Abstract
The protein Eater plays an important role in microbial recognition and defensive phagocytosis in Drosophila melanogaster. We sequenced multiple alleles of the eater gene from an African and a North American population of D. melanogaster and found signatures of a partial selective sweep in North America that is localized around the second intron. This pattern is consistent with local adaptation to novel selective pressures during range expansion out of Africa. The North American sample is divided into two predominant haplotype groups, and the putatively selected haplotype is associated with a significantly higher gene expression level, suggesting that gene regulation is a possible target of selection. The eater alleles contain from 22 to 40 repeat units that are characterized by the presence of a cysteine-rich NIM motif. NIM repeats in the structural stalk of the protein exhibit concerted evolution as a function of physical location in the repeat array. Several NIM repeats within eater have previously been implicated in binding to microbial ligands, a function which in principle might subject them to special evolutionary pressures. However, we find no evidence of elevated positive selection on these pathogen-interacting units. Our study presents an instance where gene expression rather than protein structure is thought to drive the adaptive evolution of a pathogen recognition molecule in the immune system.
Keywords: Drosophila, immunity, phagocytosis, population genetics, evolution, receptor
Introduction
All living organisms have a vital need to protect themselves against pathogenesis, and hosts are thus constantly being forced to adapt their defenses to novel and reciprocally evolving pathogens and parasites (Ebert 2000). Population genetic analyses can answer questions about the role of local adaptation in driving rapid evolution and the geographic distribution of selected alleles, as well as help determine the relative importance of selection on standing genetic variation versus on novel variants introduced by mutation. Additionally, population geneticists studying host–pathogen relationships can localize the specific targets of selection within proteins and determine whether these correspond to domains that interact directly with pathogens. In the present paper, we address these questions with respect to the evolution of the eater gene of Drosophila melanogaster.
The gene eater encodes a recognition receptor that is critical for defensive phagocytosis (Kocks et al. 2005), an important first line of protection against invading microbes. In D. melanogaster, eater is expressed solely in hemocytes and is thought to be a cell–surface–bound molecule that binds to microbial compounds and stimulates phagocytosis (Kocks et al. 2005). Ablation of this single gene with RNAi knockdown can decrease phagocytosis by 55–70% (Kocks et al. 2005). eater is part of the recently described nimrod superfamily of cellular recognition molecules that also includes multiple nimrod homologues and draper (Kocks et al. 2005; Kurucz et al. 2007; Somogyi et al. 2008). Proteins in the nimrod superfamily all have similar compositions, each containing a signal peptide, a CCxGY amino acid motif, and at least one cysteine-rich NIM domain (Somogyi et al. 2008). NIM domains are defined by a consensus sequence motif (CxPxCxxxCxNGxCxxPxxCxCxxGY), which is closely related to the epidermal growth factor consensus motif (xxxxCx2–7Cx1–4(G/A)xCx1–13ttaxCx-CxxGax1–6GxxCx) (Kurucz et al. 2007). Genes in the nimrod superfamily are found in syntenic clusters in D. melanogaster as well as in other Drosophila species, the honey bee (Apis mellifera), a mosquito (Anopheles gambiae), and the red flour beetle (Tribolium castaneum) (Kurucz et al. 2007; Somogyi et al. 2008).
NIM units occur as tandem repeats in some members of the nimrod superfamily, including eater. Repeated motifs of highly similar sequence often exhibit concerted evolution due to mispairing and unequal crossing over between homologous chromosomes and to gene conversion between nonhomologous repeats. This type of evolution results in repeat arrays where paralogous repeat units are more similar to each other within species than they are to homologous units among species (Charlesworth et al. 1994). Of genes in the nimrod superfamily, eater is the only member whose NIM repeats show evidence of concerted evolution (Somogyi et al. 2008). NIM repeats in the interior of the gene appear to be evolving concertedly (Somogyi et al. 2008) and are thought to provide a structural “stalk” between the microbe binding units and the hemocyte cell membrane (Kocks et al. 2005). The first four NIM repeat units in eater, which have been shown to be necessary for microbial binding (Kocks et al. 2005), show no signs of concerted evolution (Somogyi et al. 2008).
In a molecular evolutionary comparison among Drosophila species, eater and three nimrod family genes were found to be evolving under positive selection (Sackton et al. 2007). In one nimrod gene, nimC1, the positively selected sites are clustered within putative microbial binding domains, which suggests that pathogen interactions drive this rapid evolution. In contrast, adaptive mutations in eater are scattered throughout the gene, including outside domains known to interact directly with pathogens (Sackton et al. 2007). Selective pressures on the immune system are geographically variable, corresponding to heterogeneity in pathogen identity or abundance and other environmental factors. Immune system genes therefore may show evidence of local adaptation that can be detected with population genetic statistics. For instance, immune system genes display elevated differences in allele frequencies among populations relative to the genome average (Ryan et al. 2006; McEvoy et al. 2009). Recent selection can also be detected by examining patterns of genetic variation within populations. Strong positive selection leads to a rapid rise in the frequency of an adaptive mutation, incidentally dragging neutral variants linked to the target of selection upward in frequency. This leads to excess linkage disequilibrium (Kelly 1997; Sabeti et al. 2002), decreased nucleotide diversity (Maynard Smith and Haigh 1974), and too many high- and low-frequency polymorphisms (Tajima 1989; Fu 1997; Fay and Wu 2000) relative to expectations under selective neutrality. Analyses of these properties can easily be applied to coding and noncoding regions, allowing us to detect selection on regulatory gene regions. We can potentially also identify the specific trait on which selection acts by linking genetic diversity patterns and phenotypes.
In the current work, we have sequenced the complete upstream and nonrepetitive coding region of eater in a North American and an African population of D. melanogaster. We find that both populations harbor substantial polymorphism in the number of NIM repeats and therefore for the overall size of the protein. We confirm the patterns of concerted evolution in NIM repeats that have been previously reported but also find evidence for varying degrees of concerted evolution between units. There is extensive linkage disequilibrium in the second intron of eater that extends through the upstream and 5′, nonrepetitive gene region in the North American population, with the major haplotypes at the second intron significantly associated with gene expression level. Additional analysis suggests that one of these haplotypes has recently risen to high frequency in North America, which we interpret to reflect adaptation of the immune response to the novel pathogen environment that was encountered after emigration from Africa.
Materials and Methods
Fly Strains
Drosophila melanogaster strains used for DNA sequence analysis in this study came from Zimbabwe or the United States. Strains ZW09, ZW139, ZW140, ZW142, ZW144, ZW149, ZW155, ZW184, ZW185, and ZW190 were originally collected in 2002 by J.W.O. Ballard from Victoria Falls, Zimbabwe. Strains I01, I03, I04, I06, I07, I13, I16, I17, I22, I23, I24, I26, I29, I31, I33, I34, I35, and I38 were originally collected in 2004 by E. M. Hill-Burns and B. P. Lazzaro from Ithaca (NY). Additional collections from China (Beijing, courtesy of X. Huang and R. Roush via A. G. Clark; Begun and Aquadro 1995), the Netherlands (Houten, courtesy of Z. Bochdanovits via A. G. Clark; Bochdanovits and de Jong 2003), Australia (Tasmania, courtesy of A. A. Hoffman, via A. G. Clark), and the United States (Athens and Blairsville, Georgia, courtesy of V. Corby-Harris and D. Promislow; Lazzaro et al. 2008) were used for polymerase chain reaction (PCR) to measure the size of eater. Each line was initiated by intercrossing the progeny of a single, field-inseminated female and has been maintained by mass sib mating in the laboratory since collection. The African lines in particular still segregate for residual heterozygosity.
eater is located on the right arm of chromosome 3 at cytological band 97E2. To isolate single eater alleles for sequencing, an individual male from each stock was crossed to virgin females from the deficiency line Df(3R)Tl-P, e1 ca1/TM3, Ser1 (Bloomington Drosophila Stock Center stock number 1910). Single male progeny from this cross with the genotype Df(3R)Tl-P, e1 ca1/+ were crossed to virgin females from the original deficiency line. Males and virgin females from the second cross that had the genotype Df(3R)Tl-P, e1 ca1/+ were crossed to each other to isolate a single wild-type allele from the original isofemale line along with the deficiency chromosome. Only flies that were either homozygous for a single wild-type allele or hemizygous over the deficiency were sequenced.
PCR and DNA Sequencing
PCR amplifications of genomic DNA were performed using iProof high-fidelity polymerase (BioRad) or Taq polymerase (New England Biolabs). iProof-derived products were prepared for sequencing using PCR purification columns (Invitrogen). Taq-derived products were prepared for sequencing using Exonuclease I (USB Corp.) and shrimp alkaline phosphatase (USB Corp.). PCR products were then directly sequenced. DNA sequences for the US and Zimbabwe populations were collected for all nonrepetitive eater coding regions, all introns, 5′ and 3′ untranslated regions, and an approximately 2 kb region upstream of the transcriptional start site. Complete sequence could not be obtained for some alleles with large numbers of repetitive internal repeats. In these cases, the length of the repetitive regions was determined by amplifying the repeat region using primers that anneal to the flanking nonrepetitive regions and sizing the products on 1% (1–2.4 kb) or 0.6% (>2.4 kb) agarose gels. This genotyping of repeat region length was done for all populations. All primers are available by request. Nucleotide sequences have been deposited in GenBank (HM165155–HM165182). Outgroup sequence were obtained from the reference genomes of D. simulans (Release 1.0) and D. yakuba (Release 2.0) (Begun et al. 2007).
