Abstract
This review article presents a general view of the recent progress in the fast developing area of surface-enhanced Raman scattering spectroscopy as an analytical tool for the detection and identification of molecular species in very small concentrations, with a particular focus on potential applications in the biomedical area. We start with a brief overview of the relevant concepts related to the choice of plasmonic nanostructures for the design of suitable substrates, their implementation into more complex materials that allow generalization of the method and detection of a wide variety of (bio)molecules and the strategies that can be used for both direct and indirect sensing. In relation to indirect sensing, we devote the final section to a description of SERS-encoded particles, which have found wide application in biomedicine (among other fields), since they are expected to face challenges such as multiplexing and high-throughput screening.
Keywords: surface-enhanced Raman scattering, sensors, metal nanoparticles, plasmon resonances
1. Introduction
Surface-enhanced Raman scattering (SERS) spectroscopy is currently a well-established analytical technique (Moskovits 2005; Kneipp 2007; Brus 2008; Stiles et al. 2008), as it offers many advantages over other spectroscopic or spectrometric techniques such as Fourier transform infrared (IR) spectroscopy, near infrared (NIR) absorption, UV-vis absorption, fluorescence, nuclear magnetic resonance (NMR), X-ray diffraction, X-ray photoelectron spectroscopy or mass spectrometry (Petry et al. 2003; Baena & Lendl 2004; Alvarez-Puebla & Liz-Marzan 2010). The implementation of Raman scattering-based techniques for life science applications is becoming very popular as they can extract a significant amount of information directly (with no need for prior sample preparation) from complex environments such as biological fluids, living tissues and cells (including sensitivity for small structural changes in macromolecules, non-invasive sampling capability, minimum sample preparation and high spatial resolution). In addition, the most significant drawback of normal Raman scattering (RS) spectroscopy for analytical applications, i.e. the inherently weak cross section, is overcome in SERS by exciting the sample in contact with a ‘plasmonic’ surface with an appropriate laser line. Under such conditions, the Raman cross section and, in turn, the signal intensity are extraordinarily increased so that levels of detection down to the single molecule can be reached, while retaining all the structural information provided by RS (Kneipp et al. 2008; Pieczonka & Aroca 2008). Thus, it is clear that the advancement in SERS detection is linked to the progress in synthesis and optical characterization of new nanostructured materials.
SERS can be achieved and maximized by carefully controlling both electronic and chemical effects, mainly through careful design of the optical substrates, but also by improving the adsorption of the analytes of interest. Therefore, the preparation of optical substrates with optimized properties is a very dynamic field of research, and, as there is no universal ‘best’ SERS platform, careful consideration of the analytical problem is required before choosing/designing a SERS sensor platform. Regarding the material, SERS has been obtained on electrodes, solid thin films and colloidal dispersions (Aroca et al. 2005a; Ko et al. 2008; Tripp et al. 2008). Metal colloids are advantageous for many reasons. First, single particles have served as a testing model ground for the most thorough theoretical studies. Second, they permit direct SERS analysis within the analyte natural solution medium. Further, their large surface area and their dispersion in liquids allow for a close adsorbent–adsorbate interaction, so that the analyte can be naturally retained onto the nanoparticles' surface. The presence of the solvent and the Brownian motion of the analyte–particle complexes minimize damage to the sample, even when using more energetic laser lines and higher power at the sample for excitation. On the other hand, the colloidal and optical stability, which are typically compromised over time, or the lack of hot spots when the different particles dispersed in the solvent do not interact with each other can be improved and controlled by means of the rational design of colloidal hybrid materials. Finally, colloidal metals can also be used for the preparation of thin films, which add portability and versatility to ‘on-field’ SERS analysis, over the regular physical fabrication techniques such as sputtering, physical vapour deposition or electron beam lithography, which are extremely difficult to find in conventional laboratories. In this review, we present the most recent advances in the optimization of metal nanoparticles for SERS, their integration into advanced hybrid systems and their use for direct SERS and for the design of encoded particles, which are gaining popularity in bio-related fields such as chemical and molecular biology, diagnosis, biodetection or bioimaging.
2. Dependence of SERS intensity on composition, size and shape
Since the first report dealing with SERS on colloids (Creighton et al. 1979), it has become clear that the composition of the metallic particles deeply affects the intensity of the SERS signal. In this respect, the most important SERS platforms so far have been made of silver and gold (Banholzer et al. 2008), while research using other metals (Cu, Pd, Pt, Rh and others) is still marginal (Tian et al. 2002). In general, it can be stated that silver is a much more efficient optical material than gold, giving rise to SERS signals 10- to 100-fold higher than similar gold nanostructures (Garcia de Abajo 2007). Additionally, silver can be excited from the UV to the IR while gold is restricted to the red or IR owing to damping by the interband transitions (Zhao et al. 2008). As a consequence, silver nanoparticles are preferred when dealing with practical applications (Cao et al. 2002; Graham et al. 2006). However, basic structural studies or applications involving living organisms are usually carried out using gold (Kneipp et al. 2006; Qian et al. 2008; Sha et al. 2008), owing to better control of its particle size and shape as well as its significantly higher biocompatibility (Murphy et al. 2008). For example, gold nanostructures have been widely used as SERS labels for in vivo detection or as nanosensors for the study of interior organelles and the composition of prokaryotic and eukaryotic micro-organisms (Kneipp et al. 2006; Tang et al. 2007).
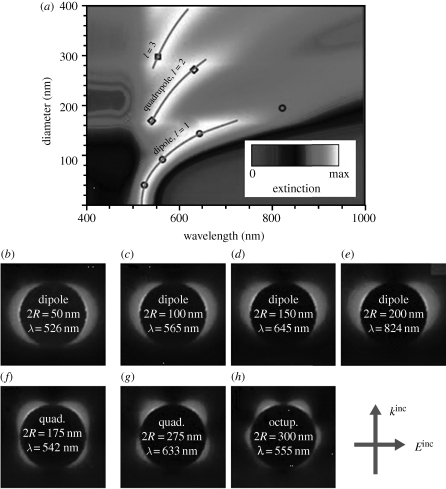
Other key factors affecting SERS intensity are the size and shape of the nanostructures. Several authors have focused on the study of the dependence of the SERS signal on size in both silver (Emory et al. 1998; Seney et al. 2009) and gold (Talley et al. 2005; Njoki et al. 2007) nanoparticles. As a rule of thumb, it has been demonstrated that SERS intensity increases with particle size. This can be explained by taking into account that the intensity of the electromagnetic field generated upon excitation with the appropriate light (localized surface plasmon resonances; LSPRs) is strongly dependent on the number of electrons excited and, thus, on the volume of the nanostructure (figure 1). Notwithstanding, the increase in the enhancement factor as a function of size is limited by the radiative damping effects, which become increasingly significant as particle size is increased. Although for many years the optimum size was thought to be around 30–100 nm (Moskovits 2005), it has been recently demonstrated that strong SERS signals can be obtained from larger particles up to 200 nm (Rodriguez-Fernandez et al. 2009). It should be noted that the nanosized gaps between NPs or within nanosized cavities (so-called hot spots, see below) are not subject to size-dependent radiative damping (Genov et al. 2004).
Figure 1.
(a) Evolution of extinction spectra over a wide range of particle sizes for gold nanospheres in water, normalized to the maximum extinction for each value of the diameter, 2R. (b–h) Near-field enhancement maps for dipolar, quadrupolar and octupolar modes in particles of different diameters, corresponding to the symbols superimposed in the upper contour plot. Adapted from Myroshnychenko et al. (2008). Copyright © RSC Publishing (2008).
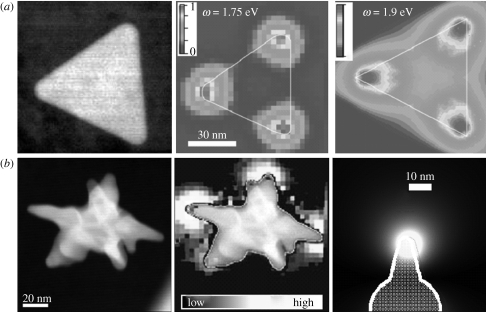
The third main factor affecting SERS intensity, which is nowadays regarded as the most important one, is nanoparticle shape (Orenforff et al. 2005; Jana & Pal 2007). Controlling the morphology provides a method to tune the optical and spectroscopic response of nanomaterials (Grzelczak et al. 2008; Lu et al. 2009; Pastoriza-Santos & Liz-Marzan 2009; Sepúlveda et al. 2009), which is an essential requirement for a wide range of applications like genetic diagnostics, immunoassay labelling and trace amount detection of drugs, biomolecules and pesticides. The tremendous progress that has been achieved in both colloid chemistry and lithographic methods resulted in the possibility of tuning the optical properties within the whole range from the UV to the NIR, through careful control over the morphology of the nanoparticles (Banholzer et al. 2008; Myroshnychenko et al. 2008; Pastoriza-Santos & Liz-Marzan 2009). This has paved the way towards the generation of new families of advanced platforms for SERS ultrasensitive analysis, in particular those related to biomedicine (Ochsenkühn et al. 2009; Alvarez-Puebla & Liz-Marzan 2010). The importance of tailoring the morphology of the nanostructures is also related to the recently demonstrated observation that, in anisotropic particles, LSPRs are not homogeneously distributed throughout the whole particle surface but give rise to a concentration of electromagnetic fields in several specific regions within the nanoparticle. Such an electromagnetic field concentration has been observed at the corners of triangular platelets (Nelayah et al. 2007; figure 2a), the ends of nanorods (Aizpurua et al. 2005; Bryant et al. 2008; Chen et al. 2009) and the edges and corners of nanobars and nanocubes (Cobley et al. 2008). In this respect, it has been recently reported that the electromagnetic field can be strongly focalized at sharp apexes such as those contained in gold ‘nanostars’. Both theoretical (boundary element method) and experimental (electron energy loss spectroscopy) results (Garcia de Abajo 2010) clearly demonstrate such field concentration (figure 2b), which results in substantially higher SERS enhancement for stars than for spheres of similar dimensions (Rodriguez-Lorenzo et al. 2009, 2010). The role played by focalization at the tips, regardless of the morphology of the entire nanostructure, has been shown through studies on more complex nanostructures, such as thorn-shaped nanowires (Pazos-Perez et al. 2010a).
Figure 2.
High-resolution scanning transmission electron microscopy (STEM) dark-field image, electron energy loss spectroscopy (EELS) intensity mapping and calculated EELS intensity map of (a) a single Ag triangle and (b) an Au nanostar. Adapted from (a) Nelayah et al. (2007) and (b) Rodriguez-Lorenzo et al. (2009). Copyright © Nature Publishing Group (2007) and ACS Publishing (2009).
Complementary to these latest approaches for electromagnetic field concentration, quite impressive progress has been directed to the controlled fabrication of so-called hot spots. Hot spots are defined as specific gaps between particles where the electromagnetic field is extremely high owing to coupling between their plasmon resonances (Brus 2008). Until very recently, hot spots were obtained only by uncontrolled aggregation of colloids; for example, by increasing the ionic strength of a colloidal suspension (Yaffe et al. in press). Carefully designed hot spots can become much more active and even enable the possibility of single molecule spectroscopy (Kneipp et al. 1997). Recent theoretical and experimental results indicate that engineered aggregates composed of dimers or trimers offer consistently higher optical enhancement than fractal aggregates (Chu et al. 2009; Laurence et al. 2009). Several approaches have been reported for the controlled and homogeneous fabrication of such sphere dimers (Li et al. 2009; Pallaoro et al. 2010) and trimers (Chen et al. 2010), not only made of spheres but also comprising nanoparticles with other geometries such as rods (Li et al. in press) or cages (Rycenga et al. 2010).
