SUMMARY
Two abundant classes of mobile elements, namely Alu and L1 elements, continue to generate new retrotransposon insertions in human genomes. Estimates suggest that these elements have generated millions of new germline insertions in individual human genomes worldwide. Unfortunately, current technologies are not capable of detecting most of these young insertions, and the true extent of germline mutagenesis by endogenous human retrotransposons has been difficult to examine. Here, we describe new technologies for detecting these young retrotransposon insertions and demonstrate that such insertions indeed are abundant in human populations. We also found that new somatic L1 insertions occur at high frequencies in human lung cancer genomes. Genome-wide analysis suggests that altered DNA methylation may be responsible for the high levels of L1 mobilization observed in these tumors. Our data indicate that transposon-mediated mutagenesis is extensive in human genomes, and is likely to have a major impact on human biology and diseases.
INTRODUCTION
Alu and L1 retrotransposons are two abundant classes of human mobile elements. With each passing generation, Alu and L1 elements “jump” to new genomic locations through the process of germline retrotransposition, thereby expanding the number of element copies in our genomes (Ostertag and Kazazian, 2001; Batzer and Deininger, 2002). Some Alu and L1 insertions, termed “private” insertions, have been generated so recently that they are found in only a single individual (Mills et al., 2007). Estimates suggest that up to 600 million of these private germline insertions have been generated in personal human genomes on a world-wide basis (Kazazian, 1999; Li et al., 2001; Brouha et al., 2003; Cordaux et al., 2006; Mills et al., 2007; Xing et al., 2009). This is an impressive mutagenesis of the human genome, and is collectively equivalent to one insertion for every 5 bp of chromosomal DNA.
Although transposon mutagenesis is not expected to be a totally random process in the human genome, this level of mutagenesis clearly would have a major impact on human biology. However, it is presently unclear whether new insertions are indeed produced in the germline as frequently as these estimates suggest. Unfortunately, transposon mutagenesis has been difficult to study in humans because we lack efficient assays for detecting new and otherwise young retrotransposon insertions. As a consequence, most of the transposon insertions that have been detected to date are common or fixed insertions that have been discovered by genome sequencing projects (Lander et al. 2001; Venter et al., 2001), and relatively few rare insertions have been discovered (Wang et al., 2006). Arguably, new and otherwise young insertions are much more interesting than common alleles, since purifying selection has not yet fully acted upon these young alleles and they are much more likely to disrupt genes. Thus, the full extent of germline mutagenesis by endogenous retrotransposons remains relatively unexplored in humans.
For the same reason, it has been difficult to determine whether human retrotransposons are actively “jumping” in somatic tumor genomes. Such retrotransposons are, for the most part, silent in normal somatic tissues (Kano et al., 2009). Thus, if they were reanimated, these elements might help to drive tumorigenesis or further destabilize an already unstable cancer genome. Several lines of evidence suggest that endogenous retrotransposons might be de-repressed in human somatic tumors as a consequence of altered DNA methylation (Bestor, 1998; Borc’his and Bestor, 2004; Howard et al., 2008). However, it remains unclear whether these elements are, in fact, mobilized at elevated frequencies in tumors. Three L1 insertion candidates have been reported in human tumors (Morse et al., 1988; Miki et al., 1992; Liu et al., 1997; Figure S1), but only one of these insertions has all of the hallmark features of a true somatic retrotransposition event (Miki et al., 1992; Figure S1). Thus, as above, the extent of transposon mutagenesis in human somatic tumors remains unclear.
As a step towards investigating transposon mutagenesis in normal and cancer genomes, we have developed novel “transposon-seq” technologies that can detect new and otherwise young retrotransposon insertions in humans. Our experiments reveal that young retrotransposon insertions indeed are highly abundant in personal human genomes and extend far beyond the number of insertions that have been detected previously. We also found that de novo L1 insertions occur at high frequencies in human lung tumor genomes. Thus, transposon-mediated mutagenesis is extensive in human genomes and is likely to have a major impact on human biology and diseases.
RESULTS
“LgL1-seq” assays for detecting germline L1 insertions in personal genomes
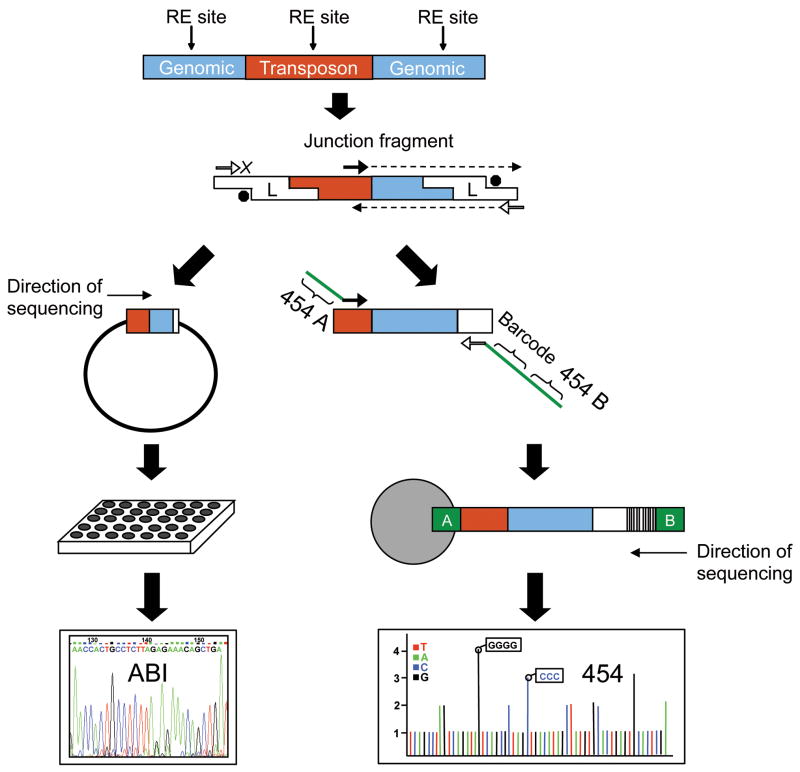
Our goal was to develop an efficient assay that, ideally, could detect a new retrotransposon insertion anywhere in the 3 billion bp haploid human genome. We began by examining traditional gel-based L1 display assays with the hope of deriving an improved assay that could tackle this challenge (Sheen et al., 2000; Ovchinnokov et al., 2001; Badge et al., 2003; Figure 1). These linker-mediated PCR assays take advantage of specific sequence changes that are present in the youngest L1 elements (the Ta elements; Skowronski et al., 1988; Boissinot et al., 2000) to amplify large collections of L1-Ta junction fragments from the human genome (Figure 1). Although multiple laboratories have successfully used L1 display to identify novel L1 insertions, such assays are inherently low-throughput and also have high false positive rates (Sheen et al., 2000; Ovchinnokov et al., 2001; Badge et al., 2003). After attempting several modifications of this approach, we found that L1 display could be significantly improved by applying high-throughput DNA sequencing directly to the initial collections of L1-Ta junction fragments that are generated by these assays. We then applied informatics filtering to the resulting data sets to eliminate false positives and to identify high probability L1-Ta insertion candidates.