DNA Sequence Analysis
DNA sequences were assembled in CodonCode Aligner (CodonCode Corp.). NIM repeat units were identified by the 26 amino acid consensus sequence CxPxCxxxCxNGxCxxPxxCxCxxGY (Somogyi et al. 2008). An alignment was built of NIM repeats using this conserved motif as in Kurucz et al. (2007) and Somogyi et al. (2008) because the nucleotides within this sequence could be aligned for all NIM repeat units from all sampled alleles and both outgroups. Alignments based on the NIM consensus sequence were used to build neighbor-joining unrooted trees. Trees were constructed in MEGA 4.0 (Tamura et al. 2007) using an amino acid model with a Poisson correction and uniform substitution rates among all sites. Five hundred bootstrap replicates were performed to indicate support of each node. In agreement with Somogyi et al. (2008), we will refer to a repeat as “independently evolving” if its sequence is found just once per individual allele and it is more closely related to homologous units in D. yakuba and D. simulans than to repeat units at nonhomologous positions within the D. melanogaster eater gene. Independently evolving units were numbered 1–11 (fig. 1).
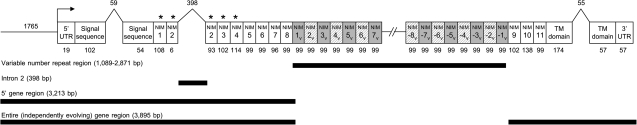
FIG. 1.
Gene structure and survey region. Variable number NIM repeat units were excluded when calculating population genetic statistics. The signal sequence, NIM 2, and the transmembrane (TM) domain are interrupted by introns, which are indicated by up-carats with the size of the intron (in base pairs) given above the carat. The numbers below sequence domains indicate their size in base pairs. The forward pointing arrow indicates the transcriptional start site. The 1765 base pair region immediately upstream of the transcriptional start site is indicated and includes the minimal enhancer region (Tokusumi et al. 2009). The boxes labeled 5′ and 3′ UTR are untranslated regions. “*” Indicates NIM repeats that have previously been implicated in microbial binding (Kocks et al. 2005). Dark lines below the gene schematic indicate various survey regions considered in different components of this article. Variable number repeat units are indicated with a ‘v’ subscript. “NIM 8–like” repeats are shown with gray and white diagonal lines, and “alternate” repeats are shown in gray. (NIM 8–like consensus motif: CKPICSxxCENGxCxAPEKCSCNGY, “alternate” consensus motif: CxxVCxxGCKNGFCxAPxKCSCxxxx.) Between 0 and 15 repeats were not sequenced in the interior of the gene (shown with hatched lines).
We compared the evolutionary patterns of the four NIM repeats that have previously been shown to be important for microbial binding (Kocks et al. 2005) to those where no such functional assignment has been made. We calculated KA, the rate of amino acid substitution, and KS, the rate of silent substitution for each NIM unit 1–11 independently (Nei and Gojobori 1986). Fixations were polarized using D. yakuba and D. simulans as outgroups and only fixations that occurred along the D. melanogaster lineage were considered. Wilcoxon rank sum tests were used to test for differences in substitution rates between microbial binding versus all other NIM repeats.
We calculated population genetic statistics on all gene regions that were not evolving concertedly. Nucleotide diversity, Tajima's D, and linkage disequilibrium were calculated using DnaSP v. 5.0 (Librado and Rozas 2009) and scripts written in the programming language R (R Development Core Team 2006). Nucleotide diversity (π) was measured both as the average pairwise differences between sequences per locus and per site with a Jukes–Cantor correction applied. Tajima's D (Tajima 1989) was calculated using all mutations. Tajima's D measures the difference between two different estimates of the population genetics parameter (4Neμ), one of which measures nucleotide diversity (θπ) and the other which relies on the number of segregating sites (θw). The f statistic, which is the nucleotide diversity in a putatively selected allele divided by the total nucleotide diversity (Macpherson et al. 2008), was calculated in R. Linkage disequilibrium was measured using the ZnS statistic (Kelly 1997), which is a standardized average of all calculations of the D statistic between all pairs of segregating sites. The standardized measure of linkage disequilibrium between pairs of sites, r2, was plotted using the LDheatmap package in R (Shin et al. 2006). Extended haplotype homozygosity (EHH), another measure of linkage disequilibrium, is the probability at a given position that two sampled alleles with the same predefined core genotype are identical by descent (Sabeti et al. 2002). EHH was calculated using a script written in R with the core genotype defined at the center of the second intron because linkage disequilibrium was most extreme in this region (see Results). We calculated ZnS, EHH, nucleotide diversity (π), Tajima's D, and f on the entire eater gene region (3,895 bp) with the variable number repeat units excluded to look for selection over the entire locus. We also calculated these statistics on the second intron (398 bp) because the most extreme values of the population genetic statistics should be near the site of selection. Lastly, we calculated the above set of statistics on the 5′ gene region (3,213 bp), which included the entire upstream region, 5′ untranslated region (UTR), NIM 1–8, and two introns because this is the region used for simulations (see Coalescent Simulations).
Coalescent Simulations
We used coalescent simulations run in the ms program (Hudson 2002) to build null distributions of our test statistics under various neutral demographic scenarios. Our empirically determined test statistics were then compared with the null distributions to test for deviations from neutrality that could be attributed to selection and to assess statistical significance of empirically observed patterns. Polymorphism data were simulated for a recombining, neutrally evolving locus of length 3,106 base pairs intended to represent the majority of the nonrepetitive coding and noncoding 5′ end of the gene with insertion or deletion events considered as single base pair mutations. The 682 base pairs at the 3′ end of the gene were not included in the simulations because recombination distance across the intervening repetitive region could not be accurately incorporated into the simulations. For these simulations, a fixed number of 86 segregating sites (our empirical observation at eater) was assumed. We also simulated patterns of polymorphism at the second intron, which was modeled as a recombining locus that was 398 base pairs long with 15 segregating sites. The local recombination rate was estimated using the D. melanogaster Recombination Rate Calculator (Singh et al. 2005) and was estimated to be 1.77 cM/Mbp for this locus. The effective population size was assumed to be 106, and the mutation rate was assumed to be 1.5 × 10−9/bp/generation (Li 1997).
Our simplest demographic scenario assumes a panmictic population of constant size. Our two other scenarios account for the bottleneck that the North American population underwent when it was founded from an ancestral African population (David and Capy 1988). The details of this bottleneck have recently been inferred in detail by two separate analyses of data sets from the Netherlands and East Africa (Li and Stephan 2006; Thornton and Andolfatto 2006). Previous work has shown that all non-African populations were derived from a single colonization event (Baudry et al. 2004; Schlötterer et al. 2006), so it is appropriate to apply the parameters inferred from these data to North American populations (Macpherson et al. 2008). The exact parameters of the bottleneck that were inferred differ between studies (Macpherson et al. 2008). Thornton and Andolfatto (2006) estimated three parameters of the bottleneck (referred to as the TA scenario from this point forward): the timing of the population size reduction (Tb), the timing of recovery (Tr), and the ratio of the population size during the bottleneck to the size before and after (Rb). The best estimate from their approximate Bayesian methods suggests that Tb was 16,000 years ago, Tr was 3,000 years ago, and that Rb was 0.029. Assuming 10 generations a year (Thornton and Andolfatto 2006), this corresponds to a Tb of 0.022 × 4Ne generations ago and a Tr of 0.0042 × 4Ne generations ago. Li and Stephan (2006) used a maximum likelihood procedure and estimated that Tb was 15,800 years (0.0367 × 4Ne generations ago) and lasted only a few hundred years (Tr equal to 0.0360 × 4Ne generations ago) and that Rb was 0.002. In addition, they estimate that previous to the bottleneck out of Africa, the African population underwent an expansion in population size. They estimated that the expansion (Te) occurred 60,000 years ago (0.1395 × 4Ne generations ago) and that the ratio of the expansion size to the current population size (Re) was 8.0. We will refer to these parameters as the LS scenario. Simulations under the LS scenario that incorporate this expansion of the ancestral population are a significantly better fit to overall genomic patterns of polymorphism than simulations under the TA scenario (Li and Stephan 2006), and an ancient expansion explains the excess of rare derived mutations that are observed within African populations.
