 | 8.26 | 2.12E–05 | 3.61E–02 |
| Top cluster for the set of 645 inducible target genes (Geometric mean: 1.03E–3) | |||||
| KEGG_PATHWAY | mmu00480:Glutathione metabolism | 10 | 1.55 | 6.85E–05 | 8.54E–02 |
| INTERPRO | IPR003081:Glutathione S-transferase, Mu class | 5 | 0.78 | 1.07E–04 | 2.07E–01 |
| PIR_SUPERFAMILY | PIRSF000503:glutathione transferase | 7 | 1.09 | 1.56E–04 | 2.82E–01 |
| GOTERM_MF_4 | GO:0004364∼glutathione transferase activity | 7 | 1.09 | 2.80E–04 | 4.02E–01 |
| INTERPRO | IPR012335:Thioredoxin fold | 11 | 1.71 | 5.19E–04 | 1.00 |
| INTERPRO | IPR004045:Glutathione S-transferase, N-terminal | 6 | 0.93 | 1.37E–03 | 2.62 |
| INTERPRO | IPR004046:Glutathione S-transferase, C-terminal | 6 | 0.93 | 1.37E–03 | 2.62 |
| SP_PIR_KEYWORDS | Dimer | 4 | 0.62 | 3.57E–03 | 5.39 |
| KEGG_PATHWAY | mmu00980:Metabolism of xenobiotics by cytochrome P450 | 9 | 1.40 | 1.06E–02 | 12.4 |
| COG_ONTOLOGY | Posttranslational modification, protein turnover, chaperones | 11 | 1.71 | 1.17E–01 | 73.2 |
Under the hypothesis that Nrf2 is part of a broader regulatory network and needs the cooperation of other TFs to regulate the expression of its target genes, we performed TF-binding site (TFBS) over-representation analyses of the Nrf2 ChIP-Seq peak sequences associated with the basal and inducible target gene sets (Table 4). We first masked out in the sequences all Nrf2 predicted sites with a score ≥0.8 to focus our search on the other motifs present in the sequences. In both the basal and inducible datasets, we identified two significant signatures from a homeodomain and a MADS TF (Table 4). Interestingly, many homeodomain TFs are part of the Nrf2 basal and/or inducible target gene sets, three of them, Dlx2, Pbx1 and Meis1, being present in both and therefore candidates to participate in the Nrf2 regulatory network.
Table 4.
TFBS over-representation in subsets of Nrf2 ChIP-Seq peaks associated with basal and inducible target genes using the oPOSSUM system and the vertebrate JASPAR profiles
| TF | TF class | Background gene hits | Target gene hits | Background TFBS rate | Target TFBS rate | Z-score | Fisher score |
|---|---|---|---|---|---|---|---|
| ChIP-Seq peaks associated with the Nrf2 basal target genes | |||||||
| Lhx3 | HOMEO | 2039 | 333 | 0.0218 | 0.0341 | 30.81 | 2.59E–05 |
| MEF2A | MADS | 1179 | 196 | 0.0125 | 0.0182 | 18.68 | 1.56E–03 |
| HNF1A | HOMEO | 424 | 81 | 0.006 | 0.009 | 14.49 | 2.41E–03 |
| NFIL3 | bZIP | 1009 | 168 | 0.0118 | 0.0155 | 12.72 | 3.57E–03 |
| ChIP-Seq peaks associated with the Nrf2 inducible target genes | |||||||
| Lhx3 | HOMEO | 2039 | 306 | 0.0218 | 0.0321 | 25.15 | 1.81E–03 |
| MEF2A | MADS | 1179 | 185 | 0.0125 | 0.0179 | 17.28 | 4.72E–03 |
All NRF2 predicted sites >80% are masked in foreground and background.
Antioxidant and detoxification networks regulated by Nrf2
For validation analysis we focused on the 244 genes, representing the Nrf2 targets regulated under basal as well as inducible conditions. Our results demonstrate an upregulation of many antioxidant and xenobiotic detoxification genes in response to the de-repression of Nrf2, the most commonly described function of this TF (Figure 4A and B). These results expand our knowledge of Nrf2 regulation of these pathways by inclusion of novel direct targets as well as by identification of the DNA-binding sites in the respective promoters. Members of the glutathione system that function to scavenge reactive oxygen species and preserve cellular redox balance (such as Gsr and Gpx2) (52,53) are direct transcriptional targets of Nrf2.
Figure 4.
