Abstract
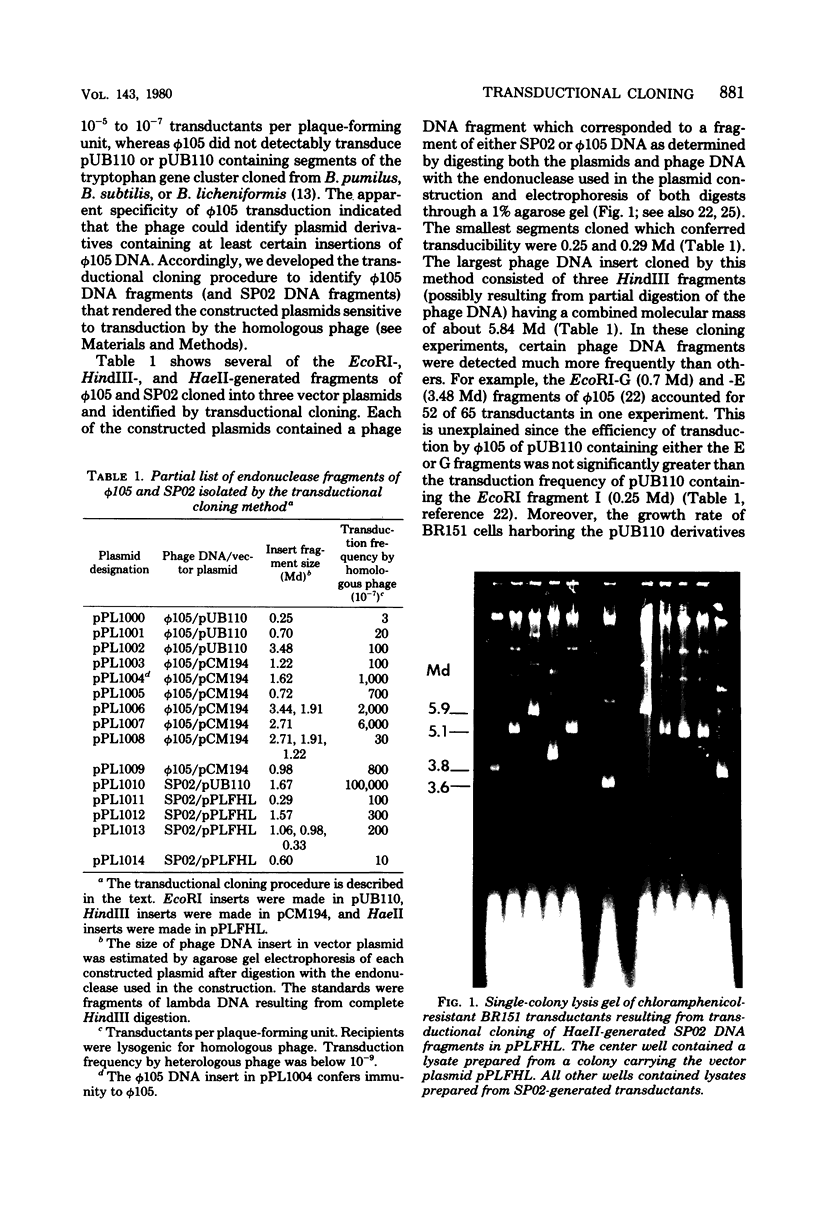
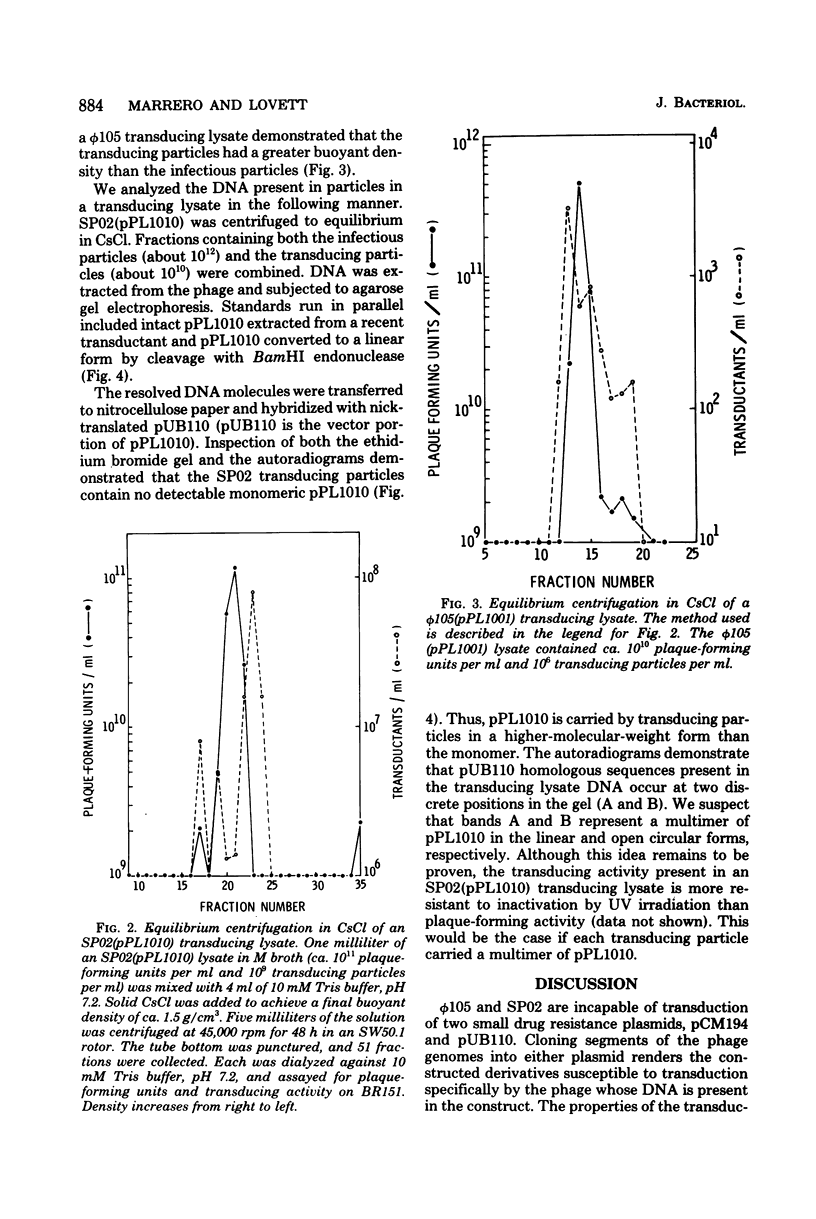
The Bacillus subtilis temperate bacteriophages phi 105 and SP02 are incapable of transduction of the small, multicopy drug resistance plasmids pUB110 and pCM194. Cloning endonuclease-generated fragments of phi 105 or SP02 DNA into each of the plasmids renders the chimeric derivatives susceptible to transduction specifically by the phage whose deoxyribonucleic acid is present in the chimera. The majority of phage deoxyribonucleic acid fragments identified that render plasmids transducible by phi 105 or SP02 appear to be internal fragments, not fragments containing the cohesive ends. However, the highest overall transduction frequency was observed in SP02-mediated transduction of a derivative of pUB110 containing a 1.6-megadalton EcoRI fragment that likely contains the SP02 cohesive ends (plasmid pPL1010). The transducing activity present in a phi 105 transducing lysate had a buoyant density slightly greater than infectious particles, whereas the majority of transducing particles in an SP02(pPL1010) transducing lysate had a buoyant density slightly less than infectious particles. Although no detectable change in plasmid structure resulted from transduction by phi 105 or SP02, deoxyribonucleic acid isolated from a purified SP02(pPL1010) transducing lysate contained no detectable monomeric pPL1010, but did contain a form of pPL1010 of higher molecular weight than the monomer.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boice L., Eiserling F. A., Romig W. R. Structure of bacillus subtilis phage SPO2 and its DNA: similarity of Bacillus subtilis phages SPO2, phi 1O5 and SPP1. Biochem Biophys Res Commun. 1969 Feb 21;34(4):398–403. doi: 10.1016/0006-291x(69)90395-7. [DOI] [PubMed] [Google Scholar]