Our eater sequences were divided into two distinct clades that we hypothesize may have adaptive significance, so we only retained simulations that matched the empirically observed topology (see Results). Specifically, we required that the final coalescence event occur at the span representing the second intron and divide the data into two clades of sizes 8 and 10, where the clade of size 8 represents an allele experiencing a partial selective sweep. In this way, we simulated coalescent trees with the same topology as seen in our data set and generated a distribution of each test statistic that would be expected for this given topology in the absence of selection. Population genetic statistics were calculated for each simulated data set with the entire population included, as well as separately with only individuals containing the putatively adaptive allele included. The distribution of each simulated test statistic was determined, and statistical significance was defined as the number of simulated data sets that had a value of the test statistic equal to or more extreme than that observed for eater. We conducted two-tailed tests on empirical estimates of eater from the entire population to test for deviations from neutrality. Empirical estimates at eater were considered significant if they fell into the 2.5% tails of the simulated distributions. We conducted one-tailed tests on the putatively adaptive haplotype group to test for a selective sweep in this class. Haplotype number, nucleotide diversity, Tajima's D, and f were considered extreme if the observed value was in the lower tail of the simulated values. ZnS was considered extreme if the observed value was in the upper tail of the simulated values.
Gene Expression
We sequenced a set of third chromosome substitution lines from a Pennsylvania, United States, collection of D. melanogaster (Fiumera et al. 2007) at the second intron of the eater locus and measured eater gene expression in the 19 lines that were a perfect match to either the “A” or “B” haplotype between base pairs 390 and 488 (see Results). These substitution lines had been previously backcrossed for eight generations to remove variation on the second, fourth, and sex chromosomes and thus only vary at the third chromosome (Fiumera et al. 2007). This should reduce the amount of transregulatory variation in eater expression. We designed primers specific to eater and to a housekeeping gene, rp49, which was used to control for variation in the efficiency of RNA extraction and cDNA synthesis. Transcript abundance was measured using Power SYBR Green (Applied BioSystems). Replicate samples of 10 males aged 3–5 days posteclosion were taken from each of two individual fly vials per line, RNA was extracted using a modified Trizol protocol (Invitrogen), and all quantitative PCR reactions were run in duplicate. This procedure was done twice, on separate days and for different fly generations. Significance of the “A” or “B” haplotype to predict eater transcript abundance was assessed using Proc Mixed in SAS (SAS Institute, Cary, NC) after accounting for the random effect of experiment day, line nested within genotype, vial nested within line and genotype, and random variance among replicate samples drawn from the same vial, as well as the fixed effect of the estimated abundance of rp49 transcripts.
Analysis of Variable Number Repeat Units
Repeat units between NIM 8 and NIM 9 have high sequence similarity at the nucleotide and amino acid level and have previously been shown to be evolving concertedly (Somogyi et al. 2008). We found individual D. melanogaster to be polymorphic for the number of repeats of this type (see Results). These variable number repeat units, which are 99 base pairs in length, cluster together into two types (“NIM 8–like” core consensus motif [78 base pairs/26 amino acids]: CKPICSxxCENGxCxAPEKCSCNGY; “alternate” core consensus motif: CxxVCxxGCKNGFCxAPxKCSCxxxx), which are always found in tandem (fig. 1; Somogyi et al. 2008). We labeled these units starting with the ones closest to NIM 8 or NIM 9 and counting inward toward the center of the array. Units immediately 3′ of NIM 8 were numbered starting with 1v and units immediate 5′ of NIM 9 were numbered starting with −1v (fig. 1).
We obtained an average of 1,393 bp (∼14 units) of sequence across the variable number repeat units per individual and measured the physical size of PCR products in this region for all individuals. The software package RST calc (Goodman 2008) was used to calculate genetic differentiation between populations. Because RST calc requires diploid samples and our fly lines were artificially made haploid, we randomly assigned alleles into diploid combinations to create artificial “genotypes.” Statistical significance of pairwise comparisons between populations was determined by permuting the sequenced alleles among subpopulations and recalculating RST 10,000 times to determine an empirical null distribution. To determine the statistical significance of the worldwide value of RST, we ran 10,000 bootstrap simulations to determine a confidence interval of our observed RST value. We measured genetic distance between all pairs of variable number repeat units using Kimura's two-parameter model in the ape package in R (Paradis et al. 2004).
Results
Summary Population Genetic Statistics
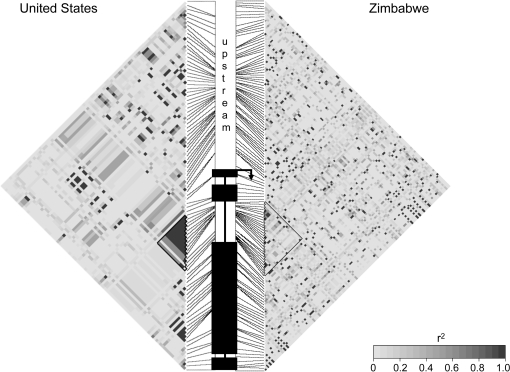
We sought to determine whether the eater gene, which is required for immunological phagocytosis, shows signs of recent adaptation at the molecular population genetic level. We sequenced multiple eater alleles from a Zimbabwe and a US population of D. melanogaster (fig. 2) and estimated sequence diversity and linkage disequilibrium for each population (table 1 and fig. 3). Linkage disequilibrium (fig. 3; second intron is outlined with a black triangle) and diversity (fig. 4) are highest in and around the second intron in the US population but not the Zimbabwe population. We simulated 1000 coalescent genealogies of a neutrally evolving equilibrium population under the estimated recombination rate, none of which showed linkage disequilibrium at the second intron that was as high as we observed in the US population (P < 10−3; table 2). These patterns reflect the presence of two high-frequency haplotype groups that are substantially diverged from each other. The presence of two high-frequency haplotypes could in principle be explained by a partial selective sweep, balancing selection, or some nonequilibrium demographic scenarios. However, the lack of variation within each haplotype class is contrary to the expectation for an ancient balanced polymorphism (e.g., Hudson and Kaplan 1988), rendering a partial sweep or nonequilibrium demography as more plausible explanations.
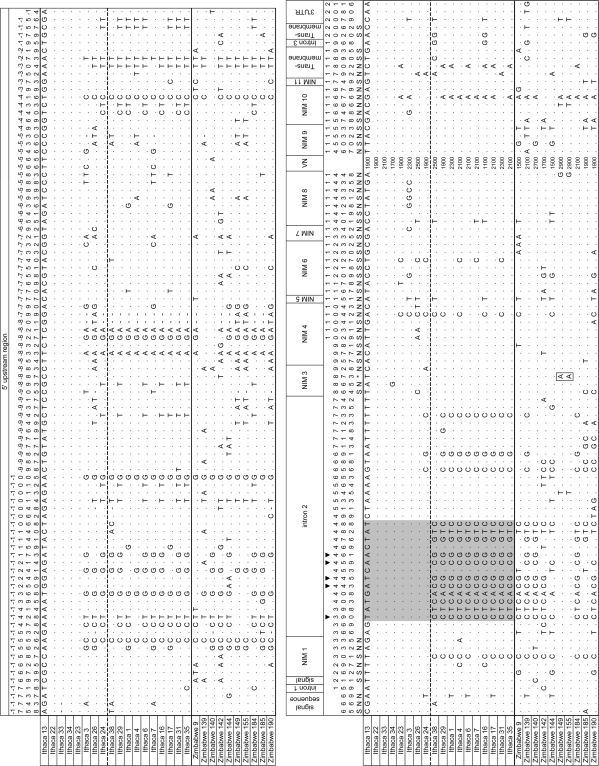
FIG. 2.
Polymorphic sites for the eater locus. The US population (Ithaca, NY) is divided into “A”- and “B”-type haplotypes based on sequence between base pairs 390 and 488 (highlighted in gray). “A” haplotypes are above the dotted line and “B” haplotypes are below. Nonsynonymous (N) and synonymous polymorphisms (S) in coding regions are indicated. Stop codons were found segregating in two individuals from Zimbabwe (boxed). CF2-II motif polymorphisms are shown (▾; see fig. 5). Variable number repeat units between NIM 8 and NIM 9 could not be aligned with confidence and are not shown, but the approximate length of that region is shown in base pairs (VN). Sites with alignment gaps were considered if there was a polymorphism. Base pair position within the gene corresponds with Figure 1 with the first position being the transcriptional start site and the 5′ upstream region indicated as −1765 to −1.
Table 1.