3. Advanced SERS sensor platforms
The design of efficient and flexible nanostructured substrates for SERS detection is one of the main challenges to be achieved before the technique can be widely applied. Although colloidal dispersions of plasmonic nanoparticles may be (and actually are) used as prepared, their integration into advanced materials for generating such enhancing platforms, which may offer flexibility and functionality, is a key aspect in sensor engineering. One of the main restrictions arises from the need for the probe molecules to be in close contact with the metallic surface, so that they ‘feel’ the high electromagnetic field due to the LSPR. Unfortunately, many molecular families show very low affinity towards coinage metal surfaces, and therefore their detection has not been possible for a long time. Novel strategies are thus required to retain such molecular species close enough to the nanostructured metal, which requires the rational design of materials for which specific properties such as electrostatic charge, chemical affinity and mechanical response can be tailored to facilitate or even force the retention of the desired analyte onto the optical enhancer. We describe in this section some of the main directions in which this line of research has evolved.
The use of electrostatic attraction is probably one of the simplest approaches that can be exploited, for example, by tuning the nanoparticle charge as a function of solution pH (Alvarez-Puebla et al. 2005). This strategy has been employed with standard, citrate-reduced gold or silver colloids, but applicability has been demonstrated only for the analysis of positively charged analytes, mainly because of the difficulty in obtaining metal colloids with a highly positive surface charge. However, this drawback can be solved by rational choice of a suitable capping agent, for example using appropriate amino acids as stabilizers. Thus, as a function of the amino acid nature (number of amino groups) and the pH of the medium, the surface charge on the particles can be tuned from 30 to −50 mV (Alvarez-Puebla & Aroca 2009). It is, however, important to realize that not all amino acids can be used, since those containing thiol groups can passivate the gold or silver surface, thereby inhibiting the retention of the molecules under study. Alternatively, tuning the dielectric properties (hydrophilic/hydrophobic nature) of the surface can be a suitable strategy to enhance the retention of non-polar molecules. In this case, the metal nanostructures are capped/functionalized with a hydrophobic monolayer that produces a high affinity for the analyte. Examples include the deposition of an aliphatic self-assembled monolayer (SAM) as a partition interface, which has been successfully applied to the analysis of polycyclic aromatic hydrocarbons (PAHs) (Jones et al. 2009) or polychlorinated biphenyls (Bantz & Haynes 2009). Other hydrophobic molecules, such as calixarenes (Guerrini et al. 2006), viologen (Guerrini et al. 2009a) or dithiocarbamates (Guerrini et al. 2009b), have been extensively used for the detection of PAHs. Unfortunately, many other types of analytes still cannot be detected using these strategies. For example, metallic ions cannot be directly detected by Raman, since this is a molecular spectroscopy. Thus, indirect detection approaches must be devised, which involve coupling an appropriate chelating molecule to the gold or silver surfaces, so that detection of the metal ions can be achieved through vibrational changes induced on the ligand upon complexation (Zhao et al. 2009).
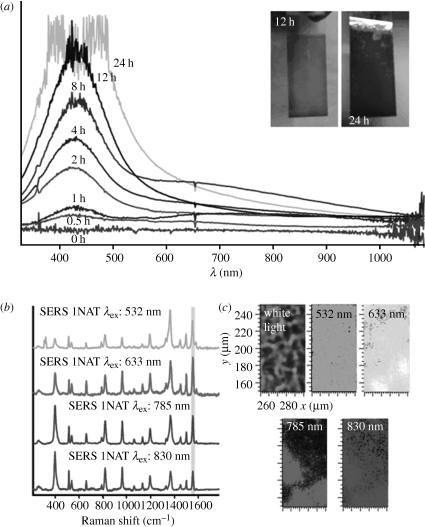
Recent and more general methods have been proposed through the design of hybrid materials that can unspecifically trap molecules from a solution. Mechanical retention and identification of analytes have been reported using polymers that are responsive to external stimuli, such as temperature (e.g. poly-N-isopropylacrylamide; Alvarez-Puebla et al. 2009) or pH (i.e. lipoic acid–polyethylene glycol (PEG)–polymethacrylic acid block copolymer; Qian et al. 2009). Although these materials do allow detection of otherwise elusive compounds (such as naphthol), the intensities that can be registered are not very high because they usually comprise single nanoparticles coated with the responsive material, and this coating hinders the interaction between particles and hot spots (Markel et al. 1999). An additional advantage of using colloidal substrates is that they can be used as prepared or assembled into solid thin films or even in bulk materials, thereby providing more flexibility to the sensing process. In line with this trapping concept, other strategies have been reported involving the incorporation of colloidal silver nanoparticles inside exponentially grown layer-by-layer (LBL) films (polyelectrolyte multilayer films with thickness exponentially increasing with the number of deposited layers; Podsiadlo et al. 2008; Srivastava et al. 2008), generating a high density of hot spots and allowing the direct, ultrasensitive analysis of molecules that could not be retained by other methods (Abalde-Cela et al. 2009). These substrates were found to provide intense and uniform signals, even when excited with different laser lines (figure 3). Trapping substrates can also be prepared by in situ reduction of silver particles inside bulk polymer gels, which brings together the benefits of molecular trapping, generation of dynamic hot spots and reusability (Aldeanueva-Potel et al. 2009).
Figure 3.
(a) UV/vis spectra of e-LBL films infiltrated with Ag nanoparticles, as a function of immersion time of the e-LBL film in a silver colloid. Inset: digital photographs of the films after 12 and 24 h of immersion time. (b) SERS spectra of 1-naphthalenethiol (1NAT) from an LBL Ag film using different excitation laser lines. (c) SERS mapping of 1NAT (1553 cm−1) on the films with different excitation laser lines. Adapted from Abalde-Cela et al. (2009). Copyright © Wiley Interscience (2009).
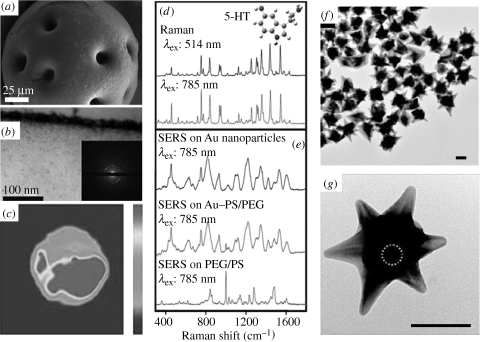
With regard to plasmonic platforms in solid thin-film format, research interest is usually related to their capability of providing a portable substrate, which is easy to use in the field, with a high enhancing efficiency owing to containing a dense collection of hot spots. Such nanostructured substrates can be easily fabricated by means of physical methods such as sputtering/physical vapour deposition (Volpati et al. 2008) or e-beam (Xia et al. 1999), block copolymer micelle (Sanchez-Iglesias et al. 2010) and dip-pen (Piner et al. 1999) lithographies. However, these techniques often require the use of expensive equipment commonly available in surface or materials science research laboratories, but very rarely in spectroscopy or analytical chemistry laboratories. Alternatively, colloidal particles can be easily prepared and engineered into a thin-film format. The simplest technique for thin-film fabrication undoubtedly comprises casting and air-drying a polymeric solution containing the relevant metal nanoparticles (dos Santos et al. 2004). However, this method can significantly reduce the enhancing efficiency of the particles because the concentration of scattering centres is often too low and most of them are located inside the polymer film rather than on the surface. Solutions to overcome this problem involve the application of modern thin-film fabrication methods such as SAMs (Gellner et al. 2009), Langmuir–Blodgett deposition (Lu et al. 2004), or LBL assembly (Goulet et al. 2005; Jiang & Tsukruk 2006). Interestingly, similar fabrication approaches can be exploited for the fabrication of ‘optical accumulators’, to be used for monitoring large sample volumes in continuous flows, or even for making discrete hybrid particles with sizes ranging from below micrometre level to several micrometres. Although different in nature, both substrates can have the same function: increasing the detection limit. The concept of optical accumulators is based on the retention of the analyte of interest during the continuous flow of the sample, until sufficient concentration is reached to enable SERS detection (Ko & Tsukruk 2008). This method has been applied for the simultaneous detection of several analytes with concentrations in the sample fluid as low as 10−18 M (Aldeanueva-Potel et al. 2010). On the other hand, SAMs, LBL assembly or simply in situ reduction of adsorbed metal ions can be carried out on the surface of small particles, typically polymer or silica spheres (Jiang & Tsukruk 2006; Braun et al. 2007a; Farah et al. 2009), but also other substrates such as carbon nanotubes (Sanles-Sobrido et al. 2009b; Taladriz-Blanco et al. 2009), which provide both optical and colloidal stability to the supported gold or silver nanoparticles. Controlled aggregation of the metal nanoparticles on the microparticle templates leads to the formation of stable and reproducible hot spots. Additionally, if the size of these particles is big enough to be observed under an optical microscope, the amount of optical enhancer required for the SERS analysis can be reduced, with a subsequent increase in the detection limit. Unfortunately, ‘looking for the bead’ under the microscope is a time-consuming process. Progress in colloidal synthesis has permitted the generation of bifunctional materials, in either the nanometre (Wei et al. 2009; Pazos-Perez et al. 2010b) or submicrometre (Spuch-Calvar et al. 2009) range, which combine optical and magnetic properties within a single entity, thereby opening up the possibility of decreasing the required amount of optical substrate, since it can be concentrated upon application of a magnetic field (figure 4).
Figure 4.
(a) SEM image of Au–PEG/PS bead. (b) Cross-sectional TEM image and electron diffraction pattern of the bead crust. (c) SERS mapping of one Au–PEG/PS microbead, showing the distribution of serotonin (5-HT) (at 1531 cm−1). (d) Raman spectra of 5-HT obtained with different excitation wavelengths. (e) SERS spectra of 5-HT on Au colloids and on Au–PEG/PS microbeads (Raman scattering spectrum of PEG/PS with the same concentration of 5-HT is also shown). Adapted from Farah et al. (2009). (f,g) TEM images of Au nanostars with superparamagnetic cores (bar = 50 nm), with the approximate position of the Fe3O4 core outlined by a dashed circle. Adapted from Wei et al. (2009). Copyright © Wiley Interscience (2009) and ACS Publishing (2009).