Figure 1. Strategy to sequence retrotransposon insertion junctions.
The approaches that were used to sequence retrotransposon insertion junctions are depicted. ABI (left side) and pyrosequencing (right side). The “transposon” is either L1-Ta or Alu. Human genomic DNA is digested with a restriction endonuclease and ligated to a linker. The linker is partially double-stranded with a 3′ amine group on the short strand. This prevents amplification of random genomic DNA from the linker primers. Amplification only occurs if there is extension from the transposon-specific primer. This completes the double-stranded linker and creates the sequence for the linker-specific primer to anneal to, thus allowing the PCR reaction to proceed. Left side-ABI sequencing. After an initial PCR amplification, a second round of PCR is performed with nested primers. Second round PCR products are cloned into pBLUESCRIPT using a restriction site in the nested retrotransposon primer and another restriction site in the linker. Resulting colonies are sent for ABI Sanger capillary sequencing. Right side-pyrosequencing. After an initial PCR amplification, a second PCR is performed with nested primers. The retrotransposon nested primer contains the “A” adapter sequence for 454 sequencing whereas the linker-specific primer contains an 8 bp unique barcode for each sample and the “B” adapter sequence. Samples are pooled in equal molar ratios for emulsion PCR with beads binding only the “A” end. Thus sequencing occurs from the “B” end only, avoiding possible problems with sequencing through the poly (A) tail of L1. A similar approach was used with Alu except the 5’ junctions were amplified and sequenced. Please see Experimental Procedures and Extended Experimental Procedures for primers and additional details.
We explored both ABI capillary sequencing and 454 pyrosequencing to recover large collections of L1 insertion junctions from our linker-mediated PCR experiments (Figure 1). In our first “L1-seq” experiment, we examined 4,600 PCR junction fragments by cloning them into plasmids and sequencing them with ABI Sanger capillary sequencing (Figure 1-left and Table 1). The PCR junction fragments for this experiment were generated from the genomes of: i) 24 ethnically diverse humans (the Coriell Polymorphism Discovery Panel; Collins et al., 1999), ii) 14 additional diverse humans from the Coriell repository, and iii) eight ATCC cell lines that were derived from human tumors (Please see Experimental Procedures and Extended Experimental Procedures). This approach was highly successful, and yielded 785 distinct L1 insertions from the genomes examined (Table 1 and Table S1). After comparing these insertions to the reference human genome sequence and to previously-discovered retrotransposon insertion polymorphisms (deposited to dbRIP, a database of human retrotransposon insertion polymorphisms; Wang et al., 2006), 152 novel L1 insertion polymorphisms were identified (Table 1 and Table S1).
Table 1. Transposon-seq results.
The Alu and L1 transposon-seq data for this study are summarized. Please see Tables S1–S4, Experimental Procedures, and Extended Experimental Procedures for supporting data and details.
| Sequencing Strategy | Sample Description | Retrotransposon | Reads | Mapped | Distinct Retrotransposons | Previously Unknown | PCR Validated |
|---|---|---|---|---|---|---|---|
| ABI capillary sequencing | Pools of diverse human DNA and tumor-derived cell line DNA. | L1 | 4,600 | 3,795 | 785 | 152 | 64/66 (97%) |
| Pyrosequencing | Lung tumor and adjacent normal lung DNA. Brain tumor and matched normal blood DNA. | L1 | 286,126 | 50,532 | 1,389 | 650 | 162/182 (89%) |
| Pyrosequencing | Brain tumor and matched normal blood DNA. | Alu | 35,022 | 22,338 | 3,799 | 403 | 53/56 (95%) |
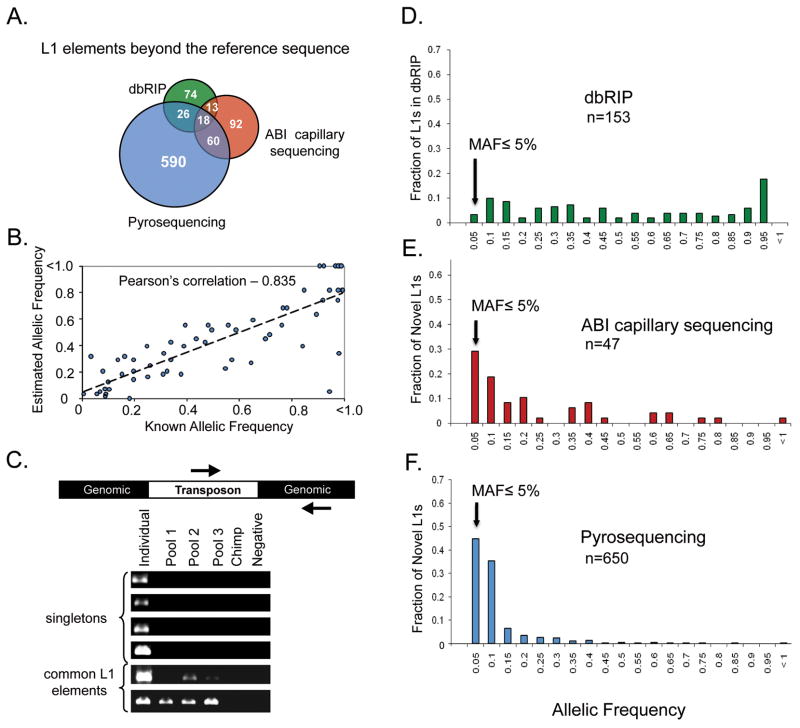
To investigate these L1 insertions further, we performed a series of PCR validation studies (Table 1and Table S1). These studies confirmed that our trace data were highly accurate (with a validation rate of 97%) and also confirmed that our collections include both common and rare L1 alleles (Table S1 and Figure 2). In contrast to dbRIP, which predominantly contains common L1 alleles (Wang et al., 2006), 30% of the L1 insertions discovered from our ABI sequencing experiments had minor allelic frequencies (MAFs) of 5% or below (compare Figures 2D and 2E). Likewise, follow-up PCR experiments revealed that 9/47 (19.1%) of the rare insertions were found only in a single human in our study (Figure 2C; MAF <1.1%). Similar results were obtained with a second set of experiments that were conducted with pyrosequencing (Figure 2F, see below). Thus, although rare L1 insertions have gone largely undetected previously, our data clearly indicate that such insertions are abundant in human populations.