Population Genetic Summary Statistics for Independently Evolving Regions of eater.
| n | h | S | bp | π | πns | πs | Tajima's D | ZnS | |
| Zimbabwe | |||||||||
| Entire gene | 10 | 9 | 142 | 3895 | 0.01188 | 0.00367 | 0.02075 | −0.5199 | 0.1321 |
| 5' gene region | 10 | 9 | 123 | 3213 | 0.01271 | 0.00356 | 0.01934 | −0.4621 | 0.1341 |
| Intron 2 | 10 | 8 | 30 | 398 | 0.02903 | n/a | n/a | 0.2622 | 0.1796 |
| New York | |||||||||
| Entire gene | 18 | 16 | 94 | 3895 | 0.00768 | 0.00343 | 0.00654 | 0.2665 | 0.1462 |
| 5' gene region | 18 | 16 | 86 | 3213 | 0.00861 | 0.00345 | 0.00862 | 0.2906 | 0.1560*** |
| Intron 2 | 18 | 5*** | 15 | 398 | 0.01867*** | n/a | n/a | 2.6697*** | 0.7474* |
| New York (“A” group) | |||||||||
| Entire gene | 8 | 6 | 64 | 3895 | 0.00564 | 0.00386 | 0.00589 | −0.7286 | 0.2987 |
| 5' gene region | 8 | 6** | 59 | 3213 | 0.00635 | 0.00434 | 0.00913 | −0.7129*** | 0.312*** |
| Intron 2 | 8 | 2* | 3 | 398 | 0.00188** | n/a | n/a | −1.4475* | 1* |
| New York (“B” group) | |||||||||
| Entire gene | 10 | 10 | 56 | 3895 | 0.00477 | 0.00284 | 0.00493 | −0.3730 | 0.187 |
| 5' gene region | 10 | 10 | 49 | 3213 | 0.00499 | 0.00285 | 0.00509 | −0.4352 | 0.1869 |
| Intron 2 | 10 | 3 | 4 | 398 | 0.00201 | n/a | n/a | −1.6671 | 0.5062 |
NOTE.—n/a, not applicable; n, number of alleles sampled; h, number of haplotypes; S, segregating sites; bp, sequence length in base pairs; π is nucleotide diversity per base pair; ns, nonsynonymous; s, synonymous. The US population was both analyzed as a whole and split into haplotype groups. Tajima's D (Tajima 1989) includes all mutations. ZnS (Kelly 1997) is linkage disequilibrium based on segregating sites. Statistically significant values (*0.01 ≤ P < 0.05; **0.001 ≤ P < 0.01; ***P < 0.001) under a null demographic model are indicated (*) for the 5′ gene region and intron 2 in the New York population and the “A” haplotype group (see table 2).
FIG. 3.
Linkage disequilibrium (r2) plotted across the concatenated gene region. Each pixel represents r2 plotted between a pair of segregating sites. Exons are shown with black boxes, with the transcriptional start site indicated with an arrow. Introns are shown as lines between the exons. The black triangle indicates the block of high linkage disequilibrium in the second intron of the US (Ithaca, NY) population and is indicated in the Zimbabwe population for comparison.
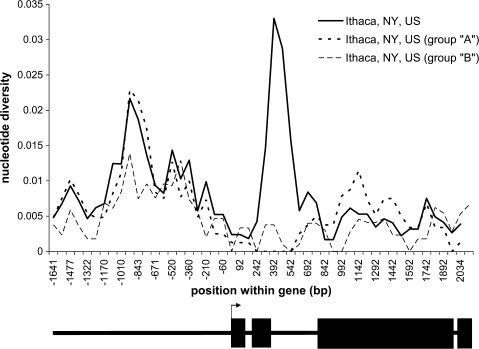
FIG. 4.
Plot of nucleotide diversity in the North American population by haplotype group. Nucleotide diversity is plotted for sliding windows with a window length of 200 sites and a step size of 75 sites. A schematic of the gene is shown below the graph with exons indicated as black boxes and the transcription start site indicated with an arrow. A spike in apparent diversity is seen over intron 2, where two divergent haplotypes (groups “A” and “B”) are segregating. There is no excess diversity within either haplotype.
Table 2.
Distribution of Summary Statistics Based on Simulations under Three Different Demographic Models.
| h | hA | π | πA | f | D | DA | ZnS | ZnSA | |
| Second intron | |||||||||
| Observed: | |||||||||
| eater (US) | 5 | 2 | 7.431 | 0.750 | 0.1009 | 2.6697 | −1.4475 | 0.7474 | 1.0 |
| Simulations | |||||||||
| Null model | 11.31 (9, 14) | 4.91 (3, 8) | 4.56 (3.24, 5.85) | 2.94 (0.93, 5.5) | 0.64 (0.21, 1.15) | 0.18 (−0.68, 1.29) | 0.08 (−1.47, 1.43) | 0.14 (0.09, 0.26) | 0.33 (0.08 ,0.81) |
| P value | 0* | 0.017* | 0* | 0.001* | 0* | 0* | 0.031* | 0* | 0.016* |
| TA | 5.74 (2, 12) | 2.56 (1, 6) | 6.34 (2.92, 7.84) | 1.06 (0, 4.64) | 0.19 (0, 0.75) | 1.72 −1.26, 3.03) | −0.53 (−1.74, 1.91) | 0.55 (0.06, 1) | 0.54 (0.02, 1) |
| P value | 0.547 | 0.534 | 0.872 | 0.549 | 0.5 | 0.1 | 0.272 | 0.235 | 0.261 |
| LS | 9.01 (5, 13) | 3.88 (1, 7) | 5.92 (4.01, 7.18) | 2.3 (0, 5.46) | 0.39 (0, 0.88) | 1.36 (−0.31, 2.45) | −0.31 (−1.64, 1.44) | 0.34 (0.17, 0.65) | 0.43 (0.04, 1) |
| P value | 0.076 | 0.182 | 0* | 0.201 | 0.156 | 0* | 0.091 | 0.018* | 0.077 |
| 5′ gene region | |||||||||
| Observed | |||||||||
| eater (US) | 16 | 6 | 26.74 | 19.75 | 0.74 | 0.2906 | −0.7129 | 0.1560 | 0.312 |
| Simulations | |||||||||
| Null model | 17.39 (15, 18) | 7.8 (7, 8) | 25.47 (22.46, 28.75) | 23.27 (17.64, 28.32) | 0.91 (0.77, 1.06) | 0.08 (−0.43, 0.63) | 0.1 (−0.51, 0.91) | 0.08 (0.07, 0.1) | 0.18 (0.15, 0.24) |
| P value | 0.107 | 0.005* | 0.782 | 0.089 | 0.018* | 0.218 | 0* | 0* | 0* |
| TA | 14.94 (12, 17) | 6.59 (4, 8) | 34.01 (22.6, 40.13) | 19.45 (3.75, 32.46) | 0.58 (0.11, 1.09) | 1.51 (−0.4, 2.53) | −0.01 (−1.87, 1.53) | 0.31 (0.19, 0.52) | 0.44 (0.22, 0.85) |
| P value | 0.851 | 0.396 | 0.059 | 0.495 | 0.713 | 0.941 | 0.238 | 1 | 0.851 |
| LS | 17.28 (14, 18) | 7.58 (6, 8) | 31.46 (26.35, 36.38) | 26.1 (17.18, 32.68) | 0.83 (0.6, 1.07) | 1.08 (0.22, 1.9) | 0.36 (−0.56, 1.22) | 0.12 (0.09, 0.15) | 0.23 (0.17, 0.29) |
| P value | 0.139 | 0.04* | 0.06 | 0.069 | 0.188 | 0.97 | 0.02* | 0.06 | 0* |
NOTE.—Empirical estimates of population genetic statistics are given for the US population and the “A” haplotype group. For the simulated distributions, the means and 95% confidence intervals (in parentheses) under three different demographic models are indicated. TA refers to the model based on Thornton and Andolfatto (2006), and LS refers to the model based on Li and Stephan (2006). π is nucleotide diversity, or average pairwise differences, per locus. f is nucleotide diversity in the putatively selected allele (“A”) divided by the total nucleotide diversity. D is Tajima's D including all mutations. P values indicate the proportion of simulations where the simulated values were more extreme than the empirical estimate at eater. Tests of h, π, D, ZnS were two tailed, and the means were standardized around 0 to calculated P values. f, hA, πA, DA, and ZnSA were used to test for a partial selective sweep, and simulated values were considered extreme if they were less than empirical estimates of f, hA, πA, and DA or greater than empirical estimates of ZnSA.
*p values less than 0.05.
To see if nonequilibrium patterns extended beyond the second intron, we compared patterns of nucleotide diversity and the site frequency spectrum at the second intron with those across the rest of the gene region. When interpreting our results, we considered only the 5′ end of the gene (3,213 bp) because simulations could not be performed across the entire gene region (3,895 bp) due to the variable number repeat units (see Materials and Methods). Tajima's D (Tajima 1989), nucleotide diversity, and linkage disequilibrium (ZnS; Kelly 1997) are all elevated in the second intron relative to the rest of the 5′ gene region in the North American population (second intron: Tajima's D = +2.6697, π = 0.01867, ZnS = 0.7474; 5′ gene region: Tajima's D = +0.2906, π = 0.00861, ZnS = 0.1560; table 1). These patterns of diversity are extremely unlikely under the standard neutral null model (table 2). Tajima's D is significantly positive at the second intron (P < 0.01), and linkage disequilibrium is significantly high at both the second intron (P < 0.001) and in the 5′ gene region (P < 0.001).