- Bott K. F., Wilson G. A. Development of competence in the Bacillus subtilis transformation system. J Bacteriol. 1967 Sep;94(3):562–570. doi: 10.1128/jb.94.3.562-570.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramucci M. G., Lovett P. S. Selective plasmid transduction in Bacillus pumilus. J Bacteriol. 1977 Sep;131(3):1029–1032. doi: 10.1128/jb.131.3.1029-1032.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramucci M. G., Lovett P. S. Temperate bacteriophage infectious for asporogenic variants of Bacillus pumilus. J Virol. 1974 Nov;14(5):1281–1287. doi: 10.1128/jvi.14.5.1281-1287.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow L. T., Boice L., Davidson N. Map of the partial sequence homology between DNA molecules of Bacillus subtilis bacteriophages SPO2 and phi105. J Mol Biol. 1972 Jul 28;68(3):391–400. doi: 10.1016/0022-2836(72)90093-9. [DOI] [PubMed] [Google Scholar]
- Chow L. T., Davidson N. Electron microscope study of the structures of the Bacillus subtilis prophages, SPO2 and phi105. J Mol Biol. 1973 Apr 5;75(2):257–264. doi: 10.1016/0022-2836(73)90019-3. [DOI] [PubMed] [Google Scholar]
- Collins J., Hohn B. Cosmids: a type of plasmid gene-cloning vector that is packageable in vitro in bacteriophage lambda heads. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4242–4246. doi: 10.1073/pnas.75.9.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Garro A. J., Leffert H., Marmur J. Genetic mapping of a defective bacteriophage on the chromosome of Bacillus subtilis 168. J Virol. 1970 Sep;6(3):340–343. doi: 10.1128/jvi.6.3.340-343.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Contente S., Dubnau D. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J Bacteriol. 1978 Apr;134(1):318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemphill H. E., Whiteley H. R. Bacteriophages of Bacillus subtilis. Bacteriol Rev. 1975 Sep;39(3):257–315. doi: 10.1128/br.39.3.257-315.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keggins K. M., Lovett P. S., Duvall E. J. Molecular cloning of genetically active fragments of Bacillus DNA in Bacillus subtilis and properties of the vector plasmid pUB110. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1423–1427. doi: 10.1073/pnas.75.3.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keggins K. M., Lovett P. S., Marrero R., Hoch S. O. Insertional inactivation of trpC in cloned Bacillus trp segments: evidence for a polar effect on trpF. J Bacteriol. 