4. Direct SERS sensing
The most common way of using SERS is the direct detection of the target analyte, i.e. the identification of the specific SERS spectral fingerprint of the probe molecule upon direct binding onto a plasmonic metal nanostructure (Alvarez-Puebla & Liz-Marzan 2010). Numerous examples have been reported on the application of colloidal suspensions for the ultrasensitive detection and characterization of both small and macro-biomolecules (nucleic acids or proteins), in vivo monitoring of metabolites or pathogens or the classification of living organisms. Although small biomolecules can be detected by means of other techniques such as fluorescence, NMR, IR spectroscopy or mass spectrometry, SERS offers higher detection limits in conjunction with a complete structural characterization of the sample, it does not require complex separation steps and it can be applied in situ under environmental/biological conditions. SERS ultradetection on colloids has been successfully reported for various substances of interest including highly toxic metal ions such as mercury (Wang et al. 2009), illegal drugs (Sägmüller et al. 2001; Faulds et al. 2002) and therapeutic compounds (Farquharson et al. 2008) or relevant metabolites, such as cancer biomarkers (Seballos et al. 2005), in different biological fluids. The versatility of SERS acquisition (i.e. backscattering through a glass or quartz window) can also be exploited for coupling with other techniques for online measurements such as flow cytometry (Watson et al. 2008; Sebba et al. 2009) or microfluidics. Recent improvements in microfluidic technology (Christopher & Anna 2007) paved the way for a variety of chemical and biochemical analyses using SERS as a transducer (Yea et al. 2005; Chen & Choo 2008). Direct detection in microfluidic channels has several advantages over SERS detection on colloids or hybrid materials, including the use of small sample volumes, with the subsequent reduction in waste production, simplicity of use and continuous flow monitoring in real time. SERS combined with microfluidics has been, for example, reported for the online monitoring of promethazine (antihistamine and sedative drug) or mitoxantrone (anti-cancer agent) (Ackermann et al. 2007).
Although we have described above the design of hybrid functional materials to expand the applicability of SERS as a detection technique, the use of specific molecular receptors is worth special consideration. This approach is based on the functionalization of the metal surface with a selective antibody for the analyte of interest (the antigen). The hybrid biosensor can thus be exposed to the sample and SERS spectra are acquired before and after the formation of the antibody–antigen complex, so that the changes induced in the vibrational pattern of the antibody are registered. Since certain vibrational peaks remain unaltered, these can be used as an internal standard and quantitative data can be extracted upon calibration. Another advantage related to the selectivity of the antibody is the ability to carry out the test directly in complex fluids. This method has been successfully applied to the detection of the cocaine metabolite benzoylecgonine (Sanles-Sobrido et al. 2009b).
However, SERS spectroscopy is not restricted to the determination of small analytes, but can also be used for both characterization and ultradetection of biomacromolecules. Methods for biomacromolecule analysis usually rely on the use of secondary detection labels, which requires the use of two different selective linkers (antibodies), one for capture and one for detection. This not only increases the analysis cost but also makes it rather time consuming, and thus the development of label-free methods has become a matter of extensive research, in which SERS has become a viable alternative. Nucleic acids (DNA and RNA) are the easiest macromolecules to detect by SERS owing to their molecular structure. They are constituted by nitrogenous aromatic heterocyclic bases (i.e. purines or pyrimidines) that act as Lewis bases and can easily coordinate with silver and gold surfaces while retaining a large SERS cross section. While SERS analysis takes advantage of these structural properties, the large number of nitrogenous functional groups, with potential to bind the metallic surface, results in a random orientation of the macromolecule. Therefore, DNA or RNA SERS spectra are characterized by strong fluctuations in the relative intensity of their bands, in agreement with surface selection rules (Moskovits & Suh 1984). To solve this situation, single- and double-stranded thiolated DNA or RNA oligomers have been used, which can direct the adsorption of the macromolecule to be perpendicular to the surface (Barhoumi et al. 2008). Further, other recent approaches include the combination of SERS with microfluidic systems for sequence-specific detection of DNA (Strelau et al. 2010).
On the other hand, although proteins and peptides also contain amino groups, the SERS analysis of proteins is not so advanced as that of nucleic acids because their aromatic character is considerably lower. However, a number of reports have been recently published regarding the detection, conformation elucidation and bioreactivity of proteins by means of direct SERS analysis. For example, single-molecule detection has been reported for the green fluorescent protein (Habuchi et al. 2003) and the photoactive yellow protein (Singhal & Kalkan 2009), as well as ultradetection of human insulin, insulin lispro (Drachev et al. 2004), glutathione (Huang et al. 2009) and various others in purified samples. Notably, Han et al. (2008) recently devised a new analytical procedure for label-free, multiplex protein detection comprising electrophoretic sequential protein separation. Using this method, the detection of myoglobin was possible within a bovine serum albumin complex solution. Regarding the conformational elucidation of proteins and peptides, SERS can be considered as a good alternative to resonance Raman (Spiro & Czernuszewicz 1995) for several reasons. First, SERS is usually carried out with excitation in the visible–NIR range, minimizing sample damage. Second, SERS cross sections are much higher than those for RRS, permitting conformational characterization at very low concentrations and thus removing possible solid-state artefacts. Further, SERS permits the study of the native conformation and its natural fate under biological conditions, giving rise to direct information that is easy to extrapolate to real problems (Iafisco et al. 2008; Sengupta et al. 2008). One of the main challenges that Raman (and also SERS) spectroscopy faces when dealing with proteins and peptides is the complexity involved in complete band assignment, which is essential for structural characterization. However, advances in theoretical modelling have made possible the calculation of accurate vibrational patterns for small- and medium-sized peptides. For example, the theoretical Raman and SERS spectra of ‘penetratin’ were accurately reproduced and compared with experimental data (figure 5). Proper reproduction of the SERS spectrum involved using the SERS spectra of three aromatic dipeptides (phenylalanine–cysteine, tyrosine–cysteine and tryptophan–cysteine) as an empirical ‘basis set’ (Wei et al. 2008). Other alternative approaches for the detection of biomacromolecules involve the incorporation of Raman dye labels into the metal nanoparticle–DNA (or protein) conjugates, so that, by detecting the SERS signals of the Raman dye, the DNA or protein analytes can be readily reported, even at very low concentrations (Braun et al. 2007b; Fabris et al. 2007).
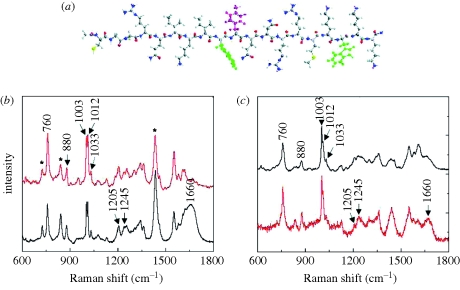
Figure 5.
Comparison of empirically predicted (red) and directly measured experimental (black) spectra of penetratin. (a) Molecular model of penetratin peptide, including one phenylalanine (purple) and two tryptophans (green). (b) Raman spectra. (c) SERS spectra. (b) Red lines, predicted Raman; black lines, experimental Raman. (c) Black lines, predicted SERS; red lines, experimental SERS. Adapted from Wei et al. (2008). Copyright © ACS Publishing (2008).
Increasing in complexity, the detection and classification of pathogenic micro-organisms (viruses, bacteria, yeast, fungi and protozoa) (Alexander 2008; Jarvis & Goodacre 2008; Tripp et al. 2008) and bacteria (Premasiri et al. 2005; Evanoff et al. 2006), as well as the study of complex tissues (Pinzaru et al. 2008), have been largely improved through improvements in optical spectroscopy (i.e. spectral spatial resolution) and nanoparticle synthesis. SERS has proved useful for the identification of micro-organisms in complex mixtures (so that isolation of single strains is not required), within a few seconds (Shanmukh et al. 2006). This is due to the powerful combination of confocal optical microscopy and the unique vibrational fingerprints obtained in SERS measurements. The most common strategy comprises the addition of colloidal suspensions to the sample where the micro-organisms are, so that the nanoparticles adhere to the bacterial cell walls (figure 6a,b), allowing their identification through the specific composition of the membrane (figure 6c–e). This approach can be improved by combination of the SERS vibrational spectra with statistical classification methods, such as principal component analysis, partial least squares or hierarchical clustering analysis. By using these chemometric pattern recognition algorithms, one can identify individual species, including the strain, in a populated sample of different micro-organisms (Pearman & Fountain 2006; Patel et al. 2008). Equivalent approaches have been developed for the identification of eukaryotic cells (Sayin et al. 2009; Sujith et al. 2009).
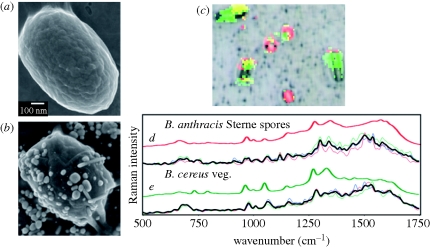
Figure 6.
Bacillus anthracis Sterne spore (a) without and (b) with silver nanoparticle coating. (c) SERS-processed RGB image of a complex bacterial mixture overlaid on BFI. (d) Comparison of extracted Raman spectra from single spores (thin coloured lines), average spectrum (black line) and a library spectrum of BASP (thick red line); (e) extracted Raman spectra from single cells (thin coloured lines), average spectrum (black line) and a library spectrum of BCVG (thick green line). Adapted from Guicheteau et al. (in press). Copyright © Wiley Interscience (2010).
Two excellent recent reviews by Willets (2009) and Chourpa et al. (2008) describe the improvements and challenges in the application of SERS to studies inside living cells. To date, most of the SERS applications inside cells have been related to the incorporation of plasmonic particles inside micro-organisms, usually by the addition of the nanoparticles directly to the growth medium. However, this technique provides only limited information because the particles mostly accumulate in the cytosol and the nucleus membrane. Additionally, the SERS spectra acquired from these particles are ‘contaminated’ by the vibrational bands of their coating molecules (serum albumin), which makes the interpretation of the obtained vibrational pattern extremely difficult. A final difficulty is related to the need for a large amount of single particles within the probe area to obtain high-quality measurements, which has been partially solved by using controlled aggregates containing hot spots, but the larger size of the aggregates subsequently increases the difficulty in particle internalization. Therefore, alternative approaches are required, such as the functionalization of the plasmonic materials with appropriate biointerfaces or the use of encoded particles, as described in the following section.
5. SERS-encoded nanoparticles
Rapid and sensitive diagnostic techniques are central to human health. All the direct methods described above exploit an intrinsic spectroscopic property of the analyte of interest or a specific molecular recognition event such as antibody–antigen, DNA–DNA or receptor–ligand interactions to carry out the detection (Edelstein et al. 2000). However, the simultaneous detection of several components (multiplexing) is difficult to achieve using direct SERS, owing to the inherent complexity of biological fluids. These limitations might, however, be overcome using the so-called encoded particles, which are labelled materials directly bound to a biointerface, providing it with a specific signature for indentifying a recognition event. These detection schemes can be termed indirect because the signal does not originate from the analyte, but from the labelled material instead (Bake & Walt 2008).
Encoded particles are cost-efficient platforms that address some of the limitations posed by more conventional substrates, such as: (i) amenability to high-throughput screening and multiplexing (Raez et al. 2007), (ii) larger surface area for receptor conjugation or solid-phase synthesis, (iii) better accessibility of the analytes to the entire sample volume for interaction with bead-conjugated receptors, and (iv) greater versatility for sample analysis and data acquisition. In particular, encoded nanoparticles (Bake & Walt 2008) appear as a fast, reliable and sensitive tool for multiplexed high-throughput screening (Fenniri & Alvarez-Puebla 2007), biodiagnosis (Xue et al. 2009) and bioimaging (Qian et al. 2008).