To test whether this reflects the pooling of two intermediate frequencies, disparate allelic classes, we calculated the statistics separately for each haplotype group in the North American population (table 1). Only two sequence haplotypes were observed between base pairs 390 and 488 (99 base pairs) within the second intron (398 base pairs), so the alleles were divided into group “A” or group “B” based on this sequence (fig. 2, region in gray). The “A” group is named because it is a perfect match to the reference genome of D. melanogaster (Adams et al. 2000). Across the remainder of the second intron, these haplotypes each have very little variation within haplotype group (πA = 0.00188; πB = 0.00201), but 11 of 15 segregating sites in this 398 bp window are fixed differences between the two groups (πcombined = 0.01867; table 1 and fig. 2). The reduction in nucleotide diversity extends approximately 800 base pairs in the “A” haplotype (fig. 4). Across the 5′ gene region, the levels of nucleotide diversity are similar in each haplotype group (πA: 0.00635, πB: 0.00499). The “A” haplotype has a higher level of linkage disequilibrium and a more negative value of Tajima's D than the “B” haplotype does (ZnS A: 0.312, B: 0.1869; Tajima's D A: −0.7129, B: −0.4352). Compared with the standard null neutral model, the “A” haplotype group has a significantly low value of Tajima's D (P < 0.001), too few haplotypes (P = 0.005), excess linkage disequilibrium (P < 0.001), and a low value of f (P = 0.018), indicating a deficit in nucleotide diversity in haplotype “A” relative to the entire population (tables 1 and 2). The observed reduction in nucleotide diversity and excess linkage disequilibrium in the “A” haplotype is consistent with a recent rise to high frequency due to a partial selective sweep.
The Zimbabwe population, in contrast, did not show evidence for haplotype structuring or a recent selective sweep around the second intron. The patterns of diversity are compatible with our expectations for a neutrally evolving African population. The Zimbabwe population harbors substantially more diversity than the US population (πZimbabwe: 0.01188, πUS: 0.00768; table 1), reflecting the larger effective size of this population, which is presumed to be ancestral to the US population (David and Capy 1988). Two individuals in the Zimbabwe population have a stop codon in the third NIM repeat that presumably results in a truncated version of Eater (fig. 2). Such potentially deleterious mutations are expected to occur at low frequencies in populations that are in mutation–selection balance. For both populations, the diversity at synonymous sites exceeded that at nonsynonymous sites in eater (table 1), as would be expected if purifying selection acts to remove deleterious amino acid variation.
Standard Neutral and Bottleneck Simulations
The observed population genetic statistics are highly suggestive of the haplotype “A” having recently risen to high frequency in the US population due to positive selection. These patterns are not compatible with a standard neutral null model of evolution. However, selectively neutral demographic processes such as population expansions or bottlenecks can often lead to patterns that mimic those expected under natural selection. It is believed that all non-African populations have only recently been founded from Africa (David and Capy 1988; Baudry et al. 2004). Models that incorporate bottlenecks similar to what these populations underwent as they expanded their population range are a better fit to genomewide patterns of diversity (Li and Stephan 2006) than the standard neutral null model. Two recent analyses have described bottleneck models that can be applied to the North American population (Li and Stephan 2006; Thornton and Andolfatto 2006), one with a prolonged bottleneck that ended recently and the other with a short, ancient bottleneck that was preceded by a population expansion. Simulations under these models can give us a mean and range of values for the number of haplotypes, nucleotide diversity, linkage disequilibrium, and Tajima's D that we can expect to observe at the eater locus in the absence of selection. If our empirically observed statistics fall into the tails of the null distributions (see Methods), then we infer that selection may have occurred.
The TA model (Thornton and Andolfatto 2006) describes a hypothesized prolonged bottleneck that ended recently. The population genetic statistics that we observed at the eater locus are all consistent with the TA demographic scenario (table 2; supplementary fig. 1a and b, Supplementary Material online). The LS model (Li and Stephan 2006) presumes a bottleneck that was ancient, brief, and was preceded by a population expansion in the ancestral African population. Using this model, there are expected to be significantly more haplotypes (P = 0.04), a higher value of Tajima's D (P = 0.02), less linkage disequilibrium (P < 0.001), and a shorter extent of EHH in the “A” group than is actually observed at the eater locus (table 2 and supplementary fig. 1c and d, Supplementary Material online). This indicates that the values observed at the eater locus cannot be explained by an ancient and brief bottleneck such as the one proposed by Li and Stephan (2006). The ranges of the distributions of most test statistics are much wider when modeling the TA scenario than under the LS scenario (table 2) and would attribute demographic explanations to all but the most extreme instances of positive selection. The TA scenario is likely to be too conservative for the detection of less radical selective pressures. Of the three neutral models presented, the LS scenario best explains genomic patterns of polymorphism in derived populations of D. melanogaster (Li and Stephan 2006), and thus, we favor interpretation of our results in light of this scenario.
Gene Expression Differences between Haplotypes and Potential Targets of Selection
If natural selection has indeed shaped patterns of variation at eater, then we might expect to see a phenotypic difference associated with the high-frequency haplotypes that could be the target of selection. We therefore examined the sequences of the “A” and “B” groups to find candidate sequence differences that could give us insight into the nature of a potential phenotypic difference. The excess linkage disequilibrium in the “A” haplotype extends through NIM 1 and NIM 2, two repeat units that are implicated in microbial binding (Kocks et al. 2005). However, no fixed nonsynonymous differences were found between the haplotype groups in these NIM repeats (fig. 2). Because the population genetic statistics were most extreme at the second intron of eater, we evaluated group “A” and “B” sequences at this intron with a sequence motif finder against insect motifs within the library TRANSFAC at GenomeNet (http://motif.genome.jp/). Four putative chorion transcription factor 2 (CF2-II)–binding regions were found in the “A” haplotype (fig. 5) using the search motif sequence GTATATATA. All four regions had polymorphisms in the “B” haplotype that made them poorer matches to the consensus motif. This sequence motif can be either an enhancer or a suppressor during D. melanogaster oogenesis and embryonic muscle development (Hsu et al. 1996; García-Zaragoza et al. 2008) and is a suppressor of expression of the antimicrobial peptide gloverin in the silkworm Bombyx mori (Mrinal and Nagaraju 2008). We therefore hypothesized that the second intron might contain one or more regulatory sequences and that transcriptional differences between the alleles is the target of selection.
FIG. 5.
Polymorphisms in putative CF2-II transcription factor recognition motifs in positions within the second intron of North American haplotypes “A” and “B.” Motif GTATATATA is considered a perfect match. The score indicates how well the input sequence matches the motif. The position within the gene region is indicated above each nucleotide. Haplotype group “A” has four high score matches to the CF2-II motif.
To test the hypothesis that sequence variation between the “A” and “B” haplotype groups results in differing expression levels of the eater gene, we measured constitutive expression of eater in adult males in 19 D. melanogaster genetic lines that were homozygous for either the “A” or “B” haplotype. We found that lines bearing the “A” haplotype express significantly more eater than “B” haplotype lines (P = 0.0417), exhibiting an average of 69% higher expression (table 3). Although this observation does not directly test the function of the putative CF2-II–binding sequences that are present in the “A” haplotype but absent in the “B” haplotype, it is consistent with the hypothesis that these or other unidentified regulatory sequences cause functional differentiation between the two haplotypes and may be targets of selection.
Table 3.
Factors and P Values of a Linear Model Used to Show that US Haplotype Group “A” Expresses eater at a Higher Level than group “B”.
| Factor Name | Effect Type | df | Z Value or f Value | P Value |
| Line (Haplotype)a | Random | 2.34 | 0.0097 | |
| Vial (Line × Haplotype)b | Random | 0.35 | 0.3634 | |
| Sample (Line × Haplotype × Vial)c | Random | 2.12 | 0.0170 | |
| Dayd | Random | 0.69 | 0.2448 | |
| Extraction (Day)e | Random | 0.95 | 0.1711 | |
| Residual | Random | 9.96 | <0.0001 | |
| Rp49f | 1 | 1508.9 | <0.0001 | |
| Haplotypeg | Fixed | 1 | 4.83 | 0.0417 |
NOTE.—df, degrees of freedom.
Background variation due to genetic line.
Variation due to rearing vial.
Random variation among replicate samples of flies within the vial.
Variation due to day of experiment.
Variation due to RNA extraction.
Variation due to amount of RNA, measured as expression of housekeeping gene.
Variation due to haplotype.
Evolutionary Patterns of NIM Repeats
The first four NIM repeat units (NIM 1–4) have previously been implicated in microbial binding (Kocks et al. 2005). We considered that these repeats specifically might participate in host–pathogen coevolutionary interactions that the other NIM repeats would not. To test this hypothesis, we compared the rate of nonsynonymous substitution (KA) and of synonymous substitution (KS) between NIM 1–4 and the remaining independently evolving NIM repeats (NIM 5–11) (supplementary table 1, Supplementary Material online). We found no evidence for any difference in the evolutionary patterns between the two sets of repeats, with KA and KS not significantly differing between the two groups (Wilcoxon signed rank test, KA P value = 0.6202, KS P value = 0.2183; supplementary table 1, Supplementary Material online). We also examined the phylogenetic relationship of each NIM repeat among D. melanogaster, D. simulans, and D. yakuba. The accepted relationship among these species places D. melanogaster and D. simulans as sister species and D. yakuba as the outgroup (Begun et al. 2007). Six NIM repeats had phylogenetic relationships that deviated from this pattern, which could indicate elevated selective pressures along particular branches. However, these repeats were evenly distributed between NIM 1–4 (microbial binding) and NIM 5–11 (unknown function) (supplementary table 1, Supplementary Material online). Nucleotide diversity levels were not different between the two sets of functionally distinct repeats in either Zimbabwe or US populations (πZimbabwe P value = 0.7879, πUS P value = 0.7748). NIM 2, a unit with a putative role in microbial binding, had no polymorphism in either the Zimbabwe and US populations (supplementary table 1, Supplementary Material online) or in additional populations sampled from Australia, the Netherlands, or China (data not shown). The second intron lies within this NIM repeat, so the deficit in diversity of NIM 2 may be linked with the unusual evolutionary patterns of the intron.
