Abstract
Signatures of balancing selection operating on specific gene loci in endemic pathogens can identify candidate targets of naturally acquired immunity. In malaria parasites, several leading vaccine candidates convincingly show such signatures when subjected to several tests of neutrality, but the discovery of new targets affected by selection to a similar extent has been slow. A small minority of all genes are under such selection, as indicated by a recent study of 26 Plasmodium falciparum merozoite-stage genes that were not previously prioritized as vaccine candidates, of which only one (locus PF10_0348) showed a strong signature. Therefore, to focus discovery efforts on genes that are polymorphic, we scanned all available shotgun genome sequence data from laboratory lines of P. falciparum and chose six loci with more than five single nucleotide polymorphisms per kilobase (including PF10_0348) for in-depth frequency–based analyses in a Kenyan population (allele sample sizes >50 for each locus) and comparison of Hudson–Kreitman–Aguade (HKA) ratios of population diversity (π) to interspecific divergence (K) from the chimpanzee parasite Plasmodium reichenowi. Three of these (the msp3/6-like genes PF10_0348 and PF10_0355 and the surf4.1 gene PFD1160w) showed exceptionally high positive values of Tajima's D and Fu and Li's F indices and have the highest HKA ratios, indicating that they are under balancing selection and should be prioritized for studies of their protein products as candidate targets of immunity. Combined with earlier results, there is now strong evidence that high HKA ratio (as well as the frequency-independent ratio of Watterson's θ/K) is predictive of high values of Tajima's D. Thus, the former offers value for use in genome-wide screening when numbers of genome sequences within a species are low or in combination with Tajima's D as a 2D test on large population genomic samples.
Keywords: tests of neutrality, allele frequency, polymorphism and divergence, immunity, antigens
Introduction
Identifying the most important targets of immunity expressed by large and complex eukaryotic pathogens is difficult but can benefit from studies of pathogen genetics and evolution. Experimental blood-stage infection and challenge studies on genetically crossed rodent malaria parasites in mice have mapped important targets of parasite strain-specific immunity to the loci encoding merozoite surface protein 1 (MSP1) (Martinelli et al. 2005; Cheesman et al. 2009) and apical membrane antigen 1 (AMA1) (Pattaradilokrat et al. 2007), with results indicating there may be few other such important polymorphic targets in that experimental system. However, human malaria parasites such as Plasmodium falciparum encode many proteins that are not present in rodent malaria parasites, the importance of which must be largely investigated without genetic crossing and infection experiments, as a relatively inaccessible experimental primate model makes such approaches difficult (Hayton et al. 2008).
A population genetic approach is to consider the effects of frequency-dependent selection on pathogens due to the memory component of acquired immune responses, which will generally lead to balancing selection maintaining polymorphism of the genes encoding immune targets in endemic pathogens (Conway and Polley 2002). A series of initial studies on the statistical distribution of DNA sequence polymorphism within and among populations has indicated strong signatures of balancing selection on particular vaccine candidate antigens of the malaria parasite P. falciparum, including AMA1 (Polley and Conway 2001; Cortes et al. 2003; Polley et al. 2003), MSP1 (Conway et al. 2000), MSP2 (Conway 1997; Ferreira and Hartl 2007), MSP3 (Polley et al. 2007), erythrocyte-binding antigen 175 (Baum et al. 2003; Verra et al. 2006), and thrombospondin-related adhesive protein (Weedall et al. 2007).
To identify new candidate targets of naturally acquired immunity in P. falciparum, we previously made a prospective search for signatures of balancing selection in less-studied merozoite-stage protein genes (Tetteh et al. 2009). Out of a panel of 26 genes screened, 1 had a strong signature (PF10_0348, a gene predicted to encode an MSP3/6-like protein) (Tetteh et al. 2009), for which the protein has been recently characterized independently and named DBLMSP (merozoite surface protein containing a duffy binding–like [DBL] domain) (Wickramarachchi et al. 2009). This hit rate is low, consistent with expectations that balancing selection generally operates on only a small minority of genes (Bubb et al. 2006; Charlesworth 2006; Andres et al. 2009) and prompts consideration of modifications that could increase efficiency in screening for such genes. We had selected that initial panel for study regardless of any preexisting information on polymorphism as such data were scant and had taken a two-step approach to 1) first identify genes with a high ratio of polymorphism (π) to interspecific divergence (K, from the chimpanzee parasite Plasmodium reichenowi) in the Hudson–Kreitman–Aguade (HKA) test or a skew in the intraspecific versus interspecific nonsynonymous (NS) to synonymous (S) ratios in the McDonald–Kreitman (MK) test, by sequencing the genes from a global sample of P. falciparum laboratory isolates and P. reichenowi, and 2)select the genes with highest positive HKA or MK ratios for endemic population–based analysis of allele frequency distributions with Tajima's D and Fu and Li's F indices.