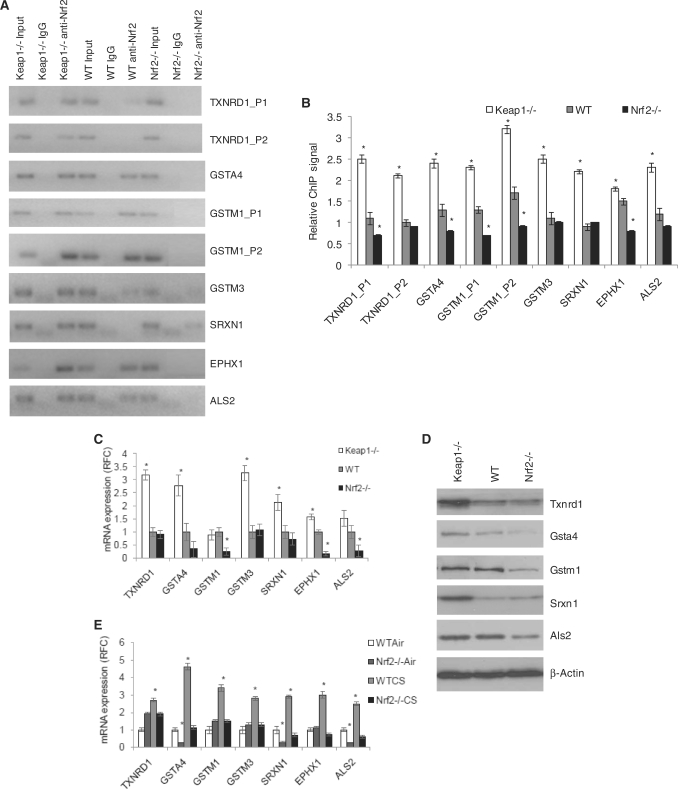
Antioxidant and xenobiotic detoxification genes are validated as Nrf2 direct targets. (A) and (B) The panels represent ChIP–PCR analysis and densitometry quantification for selected known Nrf2 target gene-binding sites—Txnrd1_P1 and its novel binding site Txnrd1_P2, novel binding sites in Gsta4, three Gstm1-binding sites (Gstm1_P1, Gstm1_P2, Gstm1_P3), Gstm3, Srxn1, Ephx1 and Als2. (C) qRT-PCR mRNA expression analysis in MEFs. (D) Immunoblot analysis for Txnrd1, Gsta4, Gstm1, Srxn1 and Als2 in MEFs. β-Actin was used as the loading control. (E) qRT-PCR for mRNA expression in lung lysates from WT and Nrf2−/− Air and CS exposed mice, corroborating with the in vitro Nrf2 targets. *P-value < 0.01, as analyzed by ANOVA analysis.
We validated the known binding sites for Nqo1, Gsta3 and Txnrd1 and an additional upstream site as direct Nrf2-binding site (Txnrd1_P2) [Figures 1A(ii, iii), 4A and B]. In addition, we identified and validated Nrf2-binding sites in promoters of known cytoprotective Nrf2 target genes such as Gsta4, Gstm1, Gstm3 and Srxn1 along with the less known targets Ephx1 and Als2 (Figure 4A and B). DNA-binding results as analyzed by ChIP–PCR directly corresponded with greater mRNA expression of the respective genes in Keap1−/− and/or WT MEFs as compared with Nrf2−/− MEFs (Figure 4C). However, in the case of Gstm1 and Als2, the microarray and ChIP–PCR results were not entirely validated by the qRT-PCR as the difference in Nrf2 binding between Keap1−/− and WT MEFs did translate into a significant difference in gene expression in the microarray but not in the qRT-PCR. This discrepancy between our high-throughput and PCR gene-expression data may be a result of the differential sensitivities of the techniques involved.
As a means for further confirmation of target genes, we assessed the protein expression levels of selected genes by immunoblot (Figure 4D). Immunoblots of Txnrd1 and Srxn1corroborate them as inducible targets while Gstm1 and Als2 were validated as basal-only targets and Gsta4 was confirmed as a common- target (Figure 4D).
To confirm our results in an in vivo stress-regulated model, we analyzed the expression patterns of these genes in the lungs of 1-day CS-exposed mice from WT and Nrf2−/− (Nrf2−/−CS) mice compared with their air controls (WTAir and Nrf2−/−Air). In this model, Nrf2 basal targets are genes downregulated in Nrf2−/− air-exposed cells compared with WT air-exposed cells, and inducible targets are genes upregulated in CS-exposed WT cells compared with air-exposed WT cells. The antioxidant and xenobiotic detoxification genes identified as Nrf2 direct targets in the MEF genetic model were validated in qRT-PCR assays to be Nrf2 targets in mouse lungs exposed to CS-induced stress, thereby highlighting the physiological importance of the identified targets (Figure 4E).