Properties of the Variable Number Repeat Units
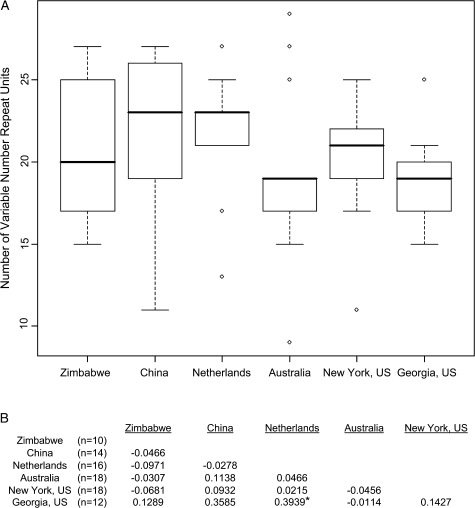
The number of repeats in the region between NIM 8 and NIM 9 is polymorphic and ranges between 11 and 29 (fig. 6a). The Zimbabwe and China populations have the highest variation in the number of repeat units. Worldwide RST, a measure of genetic differentiation that ranges from 0 for completely undifferentiated to 1 for complete isolation, was 0.00388 (95% confidence interval: −0.0488, 0.4082) that indicates a lack of differentiation between populations. Pairwise comparisons between individual populations ranged from −0.0971 to 0.3939 (fig. 6b), and all were nonsignificant (P > 0.05) after a Bonferroni correction for multiple tests. We therefore find no evidence that the overall length of the variable number repeat region is geographically differentiated or locally adapted.
FIG. 6.
Absence of genetic differentiation (RST) between populations in variable number repeat sizes. (a) Box plots of the distribution of sizes of variable number repeat region by population. (b) Pairwise RST values between populations. *P = 0.0287 (not significant after a Bonferroni correction); P > 0.05 for all other pairwise comparisons.
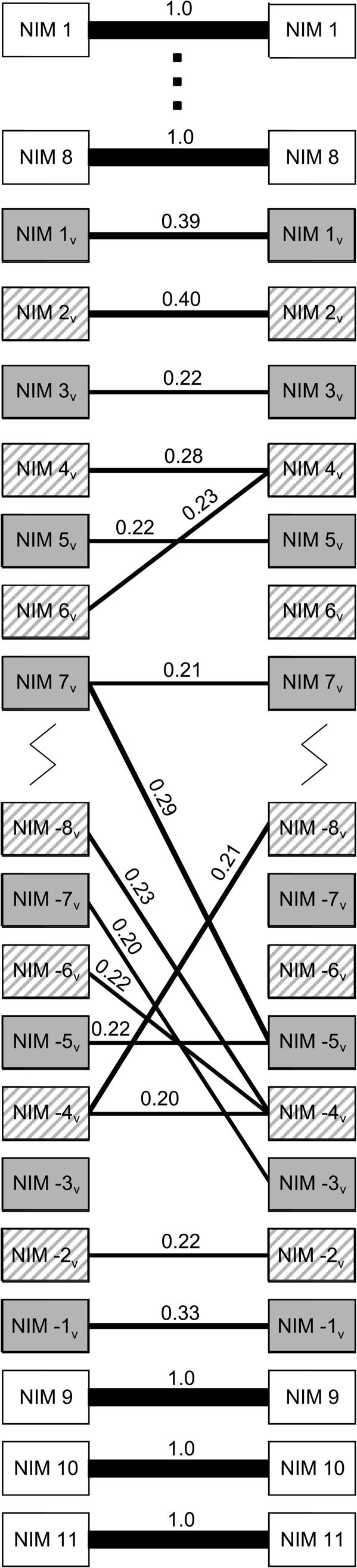
At the nucleotide sequence level, the variable number repeat units do not tightly cluster phylogenetically based on physical location in the array, in contrast with the conserved number NIM 1–11, whose nearest phylogenetic neighbors are always physically homologous repeats in alleles isolated from different individuals and from the outgroup species (Somogyi et al. 2008). This suggests that the variable number repeat units are evolving concertedly by birth and death of repeat units and gene conversion across paralogous units within the variable number repeat array. This is in contrast with the conserved number repeat units, which are evolving independently. We find evidence that there is also variation in the evolutionary patterns within the variable number repeat region. For units on the periphery of the variable number repeat region, the lowest genetic distances between units was generally found when comparing between units at the same homologous position (fig. 7). No such pattern existed for units in the interior of the array. This indicates a higher degree of independent evolution in units in the periphery than in interior units and suggests that the birth-and-death process that gives rise to new alleles is most likely to occur in the interior of the gene.
FIG. 7.
Nearest genetic neighbors between NIM repeat units. Genetic distances were calculated between all pairwise combinations of NIM repeats from different individuals. The thickness of the connecting lines and the number on the line indicate the proportion of times that the nearest neighbor of a particular repeat unit was the indicated NIM repeat. Genetic distances were calculated with the Kimura 2-parameter model using an alignment of the 78 base pair NIM consensus motif that is conserved between all repeat units. NIM 1 through NIM 8 all showed the same pattern, so the intervening repeats are not shown (region indicated with dots). Variable number repeat units are shaded (“NIM 8 like” = gray and white stripes and “alternate” = gray). Some variable number repeats units were not sequenced (region indicated with a jagged line).
Discussion
The patterns of genetic diversity and the divergence in gene expression between two high-frequency haplotypes give strong support for a partial selective sweep at the eater locus in a North American population. One haplotype group, labeled the “A” group, has a high level of linkage disequilibrium, EHH that reaches over a long genomic distance, and a negative value of Tajima's D. These extreme values reach statistical significance under two of three previously described demographic null models (neutral and LS models) of selective neutrality. The model to which the eater data can be fit (the TA model) is so general that it provides little resolution between selective and neutral scenarios. Overall, our data are consistent with the “A” haplotype having recently risen to high frequency in North America due to an incomplete selective sweep. Notably, the expression of eater in isogenic lines with the “A” allele is on average 69% higher than in lines with the “B” allele. This expression difference offers a phenotypic basis upon which selection could act.
The “A” group does not display the strong deficit in nucleotide diversity that could be expected if it had recently and rapidly reached high frequency. This may, however, be a consequence of our assignment of individual alleles to haplotype groups. Of the eight alleles in the “A” group, seven are identical across the entire second intron and five have an average of 2.4 pairwise differences among them for the entire length of the gene, compared with the average of 21.4 pairwise differences among alleles in the “A” group as a whole. The eighth “A” allele brings in the majority of sites segregating in that haplotype group and appears to be a recombinant between the “A” and “B” haplotype groups. A less conservative assignment that excluded this eighth allele from haplotype group “A” would have led to a much more extreme deficit of nucleotide diversity in the “A” group. The 99 base pairs that we used to define the “A” haplotype are perfectly conserved in two lines from Zimbabwe, suggesting that this allele was present in the ancestral population prior to founding of the North American population (cf., Pool et al. 2006), although we cannot exclude the possibility that the haplotype was reintroduced back into the African population by back migration. Selective events that act on standing genetic variation leave much less dramatic signatures than those seen when selection strongly favors novel mutations (Przeworski et al. 2005). The fact that we are able to see any distortions to the site frequency spectrum at all suggests strong positive selection at this locus.
It is striking that the two “A” haplotypes found in the Zimbabwe population are identical across the entire 4-kb region in those two individuals (fig. 2). This sample size is too small to do simulations similar to those we did with the US population, but this observation begs the question of whether or not selective sweeps involving these two haplotypes are happening in other populations or if this selective sweep is unique to the US population. Geographically restricted selective sweeps could potentially stem from adaptation of populations to their local environments (Aminetzach et al. 2005; Macpherson et al. 2008). We surveyed eight alleles from each of two additional populations from the Netherlands and China at the second intron (data not shown) but found no evidence of the “A” haplotype being present in either of these populations. This suggests that, of the derived populations, the selective sweep involving the “A” haplotype is a local phenomenon restricted to the North American population. In contrast, we find no evidence of genetic differentiation (RST = 0.00388) in the total number of NIM repeat units among populations around the world. The lack of differentiation indicated by this RST value suggests that the number of repeats is not free to drift to different frequencies in individual populations, and certainly is not adaptively diverging among subpopulations, but instead that the number of repeats is subject to purifying selection.