Figure 2. Characterization of novel L1 elements.
A) For both ABI sequencing and pyrosequencing experiments, L1 elements that were not present in the human reference sequence (hg18) were compared with L1 polymorphisms that have been deposited to dbRIP (Wang et al., 2006). The Venn diagram depicts the relationships between our candidates and L1 insertions that have been deposited to dbRIP but are absent from hg18. Note the minimal overlap between our data sets and the L1 polymorphisms in dbRIP, indicating that our L1-seq method has recovered many novel insertions. Our assays predominantly detected L1-Ta and pre-Ta elements (Table S5). B) Correlation of estimated and previously determined allelic frequencies. For the pyrosequencing experiment, allelic frequencies were estimated using sequence data and were compared to known allelic frequencies of polymorphic L1 elements (n = 63). C) A subset of L1 elements that appeared to be "singletons" based on their presence in a single individual (from sequence data) and absence from hg18 and dbRIP were verified by PCR in pools of diverse human DNA. The Individual lane is the individual from whom the L1 was initially sequenced. Pool lanes contain DNA from 15 diverse humans. Chimp is Coriell #NA03448A and Negative is a control PCR with no template. D) Histogram of allelic frequencies for polymorphic L1s in dbRIP (Wang et al., 2006). Allelic frequency data was extracted from dbRIP (n = 153). E) Allelic frequencies of L1’s identified by ABI Sanger dideoxy sequencing. Allelic frequencies were determined by PCR in a panel of 46 diverse individuals (n = 46). F) Allelic frequencies of L1’s identified by pyrosequencing. Allelic frequencies were determined as depicted in B (n = 650).
Frequent somatic L1 insertions in lung tumor genomes
Despite the fact that we examined only eight cell lines from human tumors in this initial experiment (leukemias, breast, and lung cancers), we identified what appeared to be a somatic L1 insertion in one of these cell lines. In particular, we identified an L1 insertion in the lung-tumor derived cell line NCI H1395 that was absent from the matched normal DNA from the same patient’s blood (Figure 3A). These results suggested the possibility that lung tumor genomes might support high levels of L1 mobilization. To explore this question further, we examined in parallel from the same tissue sources, DNA samples isolated from 20 primary non-small cell lung cancers and 20 matched normal adjacent tissues. Since L1 is highly active in both the mouse brain (Moutri et al., 2005) and human neural stem cells (Coufal et al., 2009), we also examined 10 human brain tumors (5 glioblastoma and 5 medulloblastoma) and 10 matched blood leukocyte controls to determine whether somatic L1 retrotransposition might be occurring in these tumors as well.
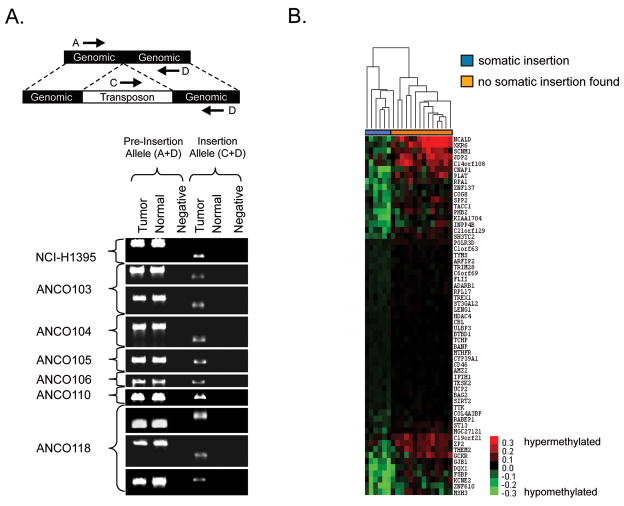
Figure 3. PCR validation of somatic insertions and identification of a hypomethylation signature in tumors with new L1 insertions.
A) Nine somatic L1 insertions were identified by screening low frequency L1 insertions that were identified initially from our pyrosequencing data (Table S2) with PCR assays. Shown here are those verified as somatic insertions by their presence in the tumor tissue and absence from adjacent normal tissue. Negative lanes are control PCRs with no template. Primers flanking the putative insertion sites were used to amplify the pre-insertion alleles. Primers within the 3′ end of an L1 consensus sequence and downstream of the putative insertion sites were used to verify the presence of each L1. Anonymous patient identifiers are on the left. NCI-H1395 is a tumor-derived cell line matched with its normal B-lymphocyte-control (both from ATCC). The three somatic L1 insertion candidates that have been reported previously in the literature are shown in Figure S1. B) The methylation status of the 20 lung tumor and normal adjacent tissues used in the pyrosequencing experiment were analyzed by Illumina Infinium analysis. Fifty-nine probes were identified whose changes in methylation status in the tumor specimens relative to matched normal tissues were tightly correlated with somatic L1 retrotransposition (Table S6). The six tumors that were positive for somatic L1 insertions (blue) clustered together along with one of the remaining tumor samples (i.e., sample 119), which did not have a somatic L1 insertion (orange). Tumor ANCO119 (seventh from the left) clustered with the L1-positive tumors but lacked an L1 insertion, suggesting that it might also have an L1-permissive state. This signature expands to 1,928 correlated probes if tumor 119 is included in the L1 permissive class and the least correlated tumor of the six L1-positive tumors (ANCO106) is shifted to the L1-negative class (see Experimental Procedures).