1979 Sep;139(3):1001–1006. doi: 10.1128/jb.139.3.1001-1006.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett P. S., Duvall E. J., Keggins K. M. Bacillus pumilus plasmid pPL10: properties and insertion into Bacillus subtilis 168 by transformation. J Bacteriol. 1976 Aug;127(2):817–828. doi: 10.1128/jb.127.2.817-828.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett P. S., Keggins K. M. Bacillus subtilis as a host for molecular cloning. Methods Enzymol. 1979;68:342–357. doi: 10.1016/0076-6879(79)68025-4. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murialdo H., Becker A. Head morphogenesis of complex double-stranded deoxyribonucleic acid bacteriophages. Microbiol Rev. 1978 Sep;42(3):529–576. doi: 10.1128/mr.42.3.529-576.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palefski S., Hemphill H. E., Kolenbrander P. E., Whiteley H. R. Dominance relationships in mixedly infected Bacillus subtilis. J Virol. 1972 Apr;9(4):594–601. doi: 10.1128/jvi.9.4.594-601.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins J. B., Zarley C. D., Dean D. H. Restriction endonuclease mapping of bacteriophage phi105 and closely related temperate Bacillus subtilis bacteriophages rho10 and rho14. J Virol. 1978 Oct;28(1):403–407. doi: 10.1128/jvi.28.1.403-407.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutberg L., Armentrout R. W., Jonasson J. Unrelatedness of temperate Bacillus subtilis bacteriophages SP02 and phi105. J Virol. 1972 May;9(5):732–737. doi: 10.1128/jvi.9.5.732-737.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher B. M., Law M. F., Garro A. J. Correlated genetic and EcoRI cleavage map of Bacillus subtilis bacteriophage phi105 DNA. J Virol. 1978 Oct;28(1):395–402. doi: 10.1128/jvi.28.1.395-402.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Yoneda Y., Graham S., Young F. E. Restriction-fragment map of the temperate Bacillus subtilis bacteriophage SPO2. Gene. 1979 Sep;7(1):51–68. doi: 10.1016/0378-1119(79)90042-8. [DOI] [PubMed] [Google Scholar]