We have hypothesized that enhancer motifs present in the second intron of “A” group haplotypes but absent in “B” group alleles result in higher expression of eater “A” haplotypes and have noted polymorphism in putative CF2-II–binding sites as candidates for responsibility. The CF2-II zinc finger transcription factor is an alternatively spliced variant of the CF2 transcription factor that was first identified in D. melanogaster and has been shown to be important during oogenesis and in embryonic muscle tissue development (Hsu et al. 1996; García-Zaragoza et al. 2008), where it can act as either an enhancer (García-Zaragoza et al. 2008) or repressor (Hsu et al. 1996). In the silkworm B. mori, CF2 was found to act as a repressor of expression of the antimicrobial peptide gloverin (Mrinal and Nagaraju 2008). The ancestral member of the gloverin family has a CF2 motif in an intron in the 3′ UTR, and a deletion of this intron in other members of the gene family has been associated with the gain of expression of gloverin in embryos. Although the haplotype structuring and expression association we identified was centered around CF2-II motifs in the second intron of eater, this does not prove that the CF2-II sites are responsible for the expression difference and does not preclude the role of a different sequence motif either inside or outside this intron. Sequence important for eater expression has been identified in the 5′ upstream region of the gene (Tokusumi et al. 2009), and it is possible that a still unidentified region of the gene is responsible for the expression differences between haplotypes.
Increased expression of eater and other genes involved in cellular and humoral immunity has previously been reported in D. melanogaster selected for increased resistance to the bacterial pathogen Pseudomonas aeruginosa (Ye et al. 2009). This supports the hypothesis that higher expression of eater is beneficial in the face of pathogen pressure. Artificially selected lines rapidly lost resistance when the selective pressure was removed, suggesting that resistance is costly to maintain. We report evidence of a partial selective sweep at the eater locus in a North American population but not in an African population. Drosophila melanogaster was likely to have encountered novel pathogens as the population range expanded out of Africa. Geographically restricted selective sweeps can occur if selective pressures such as bacterial species and frequencies vary across different areas. The selective sweep may be ongoing which is why the allele associated with higher eater expression is not fixed in the population, or the costs related to increased expression may inhibit the fixation of this allele.
eater is a cellular recognition gene, a class which shows evidence of rapid evolution between species (Sackton et al. 2007). Like eater, other genes in this class have previously shown evidence of selection at the population level. Thioester-containing proteins, which are thought to function as opsonins and label microbes for phagocytosis, show evidence of adaptive evolution in Drosophila, Anopheles mosquitoes, and the crustacean Daphnia (Little et al. 2004; Little and Cobbe 2005; Jiggins and Kim 2006). In Tep genes, positively selected sites are often clustered around putative sites of interaction between host and pathogen, suggesting that coevolutionary arms races drive their rapid evolution. Single Tep genes show evidence of recent selection within an African population of D. melanogaster (Jiggins and Kim 2006) and divergence in gene expression levels between populations (Hutter et al. 2008). Class C scavenger receptor (SR-Cs) proteins are implicated in the internalization of microbial compounds (Rämet et al. 2001), and some members of this family display evidence of adaptive amino acid replacement between species of Drosophila (Lazzaro 2005). SR-Cs show unusual patterns of nucleotide diversity and haplotype structuring within one North American population of D. simulans that suggests a recent and rapid rise to high frequency of putatively selected haplotypes (Lazzaro 2005; Schlenke and Begun 2005). These previous studies suggest that, although cellular recognition molecules evolve rapidly as a class, unique evolutionary patterns and pressures drive the evolution of individual genes.
Partial selective sweeps have been invoked to explain the presence of high-frequency haplotypes with low genetic diversity, and previous studies have identified D. melanogaster loci with similar patterns of genetic variation as we see at eater (Hudson et al. 1997; Aminetzach et al. 2005). The Doc1420 long interspersed element–like transposon is a polymorphic insertion in D. melanogaster that results in a truncated version of a protein and confers organophosphate pesticide resistance (Aminetzach et al. 2005). There are fewer haplotypes, reduced variation, and excess linkage disequilibrium in the group of alleles containing the element, and the transposon insertion is found in high frequency in derived populations but only low frequency in ancestral African populations (Aminetzach et al. 2005). At the Sod locus, two haplotype groups, one within a fast electromorph group and one containing all slow electromorphs, each have very little or no nucleotide diversity (Lee et al. 1981). A complex pattern of selection where the fast haplotype group underwent a partial selective sweep and then a subsequent mutation led to the slow haplotype, which is different by only one amino acid, is the most likely explanation for patterns of variation at this locus (Hudson et al. 1997). It should be noted that in both these examples, the excess linkage disequilibrium and reduced genetic diversity extended as far away as 10 kb, and therefore, these loci may have been subject to stronger or more recent selection.
The coding regions of eater are largely composed of NIM repeat units. These repeats in eater have been previously identified as evolving either independently or concertedly (Somogyi et al. 2008). Four of the eleven independently evolving repeats have been implicated in microbial binding, and it has been hypothesized that repeats evolving concertedly compose a structural “stalk” between the ligand-binding NIM repeats and the phagocyte membrane (Kocks et al. 2005). Coevolutionary arms races between pathogens and the host immune response can drive unusual patterns such as accelerated rates of amino acid substitution, selective sweeps, or balancing selection. To look for evidence of pathogen-imposed selection on NIM repeats with functional evidence of microbe binding, we compared evolutionary patterns of these four repeats with the seven other independently evolving repeats. We found no evidence of a difference in the rate of amino acid substitution, patterns of genetic diversity, or phylogenetic relationships with outgroup species. This is consistent with previous evidence that, although eater potentially shows evidence of positive selection between Drosophila species, selection is not concentrated around pathogen interaction domains (Sackton et al. 2007).
Sequence similarity between NIM repeats is especially high in the interior of the gene and has led to concerted evolution. Concerted evolution can arise because of unequal crossing over due to nonhomologous pairing during recombination or because of gene conversion (Charlesworth et al. 1994). We present evidence that the repeat units in the periphery of the variable number repeat region show signs of independent evolution and that the internal repeats are truly evolving concertedly. This is indicated by the observation that units on the periphery are more likely to be most closely related to units in the same physical location in different individuals, whereas units in the interior show no such concordance between physical location and genetic distance. This is also strong evidence that the duplication and deletion of repeat units is more likely to occur in the internal repeats than in the external repeats, in part because nonhomologous pairing becomes less likely as the genetic distance between sequences increases (Stephan 1989). Polymorphism in repeat number like we observe at eater can only be caused by unequal crossing over (Smith 1976). Gene conversion is likely also driving concerted evolution in this region. In one instance, we observed patterns of concerted and independent evolution within a single NIM repeat unit. The 78 base pairs that define the core consensus “NIM” motif are evolving concertedly in NIM −1v (fig. 7). In contrast, the last 15 base pairs of this repeat are evolving independently (data not shown). This pattern can only be driven by gene conversion and indicates that multiple factors contribute to concerted evolution within eater.
The data we have collected at the eater locus support a model wherein this recognition molecule, which is critical in the cellular immune response of D. melanogaster, is subject to distinctive evolutionary pressures. However, unlike observations for other genes and contrary to our expectations, this selection is not centered around pathogen interaction domains. Instead, selection appears to be acting on gene expression level in a geographically restricted subpopulation. Further experimentation will be required to determine the organismal fitness consequences of variation in eater expression. Novel mutations that are selectively advantageous in local environments have a chance to rapidly rise to high frequency and may eventually serve as the basis for between-species divergences. Unlike comparisons between species that have found evidence of amino acid adaptation in cellular immune response genes, our data implicate noncoding regulatory changes as playing an important role in the evolution of eater.
Supplementary Material
Supplementary figure 1 and table 1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We thank Ann Hajek, Andy Clark, Sarah Short, Jacob Crawford, Jennifer Comstock, Madeline Galac, and two anonymous reviewers for comments on the manuscript. We thank Sean Hackett and Andy Clark for providing some of the lines used in this study. Part of this work was carried out by using the resources of the Computational Biology Service Unit from Cornell University which is partially funded by Microsoft Corporation. This work was supported by grants from the National Science Foundation (DEB-0415851) and the National Institutes of Health (AI064950).