Regulators of cell proliferation, cell cycle and survival networks as transcriptional targets of Nrf2
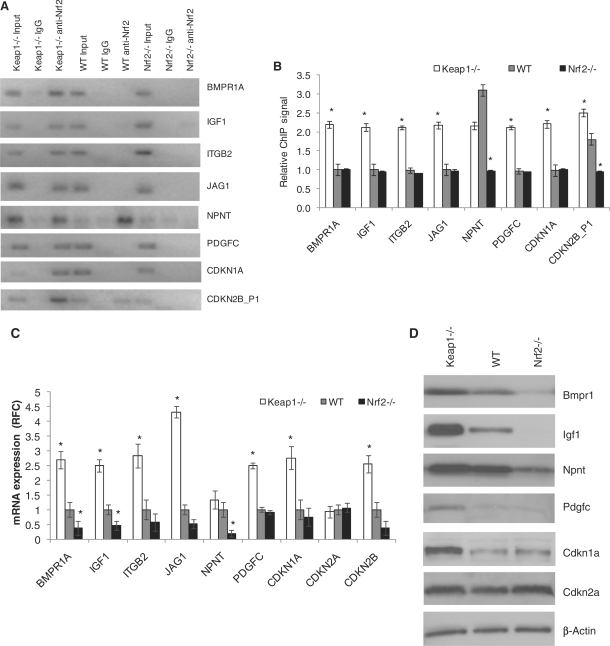
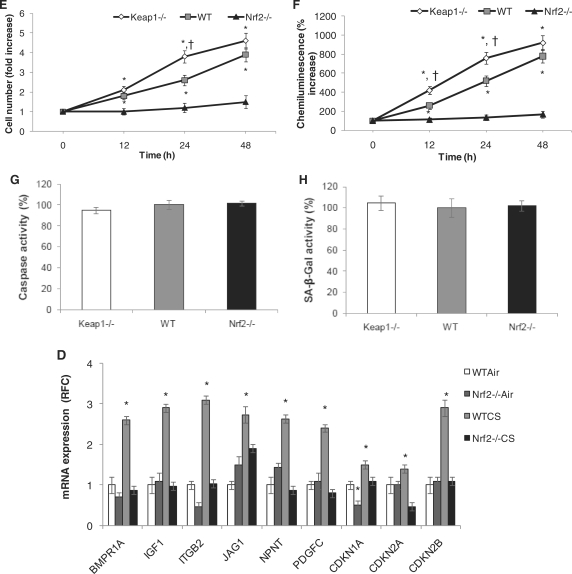
The compilation of the ChIP-Seq and gene-expression profiling analyses identified component genes of the cell proliferation/survival pathways as well as cell-cycle regulators such as Cdkn2b. Using ChIP–PCR and qRT-PCR analyses for DNA binding on the identified sites and mRNA probes respectively, we validated several of these genes as direct transcriptional targets of Nrf2 (Figure 5A, B and C). Cell proliferation regulators such as Als2 (Figure 4) and Npnt (Figure 5A and B) were found to be basal- targets of Nrf2 by ChIP–PCR and were confirmed as basal- only targets of Nrf2 by qRT-PCR and immunoblot assays (Figures 4, Figure 5C and D). Other cell proliferation associated genes as Bmpr1a, Igf1, Itgb2, Jag1 and Pdgfc were found to be inducible targets of Nrf2 as assessed by ChIP–PCR (Figure 5A and B). In contrast, the microarray analyses (as well as the qRT-PCR for Bmpr1a and Igf1) defined these genes as common Nrf2 targets. The gene-expression change is much greater between Keap1−/− and WT MEFs than between Nrf2−/− and WT MEFs and the latter might thus be due to secondary effects rather than direct Nrf2-binding changes. Additional evaluation of target gene expression using protein expression analysis confirms Bmpr1a, Igf1 and Vegfc as common targets, Pdgfc and Cdkn1a as inducible targets and Npnt as a basal-only target (Figure 5D). Next, as Cdkn2b is a part of this integrated gene set, we looked through all the Nrf2 ChIP-Seq dataset for additional cell-cycle regulators and identified Cdkn1a and Cdkn2a, none of them being differentially regulated in the microarray analysis. Using ChIP–PCR and qRT-PCR assays, Cdkn1a (p21) and Cdkn2b (p15) were found to be direct inducible targets of Nrf2, on the other hand, Cdkn2a (p16) does not appear to be a direct Nrf2 target (Figure 5A, B and C). Immunoblot analysis confirms Cdkn1a as an inducible target of Nrf2 while no change in protein expression of Cdkn2a was observed similar to the qRT-PCR results (Figure 5C and D). We checked whether the changes in the gene expression of these cell proliferation and survival promoting genes translated into increased proliferation of Keap1−/− and WT cells when compared with Nrf2−/− cells. Corroborating the gene-expression results, we found a significant increase in proliferation of Keap1−/− as well as WT cells as compared with Nrf2−/− cells under normal conditions as analyzed by increased DNA content quantified by QuantosTM cell proliferation assay and increased BrdU uptake assessed by BrDU chemiluminescence assay respectively (Figure 5E and F). Recent publications have emphasized the role of Nrf2 in promoting cell proliferation and cell-cycle progression (17,41), but this report is the first to broadly characterize the direct role of Nrf2 in executing the regulatory program governing proliferation. Since, Cdkn1a and Cdkn2b are traditionally considered as cell-cycle inhibitors and inducers of apoptosis or senescence; we sought to distinguish if their induction promoted inhibition of cell proliferation or induction of cell death or senescence in Keap1−/− cells. We found no differential induction of apoptosis or senescence, while there was a relative increase in proliferation in Keap1−/− cells when compared with WT or Nrf2−/− cells (Figure 5G and H). Regulation of Cdkns as direct targets and components of Nrf2 stress response remains a very interesting finding and its implications on cellular physiology remain to be unraveled. These genes were also validated as Nrf2 targets in vivo in mice lungs under CS-induced stress (Figure 5I). These two observations implicate novel probable roles for Cdkn1a and Cdkn2b in Nrf2-regulated cell survival and/or proliferation.