The expanding availability of data from shotgun genome sequences of different P. falciparum isolates (Jeffares et al. 2007; Mu et al. 2007; Volkman et al. 2007) together with highly accessible tools for browsing such data (Aurrecoechea et al. 2009) now allow easier and rapid screening for genes showing an unusually high level of polymorphism, so these can be immediately prioritized for population-based studies to test for signatures of selection. Here, to select candidate loci for population-based analysis in an endemic Kenyan site, we first identified six genes with an exceptionally high number of single nucleotide polymorphisms (SNPs) per kilobase, including PF10_0348 and five others that had not been previously tested (one other msp3/6-like paralogue and four unrelated genes expressed at the schizont and merozoite erythrocytic stage of infection). In the population analysis, the PF10_0348 gene gave a highly positive value of Tajima's D, as did two of the other five genes (the msp3/6-like PF10_0355 and the surf4.2 gene PFD1160w), whereas the remaining three were negative. Thus, the positive screening for polymorphic genes gave an enhanced hit rate, enabling us to confirm one and identify two new genes that have signatures of balancing selection. The protein products of allelic forms of these three genes are now prioritized for study as candidate targets of immunity. Using these and previous data on other genes, we show that allele frequency–based and polymorphism-versus-divergence analyses give independent signatures that are useful in screening for balancing selection, either separately or in 2D tests, and recommend the application of both in large-scale discovery approaches.
Materials and Methods
Screening for Highly Polymorphic P. falciparum Genes in PlasmoDB
We accessed available SNP and stage-specific transcript profile data on the PlasmoDB website (www.plasmodb.org) (Aurrecoechea et al. 2009) in November 2007 (PlasmoDB release 5.4) to screen for highly polymorphic genes expressed in the replicating asexual blood stage of P. falciparum. We found 490 annotated genes with evidence of maximum relative transcription at the later stages of the ∼48-h asexual cycle (schizonts and merozoites or >30 h into the cycle up until the stage of newly invaded “ring” stages) (Bozdech et al. 2003; Llinas et al. 2006), and on each of these we performed SNP density screening for all possible pairwise comparisons of alleles in available data from 13 laboratory P. falciparum isolates of diverse geographical sources: 3D7, HB3, Dd2, D10, V1_S, 7G8, RO33, K1, D6, FCC2, FCB, FCR3, and IT. Genes greater than 1.0 kb in length with density of at least around five SNPs per kilobase were identified as suitable for analysis, excluding those that had been previously focused on elsewhere as they encode vaccine candidates, or as members of the well-studied eba, Rh, and RhopH1/clag gene families. Six highly polymorphic genes with peak transcription during the schizont/merozoite stage were thereby selected for population-based analysis: the surf4.1 gene PFD0100c (on chromosome chr 4), surf4.2 gene PFD1160w (chr 4), PF07_0004 (chr 7), PF10_0342 (chr 10), the msp3/6-like PF10_0348 (chr 10), and surf 13.1 gene PF13_0075 (chr 13). As we encountered inconsistent amplification of PF13_0075 from field isolates in initial attempts, for the purpose of this study we replaced it with another highly polymorphic gene PF10_0355 (chr 10), which is more constitutively transcribed but that might function in schizonts and merozoites as it is MSP3/6-like in structure and paralogous to PF10_0348.