We began by re-tooling the L1-seq assay for 454 pyrosequencing, since this would greatly increase the throughput of the assay and allow us to examine these 60 samples with greater sequencing depth and efficiency (Figure 1). The standard 454 A and B primers were incorporated into the assay, and the PCR products were sequenced solely from the B end to avoid possible artifacts of sequencing through the L1 poly (A) tails (Figure 1-right). A similar Alu-seq assay was developed to detect young Alu insertions. For this assay, sequencing was performed from the 5’ end of Alu. In each case, 20 samples were combined into a single sequencing experiment (10 tumor and 10 matched control), using a bar-coding strategy (Hamady et al., 2008) that allowed us to assign a given sequence to a specific sample from the pool (Figure 1-right). Like the ABI L1-seq assay discussed above, this approach was highly successful, and yielded 1,389 distinct L1 insertions in the 60 samples that were examined (Table 1 and Table S2). After comparing these L1 candidates to the reference human genome and to dbRIP (Wang et al., 2006), 650 novel L1 insertions were identified (Table 1 and Table S2). Forty-five percent of these L1 alleles had MAFs <5% (Figures 2B, F). Moreover, 93% of the genomes had at least one rare L1 insertion that was present in only a single human in our study. An additional 403 novel Alu insertions were identified from the Alu assay, including both common and rare Alu alleles (Table 1 and Table S3). Like our ABI results outlined above, these pyrosequencing data clearly demonstrate an abundance of rare retrotransposon insertions in the genomes examined.
We hypothesized that some of these low frequency alleles might have been caused by new somatic retrotransposition events in the tumor genomes. Thus, we screened low frequency alleles with PCR assays and identified nine tumor-specific somatic L1 insertions (Figure 3A). Most of the remaining insertions were considered to be germline insertions, since they were found in just the normal tissue (or both tissues) of a patient or were found in multiple humans. All nine of the somatic insertions were present in the tumor tissues but were completely absent from the adjacent normal tissues (Figure 3A). Eight of the nine somatic insertions were further confirmed by cloning and sequencing the junction fragments that were amplified from the tumor DNAs (the ninth could not be sequenced because both the original DNA and the PCR products from the tumor tissue became depleted; data not shown). All of the somatic L1 insertions were found in the lung tumors, and no somatic insertions were identified in the brain tumors that were examined (data not shown). Likewise, no somatic Alu insertions were identified in the brain tumors that were examined (data not shown).
Although additional studies will be necessary to determine the full spectrum of cancer genomes that are permissive for Alu and L1 mobilization, these data indicate that lung tumor genomes are highly permissive for L1 mobilization. The nine somatic L1 insertions described above (Figure 3A) were found in six of the 20 lung tumors that were examined. Thus, an impressive 30% of the lung tumors had at least one new L1 insertion and some tumors had two or even three new L1 insertions. Given that our assay recovers insertions from only a fraction of the genome, this 30% measurement is likely to represent the lower boundary of somatic L1 insertion frequencies in lung tumor genomes (see Experimental Procedures). Since L1 is mostly silent in normal somatic tissues, this rate suggests that lung tumors behave like germline or early embryonic tissues with respect to L1 mobilization (Borc’his and Bestor, 2004; Moutri et al., 2005; Coufal et al., 2009; Kano et al., 2009).
A methylation signature in L1-permissive tumors
Genomic DNA methylation is often altered in human lung tumors (Daskalos et al., 2009) and is also thought to play a role in suppressing retroelements (Bestor, 1998; Borc’his and Bestor, 2004; Howard et al., 2008). Thus, we next examined the genome-wide methylation status of 27,578 CpG dinucleotides in our lung tumor specimens using the Illumina Infinium platform. In order to study the relationship between DNA methylation and somatic L1 retrotransposition, patient samples were divided into two groups: i) those with tumor-specific somatic L1 insertions and ii) those without such insertions (Figure 3B). On the basis of this classification, 59 probes were identified for which the change in DNA methylation status between the tumor and adjacent normal tissue from the same patient was significantly correlated with the L1 retrotransposition potential of the tumors (Table S6; randomly permuted datasets averaged only 1.5 significantly correlated probes). Unsupervised hierarchical clustering of the samples based on these 59 probes revealed a dendrogram consisting of two distinct sample groups (Figure 3B). All six patients exhibiting somatic L1 insertions were clustered together into one branch whereas 13/14 of the patients that lacked somatic insertions were clustered into a separate branch. Even though L1 insertions were not detected in tumor DNA from patient 119, this DNA was more related to the insertion-positive group on the basis of the differential methylation status of the 59 probes (see Experimental Procedures). This observation suggests i) that the tumor DNA from patient 119 harbors an existing insertion that was not detected, or ii) that this patient’s tumor exhibited an environment that was permissive for L1 retrotransposition but had not yet produced a new L1 insertion. Overall, these data reveal a methylation signature that distinguishes L1-permissive lung tumors from non-L1 permissive tumors. Interestingly, all correlated probes were hypomethylated to varying degrees in the tumors relative to the matched normal tissues.