Figure 5.
Cell proliferation genes and cell-cycle regulators are direct transcriptional targets of Nrf2. (A) and (B) ChIP–PCR and densitometry quantification analysis for cell proliferation genes and Cdkn genes—Cdkn1a, Cdkn2b-binding site (Cdkn2b_P1), comparing Keap1−/−, WT and Nrf2−/− MEFs. (C) qRT-PCR for mRNA expression analysis of proliferation genes and Cdkns respectively. (D) Immunoblot analysis for Bmpr1a, Igf1, Npnt, Pdgfc, Vegfc CDKN1A and CDKN2A in MEFs. β-Actin was used as the loading control. (E) QuantosTM Cell proliferation assay. (F) BrdU chemiluminescence assay. (G) Caspase activity (apoptosis) assay. (H) Cell senescence assay. (I) qRT-PCR for mRNA expression in lung lysates from WT and Nrf2−/− Air and CS exposed mice corroborating with the in vitro Nrf2 targets. *P-value < 0.01 as compared with control and †P-value < 0.01 as compared with WT at the same time point, as analyzed by ANOVA analysis.
DISCUSSION
We have described a genome-scale analysis of the regulatory network governed by the Nrf2 TF using a combination of high-throughput sequencing for ChIP and microarray-based gene-expression profiling. The genome-scale experiments were performed with MEFs isolated from mice lacking Keap1, the key mediator of Nrf2 degradation, and mice lacking Nrf2 as well as WT mice. The results agree with the recognized role of Nrf2 in regulating the expression of protective genes that attenuate cytotoxicity in response to chemical toxins, and reveal a strong role for Nrf2 in the direct regulation of cellular proliferation. The segregation of ‘cell proliferation’ gene enrichment to a basal gene-expression cluster contrasts with ‘stress response’ enrichment in an inducible cluster. An unbiased binding profile derived from the experimental data extends a previous model for the binding specificity of Nrf2. The validity of the high-throughput results were confirmed by independent ChIP and qRT-PCR assays. To confirm the functional relevance of the findings in the transgenic cells, the results were confirmed using lung samples from mice exposed to concentrated CS in comparison to air controls. These findings represent the first comprehensive genome-wide study of Nrf2 binding and, coupled with the expression profiling, reveal the broad role of Nrf2 in protecting cells against toxic conditions.
The central role of Nrf2 in activating protective gene expression is long recognized, but its role in the regulation of cell proliferation has received less attention. Studies on the control of cellular differentiation in both adipocytes (54) and osteoblasts (55) have indicated that activation of the Nrf2 pathway can inhibit differentiation. Given the relationship between the maintenance of a proliferative state and the cell cycle, these studies are suggestive of a link between Nrf2 and the cell cycle. Our own studies have shown a potential link between Nrf2 and the oxidative-stress induced inhibition of proliferation (56,57). The current study also allows for speculation about novel roles of Nrf2 pathway in regulation of diseases. For example, Nrf2 target gene, Pdgf is involved in a prenatal placental disorder, preeclampsia, because of increasing levels of their antagonist, fms-like tyrosine kinase 1 (Flt1) (58,59). In addition, Nrf2 regulated gene expression has been linked previously with vascular homeostasis and oxidative stress response in endothelial and smooth muscle cells in protection against diseases such as atherosclerosis and preeclampsia (60). These observations along with our data on positive regulation of Pdgf by Nrf2 indicate a more important role of Nrf2 pathway in developmental diseases such as preeclampsia.
The segregation of ‘proliferation’ targets and ‘stress’ targets to genetically distinct expression groups is noteworthy. The distinction suggests a potential complexity to the Nrf2 regulatory network (Figure 6). The analysis of transcription factor motif over-representation did not reveal an enriched binding site for any TF that segregates between the sets. It is possible that the distinction results from an alternative mechanism such as a microRNA activity.
Figure 6.