The challenges associated with multiplex analysis using encoded particles are related to the encoding process, which should retain the assay sensitivity, specificity and reproducibility. Regarding the fabrication of encoded particles, several alternatives have been reported for the introduction of the unique code into each batch of single particles. For example, the particles may be intrinsically labelled through shape (Qin et al. 2007), composition (Raez et al. 2007) or lithographic marks (Pregibon et al. 2007), but they may also be externally labelled by introducing into their structures quantum dots (Bruchez et al. 1998; Han et al. 2001), fluorescent dyes (Stoermer et al. 2006) or SERS molecular codes (Doering et al. 2007). Since fluorescence-encoded microbeads (Battersby et al. 2000) can be rapidly processed by conventional flow cytometry, they have become popular platforms for multiplexing assays. However, several disadvantages of using multiple fluorescent signals as encoding tools include both a limited number of barcodes and possible interference from the native fluorescence of the analyte. Notably, SERS-encoded particles can overcome this drawback because the unique vibrational fingerprints of each molecular entity (Aroca et al. 2005b) allow for an infinite number of encoding molecules that can be used simultaneously (Faulds et al. 2002). Additionally, the extremely high SERS cross sections, which may even permit single-molecule detection, allow for the identification of the different codes at very low concentration in a time-effective manner (typically ms).
The first generation of SERS tags was reported by Porter's group (Rohr et al. 1989; Ni et al. 1999), based on the co-adsorption of reporter molecules and targeting ligands onto metal nanoparticles; however, there were several limitations, such as: (i) the susceptibility of the encoded signal to dramatic changes in the intensity owing to colloidal aggregation, (ii) possible side reactions owing to the catalytic activity of the metal nanoparticles, (iii) signal enhancement of the analyte overlapping the microparticle barcode, (iv) difficulty in the functionalization of the particle surface for biomolecule immobilization, and (v) leaching of the vibrational label, decreasing the particle signal and promoting toxic reactions. To overcome all these problems, the optical enhancer needs to be coated with a suitable protective shell, usually silica (Freeman et al. 2005) or PEG, so that the Raman code cannot leach out (Qian et al. 2008) while providing a suitable surface for biofunctionalization (figure 7a). Several methods have been reported for controlled silica coating of different kinds of nanoparticles (Guerrero-Martínez et al. 2010), mainly based on the use of coupling agents to induce the polymerization of silica by hydrolysis and condensation of organosilanes on the nanoparticle surface (Stöber et al. 1968; Liz-Marzan et al. 1996). The silica shell not only circumvents the problems discussed above but also hinders the adsorption of other molecular systems onto the metallic surfaces (which could interfere with the vibrational code) while providing high colloidal stability and allowing conjugation with antibodies or nucleic acids and stabilization in biological media (Cao et al. 2002). Further research along this direction has led to the fabrication of more efficient encoded nanoparticles using hybrid materials composed of plasmonic nanoparticles protected with either polymers or silica shells (Mulvaney et al. 2003; Fernández-López et al. 2009). However, a major limitation of this approach is that single particles usually do not provide sufficient field enhancement to raise the SERS signal up to detectable levels. This problem can however be overcome by encapsulation of controlled aggregates with either silica or polymer shells (which leads to heterogeneity in both shape and size; Lutz et al. 2008; Watson et al. 2008) or by confined epitaxial growth of gold or silver islands inside homogeneous, hollow silica capsules (Sanles-Sobrido et al. 2009a).
Figure 7.
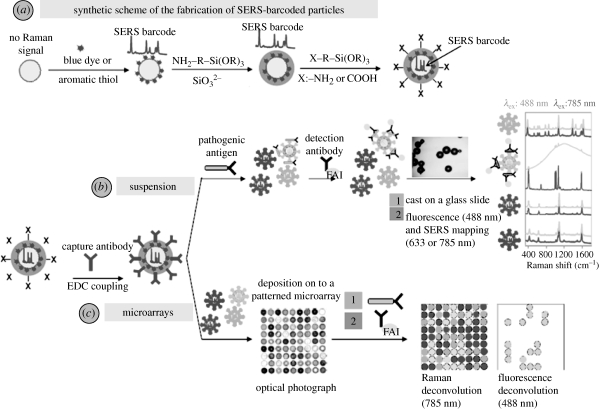
(a) Schematic image illustrating the fabrication of SERS-encoded particles; (b) strategy for the detection of analytes of interest, i.e. pathogenic antigen, with suspension experiments; (c) detection route through microarray patterning.
Regarding practical applicability, these materials show great versatility. Encoded nanoparticles can be used either in suspension assays or patterned into microchips. In a typical suspension assay (figure 7b), a mixture of different antibodies, each of them previously labelled with one particular encoded particle, is added to the fluid of interest. After reaction with their specific antigens, the particles are separated by centrifugation, washed and exposed to a detection antibody (labelled with a fluorophore), which selectively binds to those particles that were complexed to the corresponding capture antibody. After fluorophore binding, the particles are centrifuged and washed again, and a small portion is cast onto a glass slide and analysed in a micro-Raman system. Positive diagnosis of the presence of a pathogen, disease marker, etc. requires the presence of both fluorescence and SERS tags on the same particle (Cao et al. 2002). Although, in principle, this method does not require homogeneity in size or shape, if automation by flow cytometry or microfluidics is to be implemented, size homogeneity is essential to avoid interference due to different Rayleigh scattering signals (which are morphology dependent; Sebba et al. 2009). An alternative route to suspension assays is the generation of microchips (figure 7c) containing encoded particles within well-defined spatial regions and fixed to a surface, which allows fast recognition and avoids the need for washing steps such as centrifugation. On the other hand, the readout from the chip can be performed automatically. Notwithstanding, the preparation of these platforms requires particles with a high homogeneity in both size and shape to be used (Raez et al. 2007).
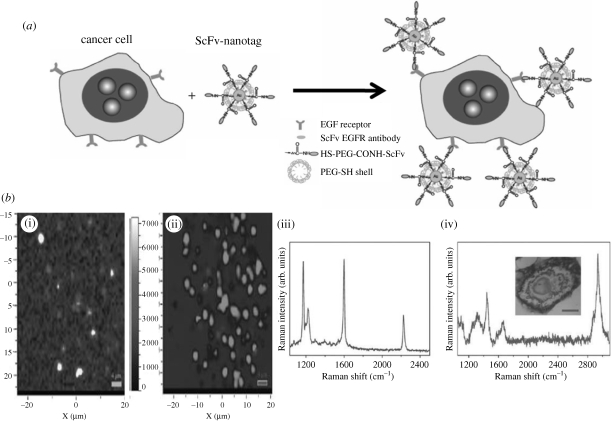
Encoded particles not only pave the way towards the design of new and fast advanced sensor devices, capable of monitoring multiple parameters in a single readout, but can also be applied in bioimaging. Nabiev et al. (1991) reported the SERS spectra of the anti-tumour drug doxorubicin, recorded from treated cancer cells incubated with citrate-reduced silver colloids. These spectra were the first feasibility data demonstrating the promising perspectives of SERS spectroscopy in bioimaging, and it has currently become a key topic of research. SERS bioimaging is based on the functionalization of encoded particles with bioligands with affinity for specific receptors of the cell membrane (figure 8). This technique can be applied to in vivo imaging of cells (Chourpa et al. 2008), tissues and organs (Lutz et al. 2008). The high intensity provided by SERS-encoded particles and the possibility of preparing extremely bright, biocompatible and small capsules (Sanles-Sobrido et al. 2009a) constitute a highly competitive alternative to quantum dots and magnetic nanoparticles. Nie's group successfully demonstrated that irradiation of tissues with NIR lasers allows the SERS fingerprint to be recorded of PEGylated gold-encoded nanoparticles, functionalized with an antibody, so as to target tumour biomarkers such as epidermal growth factor receptors on human cancer (Qian et al. 2008).
Figure 8.
(a) Preparation of targeted SERS nanoparticles; (b) SERS imaging of H9c2 cardiac myocytes. (i) SERS image of the normalized CN intensity map of a fixed H9c2 cell. (ii) Multivariate deconvolution of the MMBN SERS spectra for fixed H9c2. (iii) SERS spectrum of MMBN (4-(mercaptomethyl)benzonitrile) Raman reporter molecule. (iv) Cell Raman spectrum taken from the nucleus region of the cell shown in the inset. (a) Adapted from Qian et al. (2008); (b) Adapted from Kennedy et al. (2009). Copyright © Nature Publishing Group (2008) and ACS (2009).
6. Conclusions and outlook
We have shown in this review that SERS has been established as a solid and reliable analytical technique for the detection of extremely low amounts of a wide variety of molecular species. Although the requirement of a close contact of the analyte with the enhancing metallic surface has been a limitation, the design of novel hybrid substrates opened up the possibility of complete generalization of SERS detection. Additionally, through the use of antibodies or other selective receptors that can be bound to the nanostructured metal surface, recognition can be made specific and even quantitative. This is extremely important for biomedical applications, since it is the basis of early diagnosis of important diseases. We have also shown the great potential of using SERS-encoded particles for indirect detection and labelling, which can be implemented on a chip or even inside living cells, tissues or a variety of micro-organisms.
However, there are still challenges remaining, mainly related to the reproducibility of the methods for substrate fabrication, in particular when dealing with the formation of hot spots, which are responsible for the highest enhancement factors, but their efficiency is extremely sensitive towards small geometrical details within the nanostructure. Additionally, although portable Raman spectrometers are available, most of the published reports are based on very sophisticated instruments that will not find a place in routine analysis carried out in laboratories or hospitals. Thus, the field of SERS detection, in particular for biomedical applications, has great potential, as demonstrated by many examples, but is open to new developments that will undoubtedly continue to amaze us in the near future.
Acknowledgements
R.A.A.-P. acknowledges the RyC programme (MEC, Spain). This work has been funded by the Spanish Ministerio de Ciencia e Innovación (grants MAT2007-62696, MAT2008-05755 and Consolider Ingenio 2010-CSD2006-12) and the Xunta de Galicia (PGIDIT06TMT31402PR and 08TMT008314PR).
Footnotes
One contribution to a Theme Supplement ‘Scaling the heights–challenges in medical materials: an issue in honour of William Bonfield, Part I. Particles and drug delivery’.