DISCUSSION
Our transposon-seq methods have revealed that rare germline retrotransposon insertions are present in virtually all personal genomes in human populations and extend far beyond the insertions that have been identified in the reference human genome (Figures 2E, F). By applying our assays to the genomes of 76 diverse humans, we identified a total of 1,145 new Alu and L1 insertions that had not been reported previously (Tables S1–S3;Lander et al., 2001; Wang et al., 2006). A surprisingly large percentage of these insertions were rare. This fact is most clearly demonstrated by the relatively large number of rare insertions (with MAFs <5%) that were discovered in our study (Figures 2E, F) compared to the small number of rare insertions that are present in dbRIP (Figure 2D). Because we confirmed that our assays predominantly detect the very youngest and most active retrotransposons in the human genome (Table S5 and Experimental Procedures), most of these rare insertions are likely to have been generated very recently. Despite the fact that entirely different human populations were examined, we identified some of the very same full-length active L1 elements that were identified by Beck et al. in the accompanying manuscript. Nevertheless, most of the insertions in the two data sets were detected by only one of the groups, suggesting that many additional retrotransposon insertions will be discovered in personal human genomes. Consistent with this idea, peaks of structural variation have been noted that correlate with Alu and L1 elements in the Watson genome and in other human genomes (Mills et al., 2006; Levy et al. 2007; Korbel et al., 2007; Wheeler et al., 2008; Xing et al., 2009).
The 1,145 novel Alu and L1 insertions that were identified in our study of 76 humans were distributed throughout the human genome on all 24 chromosomes. A large fraction of these insertions (391/1,145 or 34.2%) were located within known RefSeq genes (Tables S1–S3). Ten of the 1,145 insertions (0.9%) were located within exons and the remaining 381 insertions were located within introns (Tables S1–S3). This percentage of insertions in exons (0.9%) approaches the percentage of the genome that is occupied by exons (1.5%; Lander et al., 2001), indicating that Alu and L1 elements can effectively mutagenize exons. Insertions in introns also can affect gene function by causing exon skipping or by influencing transcript elongation, splicing, or polyadenylation (Ostertag and Kazazian, 2001; Batzer and Deininger, 2002). Thus, young retrotransposon insertions in exons and introns are likely to have a major impact on gene function.
Our results provide strong support for a neutral theory (Kimura and Ota, 1971) model in which large pools of new germline insertions are continuously generated in personal genomes. Over time, the majority of these alleles, which are expected to be neutral, are likely to be eliminated by genetic drift. However, detrimental alleles also would be expected to be produced in these pools (Boissinot et al., 2001), including insertions in the exons of known genes and other functional DNA sequences. In fact, several dozen disease-causing transposon insertions have been identified in genes to date (reviewed in Ostertag and Kazazian, 2001; Batzer and Deininger, 2002; Mills et al., 2007) and our data suggest that many more will be found.
We also provide strong evidence that somatic L1 retrotransposition occurs frequently in human lung tumors. Although the hypothesis that retrotransposons might play a role in driving tumorigenesis has been around for several years (Bestor, 1998), it has been difficult to test in humans. Our transposon-seq technologies now reveal that transposons indeed are mobilized at high frequencies in at least one type of human tumors (Figure 3A). Moreover, L1-permissive tumors had a specific hypomethylation signature (Figure 3B), suggesting that altered DNA methylation might be responsible for the unusual levels of L1 mobilization that were observed in these tumors (see below). Once reactivated, L1 could be envisioned to mutate genes that are involved in cancer pathways (Dupuy et al., 2005; Collier et al., 2005) or to facilitate deletions and other large-scale chromosomal rearrangements that are commonly found in human tumors (Pearson and Rowley, 1985; Xing et al., 2009). Thus, high levels of L1 mobilization would further destabilize an already unstable cancer genome. Overall, our experiments in both normal and cancer genomes provide compelling evidence that human transposons are potent endogenous mutagens that continue to drive the evolution of human genomes.
Our genome-wide methylation analysis revealed a specific hypomethylation signature that was strongly correlated with the six L1-permissive lung tumors that were identified in our study (Figure 3, Table S6). We examined the genomic regions surrounding the 59 highly correlated probes and determined that no modern, full length L1-Ta, Pre-Ta, or L1-PA2 elements were located near these probes in the haploid reference human genome. However, this does not rule out a model in which master L1-Ta elements are de-repressed as a consequence of hypomethylation. The hypomethylation signature that is captured by our 59 probes could represent a broader hypomethylation state that releases master L1 elements from constraint at other genomic sites. Additional statistical modeling of our methylation data supports this possibility, and indicates that our 59-probe signature is likely to represent a much larger signature of 1928 probes that are correlated with L1 retrotransposition (Experimental Procedures). These results suggest that specific hypomethylation patterns may influence the mobilization of retrotransposons in human genomes. The relationship between these patterns of hypomethylation and other forms of genomic instability in human tumors is presently unclear.
EXPERIMENTAL PROCEDURES
DNA samples
The following genomic DNA samples were obtained from the Coriell Repository and were used to prepare L1-seq libraries: NA15029, NA15036, NA15213, NA15215, NA15223, NA15224, NA15236, NA15510, NA15221, NA15227, NA15385, NA15056, NA15590, NA15038, NA15245, NA15072, NA15144, NA15216, NA15226, NA15242, NA15268, NA15324, NA15386, NA15594, NA11200, NA11776, NA10970, NA10968, NA10975, NA10492, NA10469, NA13820, NA13619, NA10540, NA11377, NA11321, NA13597, and NA13607. The following genomic DNA samples were obtained from the ATCC (cancer-derived cell lines and one normal control): 45500 (HL-60), 45502 (THP-1), 45506 (K-562), 45508 (NCI-H1395), 45510 (NCI-BL1395), 45516 (MDA-MB-175), 45518 (MDA-MB-231), 45522 (BT-483), 45532 (ZR-75-30).
Patient-derived glioblastoma and medulloblastoma tumors and matched blood leukocytes were obtained from the Emory University School of Medicine Tissue Procurement and Banking Service. Samples were snap-frozen by dipping them in a container with isopentane cooled with liquid nitrogen. The samples then were transferred to cryovials and stored in liquid nitrogen. Genomic DNA was extracted from 25 mg of tissue using the DNeasy Tissue Kit (Qiagen). Twenty snap-frozen non-small cell lung cancer specimens (16 adenocarcinomas, 4 squamous cell carcinomas) and paired adjacent normal tissues were obtained from the Emory University School of Medicine Tissue Procurement and Banking Service. Genomic DNA was extracted from 15 mg of snap-frozen lung tissues using the DNeasy Tissue Kit (Qiagen).