Summary of Nrf2-regulated global transcriptomics circuit. This Nrf2 interaction network was generated using the String interaction network analysis tool (http://string.embl.de). The genes represented are the basal (black), inducible (white) and common (grey) target genes associated with cell proliferation and/or glutathione metabolism as defined by the functional clusters obtained through the DAVID resource and described in Table 3.
The Nrf2 pathway has been studied in diverse tissue contexts, due to potential roles in stroke recovery, COPD and cancer prevention in multiple organs. The profiling of gene-expression changes mediated by the Nrf2 pathway has been well documented. The interpretation of such studies, however, has been hampered by the inability to distinguish genes modulated directly by Nrf2 from genes induced secondarily by Nrf2-sensitive TFs. The availability of the ChIP-Seq technology and a high-quality antibody have provided this first opportunity to intersect the two classes of genomic data and consequently define direct gene targets of the pathway. The genomic regions directly bound by Nrf2 in the vicinity of Nrf2-responsive genes constitute a rich starting set for the study of Nrf2 regulation. The downstream analysis of such target regions could reveal heretofore under-appreciated properties of the elements that could distinguish inducible cis-regulatory elements from other sequences bound by Nrf2 with no functional outcome.
The development of a high-throughput ChIP-Seq procedure provides a direct path for future experimental studies. As the cost of ChIP-Seq experiments declines, it will be possible to perform temporal studies of binding in response to inducing conditions and to perform comparative analyses of responses between different tissues. Such studies will be an important focus as researchers seek to discern the tissue-specific properties of the pathway and identify functional targets for interventions, both to induce the protective enzymes in healthy tissue and to suppress the system in cancer cells where the pathway has become deregulated.
The identification of a well-defined Nrf2 regulatory program is a critical step in advancing translational research. Nrf2 plays an important role in the susceptibility to COPD and other diseases; moreover, it protects cells against a variety of environmental stressors and carcinogens. Thus, the full discovery of the global circuitry of Nrf2 as a master protector is a significant step in our understanding of its function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Operating support from the Canadian Institutes of Health Research; infrastructure support from Canada Foundation for Innovation (to W.W.W.); Scholar of the Michael Smith Foundation for Health Research and a New Investigator of the CIHR (to W.W.W.); National Institutes of Health (R01 HL081205, R01 CA140492-01A1, R01 GM079239, 1P50HL074945-01, 1P50ES015903, 4P50CA058184, 5R21/R33AI080541, P01ES018176, 5P30ES003819 to S.B.); (CA94076 to T.W.K.); Clinical innovator award from Flight Attendant Medical Research Institute (to S.B.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the team at the British Columbia Genome Sciences Center for assistance with the preparation and analysis of the ChIP-Seq data, with special recognition to Anthony Fejes and Richard Varhol. They would also like acknowledge Vikas Misra and Hannah Lee for their input for the microarray experiments.
REFERENCES
- 1.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 2.Chan K, Kan YW. Nrf2 is essential for protection against acute pulmonary injury in mice. Proc. Natl Acad. Sci. USA. 1999;96:12731–12736. doi: 10.1073/pnas.96.22.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho HY, Jedlicka AE, Reddy SP, Kensler TW, Yamamoto M, Zhang LY, Kleeberger SR. Role of NRF2 in protection against hyperoxic lung injury in mice. Am. J. Respir. Cell Mol. Biol. 2002;26:175–182. doi: 10.1165/ajrcmb.26.2.4501. [DOI] [PubMed] [Google Scholar]
- 4.Collins AR, Lyon CJ, Xia X, Liu JZ, Tangirala RK, Yin F, Boyadjian R, Bikineyeva A, Pratico D, Harrison DG, et al. Age-accelerated atherosclerosis correlates with failure to upregulate antioxidant genes. Circ. Res. 2009;104:e42–e54. doi: 10.1161/CIRCRESAHA.108.188771. [DOI] [PubMed] [Google Scholar]
- 5.Duan W, Zhang R, Guo Y, Jiang Y, Huang Y, Jiang H, Li C. Nrf2 activity is lost in the spinal cord and its astrocytes of aged mice. In Vitro Cell Dev. Biol. Anim. 2009;45:388–397. doi: 10.1007/s11626-009-9194-5. [DOI] [PubMed] [Google Scholar]