References
- Abalde-Cela S., Ho S., Rodriguez-Gonzalez B., Correa-Duarte M. A., Alvarez-Puebla R. A., Liz-Marzan L. M., Kotov N. A. 2009. Loading of exponentially grown LBL films with silver nanoparticles and their application to generalized SERS detection. Angew. Chem. Int. Ed. 48, 5326–5329. ( 10.1002/anie.200901807) [DOI] [PubMed] [Google Scholar]
- Ackermann K. R., Henkel T., Popp J. 2007. Quantitative online detection of low-concentrated drugs via a SERS microfluidic system. ChemPhysChem 8, 2665–2670. ( 10.1002/cphc.200700554) [DOI] [PubMed] [Google Scholar]
- Aizpurua J., Bryant G. W., Richter L. J., García De Abajo F. J., Kelley B. K., Mallouk T. 2005. Optical properties of coupled metallic nanorods for field-enhanced spectroscopy. Phys. Rev. B 71, 1–13. [Google Scholar]
- Aldeanueva-Potel P., Faoucher E., Alvarez-Puebla R. A., Liz-Marzan L. M., Brust M. 2009. Recyclable molecular trapping and SERS detection in silver-loaded agarose gels with dynamic hot spots. Anal. Chem. 81, 9233–9238. ( 10.1021/ac901333p) [DOI] [PubMed] [Google Scholar]
- Aldeanueva-Potel P., Correa-Duarte M. A., Alvarez-Puebla R., Liz-Marzan L. M. 2010. Free-standing carbon nanotube films as optical accumulators for multiplex SERRS. ACS Appl. Mater. Interfaces 2, 19–22. ( 10.1021/am9008715) [DOI] [PubMed] [Google Scholar]
- Alexander T. A. 2008. Development of methodology based on commercialized SERS-active substrates for rapid discrimination of Poxviridae virions. Anal. Chem. 80, 2817–2825. ( 10.1021/Ac702464w) [DOI] [PubMed] [Google Scholar]
- Alvarez-Puebla R. A., Aroca R. F. 2009. Synthesis of silver nanoparticles with controllable surface charge and their application to surface-enhanced Raman scattering. Anal. Chem. 81, 2280–2285. ( 10.1021/Ac8024416) [DOI] [PubMed] [Google Scholar]
- Alvarez-Puebla R. A., Liz-Marzan L. M. 2010. SERS-based diagnosis and biodetection. Small 6, 604–610. ( 10.1002/smll.200901820) [DOI] [PubMed] [Google Scholar]
- Alvarez-Puebla R. A., Arceo E., Goulet P. J. G., Garrido J. J., Aroca R. F. 2005. Role of nanoparticle surface charge in surface-enhanced Raman scattering. J. Phys. Chem. B 109, 3787–3792. ( 10.1021/Jp045015o) [DOI] [PubMed] [Google Scholar]
- Alvarez-Puebla R. A., Contreras-Caceres R., Pastoriza-Santos I., Perez-Juste J., Liz-Marzan L. M. 2009. Colloids as molecular traps for surface-enhanced, spectroscopic, ultra-sensitive analysis. Angew. Chem. Int. Ed. 48, 138–143. ( 10.1002/anie.200804059) [DOI] [PubMed] [Google Scholar]
- Aroca R. F., Alvarez-Puebla R. A., Pieczonka N., Sanchez-Cortez S., Garcia-Ramos J. V. 2005a Surface-enhanced Raman scattering on colloidal nanostructures. Adv. Colloid Interface Sci. 116, 45–61. ( 10.1016/j.cis.2005.04.007) [DOI] [PubMed] [Google Scholar]
- Aroca R. F., Goulet P. J. G., dos Santos D. S., Alvarez-Puebla R. A., Oliveira O. N. 2005b Silver nanowire layer-by-layer films as substrates for surface-enhanced Raman scattering. Anal. Chem. 77, 378–382. ( 10.1021/Ac048806v) [DOI] [PubMed] [Google Scholar]
- Baena J. R., Lendl B. 2004. Raman spectroscopy in chemical bioanalysis. Curr. Opin. Chem. Biol. 8, 534–539. ( 10.1016/j.cbpa.2004.08.014) [DOI] [PubMed] [Google Scholar]
- Bake K. D., Walt D. R. 2008. Multiplexed spectroscopic detections. Annu. Rev. Anal. Chem. 1, 515–547. ( 10.1146/annurev.anchem.1.031207.112826) [DOI] [PubMed] [Google Scholar]
- Banholzer M. J., Millstone J. E., Qin L., Mirkin C. A. 2008. Rationally designed nanostructures for surface-enhanced Raman spectroscopy. Chem. Soc. Rev. 37, 885–897. ( 10.1039/b710915f) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantz K. C., Haynes C. L. 2009. Surface-enhanced Raman scattering detection and discrimination of polychlorinated biphenyls. Vibr. Spectrosc. 50, 29–35. ( 10.1016/j.vibspec.2008.07.006) [DOI] [Google Scholar]
- Barhoumi A., Zhang D., Tam F., Halas N. J. 2008. Surface-enhanced Raman spectroscopy of DNA. J. Am. Chem. Soc. 130, 5523–5529. ( 10.1021/Ja800023j) [DOI] [PubMed] [Google Scholar]
- Battersby B. J., Bryant D., Meutermans W., Matthews D., Smythe M. L., Trau M. 2000. Toward larger chemical libraries: encoding with fluorescent colloids in combinatorial chemistry. J. Am. Chem. Soc. 122, 2138–2139. ( 10.1021/ja993634i) [DOI] [Google Scholar]
- Braun G., Pavel I., Morrill A. R., Seferos D. S., Bazan G. C., Reich N. O., Moskovits M. 2007a Chemically patterned microspheres for controlled nanoparticle assembly in the construction of SERS hot spots. J. Am. Chem. Soc. 129, 7760–7761. ( 10.1021/ja072533e) [DOI] [PubMed] [Google Scholar]
- Braun G., Lee S. J., Dante M., Nguyen T. Q., Moskovits M., Reich N. 2007b Surface-enhanced Raman spectroscopy for DNA detection by nanoparticle assembly onto smooth metal films. J. Am. Chem. Soc. 129, 6378–6379. ( 10.1021/ja070514z) [DOI] [PubMed] [Google Scholar]
- Bruchez M., Jr, Moronne M., Gin P., Weiss S., Alivisatos A. P. 1998. Semiconductor nanocrystals as fluorescent biological labels. Science 281, 2013–2016. ( 10.1126/science.281.5385.2013) [DOI] [PubMed] [Google Scholar]
- Brus L. 2008. Noble metal nanocrystals: plasmon electron transfer photochemistry and single-molecule Raman spectroscopy. Acc. Chem. Res. 41, 1742–1749. ( 10.1021/ar800121r) [DOI] [PubMed] [Google Scholar]
- Bryant G. W., García De Abajo F. J., Aizpurua J. 2008. Mapping the plasmon resonances of metallic nanoantennas. Nano Lett. 8, 631–636. ( 10.1021/nl073042v) [DOI] [PubMed] [Google Scholar]
- Cao Y. C., Jin R., Mirkin C. A. 2002. Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science 297, 1536–1540. ( 10.1126/science.297.5586.1536) [DOI] [PubMed] [Google Scholar]
- Chen L., Choo J. 2008. Recent advances in surface-enhanced Raman scattering detection technology for microfluidic chips. Electrophoresis 29, 1815–1828. ( 10.1002/elps.200700554) [DOI] [PubMed] [Google Scholar]
- Chen X., Li S., Can X., Banholzer M. J., Schatz G. C., Mirkin C. A. 2009. Plasmonic focusing in rod-sheath heteronanostructures. ACS Nano 3, 87–92. ( 10.1021/nn800695u) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Wang Y., Yang M., Xu J., Goh S. J., Pan M., Chen H. 2010. Measuring ensemble-averaged surface-enhanced Raman scattering in the hotspots of colloidal nanoparticle dimers and trimers. J. Am. Chem. Soc. 132, 3644–3645. ( 10.1021/ja9090885) [DOI] [PubMed] [Google Scholar]
- Chourpa I., Lei F. H., Dubois P., Manfait M., Sockalingum G. D. 2008. Intracellular applications of analytical SERS spectroscopy and multispectral imaging. Chem. Soc. Rev. 37, 993–1000. ( 10.1039/b714732p) [DOI] [PubMed] [Google Scholar]
- Christopher G. F., Anna S. L. 2007. Microfluidic methods for generating continuous droplet streams. J. Phys. D Appl. Phys. 40, R319–R336. ( 10.1088/0022-3727/40/19/R01) [DOI] [Google Scholar]
- Chu M. W., Myroshnychenko V., Chen C. H., Deng J. P., Mou C. Y., García de Abajo F. J. 2009. Probing bright and dark surface-plasmon modes in individual and coupled noble metal nanoparticles using an electron beam. Nano Lett. 9, 399–404. ( 10.1021/nl803270x) [DOI] [PubMed] [Google Scholar]
- Cobley C. M., Skrabalak S. E., Campbell D. J., Xia Y. 2008. Shape-controlled synthesis of silver nanoparticles for plasmonic and sensing applications. Plasmonics 4, 171–179. ( 10.1007/s11468-009-9088-0) [DOI] [Google Scholar]
- Creighton J. A., Blatchford C. G., Albrecht M. G. 1979. Plasma resonance enhancement of Raman scattering by pyridine adsorbed on silver or gold sol particles of size comparable to the excitation wavelength. J. Chem. Soc. Faraday Trans. 2 75, 790–798. ( 10.1039/F29797500790) [DOI] [Google Scholar]
- Doering W. E., Piotti M. E., Natan M. J., Freeman R. G. 2007. SERS as a foundation for nanoscale, optically detected biological labels. Adv. Mater. 19, 3100–3108. ( 10.1002/adma.200701984) [DOI] [Google Scholar]
- dos Santos D. S., Goulet P. J. G., Pieczonka N. P. W., Oliveira O. N., Aroca R. F. 2004. Gold nanoparticle embedded, self-sustained chitosan films as substrates for surface-enhanced Raman scattering. Langmuir 20, 10 273–10 277. ( 10.1021/La048328j) [DOI] [PubMed] [Google Scholar]
- Drachev V. P., Thoreson M. D., Khaliullin E. N., Davisson V. J., Shalaev V. M. 2004. Surface-enhanced Raman difference between human insulin and insulin lispro detected with adaptive nanostructures. J. Phys. Chem. B 108, 18 046–18 052. ( 10.1021/jp047254h) [DOI] [Google Scholar]
- Edelstein R. L., Tamanaha C. R., Sheehan P. E., Miller M. M., Baselt D. R., Whitman L. J., Colton R. J. 2000. The BARC biosensor applied to the detection of biological warfare agents. Biosens. Bioelectron. 14, 805–813. ( 10.1016/S0956-5663(99)00054-8) [DOI] [PubMed] [Google Scholar]
- Emory S. R., Haskins W. E., Nie S. M. 1998. Direct observation of size-dependent optical enhancement in single metal nanoparticles. J. Am. Chem. Soc. 120, 8009–8010. ( 10.1021/ja9808402) [DOI] [Google Scholar]
- Evanoff D. D., Heckel J., Caldwell T. P., Christensen K. A., Chumanov G. 2006. Monitoring DPA release from a single germinating Bacillus subtilis endospore via surface-enhanced Raman scattering microscopy. J. Am. Chem. Soc. 128, 12 618–12 619. ( 10.1021/ja0642717) [DOI] [PubMed] [Google Scholar]