Population distributions
In many cases, specific information was available regarding the ethnicities, genders, and ages of the humans that were examined in our study. A complete listing of all available population data can be found in the Extended Experimental Procedures. Lung tumor specimens were de-identified as a condition of Emory University’s Institutional Review Board approval, and we do not have further information beyond the pathology at diagnosis. Likewise, specific information for the polymorphism discovery panel (Coriell) is de-identified and we know only that the 24 humans represent a diverse human population (see Collins et al. 1999).
Sequencing platforms
We explored two different sequencing platforms to identify new transposon insertions in personal genomes. We began with traditional ABI Sanger sequencing to characterize L1-Ta insertion junctions. This approach provided high quality, long reads that originated within L1 elements and extended through the poly (A) tails into adjacent genomic sequences. We also explored 454 sequencing because it is relatively less expensive, particularly when used together with a bar coding and pooling strategy that analyzes multiple samples (Hamady et al. 2008). Despite the fact that the 454 reads are not as long as those that are produced by ABI sequencing, the full junction sequences were usually recovered with 454 reads, including poly (A) tails and other transposon sequences. Reads that contain this information generally have higher validation rates than reads that do not span insertion junctions. Finally, we also considered using the Illumina platform and other next-gen sequencers. Illumina has shorter reads than ABI and 454, and we decided not to explore this platform because it would not yield junction sequences. However, the higher read densities that are achieved with the Illumina platform may provide some advantages over the ABI and 454 approaches.
Targeted sequencing of retrotransposon insertion junctions
Linker-mediated PCR methods were adapted from Siebert et al., 1995. In brief: Genomic DNA was digested with restriction endonucleases and then ligated to linkers. Purified ligation products were used as templates for PCR with linker-specific and transposon-specific primers (please see the Extended Experimental Procedures for a list of the primers that were used for transposon-seq). A second PCR reaction was performed with nested primers to increase the specificity of the reaction. For ABI sequencing experiments, the PCR products were cloned into pBLUESCRIPT (Stratagene) using restriction sites that were incorporated into the nested primers. Individual colonies were used to inoculate LB-Amp (freezing) medium in 96-well plates and these (frozen) cultures were sent to Agencourt Bioscience (Beverly, MA) for ABI capillary sequencing. For pyrosequencing experiments, nested primers contained adapter sequences and eight bp barcodes (please see the Extended Experimental Procedures). Samples were pooled in equal molar ratios and gel-purified to remove small, uninformative products. Purified samples were then sent to the University of Florida Interdisciplinary Center for Biotechnology Research or the Genomics Resource Center at the Institute for Genome Sciences for emulsion PCR and Roche 454 pyrosequencing. Please see Supplemental Experimental Procedures for additional details.
Sequence Analysis and PCR validation
FASTA sequences were parsed into A and B adapters, linker, barcode, retrotransposon and unique genomic sequences. Custom Perl scripts were developed to compare the sequences to the human genome (May 2006 build, hg18) using BLAT (Kent et al., 2002). For L1, each non-redundant hg18 coordinate was compared to the coordinates of known L1s using Galaxy (Taylor et al., 2007) to determine whether an L1 element was located in the adjacent region of the reference sequence. For Alu, sequences flanking the insertion site in hg18 were used to query CAlu, a web-based Alu classifier (available at http://clustbu.cc.emory.edu/calu/index.cgi) to determine whether the Alu was present in the reference sequence. PCR validations were performed using standard protocols (see Supplemental Experimental Procedures). The primers that were used for PCR validation studies are listed in Table S1 (ABI sequencing), Table S4 (L1 pyrosequencing), and the Extended Experimental Procedures (Alu pyrosequencing).
Evaluation of transposon families and subfamilies
We confirmed that our assays are detecting the very youngest retrotransposon subfamilies in the human genome as follows. Our assays detect both existing copies of retrotransposons that have been documented in the reference haploid human genome as well as new insertions at other sites. When an existing copy is detected in the reference genome, we can determine the family or subfamily of that element from its genomic sequence. Thus, traces that map to existing copies in the genome serve as internal controls for our experiments, and allow us to estimate the percentage of young elements that are detected. For our 454 L1-Ta experiments, 92.1% of the traces that mapped to existing elements in the genome mapped to L1-Ta elements and another 5.2% of these traces mapped to pre-L1-Ta elements, which are also very young. Thus, our assay is highly specific for L1-Ta and Pre-Ta elements, and most of the elements detected (97.3%) belong to these two very young L1 classes (Table S5). Similarly, 93.1% of the L1 elements detected in our ABI experiments belong to these two young classes (Table S5). We also confirmed that our Alu assays are detecting young Alu elements: 94.6% of the traces that mapped to existing elements detected Alu Y elements and elements belonging to some of the very youngest Alu Y subfamilies (Table S5). Alu Ya5 (64.1%), Alu Y (11.2%), and other young Alu Ya subfamilies (16.9%) were the most abundant elements detected. Thus, our assays are highly specific for the very youngest retrotransposon families in the human genome.
Calculation of assay coverage and efficiency
Our assay uses a reduced representation approach to detect new insertions within a specific subset of restriction fragments in the human genome. For example, we estimate that our approach samples ~4.5 million of the 10.5 million Mse I restriction fragments that are generated for our 454 L1-seq assays. The same 4.5 million fragments are examined in all of the samples in the assay, providing greater depth of coverage for this set of fragments. We assume that we can detect an L1-Ta (or Alu) insertion if it occurs within 300 bp of an Mse I restriction site. The reference human genome contains 216 L1-Ta elements that fall within 300 bp of an Mse I site. This represents approximately 43% of the 508 L1-Ta elements in the reference human genome sequence. On this basis, we estimate that we can detect new insertions from approximately 43% of the genome. Using these estimates, we determined that we achieved an average of 3.1X coverage of the detectable L1-Ta insertion junction fragments in our initial 454 experiments (please see Extended Experimental Procedures).