- 6.Okawa H, Motohashi H, Kobayashi A, Aburatani H, Kensler TW, Yamamoto M. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem. Biophys. Res. Commun. 2006;339:79–88. doi: 10.1016/j.bbrc.2005.10.185. [DOI] [PubMed] [Google Scholar]
- 7.Papaiahgari S, Kleeberger SR, Cho HY, Kalvakolanu DV, Reddy SP. NADPH oxidase and ERK signaling regulates hyperoxia-induced Nrf2-ARE transcriptional response in pulmonary epithelial cells. J. Biol. Chem. 2004;279:42302–42312. doi: 10.1074/jbc.M408275200. [DOI] [PubMed] [Google Scholar]
- 8.Rangasamy T, Cho CY, Thimmulappa RK, Zhen L, Srisuma SS, Kensler TW, Yamamoto M, Petrache I, Tuder RM, Biswal S. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J. Clin. Invest. 2004;114:1248–1259. doi: 10.1172/JCI21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rangasamy T, Guo J, Mitzner WA, Roman J, Singh A, Fryer AD, Yamamoto M, Kensler TW, Tuder RM, Georas SN, et al. Disruption of Nrf2 enhances susceptibility to severe airway inflammation and asthma in mice. J. Exp. Med. 2005;202:47–59. doi: 10.1084/jem.20050538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thimmulappa RK, Lee H, Rangasamy T, Reddy SP, Yamamoto M, Kensler TW, Biswal S. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J. Clin. Invest. 2006;116:984–995. doi: 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Fields J, Zhao C, Langer J, Thimmulappa RK, Kensler TW, Yamamoto M, Biswal S, Dore S. Role of Nrf2 in protection against intracerebral hemorrhage injury in mice. Free Radic. Biol. Med. 2007;43:408–414. doi: 10.1016/j.freeradbiomed.2007.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goven D, Boutten A, Lecon-Malas V, Marchal-Somme J, Amara N, Crestani B, Fournier M, Leseche G, Soler P, Boczkowski J, et al. Altered Nrf2/Keap1-Bach1 equilibrium in pulmonary emphysema. Thorax. 2008;63:916–924. doi: 10.1136/thx.2007.091181. [DOI] [PubMed] [Google Scholar]
- 13.Malhotra D, Thimmulappa R, Navas-Acien A, Sandford A, Elliott M, Singh A, Chen L, Zhuang X, Hogg J, Pare P, et al. Decline in NRF2-regulated antioxidants in chronic obstructive pulmonary disease lungs due to loss of its positive regulator, DJ-1. Am. J. Respir. Crit. Care Med. 2008;178:592–604. doi: 10.1164/rccm.200803-380OC. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Malhotra D, Thimmulappa R, Vij N, Navas-Acien A, Sussan T, Merali S, Zhang L, Kelsen SG, Myers A, Wise R, et al. Heightened endoplasmic reticulum stress in the lungs of patients with chronic obstructive pulmonary disease: the role of Nrf2-regulated proteasomal activity. Am. J. Respir. Crit. Care Med. 2009;180:1196–1207. doi: 10.1164/rccm.200903-0324OC. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Suzuki M, Betsuyaku T, Ito Y, Nagai K, Nasuhara Y, Kaga K, Kondo S, Nishimura M. Down-regulated NF-E2-related factor 2 in pulmonary macrophages of aged smokers and patients with chronic obstructive pulmonary disease. Am. J. Respir. Cell Mol. Biol. 2008;39:673–682. doi: 10.1165/rcmb.2007-0424OC. [DOI] [PubMed] [Google Scholar]
- 16.Hu R, Xu C, Shen G, Jain MR, Khor TO, Gopalkrishnan A, Lin W, Reddy B, Chan JY, Kong AN. Gene expression profiles induced by cancer chemopreventive isothiocyanate sulforaphane in the liver of C57BL/6J mice and C57BL/6J/Nrf2 (-/-) mice. Cancer Lett. 2006;243:170–192. doi: 10.1016/j.canlet.2005.11.050. [DOI] [PubMed] [Google Scholar]
- 17.Osburn WO, Yates MS, Dolan PD, Chen S, Liby KT, Sporn MB, Taguchi K, Yamamoto M, Kensler TW. Genetic or pharmacologic amplification of nrf2 signaling inhibits acute inflammatory liver injury in mice. Toxicol Sci. 2008;104:218–227. doi: 10.1093/toxsci/kfn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Portales-Casamar E, Kirov S, Lim J, Lithwick S, Swanson MI, Ticoll A, Snoddy J, Wasserman WW. PAZAR: a framework for collection and dissemination of cis-regulatory sequence annotation. Genome Biol. 2007;8:R207. doi: 10.1186/gb-2007-8-10-r207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sussan TE, Rangasamy T, Blake DJ, Malhotra D, El-Haddad H, Bedja D, Yates MS, Kombairaju P, Yamamoto M, Liby KT, et al. Targeting Nrf2 with the triterpenoid CDDO-imidazolide attenuates cigarette smoke-induced emphysema and cardiac dysfunction in mice. Proc. Natl Acad. Sci. USA. 2009;106:250–255. doi: 10.1073/pnas.0804333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh A, Boldin-Adamsky S, Thimmulappa RK, Rath SK, Ashush H, Coulter J, Blackford A, Goodman SN, Bunz F, Watson WH, et al. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 2008;68:7975–7984. doi: 10.1158/0008-5472.CAN-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Z, Zhang S, Chan JY, Zhang DD. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol. Cell. Biol. 2007;27:6334–6349. doi: 10.1128/MCB.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto T, Suzuki T, Kobayashi A, Wakabayashi J, Maher J, Motohashi H, Yamamoto M. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol. Cell. Biol. 2008;28:2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blank V. Small Maf proteins in mammalian gene control: mere dimerization partners or dynamic transcriptional regulators? J. Mol. Biol. 2008;376:913–925. doi: 10.1016/j.jmb.2007.11.074. [DOI] [PubMed] [Google Scholar]
- 25.Katsuoka F, Motohashi H, Ishii T, Aburatani H, Engel JD, Yamamoto M. Genetic evidence that small maf proteins are essential for the activation of antioxidant response element-dependent genes. Mol. Cell. Biol. 2005;25:8044–8051. doi: 10.1128/MCB.25.18.8044-8051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho HY, Reddy SP, Kleeberger SR. Nrf2 defends the lung from oxidative stress. Antioxid. Redox. Signal. 2006;8:76–87. doi: 10.1089/ars.2006.8.76. [DOI] [PubMed] [Google Scholar]
- 27.Kwak MK, Wakabayashi N, Itoh K, Motohashi H, Yamamoto M, Kensler TW. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J. Biol. Chem. 2003;278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- 28.Lee JM, Calkins MJ, Chan K, Kan YW, Johnson JA. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J. Biol. Chem. 2003;278:12029–12038. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- 29.Lee JM, Li J, Johnson DA, Stein TD, Kraft AD, Calkins MJ, Jakel RJ, Johnson JA. Nrf2, a multi-organ protector? FASEB J. 2005;19:1061–1066. doi: 10.1096/fj.04-2591hyp. [DOI] [PubMed] [Google Scholar]
- 30.Purdom-Dickinson SE, Lin Y, Dedek M, Morrissy S, Johnson J, Chen QM. Induction of antioxidant and detoxification response by oxidants in cardiomyocytes: evidence from gene expression profiling and activation of Nrf2 transcription factor. J. Mol. Cell. Cardiol. 2007;42:159–176. doi: 10.1016/j.yjmcc.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen G, Xu C, Hu R, Jain MR, Nair S, Lin W, Yang CS, Chan JY, Kong AN. Comparison of (-)-epigallocatechin-3-gallate elicited liver and small intestine gene expression profiles between C57BL/6J mice and C57BL/6J/Nrf2 (-/-) mice. Pharm. Res. 2005;22:1805–1820. doi: 10.1007/s11095-005-7546-8. [DOI] [PubMed] [Google Scholar]
- 32.Stacy DR, Ely K, Massion PP, Yarbrough WG, Hallahan DE, Sekhar KR, Freeman ML, et al. Increased expression of nuclear factor E2 p45-related factor 2 (NRF2) in head and neck squamous cell carcinomas. Head Neck. 2006;28:813–818. doi: 10.1002/hed.20430. [DOI] [PubMed] [Google Scholar]
- 33.Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat. Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- 34.Li W, Khor TO, Xu C, Shen G, Jeong WS, Yu S, Kong AN. Activation of Nrf2-antioxidant signaling attenuates NFkappaB-inflammatory response and elicits apoptosis. Biochem. Pharmacol. 2008;76:1485–1499. doi: 10.1016/j.bcp.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pitha-Rowe I, Liby K, Royce D, Sporn M. Synthetic triterpenoids attenuate cytotoxic retinal injury: cross-talk between Nrf2 and PI3K/AKT signaling through inhibition of the lipid phosphatase PTEN. Invest. Ophthalmol. Vis. Sci. 2009;50:5339–5347. doi: 10.1167/iovs.09-3648. [DOI] [PubMed] [Google Scholar]
- 36.Shin S, Wakabayashi N, Misra V, Biswal S, Lee GH, Agoston ES, Yamamoto M, Kensler TW. NRF2 modulates aryl hydrocarbon receptor signaling: influence on adipogenesis. Mol. Cell. Biol. 2007;27:7188–71897. doi: 10.1128/MCB.00915-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Euskirchen GM, Rozowsky JS, Wei CL, Lee WH, Zhang ZD, Hartman S, Emanuelsson O, Stolc V, Weissman S, Gerstein MB, et al. Mapping of transcription factor binding regions in mammalian cells by ChIP: comparison of array- and sequencing-based technologies. Genome Res. 2007;17:898–909. doi: 10.1101/gr.5583007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 39.Fejes AP, Robertson G, Bilenky M, Varhol R, Bainbridge M, Jones SJ. FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics. 2008;24:1729–1730. doi: 10.1093/bioinformatics/btn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lenhard B, Wasserman WW. TFBS: Computational framework for transcription factor binding site analysis. Bioinformatics. 2002;18:1135–1136. doi: 10.1093/bioinformatics/18.8.1135. [DOI] [PubMed] [Google Scholar]