- Fabris L., Dante M., Braun G., Lee S. J., Reich N. O., Moskovits M., Nguyen T. Q., Bazan G. C. 2007. A heterogeneous PNA-based SERS method for DNA detection. J. Am. Chem. Soc. 129, 6086–6087. ( 10.1021/ja0705184) [DOI] [PubMed] [Google Scholar]
- Farah A. A., Bravo-Vasquez J. P., Alvarez-Puebla R. A., Cho J.-Y., Fenniri H. 2009. Robust Au-PEG/PS microbeads as optically stable platforms for SERS. Small 5, 1283–1286. ( 10.1002/smll.200801398) [DOI] [PubMed] [Google Scholar]
- Farquharson S., Gift A., Shende C., Inscore F., Ordway B., Farquharson C., Murren J. 2008. Surface-enhanced Raman spectral measurements of 5-fluorouracil in saliva. Molecules 13, 2608–2627. ( 10.3390/molecules13102608) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulds K., Smith W. E., Graham D., Lacey R. J. 2002. Assessment of silver and gold substrates for the detection of amphetamine sulfate by surface enhanced Raman scattering (SERS). Analyst 127, 282–286. ( 10.1039/b107318b) [DOI] [PubMed] [Google Scholar]
- Fenniri H., Alvarez-Puebla R. 2007. High-throughput screening flows along. Nat. Chem. Biol. 3, 247–249. ( 10.1038/nchembio0507-247) [DOI] [PubMed] [Google Scholar]
- Fernández-López C., Mateo-Mateo C., Álvarez-Puebla R. A., Pérez-Juste J., Pastoriza-Santos I., Liz-Marzán L. M. 2009. Highly controlled silica coating of PEG-capped metal nanoparticles and preparation of SERS-encoded particles. Langmuir 25, 13 894–13 899. ( 10.1021/la9016454) [DOI] [PubMed] [Google Scholar]
- Freeman R. G., Doering W. E., Walton I. D., Penn S. G., Davis G., Wong F., Natan M. J. 2005. Detection of biomolecules using nanoparticle surface enhanced Raman scattering tags. Proc. SPIE 5705, 114–122. ( 10.1117/12.591114) [DOI] [Google Scholar]
- Garcia de Abajo F. J. 2007. Colloquium: Light scattering by particle and hole arrays. Rev. Mod. Phys. 79, 1267–1290. ( 10.1103/RevModPhys.79.1267) [DOI] [Google Scholar]
- Garcia de Abajo F. J. 2010. Optical excitations in electron microscopy. Rev. Mod. Phys. 82, 209–275. ( 10.1103/RevModPhys.82.209) [DOI] [Google Scholar]
- Gellner M., Kömpe K., Schlücker S. 2009. Multiplexing with SERS labels using mixed SAMs of Raman reporter molecules. Anal. Bioanal. Chem. 394, 1839–1844. ( 10.1007/s00216-009-2868-8) [DOI] [PubMed] [Google Scholar]
- Genov D. A., Sarychev A. K., Shalaev V. M., Wei A. 2004. Resonant field enhancements from metal nanoparticle arrays. Nano Lett. 4, 153–158. ( 10.1021/nl0343710) [DOI] [Google Scholar]
- Goulet P. J. G., dos Santos D. S., Alvarez-Puebla R. N. A., Oliveira O. N., Aroca R. F. 2005. Surface-enhanced Raman scattering on dendrimer/metallic nanoparticle layer-by-layer film substrates. Langmuir 21, 5576–5581. ( 10.1021/la050202e) [DOI] [PubMed] [Google Scholar]
- Graham D., Faulds K., Smith W. E. 2006. Biosensing using silver nanoparticles and surface enhanced resonance Raman scattering. Chem. Commun. 42, 4363–4371. ( 10.1039/b60794k) [DOI] [PubMed] [Google Scholar]
- Grzelczak M., Perez-Juste J., Mulvaney P., Liz-Marzan L. M. 2008. Shape control in gold nanoparticle synthesis. Chem. Soc. Rev. 37, 1783–1791. ( 10.1039/b711490g) [DOI] [PubMed] [Google Scholar]
- Guerrero-Martínez A., Pérez-Juste J., Liz-Marzán L. M. 2010. Recent progress on silica coating of nanoparticles and related nanomaterials. Adv. Mater. 22, 1182–1195. ( 10.1002/adma.200901263) [DOI] [PubMed] [Google Scholar]
- Guerrini L., Garcia-Ramos J. V., Domingo C., Sanchez-Cortes S. 2006. Functionalization of Ag nanoparticles with dithiocarbamate calix[4]arene as an effective supramolecular host for the surface-enhanced Raman scattering detection of polycyclic aromatic hydrocarbons. Langmuir 22, 10 924–10 926. ( 10.1021/La062266a) [DOI] [PubMed] [Google Scholar]
- Guerrini L., Garcia-Ramos J. V., Domingo C., Sanchez-Cortes S. 2009a Nanosensors based on violagen functionalized silver nanoparticles: few molecules surface-enhanced Raman detection of polycyclic aromatic hydrocarbons in interparticle hot spots. Anal. Chem. 81, 1418–1425. ( 10.1021/ac8021746) [DOI] [PubMed] [Google Scholar]
- Guerrini L., Garcia-Ramos J. V., Domingo C., Sanchez-Cortes S. 2009b Self-assembly of dithiocarbamate calixarene on Ag nanoparticles and its application in the fabrication of surface-enhanced Raman scattering based nanosensors. Phys. Chem. Chem. Phys. 11, 1787–1793. ( 10.1039/b812811a) [DOI] [PubMed] [Google Scholar]
- Guicheteau J., Christesen S., Emge D., Tripathi A. In press Bacterial mixture identification using Raman and surface-enhanced Raman chemical imaging. J. Raman Spectrosc. ( 10.1002/jrs.2601) [DOI] [Google Scholar]
- Habuchi S., Cotlet M., Gronheid R., Dirix G., Michiels J., Vanderleyden J., De Schryver F. C., Hofkens J. 2003. Single-molecule surface enhanced resonance Raman spectroscopy of the enhanced green fluorescent protein. J. Am. Chem. Soc. 125, 8446–8447. ( 10.1021/ja0353311) [DOI] [PubMed] [Google Scholar]
- Han M., Gao X., Su J. Z., Nie S. 2001. Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nat. Biotech. 19, 631–635. ( 10.1038/90228) [DOI] [PubMed] [Google Scholar]
- Han X. X., Jia H. Y., Wang Y. F., Lu Z. C., Wang C. X., Xu W. Q., Zhao B., Ozaki Y. 2008. Analytical technique for label-free multi-protein detection based on Western blot and surface-enhanced Raman scattering. Anal. Chem. 80, 2799–2804. ( 10.1021/Ac702390u) [DOI] [PubMed] [Google Scholar]
- Huang G. G., Han X. X., Hossain M. K., Ozaki Y. 2009. Development of a heat-induced surface-enhanced Raman scattering sensing method for rapid detection of glutathione in aqueous solutions. Anal. Chem. 81, 5881–5888. ( 10.1021/Ac900392s) [DOI] [PubMed] [Google Scholar]
- Iafisco M., Palazzo B., Falini G., Di Foggia M., Bonora S., Nicolis S., Casella L., Roveri N. 2008. Adsorption and conformational change of myoglobin on biomimetic hydroxyapatite nanocrystals functionalized with alendronate. Langmuir 24, 4924–4930. ( 10.1021/la703381h) [DOI] [PubMed] [Google Scholar]
- Jana N. R., Pal T. 2007. Anisotropic metal nanoparticles for use as surface-enhanced Raman substrates. Adv. Mater. 19, 1761–1763. ( 10.1002/adma.200601749) [DOI] [Google Scholar]
- Jarvis R. M., Goodacre R. 2008. Characterisation and identification of bacteria using SERS. Chem. Soc. Rev. 37, 931–936. ( 10.1039/B705973f) [DOI] [PubMed] [Google Scholar]
- Jiang C., Tsukruk V. V. 2006. Freestanding nanostructures via layer-by-layer assembly. Adv. Mater. 18, 829–840. ( 10.1002/adma.200502444) [DOI] [Google Scholar]
- Jones C. L., Bantz K. C., Haynes C. L. 2009. Partition layer-modified substrates for reversible surface-enhanced Raman scattering detection of polycyclic aromatic hydrocarbons. Anal. Bioanal. Chem. 394, 303–311. ( 10.1007/s00216-009-2701-4) [DOI] [PubMed] [Google Scholar]
- Kennedy D. C., Tay L.-L., Lyn R. K., Rouleau Y., Hulse J., Pezacki J. P. 2009. Nanoscale aggregation of cellular-adrenergic receptors measured by plasmonic interactions of functionalized nanoparticles. ACS Nano 3, 2329–2339. ( 10.1021/nn900488u) [DOI] [PubMed] [Google Scholar]
- Kneipp K. 2007. Surface-enhanced Raman scattering. Phys. Today 60, 40–46. ( 10.1063/1.2812122) [DOI] [Google Scholar]
- Kneipp K., Wang Y., Kneipp H., Perelman L. T., Itzkan I., Dasari R. R., Feld M. S. 1997. Single molecule detection using surface-enhanced Raman scattering (SERS). Phys. Rev. Lett. 78, 1667–1670. ( 10.1103/PhysRevLett.78.1667) [DOI] [Google Scholar]
- Kneipp J., Kneipp H., McLaughlin M., Brown D., Kneipp K. 2006. In vivo molecular probing of cellular comportments with gold nanoparticles and nanoaggregates. Nano Lett. 6, 2225–2231. ( 10.1021/nl061517x) [DOI] [PubMed] [Google Scholar]
- Kneipp J., Kneipp H., Kneipp K. 2008. SERS—a single-molecule and nanoscale tool for bioanalytics. Chem. Soc. Rev. 37, 1052–1060. ( 10.1039/b708459p) [DOI] [PubMed] [Google Scholar]
- Ko H., Tsukruk V. V. 2008. Nanoparticle-decorated nanocanals for surface-enhanced Raman scattering. Small 4, 1980–1984. ( 10.1002/smll.200800301) [DOI] [PubMed] [Google Scholar]
- Ko H., Singamaneni S., Tsukruk V. V. 2008. Nanostructured surfaces and assemblies as SERS media. Small 4, 1576–1599. ( 10.1002/smll.200800337) [DOI] [PubMed] [Google Scholar]
- Laurence T. A., Braun G., Talley C., Schwartzberg A., Moskovits M., Reich N., Huser T. 2009. Rapid, solution-based characterization of optimized SERS nanoparticle substrates. J. Am. Chem. Soc. 131, 162–169. ( 10.1021/ja806236k) [DOI] [PubMed] [Google Scholar]
- Li W., Camargo P. H. C., Lu X., Xia Y. 2009. Dimers of silver nanospheres: facile synthesis and their use as hot spots for surface-enhanced Raman scattering. Nano Lett. 9, 485–490. ( 10.1021/nl803621x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Pedano M. L., Chang S. H., Mirkin C. A., Schatz G. C. In press Gap structure effects on surface-enhanced Raman scattering intensities for gold gapped rods. Nano Lett. ( 10.1021/nl100099g) [DOI] [PubMed] [Google Scholar]
- Liz-Marzan L. M., Giersig M., Mulvaney P. 1996. Synthesis of nanosized gold–silica core-shell particles. Langmuir 12, 4329–4335. ( 10.1021/la9601871) [DOI] [Google Scholar]
- Lu Y., Liu G. L., Lee L. P. 2004. High-density silver nanoparticle film with temperature-controllable interparticle spacing for a tunable surface enhanced Raman scattering substrate. Nano Lett. 5, 5–9. ( 10.1021/nl048965u) [DOI] [PubMed] [Google Scholar]