Sequencing Polymorphic Genes from an Endemic P. falciparum Population
Cross-sectional venous blood samples were obtained in August–September 1998 from a broad sample of children and adults (age range 0.5–80 years, median 8 years) resident in Ngerenya village in Kilifi District, Kenya, in a study of malaria reviewed and approved by the Scientific Steering Committee and the Ethics committee of the Kenya Medical Research Institute. This population had low to moderate endemic malaria transmission at the time of sampling (Mwangi et al. 2008). Parasite DNA was extracted from frozen heparinized venous blood samples from individuals that were slide positive for P. falciparum, using the QIAamp DNA Blood Mini Kit (QIAGEN, Crawley, UK). A population sample of at least 50 allele sequences of each gene was sought as optimal for frequency-based tests, so the six genes selected for study were each amplified from 90 of these individual parasite-positive DNA isolates, using oligonucleotide primers and amplification conditions listed in supplementary table S1 (part A) (Supplementary Material online). The P. reichenowi orthologues of these genes were amplified from DNA from the blood of a laboratory chimpanzee infected with P. reichenowi (CDC-1 “Oscar” strain), using primers and amplification conditions listed in supplementary table S1 (part B) (Supplementary Material online); the PF10_0348 ortholog sequence in P. reichenowi had been obtained previously (Tetteh et al. 2009). Polymerase chain reaction (PCR) products were purified with the QIAquick PCR Purification Kit (QIAGEN) and sequenced using the amplification primers and several internal sequencing primers, with ABI BIGDYE terminator v3.1 chemistry and electrophoresis on ABI 3130xl and ABI 3730 capillary sequencers (Applied Biosystems, Warrington, UK). Sequences were assembled, edited, and aligned using SeqMan and MegAlign software (Lasergene 7; DNASTAR, Madison, WI). For each locus, each isolate giving clear single-allele sequence representing the sole or predominant haploid parasite allelic type within the blood was analyzed, whereas isolates that showed mixed and electrophoretically superimposed allele sequences were not analyzed. All singleton nucleotide polymorphisms were confirmed by independent reamplification and resequencing from the relevant samples.
Tests of Neutrality and Linkage Disequilibrium
Tests for departures from neutrality were based on allele frequency distribution indices (Tajima's D, and Fu and Li's F) and comparisons of variation within and between species (HKA and MK ratios), using DnaSP5.0 (Rozas 2009). Tajima's D test takes into account the difference between average pairwise nucleotide diversity between sequences (π) and Watterson's population nucleotide diversity parameter theta (θ) expected under neutrality from the total number of segregating sites (S) (Tajima 1989c). Fu and Li's F test statistic is based on the difference between the observed number of singleton nucleotide polymorphisms and the number expected under neutrality given the total number of segregating sites and Watterson's estimate of nucleotide diversity (θ) (Fu and Li 1993). The HKA ratio is used to identify genes with exceptionally high ratios of polymorphism (π) over divergence (K) from a closely related species (in this case P. reichenowi) (Hudson et al. 1987; Innan 2006). The MK test counts the numbers of NS and S polymorphic sites within species and fixed differences between closely related species, using a Fisher's exact test on the 2 × 2 contingency table (McDonald and Kreitman 1991).
Linkage disequilibrium (LD), the association of nucleotide variants at different polymorphic sites, was assayed for all informative pairs of polymorphic sites within the genes, using DnaSP5.0. The r2 indices (square of the correlation coefficient of allelic states at each pair of loci) were calculated and tested for departures from randomness by Fisher's exact test. The value of r2 ranges from 0 to 1, although its values are constrained by the underlying allele frequencies (Hill and Robertson 1968).
Results
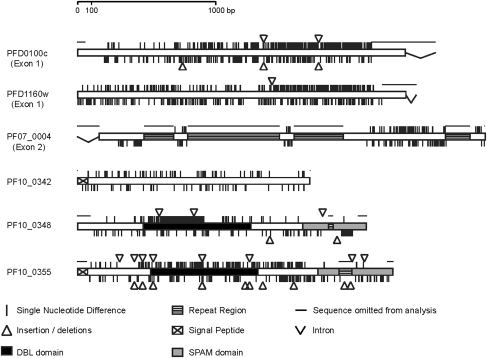
For each of the six loci, a majority of the 90 Kenyan isolates each yielded a clear single-allele sequence, representing the P. falciparum allelic type that was dominant within each blood sample. Mixed P. falciparum genotype infections in a proportion of isolates yielded electrophoretically superimposed allelic sequences, excluded from analysis here (numbers excluded generally differ among genes due to the relative amounts of sequence polymorphism, particularly disruptive effects of repeat polymorphisms, and stochastic effects of PCR). Thus, the sample size of alleles for each locus was in excess of the target number of 50 for statistical power, ranging from 51 (for PFD0100c) to 79 (for PF10_0342) (table 1). Three of the genes (PF07_0004, PF10_0348, and PF10_0355) contained regions of repeat sequences that were excluded from the alignment-based tests below (the amino acid translations of these repeats are shown separately in supplementary fig. S1, Supplementary Material online). The positions of nucleotide polymorphisms in the genes in this population and fixed differences from P. reichenowi are shown schematically in figure 1. Full alignments of all alleles for each of the genes are given in supplementary figure S2 (Supplementary Material online) and in EMBL Nucleotide Sequence Database alignment files (accession numbers are listed in supplementary table S2, Supplementary Material online).