Our approach for examining the efficiency of our assay using the reference haploid genome sequence is outlined as follows. We identified 51 fixed L1-Ta elements that are located within 300 bp of an Mse I restriction cleavage site. Since these elements are fixed in human populations (Myers et al., 2002) and are expected to be present in all of the genomes examined, these elements serve as internal controls for the efficiency of detection. We achieved an average of 72.3% detection of these 51 elements in our initial 454 L1-Ta experiments at 3.1X sequencing coverage (please see Extended Experimental Procedures). When the sequencing coverage was increased to 5.8X coverage, our efficiency increased to an average of 84% (please see Extended Experimental Procedures). Additional increases in sequencing coverage beyond 3.1X to 5.8X are predicted to yield relatively small additional increases in L1 discovery, since the redundancy of L1 detection also increases. Further increases could be achieved by combining two or three restriction enzymes in the assay, which would provide genome-wide coverage. Some insertions will be difficult or impossible to recover with our assays. For example, insertions that occur within the un-sequenced portions of the human genome will not be detected. Likewise, it can be difficult or impossible to map insertions that fall within or near other transposon repeats. Thus, a fraction of the insertions in a given genome will be refractory to detection with our methods.
Methylation analysis
Genomic DNAs from 20 lung tumors and matched normal tissues were bisulfite-modified and then used to perform genome-wide DNA methylation profiling with the Illumina Infinium Human Methylation 27 v1.0 platform. Probes were identified for which the changes in methylation status between the tumors and their matched normal adjacent tissues were correlated with somatic L1 retrotransposition using the Quantitative Response feature within the Significance Analysis of Microarrays (SAM) software package (Tusher et al., 2001; see Extended Experimental Procedures). Differential methylation data for significantly correlated probes was hierarchically clustered using Cluster 3.0 (distance metrics: genes = Euclidean, arrays = Pearson; linkage = average), and visualized with Java Treeview.
Statistical modeling of methylation data
One of the L1-negative tumors (ANCO119) appeared to cluster with the six L1-positive tumors based on the 59 differentially methylated probes, suggesting that this might represent a tumor that is in fact permissive for retrotransposition, but for which an L1 retrotransposition event had not yet been found (Figure 3B, Table S6). To test this possibility quantitatively, we moved ANCO119 into the L1-positive group and re-ran the Quantitative Response feature using SAM. This increased the number of probes detected to 139 for the expanded sample set, all of which were hypomethylated in the L1-positive group of tumors relative to their corresponding normal tissues. As a control, we ran 13 additional analyses, where, in each analysis, we similarly moved one of the 13 other L1-negative tumors into the L1-positive category. In all 13 of these cases, this resulted in a significant decrease in the number of correlated probes. Thus, unlike the other L1-negative tumors, the methylation status of sample ANCO119 is more closely related to the L1-positive tumors.
Motivated by this finding, we performed additional analyses, where, in each analysis, we moved one of these seven tumors (i.e., one of the six L1-positive tumors or the ANCO119 tumor) into the “L1-negative” group. In all but one case, this resulted in significantly fewer differentially methylated probes being correlated with the L1-positive category. However, the additional step of moving one of the L1-positive tumors, namely ANCO106, to the L1-negative category resulted in a dramatic increase in the number of correlated probes detected (to 1,928). Interestingly, again, all 1928 correlated probes were hypomethylated in the L1-positive category of tumors relative to their normal tissue counterparts.
To determine the combination of six tumors out of 20 that gave the maximal number of significantly correlated hypomethylated probes, we analyzed every possible combination (n=38,760) such that six tumors were placed in one category and 14 in the other, and re-ran the analysis. In every other combination, the number of hypomethylated probes that correlated with these arbitrary tumor partitionings was less than 1,928. Thus, the profile obtained using the five L1-positive tumors (ANCO103, ANCO104, ANCO105, ANCO110, ANCO118) plus the one L1-negative tumor (ANCO119) corresponds to the optimal (six tumor) hypomethylation signal in this data set. The correlation between this optimal hypomethylation profile and L1 retrotransposition is statistically significant: based on Fisher’s exact test the probability of having observed this in the absence of such a correlation is p = 0.0022.
Statistical analysis of lung vs. brain tumors
The fact that we detected nine new L1 insertions in 20 lung tumor samples but none in 10 brain tumor samples suggested that tumor-specific retrotransposition might be more prevalent in lung tumors compared to brain tumors. To test the hypothesis that the observed difference is merely due to chance variation, we performed the following statistical analysis. The maximum likelihood estimate for the frequency of a lung-tumor-specific L1 retrotransposition event is 1.4 × 10−7. This is based on our observation of new insertions within nine of the approximately 66 million fragments that were implicitly tested within 20 lung tumor samples. Our estimate of implicit tests performed assumes: (i) that the 2.7 billion bp sequenced portion of the human genome contains approximately 10.5 million fragments after digestion with a restriction enzyme that cuts on average every 256 bp; (ii) that we can detect insertions in ~43% of these fragments; (iii) that, among these, the average efficiency of detection is 72.3%; and (iv) that as a result, 3.3 million independent tests were performed per sample. Based on a binomial probability distribution where the total number of tests is 33 million and the number of ‘successes’ is zero, we thus obtain a probability of 0.011 that we would have observed no tumor-specific L1 retrotransposition events within the 10 brain tumor samples due to chance alone. Given a significance cutoff of 0.05, we reject the null hypothesis and conclude that the frequency of L1 retrotransposition is different in brain versus lung tumor samples. However, it is important to note that this does not necessarily indicate that L1 mobilization is absent from brain tumors. It remains possible that the rate of L1 mobilization is slightly lower in brain tumors, or that we did not sample the most active brain tumor types in our study.