- Lu X., Rycenga M., Skrabalak S. E., Wiley B., Xia Y. 2009. Chemical synthesis of novel plasmonic nanoparticles. Annu. Rev. Phys. Chem. 60, 167–192. ( 10.1146/annurev.physchem.040808.090434) [DOI] [PubMed] [Google Scholar]
- Lutz B. R., Dentinger C. E., Nguyen L. N., Sun L., Zhang J., Allen A. N., Chan S., Knudsen B. S. 2008. Spectral analysis of multiplex Raman probe signatures. ACS Nano 2, 2306–2314. ( 10.1021/nn800243g) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markel V. A., Shalaev V. M., Zhang P., Huynh W., Tay L., Haslett T. L., Moskovits M. 1999. Near-field optical spectroscopy of individual surface-plasmon modes in colloid clusters. Phys. Rev. B 59, 10903 ( 10.1103/PhysRevB.59.10903) [DOI] [Google Scholar]
- Moskovits M. 2005. Surface-enhanced Raman spectroscopy: a brief retrospective. J. Raman Spectrosc. 36, 485–496. ( 10.1002/jrs.1362) [DOI] [Google Scholar]
- Moskovits M., Suh J. S. 1984. Surface selection-rules for surface-enhanced Raman-spectroscopy—calculations and application to the surface-enhanced Raman spectrum of phthalazine on silver. J. Phys. Chem. 88, 5526–5530. ( 10.1021/j150667a013) [DOI] [Google Scholar]
- Mulvaney S. P., Musick M. D., Keating C. D., Natan M. J. 2003. Glass-coated, analyte-tagged nanoparticles. A new tagging system based on detection with surface-enhanced Raman scattering. Langmuir 19, 4784–4790. ( 10.1021/la026706j) [DOI] [Google Scholar]
- Murphy C. J., Gole A. M., Stone J. W., Sisco P. N., Alkilany A. M., Goldsmith E. C., Baxter S. C. 2008. Gold nanoparticles in biology: beyond toxicity to cellular imaging. Acc. Chem. Res. 41, 1721–1730. ( 10.1021/ar800035u) [DOI] [PubMed] [Google Scholar]
- Myroshnychenko V., Rodriguez-Fernandez J., Pastoriza-Santos I., Funston A. M., Novo C., Liz-Marzan L. M., Garcia de Abajo F. J. 2008. Modelling the optical response of gold nanoparticles. Chem. Soc. Rev. 37, 1792–1805. ( 10.1039/b711486a) [DOI] [PubMed] [Google Scholar]
- Nabiev I. R., Morjani H., Manfait M. 1991. Selective analysis of antitumor drug interaction with living cancer cells as probed by surface-enhanced Raman spectroscopy. Eur. Biophys. J. 19, 311–316. ( 10.1007/BF00183320) [DOI] [PubMed] [Google Scholar]
- Nelayah J., et al. 2007. Mapping surface plasmons on a single metallic nanoparticle. Nat. Phys. 3, 348–353. ( 10.1038/nphys575) [DOI] [Google Scholar]
- Ni J., Lipert R. J., Dawson G. B., Porter M. D. 1999. Immunoassay readout method using extrinsic Raman labels adsorbed on immunogold colloids. Anal. Chem. 71, 4903–4908. ( 10.1021/ac990616a) [DOI] [PubMed] [Google Scholar]
- Njoki P. N., Lim I. I. S., Mott D., Park H. Y., Khan B., Mishra S., Sujakumar R., Luo J., Zhong C. J. 2007. Size correlation of optical and spectroscopic properties for gold nanoparticles. J. Phys. Chem. C 111, 14 664–14 669. ( 10.1021/Jp074902z) [DOI] [Google Scholar]
- Ochsenkühn M. A., Jess P. R. T., Stoquert H., Dholakia K., Campbell C. J. 2009. Nanoshells for surface-enhanced Raman spectroscopy in eukaryotic cells: cellular response and sensor development. ACS Nano 3, 3613–3621. ( 10.1021/nn900681c) [DOI] [PubMed] [Google Scholar]
- Orenforff C. J., Gearheart L., Jana N. R., Murphy C. J. 2005. Aspect ratio dependence on surface enhanced Raman scattering using silver and gold nanorod substrates. Phys. Chem. Chem. Phys. 8, 165–170. ( 10.1039/b512573a) [DOI] [PubMed] [Google Scholar]
- Pallaoro A., Braun G. B., Reich N. O., Moskovits M. 2010. Mapping local pH in live cells using encapsulated fluorescent SERS nanotags. Small 6, 618–622. ( 10.1002/smll.200901893) [DOI] [PubMed] [Google Scholar]
- Pastoriza-Santos I., Liz-Marzan L. M. 2009. N, N-dimethylformamide as a reaction medium for metal nanoparticle synthesis. Adv. Funct. Mater. 19, 679–688. ( 10.1002/adfm.200801566) [DOI] [Google Scholar]
- Patel I. S., Premasiri W. R., Moir D. T., Ziegler L. D. 2008. Barcoding bacterial cells: a SERS-based methodology for pathogen identification. J. Raman Spectrosc. 39, 1660–1672. ( 10.1002/jrs.2064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazos-Perez N., Barbosa S., Rodriguez-Lorenzo L., Aldeanueva-Potel P., Perez-Juste J., Pastoriza-Santos I., Alvarez-Puebla R. A., Liz-Marzan L. M. 2010a Growth of sharp tips on gold nanowires leads to increased surface-enhanced Raman scattering activity. J. Phys. Chem. Lett. 1, 24–27. ( 10.1021/jz900004h) [DOI] [PubMed] [Google Scholar]
- Pazos-Perez N., Rodriguez-Gonzalez B., Hilgendorff M., Giersig M., Liz-Marzan L. M. 2010b Gold encapsulation of star-shaped FePt nanoparticles. J. Mater. Chem. 20, 61–64. ( 10.1039/b911175a) [DOI] [Google Scholar]
- Pearman W. F., Fountain A. W. 2006. Classification of chemical and biological warfare agent simulants by surface-enhanced Raman spectroscopy and multivariate statistical techniques. Appl. Spectrosc. 60, 356–365. ( 10.1366/000370206776593744) [DOI] [PubMed] [Google Scholar]
- Petry R., Schmitt M., Popp J. 2003. Raman spectroscopy—a prospective tool in the life sciences. ChemPhysChem 4, 14–30. ( 10.1002/cphc.200390004) [DOI] [PubMed] [Google Scholar]
- Pieczonka N. P., Aroca R. F. 2008. Single molecule analysis by surfaced-enhanced Raman scattering. Chem. Soc. Rev. 37, 946–954. ( 10.1039/b709739p) [DOI] [PubMed] [Google Scholar]
- Piner R. D., Zhu J., Xu F., Hong S., Mirkin C. A. 1999. Dip-pen lithography. Science 283, 661–663. ( 10.1126/science.283.5402.661) [DOI] [PubMed] [Google Scholar]
- Pinzaru S. C., Andronie L. M., Domsa I., Cozar O., Astilean S. 2008. Bridging biomolecules with nanoparticles: surface-enhanced Raman scattering from colon carcinoma and normal tissue. J. Raman Spectrosc. 39, 331–334. ( 10.1002/jrs.1907) [DOI] [Google Scholar]
- Podsiadlo P., et al. 2008. Exponential growth of LBL films with incorporated inorganic sheets. Nano Lett. 8, 1762–1770. ( 10.1021/nl8011648) [DOI] [PubMed] [Google Scholar]
- Pregibon D. C., Toner M., Doyle P. S. 2007. Multifunctional encoded particles for high-throughput biomolecule analysis. Science 315, 1393–1396. ( 10.1126/science.1134929) [DOI] [PubMed] [Google Scholar]
- Premasiri W. R., Moir D. T., Klempner M. S., Krieger N., Jones G., Ziegler L. D. 2005. Characterization of the surface enhanced Raman scattering (SERS) of bacteria. J. Phys. Chem. B 109, 312–320. ( 10.1021/jp040442n) [DOI] [PubMed] [Google Scholar]
- Qian X., et al. 2008. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat. Biotech. 26, 83–90. ( 10.1038/nbt1377) [DOI] [PubMed] [Google Scholar]
- Qian X., Li J., Nie S. 2009. Stimuli-responsive SERS nanoparticles: conformational control of plasmonic coupling and surface Raman enhancement. J. Am. Chem. Soc. 131, 7540–7541. ( 10.1021/ja902226z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L., Banholzer M. J., Millstone J. E., Mirkin C. A. 2007. Nanodisk codes. Nano Lett. 7, 3849–3853. ( 10.1021/nl072606s) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raez J., Blais D. R., Zhang Y., Alvarez-Puebla R. A., Bravo-Vasquez J. P., Pezacki J. P., Fenniri H. 2007. Spectroscopically encoded microspheres for antigen biosensing. Langmuir 23, 6482–6485. ( 10.1021/la700701x) [DOI] [PubMed] [Google Scholar]
- Rodriguez-Fernandez J., Funston A. M., Perez-Juste J., Alvarez-Puebla R. A., Liz-Marzan L. M., Mulvaney P. 2009. The effect of surface roughness on the plasmonic response of individual sub-micron gold spheres. Phys. Chem. Chem. Phys. 11, 5909–5914. ( 10.1039/b905200n) [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lorenzo L., Alvarez-Puebla R. A., Pastoriza-Santos I., Mazzucco S., Stephan O., Kociak M., Liz-Marzan L. M., de Abajo F. J. G. 2009. Zeptomol detection through controlled ultrasensitive surface-enhanced Raman scattering. J. Am. Chem. Soc. 131, 4616–4618. ( 10.1021/Ja809418t) [DOI] [PubMed] [Google Scholar]
- Rodriguez-Lorenzo L., Alvarez-Puebla R. A., Garcia de Abajo F. J., Liz-Marzan L. M. 2010. Surface enhanced raman scattering using star-shape gold colloidal nanoparticles. J. Phys. Chem. C 114, 7336–7340 ( 10.1021/jp909253w) [DOI] [Google Scholar]
- Rohr T. E., Cotton T., Fan N., Tarcha P. J. 1989. Immunoassay employing surface-enhanced Raman spectroscopy. Anal. Biochem. 182, 388–398. ( 10.1016/0003-2697(89)90613-1) [DOI] [PubMed] [Google Scholar]
- Rycenga M., Camargo P. C. H., Li W., Moran C. H., Xia Y. 2010. Understanding the SERS effects of single silver nanoparticles and their dimers, one at a time. J. Phys. Chem. Lett. 1, 696–703. ( 10.1021/jz900286a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sägmüller B., Schwarze B., Brehm G., Schneider S. 2001. Application of SERS spectroscopy to the identification of (3,4-methylenedioxy) amphetamine in forensic samples utilizing matrix stabilized silver halides. Analyst 126, 2066–2071. ( 10.1039/b105321n) [DOI] [PubMed] [Google Scholar]
- Sanchez-Iglesias A., Aldeanueva-Potel P., Ni W., Perez-Juste J., Pastoriza-Santos I., Alvarez-Puebla R., Mbenkum N. B., Liz-Marzan L. M. 2010. Chemical seeded growth of Ag nanoparticles arrays and their applications as reproducible SERS substrates. Nano Today 5, 21–27. ( 10.1016/j.nantod.2010.01.002) [DOI] [Google Scholar]