- 41.Ho Sui SJ, Mortimer JR, Arenillas DJ, Brumm J, Walsh CJ, Kennedy BP, Wasserman WW. oPOSSUM: identification of over-represented transcription factor binding sites in co-expressed genes. Nucleic Acids Res. 2005;33:3154–3164. doi: 10.1093/nar/gki624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A, et al. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teytelman L, Ozaydin B, Zill O, Lefrancois P, Snyder M, Rine J, Eisen MB. Impact of chromatin structures on DNA processing for genomic analyses. PLoS ONE. 2009;4:e6700. doi: 10.1371/journal.pone.0006700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 47.Itoh K, Igarashi K, Hayashi N, Nishizawa M, Yamamoto M. Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol. Cell Biol. 1995;15:4184–4193. doi: 10.1128/mcb.15.8.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friling RS, Bergelson S, Daniel V. Two adjacent AP-1-like binding sites form the electrophile-responsive element of the murine glutathione S-transferase Ya subunit gene. Proc. Natl Acad. Sci. USA. 1992;89:668–672. doi: 10.1073/pnas.89.2.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tompa M, Li N, Bailey TL, Church GM, De Moor B, Eskin E, Favorov AV, Frith MC, Fu Y, Kent WJ, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 50.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 51.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 52.Harvey CJ, Thimmulappa RK, Singh A, Blake DJ, Ling G, Wakabayashi N, Fujii J, Myers A, Biswal S. Nrf2-regulated glutathione recycling independent of biosynthesis is critical for cell survival during oxidative stress. Free Radic. Biol. Med. 2009;46:443–453. doi: 10.1016/j.freeradbiomed.2008.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh A, Rangasamy T, Thimmulappa RK, Lee H, Osburn WO, Brigelius-Flohe R, Kensler TW, Yamamoto M, Biswal S. Glutathione peroxidase 2, the major cigarette smoke-inducible isoform of GPX in lungs, is regulated by Nrf2. Am. J. Respir. Cell. Mol. Biol. 2006;35:639–650. doi: 10.1165/rcmb.2005-0325OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahashi T, Tabuchi T, Tamaki Y, Kosaka K, Takikawa Y, Satoh T. Carnosic acid and carnosol inhibit adipocyte differentiation in mouse 3T3-L1 cells through induction of phase2 enzymes and activation of glutathione metabolism. Biochem. Biophys. Res. Commun. 2009;382:549–554. doi: 10.1016/j.bbrc.2009.03.059. [DOI] [PubMed] [Google Scholar]
- 55.Hinoi E, Fujimori S, Wang L, Hojo H, Uno K, Yoneda Y. Nrf2 negatively regulates osteoblast differentiation via interfering with Runx2-dependent transcriptional activation. J. Biol. Chem. 2006;281:18015–18024. doi: 10.1074/jbc.M600603200. [DOI] [PubMed] [Google Scholar]
- 56.Reddy NM, Kleeberger SR, Yamamoto M, Kensler TW, Scollick C, Biswal S, Reddy SP. Genetic dissection of the Nrf2-dependent redox signaling-regulated transcriptional programs of cell proliferation and cytoprotection. Physiol. Genomics. 2007;32:74–81. doi: 10.1152/physiolgenomics.00126.2007. [DOI] [PubMed] [Google Scholar]
- 57.Reddy NM, Kleeberger SR, Bream JH, Fallon PG, Kensler TW, Yamamoto M, Reddy SP. Genetic disruption of the Nrf2 compromises cell-cycle progression by impairing GSH-induced redox signaling. Oncogene. 2008;27:5821–5832. doi: 10.1038/onc.2008.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maynard SE, Min JY, Merchan J, Lim KH, Li J, Mondal S, Libermann TA, Morgan JP, Sellke FW, Stillman IE, et al. Excess placental soluble fms-like tyrosine kinase 1 (sFlt1) may contribute to endothelial dysfunction, hypertension, and proteinuria in preeclampsia. J. Clin. Invest. 2003;111:649–658. doi: 10.1172/JCI17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luttun A, Carmeliet P. Soluble VEGF receptor Flt1: the elusive preeclampsia factor discovered? J. Clin. Invest. 2003;111:600–602. doi: 10.1172/JCI18015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mann GE, Niehueser-Saran J, Watson A, Gao L, Ishii T, de Winter P, Siow RC. Nrf2/ARE regulated antioxidant gene expression in endothelial and smooth muscle cells in oxidative stress: implications for atherosclerosis and preeclampsia. Sheng Li Xue Bao. 2007;59:117–27. [PubMed] [Google Scholar]
- 61.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.