Supplementary Material
Acknowledgments
We thank Kristie Jones and Luke Tallon for help with 454 Titanium sequencing. We also thank Shari Corin, Julienne Mullaney, Michael Zwick, Ichiro Matsumura, and Xiaodong Cheng for helpful advice, and Mark Mazaitis for help with artwork. This work was funded by grants from the American Cancer Society (PF-07-130-01-MGO to M.T.M.), SUN Microsystems (to W.S.P. and S.E.D), the National Institute for General Medical Sciences, National Insitutes of Health (NIH) (GM078541 to A.F.N.), the National Cancer Institute, NIH (R01CA086335 and R01 CA116804 to E.G.V.M.; 1R01CA132065 and 5P01CA116676 to P.M.V.), and the National Human Genome Research Institute, NIH (F32HG004207 to R.E.M. and R01HG002898 to S.E.D.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Badge RM, Alisch RS, Moran JV. ATLAS: a system to selectively identify human-specific L1 insertions. Am J Hum Genet. 2003;72:823–838. doi: 10.1086/373939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;35:501–538. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Bestor TH. The host defense function of genomic methylation patterns. Novartis Found Symp. 1998;214:187–195. doi: 10.1002/9780470515501.ch11. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Chevret P, Furano AV. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol. 2000;17:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Entezam A, Furano AV. Selection against deleterious LINE-1- containing loci in the human lineage. Mol Biol Evol. 2001;18:926–935. doi: 10.1093/oxfordjournals.molbev.a003893. [DOI] [PubMed] [Google Scholar]
- Borc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH. Hot L1s account for the bulk of retrotransposition activity in the human population. Proc Natl Acad Sci USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumors using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- Collins FS, Brooks LD, Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1999;8:1229–1231. doi: 10.1101/gr.8.12.1229. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Hedges DJ, Herke SW, Batzer MA. Estimating the retrotransposition rate of human Alu elements. Gene. 2006;373:134–137. doi: 10.1016/j.gene.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O’Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalos A, Nikolaidis G, Xinarianos G, Savvardi P, Cassidy A, Zakopoulou R, Kotsinas A, Gorgoulis V, Field JK, Liloglou T. Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int J Cancer. 2009;124:81–87. doi: 10.1002/ijc.23849. [DOI] [PubMed] [Google Scholar]
- Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- Hamady M, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–408. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, Kazazian HH. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH., Jr An estimated frequency of endogenous insertional mutations in humans. Nat Genet. 1999;22:130. doi: 10.1038/9638. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Hausler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Ota T. Protein polymorphism as a phase of molecular evolution. Nature. 1971;229:467–469. doi: 10.1038/229467a0. [DOI] [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, Fitzhugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, Walenz BP, Axelrod N, Huang J, Kirkness EF, Denisov G, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Scaringe WA, Hill KA, Roberts S, Mengos A, Careri D, Pinto MT, Kasper CK, Sommer SS. Frequency of recent retrotransposition events in the human factor IX gene. Hum Mutat. 2001;17:511–519. doi: 10.1002/humu.1134. [DOI] [PubMed] [Google Scholar]
- Liu J, Nau MM, Zucman-Rossi J, Powell JI, Allegra CJ, Wright JJ. LINE-1 element insertion at the t(11:22) translocation breakpoint of a desmoplastic small round cell tumor. Genes Chromosomes Cancer. 1997;18:232–239. doi: 10.1002/(sici)1098-2264(199703)18:3<232::aid-gcc10>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, Vogelstein B, Nakamura Y. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643–645. [PubMed] [Google Scholar]
- Mills RE, Luttig CT, Larkins CE, Beauchamp A, Tsui C, Pittard WS, Devine SE. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006;16:1182–1190. doi: 10.1101/gr.4565806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, Bennett EA, Iskow RC, Devine SE. Which transposable elements are active in the human genome? Trends Genet. 2007;23:183–189. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Morse B, Rotherg PG, South VJ, Spandorder JM, Astrin SM. Insertional mutagenesis of the myc locus by a LINE-1 sequence in a human breast carcinoma. Nature. 1988;333:87–90. doi: 10.1038/333087a0. [DOI] [PubMed] [Google Scholar]
- Moutri AR, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Myers JS, Vincent BJ, Udall H, Watkins WS, Morrish TA, Kilroy GE, Swergold GD, Henke J, Henke L, Moran JV, et al. A comprehensive analysis of recently integration human Ta L1 elements. Am J Hum Genet. 2002;71:312–326. doi: 10.1086/341718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- Ovchinnokov I, Troxel AB, Swergold GD. Genomic characterization of recent human LINE-1 insertions: evidence supporting random insertion. Genome Res. 2001;11:2050–2058. doi: 10.1101/gr.194701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson M, Rowley JD. The relation of oncogenesis and cytogenetics in leukemia and lymphoma. Annu Rev Med. 1985;36:471–483. doi: 10.1146/annurev.me.36.020185.002351. [DOI] [PubMed] [Google Scholar]
- Sheen FM, Sherry ST, Risch GM, Robichaux M, Nasidze I, Stoneking M, Batzer MA, Swergold GD. Reading between the LINEs: human genomic variation induced by LINE-1 retrotransposition. Genome Res. 2000;10:1496–1508. doi: 10.1101/gr.149400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert PD, Kellogg DE, Lukyanov KA, Lukyanov SA. An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowronski J, Fanning TG, Singer MF. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol Cell Biol. 1988;8:1385–1397. doi: 10.1128/mcb.8.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J, Schenck I, Blankenberg D, Nekrutenko A. Using galaxy to perform large-scale interactive data analyses. In: Baxevanis A, editor. Current Protocols in Bioinformatics. Unit 10.5. New York, New York: John Wiley and Sons, Inc; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Wang J, Song L, Grover D, Azrak S, Batzer MA, Liang P. dbRIP: A highly integrated database of retrotransposon insertion polymorphisms in humans. Hum Mutat. 2006;27:323–329. doi: 10.1002/humu.20307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- Xing J, Zhang Y, Han K, Salem AH, Sen SK, Huff CD, Zhou Q, Kirkness EF, Levy S, Batzer MA, et al. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009;19:1516–1526. doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.