- Sanles-Sobrido M., Exner W., Rodríguez-Lorenzo L., Rodríguez-González B., Correa-Duarte M. A., Álvarez-Puebla R. A., Liz-Marzán L. M. 2009a Design of SERS-encoded, submicron, hollow particles through confined growth of encapsulated metal nanoparticles. J. Am. Chem. Soc. 131, 2699–2705. ( 10.1021/ja8088444) [DOI] [PubMed] [Google Scholar]
- Sanles-Sobrido M., Rodríguez-Lorenzo L., Lorenzo-Abalde S., González-Fernández A., Correa-Duarte M. A., Álvarez-Puebla R. A., Liz-Marzán L. M. 2009b Label-free SERS detection of relevant bioanalytes on silver-coated carbon nanotubes: the case of cocaine. Nanoscale 1, 153–158. ( 10.1039/b9nr00059c) [DOI] [PubMed] [Google Scholar]
- Sayin I., Kahraman M., Sahin F., Yurdakul D., Culha M. 2009. Characterization of yeast species using surface-enhanced Raman scattering. Appl. Spectrosc. 63, 1276–1282. ( 10.1366/000370209789806849) [DOI] [PubMed] [Google Scholar]
- Seballos L., Zhang J. Z., Sutphen R. 2005. Surface-enhanced Raman scattering detection of lysophosphatidic acid. Anal. Bioanal. Chem. 383, 763–767. ( 10.1007/s00216-005-0097-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebba D. S., Watson D. A., Nolan J. P. 2009. High throughput single nanoparticle spectroscopy. ACS Nano 3, 1477–1484. ( 10.1021/nn9003346) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seney C. S., Gutzman B. M., Goddard R. H. 2009. Correlation of size and surface-enhanced Raman scattering activity of optical and spectroscopic properties for silver nanoparticles. J. Phys. Chem. C 113, 74–80. ( 10.1021/Jp805698e) [DOI] [Google Scholar]
- Sengupta A., Thai C. K., Sastry M. S. R., Matthaei J. F., Schwartz D. T., Davis E. J., Baneyx F. 2008. A genetic approach for controlling the binding and orientation of proteins on nanoparticles. Langmuir 24, 2000–2008. ( 10.1021/la702079e) [DOI] [PubMed] [Google Scholar]
- Sepúlveda B., Angelomé P. C., Lechuga L. M., Liz-Marzan L. M. 2009. LSPR-based nanobiosensors. Nano Today 4, 244–251. ( 10.1016/j.nantod.2009.04.001) [DOI] [Google Scholar]
- Sha M. Y., Xu H., Natan M. J., Cromer R. 2008. Surface-enhanced Raman scattering tags for rapid and homogeneous detection of circulating tumor cells in the presence of human whole blood. J. Am. Chem. Soc. 130, 17 214–17 215. ( 10.1021/ja804494m) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmukh S., Jones L., Driskell J., Zhao Y. P., Dluhy R., Tripp R. A. 2006. Rapid and sensitive detection of respiratory virus molecular signatures using a silver nanorod array SERS substrate. Nano Lett. 6, 2630–2636. ( 10.1021/nl061666f) [DOI] [PubMed] [Google Scholar]
- Singhal K., Kalkan A. K. 2009. Surface-enhanced Raman scattering captures conformational changes of single photoactive yellow protein molecules under photoexcitation. J. Am. Chem. Soc. 132, 429–431. ( 10.1021/ja9028704) [DOI] [PubMed] [Google Scholar]
- Spiro T. G., Czernuszewicz R. S. 1995. Resonance Raman-spectroscopy of metalloproteins. Biochem. Spectrosc. 246, 416–460. ( 10.1016/0076-6879(95)46020-9) [DOI] [PubMed] [Google Scholar]
- Spuch-Calvar M., Rodriguez-Lorenzo L., Morales M. P., Alvarez-Puebla R. A., Liz-Marzan L. M. 2009. Bifunctional nanocomposites with long-term stability as SERS optical accumulators for ultrasensitive analysis. J. Phys. Chem. C 113, 3373–3377. ( 10.1021/Jp810470n) [DOI] [Google Scholar]
- Srivastava S., Ball V., Podsiadlo P., Lee J., Ho P., Kotov N. A. 2008. Reversible loading and unloading of nanoparticles in exponentially growing polyelectrolyte LBL films. J. Am. Chem. Soc. 130, 3748–3749. ( 10.1021/ja7110288) [DOI] [PubMed] [Google Scholar]
- Stiles P. L., Dieringer J. A., Shah N. C., Van Duyne R. R. 2008. Surface-enhanced Raman spectroscopy. Annu. Rev. Anal. Chem. 1, 601–626. ( 10.1146/annurev.anchem.1.031207.112814) [DOI] [PubMed] [Google Scholar]
- Stöber W., Fink A., Bohn E. 1968. Controlled growth of monodisperse silica spheres in the micron size range. J. Colloid Interface Sci. 26, 62–69. ( 10.1016/0021-9797(68)90272-5) [DOI] [Google Scholar]
- Stoermer R. L., Cederquist K. B., McFarland S. K., Sha M. Y., Penn S. G., Keating C. D. 2006. Coupling molecular beacons to barcoded metal nanowires for multiplexed, sealed chamber DNA bioassays. J. Am. Chem. Soc. 128, 16892–16903. ( 10.1021/ja0658261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelau K. K., Kretschmer R., Möller R., Fritzsche W., Popp J. 2010. SERS as tool for the analysis of DNA-chips in a microfluidic platform. Anal. Bioanal. Chem. 396, 1381–1384. ( 10.1007/s00216-009-3374-8) [DOI] [PubMed] [Google Scholar]
- Sujith A., Itoh T., Abe H., Yoshida K. I., Kiran M. S., Biju V., Ishikawa M. 2009. Imaging the cell wall of living single yeast cells using surface-enhanced Raman spectroscopy. Anal. Bioanal. Chem. 394, 1803–1809. ( 10.1007/s00216-009-2883-9) [DOI] [PubMed] [Google Scholar]
- Taladriz-Blanco P., Rodriguez-Lorenzo L., Sanles-Sobrido M., Herves P., Correa-Duarte M. A., Alvarez-Puebla R. A., Liz-Marzan L. M. 2009. SERS study of the controllable release of nitric oxide from aromatic nitrosothiols on bimetallic, bifunctional nanoparticles supported on carbon nanotubes. ACS Appl. Mater. Interfaces 1, 56–59. ( 10.1021/Am800141j) [DOI] [PubMed] [Google Scholar]
- Talley C. E., Jackson J. B., Oubre C., Grady N. K., Hollars C. W., Lane S. M., Huser T. R., Nordlander P., Halas N. J. 2005. Surface-enhanced Raman scattering from individual Au nanoparticles and nanoparticle dimer substrates. Nano Lett. 5, 1569–1574. ( 10.1021/Nl050928v) [DOI] [PubMed] [Google Scholar]
- Tang H.-W., Yang X. B., Kirkham J., Smith D. A. 2007. Probing intrinsic and extrinsic components in single osteosarcoma cells by near-infrared surface-enhanced Raman scattering. Anal. Chem. 79, 3646–3653. ( 10.1021/ac062362g) [DOI] [PubMed] [Google Scholar]
- Tian Z. Q., Ren B., Wu D. Y. 2002. Surface-enhanced Raman scattering: from noble to transition metals and from rough surfaces to ordered nanostructures. J. Phys. Chem. B 106, 9463–9483. ( 10.1021/Jp0257449) [DOI] [Google Scholar]
- Tripp R. A., Dluhy R. A., Zhao Y. 2008. Novel nanostructures for SERS biosensing. Nano Today 3, 31–37. ( 10.1016/S1748-0132(08)70042-2) [DOI] [Google Scholar]
- Volpati D., Job A. E., Aroca R. F., Constantino C. J. L. 2008. Molecular and morphological characterization of bis benzimidazo perylene films and surface-enhanced phenomena. J. Phys. Chem. B 112, 3894–3902. ( 10.1021/jp077588h) [DOI] [PubMed] [Google Scholar]
- Wang G., Lim C., Chen L., Chon H., Choo J., Hong J., deMello A. J. 2009. Surface-enhanced Raman scattering in nanoliter droplets: towards high-sensitivity detection of mercury (II) ions. Anal. Bioanal. Chem. 394, 1827–1832. ( 10.1007/s00216-009-2832-7) [DOI] [PubMed] [Google Scholar]
- Watson D. A., Brown L. O., Gaskill D. F., Naivar M., Graves S. W., Doorn S. K., Nolan J. P. 2008. A flow cytometer for the measurement of Raman spectra. Cytometry Part A 73A, 119–128. ( 10.1002/cyto.a.20520) [DOI] [PubMed] [Google Scholar]
- Wei F., Zhang D., Halas N. J., Hartgerink J. D. 2008. Aromatic amino acids providing characteristic motifs in the Raman and SERS spectroscopy of peptides. J. Phys. Chem. B 112, 9158–9164. ( 10.1021/jp8025732) [DOI] [PubMed] [Google Scholar]
- Wei Q., Song H.-M., Leonov A. P., Hale J. A., Oh D., Ong Q. K., Ritchie K., Wei A. 2009. Gyromagnetic imaging: dynamic optical contrast using gold nanostars with magnetic cores. J. Am. Chem. Soc. 131, 9728–9734. ( 10.1021/ja901562j) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willets K. A. 2009. Surface-enhanced Raman scattering (SERS) for probing internal cellular structure and dynamics. Anal. Bioanal. Chem. 394, 85–94. ( 10.1007/s00216-009-2682-3) [DOI] [PubMed] [Google Scholar]
- Xia Y., Rogers J. A., Paul K. E., Whitesides G. M. 1999. Unconventional methods for fabricating and patterning nanostructures. Chem. Rev. 99, 1823–1848. ( 10.1021/cr980002q) [DOI] [PubMed] [Google Scholar]
- Xue M. Q., Zhang Z., Zhu N., Wang F. F., Zhao X. S., Cao T. B. 2009. Transfer printing of metal nanoparticles with controllable dimensions, placement, and reproducible surface-enhanced Raman scattering effects. Langmuir 25, 4347–4351. ( 10.1021/La900462f) [DOI] [PubMed] [Google Scholar]
- Yaffe N. R., Ingram A., Graham D., Blanch E. W. In press A multi-component optimisation of experimental parameters for maximising SERS enhancements. J. Raman Spectrosc. ( 10.1002/jrs.2495) [DOI] [Google Scholar]
- Yea K.-H., Lee S., Kyong J. B., Choo J., Lee E. K., Joo S.-W., Lee S. 2005. Ultra-sensitive trace analysis of cyanide water pollutant in a PDMS microfluidic channel using surface-enhanced Raman spectroscopy. Analyst 130, 1009–1011. ( 10.1039/b501980j) [DOI] [PubMed] [Google Scholar]
- Zhao J., Pinchuk A. O., Mcmahon J. M., Li S., Ausman L. K., Atkinson A. L., Schatz G. C. 2008. Methods for describing the electromagnetic properties of silver and gold nanoparticles. Acc. Chem. Res. 41, 1710–1720. ( 10.1021/ar800028j) [DOI] [PubMed] [Google Scholar]
- Zhao Y., Newton J. N., Liu J., Wei A. 2009. Dithiocarbamate-coated SERS substrates: sensitivity gain by partial surface passivation. Langmuir 25, 13 833–13 839. ( 10.1021/la902087e) [DOI] [PubMed] [Google Scholar]