References
- Adams MD, Celniker SE, Holt RA, et al. (195 co-authors) The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Aminetzach YT, Macpherson JM, Petrov DA. Pesticide resistance via transposition-mediated adaptive gene truncation in Drosophila. Science. 2005;309:764–767. doi: 10.1126/science.1112699. [DOI] [PubMed] [Google Scholar]
- Baudry E, Viginier B, Veuille M. Non-African populations of Drosophila melanogaster. Mol Biol Evol. 2004;21:1482–1491. doi: 10.1093/molbev/msh089. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Aquadro CF. Molecular variation at the vermillion locus in geographically diverse populations of D. melanogaster and D. simulans. Genetics. 1995;140:1019–1032. doi: 10.1093/genetics/140.3.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun DJ, Holloway AK, Stevens K, et al. (13 co-authors) Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochdanovits Z, de Jong G. Experimental evolution in D. melanogaster: interaction of temperature and food regime selection regimes. Evolution. 2003;57:1829–1836. doi: 10.1111/j.0014-3820.2003.tb00590.x. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- David JR, Capy P. Genetic variation of Drosophila melanogaster natural populations. Trends Genet. 1988;4:106–111. doi: 10.1016/0168-9525(88)90098-4. [DOI] [PubMed] [Google Scholar]
- Ebert D. Experimental evidence for rapid parasite adaptation and its consequences for the evolution of virulence. In: Poulin R, Morand S, Skorping A, editors. Evolutionary biology of host-parasite relationships: theory meets reality. Amsterdam: Elsevier Science.; 2000. pp. 163–184. [Google Scholar]
- Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiumera AC, Dumont BL, Clark AG. Associations between sperm competition and natural variation in male reproductive genes on the third chromosome of Drosophila melanogaster. Genetics. 2007;176:1245–1260. doi: 10.1534/genetics.106.064915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Zaragoza E, Mas JA, Vivar J, Arredondo JJ, Cervera M. CF2 activity and enhancer integration are required for proper muscle gene expression in Drosophila. Mech Dev. 2008;125:617–630. doi: 10.1016/j.mod.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Goodman SJ. RST Calc: a collection of computer programs for calculating estimates of genetic differentiation from microsatellite. Mol Ecol. 2008;6:881–885. [Google Scholar]
- Hsu T, Bagni C, Sutherland JD, Kafatos FC. The transcriptional factor CF2 is a mediator of EGF-R-activated dorsoventral patterning in Drosophila oogenesis. Genes Dev. 1996;10:1411–1421. doi: 10.1101/gad.10.11.1411. [DOI] [PubMed] [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL. The coalescent process in models with selection and recombination. Genetics. 1988;120:831–840. doi: 10.1093/genetics/120.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Sáez AG, Ayala FJ. DNA variation at the Sod locus of Drosophila melanogaster: an unfolding story of natural selection. Proc Natl Acad Sci U S A. 1997;94:7725–7729. doi: 10.1073/pnas.94.15.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter S, Saminadin-Peter SS, Stephan W, Parsch J. Gene expression variation in African and European populations of Drosophila melanogaster. Genome Biol. 2008;9:R12. doi: 10.1186/gb-2008-9-1-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins FM, Kim KW. Contrasting evolutionary patterns in Drosophila immune receptors. J Mol Evol. 2006;63:769–780. doi: 10.1007/s00239-006-0005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JK. A test of neutrality based on interlocus associations. Genetics. 1997;146:1197–1206. doi: 10.1093/genetics/146.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocks C, Cho JH, Nehme N, et al. (16 co-authors) Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell. 2005;123:335–346. doi: 10.1016/j.cell.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Kurucz E, Márkus R, Zsámboki J, et al. (13 co-authors) Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr Biol. 2007;17:649–654. doi: 10.1016/j.cub.2007.02.041. [DOI] [PubMed] [Google Scholar]
- Lazzaro BP. Elevated polymorphism and divergence in the class C scavenger receptors of Drosophila melanogaster and D. simulans. Genetics. 2005;169:2023–2034. doi: 10.1534/genetics.104.034249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro BP, Flores HA, Lorigan JG, Yourth CP. Genotype-by-environment interactions and adaptation to local temperature affect immunity and fecundity in Drosophila melanogaster. PLoS Pathog. 2008;4:e1000025. doi: 10.1371/journal.ppat.1000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YM, Misra HP, Ayala FJ. Superoxide dismutase in Drosophila melanogaster: biochemical and structural characterization of allozyme variants. Proc Natl Acad Sci U S A. 1981;78:7052–7055. doi: 10.1073/pnas.78.11.7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Stephan W. Inferring the demographic history and rate of adaptive substitution in Drosophila. PLoS Genet. 2006;2:e166. doi: 10.1371/journal.pgen.0020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH. Molecular evolution. Sunderland (MA): Sinauer Associates; 1997. [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Little TJ, Cobbe N. The evolution of immune-related genes from disease carrying mosquitoes: diversity in a peptidoglycan- and thioester-recognizing protein. Insect Mol Biol. 2005;14:599–605. doi: 10.1111/j.1365-2583.2005.00588.x. [DOI] [PubMed] [Google Scholar]
- Little TJ, Colbourne JK, Crease TJ. Molecular evolution of Daphnia immunity genes: polymorphism in a gram-negative binding protein gene and a 2-macroglobulin gene. J Mol Evol. 2004;59:498–506. doi: 10.1007/s00239-004-2641-8. [DOI] [PubMed] [Google Scholar]
- Macpherson JM, González J, Witten DM, Davis JC, Rosenberg NA, Hirsh AE, Petrov DA. Nonadaptive explanations for signatures of partial selective sweeps in Drosophila. Mol Biol Evol. 2008;25:1025–1042. doi: 10.1093/molbev/msn007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J, Haigh J. The hitch-hiking effect of a favourable gene. Genet Res. 1974;23:23–35. [PubMed] [Google Scholar]
- McEvoy BP, Montgomery GW, McRae AF, et al. (27 co-authors) Geographical structure and differential natural selection among North European populations. Genome Res. 2009;19:804–814. doi: 10.1101/gr.083394.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrinal N, Nagaraju J. Intron loss is associated with gain of function in the evolution of the gloverin family of antibacterial genes in Bombyx mori. J Biol Chem. 2008;283:23376–23387. doi: 10.1074/jbc.M801080200. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Pool JE, Bauer Du Mont V, Mueller JL, Aquadro CF. A scan of molecular variation leads to the narrow localization of a selective sweep affecting both Afrotropical and cosmopolitan populations of Drosophila melanogaster. Genetics. 2006;172:1093–1105. doi: 10.1534/genetics.105.049973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M, Coop G, Wall JD. The signature of positive selection on standing genetic variation. Evolution. 2005;59:2312–2323. [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2006. [Google Scholar]
- Rämet M, Pearson A, Manfruelli P, Li X, Koziel H, Göbel V, Chung E, Krieger M, Ezekowitz RAB. Drosophila scavenger receptor CI is a pattern recognition receptor for bacteria. Immunity. 2001;15:1027–1038. doi: 10.1016/s1074-7613(01)00249-7. [DOI] [PubMed] [Google Scholar]
- Ryan AW, Mapp J, Moyna S, Mattiangeli V, Kelleher D, Bradley DG, McManus R. Levels of interpopulation differentiation among different functional classes of immunologically important genes. Genes Immun. 2006;7:179–183. doi: 10.1038/sj.gene.6364266. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, et al. (17 co-authors) Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- Sackton TB, Lazzaro BP, Schlenke TA, Evans JD, Hultmark D, Clark AG. Dynamic evolution of the innate immune system in Drosophila. Nat Genet. 2007;39:1461–1468. doi: 10.1038/ng.2007.60. [DOI] [PubMed] [Google Scholar]
- Schlenke TA, Begun DJ. Linkage disequilibrium and recent selection at three immunity receptor loci in Drosophila simulans. Genetics. 2005;169:2013–2022. doi: 10.1534/genetics.104.035337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlötterer C, Neumeier H, Sousa C, Nolte V. Highly structured Asian Drosophila melanogaster populations: a new tool for hitchhiking mapping? Genetics. 2006;172:287–292. doi: 10.1534/genetics.105.045831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J-H, Blay S, McNeney B, Graham J. LDheatmap: an R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J Stat Soft. 2006;16 Code Snippet 3. [Google Scholar]
- Singh ND, Arndt PF, Petrov DA. Effect of recombination on patterns of substitution in Drosophila. Genetics. 2005;169:709–722. doi: 10.1534/genetics.104.032250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GP. Evolution of repeated DNA sequences by unequal crossover. Science. 1976;191:528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- Somogyi K, Sipos B, Pénzes Z, Kurucz E, Zsámboki J, Hultmark D, Andó I. Evolution of genes and repeats in the Nimrod superfamily. Mol Biol Evol. 2008;25:2337–2347. doi: 10.1093/molbev/msn180. [DOI] [PubMed] [Google Scholar]
- Stephan W. Tandem-repetitive noncoding DNA: forms and forces. Mol Biol Evol. 1989;6:198–212. doi: 10.1093/oxfordjournals.molbev.a040542. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thornton K, Andolfatto P. Approximate Bayesian inference reveals evidence for a recent, severe bottleneck in a Netherlands population of Drosophila melanogaster. Genetics. 2006;172:1607–1619. doi: 10.1534/genetics.105.048223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokusumi T, Shoue DA, Tokusumi Y, Stoller JR, Schulz RA. New hemocyte-specific enhancer-reporter transgenes for the analysis of hematopoiesis in Drosophila. Genesis. 2009;47:771–774. doi: 10.1002/dvg.20561. [DOI] [PubMed] [Google Scholar]
- Ye YH, Chenoweth SF, McGraw EA. Effective but costly, evolved mechanisms of defense against a virulent opportunistic pathogen of Drosophila melanogaster. PLoS Pathog. 2009;5:e1000358. doi: 10.1371/journal.ppat.1000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.