Table 1.
Tests of Neutrality on Polymorphisms in Six Plasmodium falciparum Genes in a Kenyan Population.
| MK |
||||||||||||
| Number of isolates (n) | π (×10−3) | K (×10−3) | HKAr (π/K) | S |
NS |
MK | Fu and Li’s F | |||||
| Locus | nt | Fixed | Poly | Fixed | Poly | P value | Tajima's D | |||||
| PFD0100c | 51 | 2,061a,b,c | 29.6 | 162.4 | 0.18 | 40 | 49 | 142 | 242 | 0.18 | −0.80 | −1.37 |
| PFD1160w | 69 | 2,199a,b,c | 43.0 | 153.8 | 0.28 | 39 | 25 | 178 | 216 | 0.02* | 1.58g | 1.78* |
| PF07_0004 | 59 | 1,152a,b,c,d | 16.6 | 157.8 | 0.11 | 27 | 23 | 90 | 64 | 0.62 | −0.48 | −0.04 |
| PF10_0342 | 79 | 1,680 | 6.5 | 40.7 | 0.16 | 10 | 13 | 39 | 44 | 0.82 | −0.16 | −0.24 |
| PF10_0348 | 56 | 1,896a,c,d,e,f | 36.3 | 70.4 | 0.52 | 18 | 62 | 57 | 133 | 0.24 | 1.83g | 2.11** |
| PF10_0355 | 53 | 2,073a,d,e,f | 33.8 | 99.2 | 0.34 | 30 | 46 | 101 | 116 | 0.35 | 3.44*** | 2.98** |
NOTE.—Nt, number of aligned nucleotide positions analyzed. Full alignments shown in supplementary figure S2 (Supplementary Material online), with P. reichenowi orthologues and the reference 3D7 sequence for comparison.
Less sequence aligned when Plasmodium reichenowi added to analysis (PF10_0348 n = 1818, PF10_0355 n = 2061, PFD0100c n = 1893, PFD1160w n = 2031, and PF07_0004 n = 996).
Specific region generated (PFD1160w exon 1, PFD0100c exon 1, and PF07_0004 exon 2).
Complex codons not analyzed by DNAsp software (PF10_0348 n = 3, PFD0100c n = 17, PFD1160w n = 17, and PF07_0004 n = 3).
Repeats removed from gene sequences for analysis.
Divergent allele in a minority of samples removed from alignment-based analysis (PF10_0348 n = 7 and PF10_0355 n = 10, included in supplementary fig. S2, Supplementary Material online).
Stop codon in eight alleles of PF10_0348, and eight stop codons in the P. reichenowi orthologue of PF10_0355 (codons removed from analysis).
Sliding window analysis shows significant regions (windows of 100 nucleotide sites, step size 50 sites).
*P < 0.05, **P < 0.02, ***P < 0.001.
FIG. 1.
Schematic diagram of each of the six gene loci studied. The positions of nucleotide polymorphisms (within Plasmodium falciparum, marked above each gene scheme) and fixed differences (between P. falciparum and Plasmodium reichenowi, marked below each gene scheme) in nonrepeat regions are indicated. Repeat regions (in three of the genes) are excluded from alignment-based analysis (their translated sequences for representative alleles are shown in supplementary fig. S1, Supplementary Material online). Alignments of the nonrepeat sequences of all alleles and P. reichenowi are shown for each of the loci in supplementary figure S2 (Supplementary Material online).
Of the six genes in this Kenyan population sample, PF10_0342 showed the lowest nucleotide polymorphism (π = 6.5 × 10−3) and PFD1160w showed the highest (π = 43.0 × 10−3) (table 1). Considering the HKA ratio, of polymorphism (π) divided by interspecific divergence from P. reichenowi (K), values ranged from 0.11 for PF07_0004 up to 0.52 for PF10_0348 (table 1). These values are higher than those indicated for the genes from a screen of shotgun genome sequence data (accessible at www.plasmodb.org) as some of the gene regions with low level of polymorphism were not studied here (e.g., the first exon of PF07_0004, the second exon of PFD1160w, and the second and third exons of PFD0100c). Only one locus (PFD1160w) had a significant MK test result showing an excess of NS versus S polymorphisms compared with fixed differences (P = 0.02). Three of the genes (PFD1160w, PF10_0348, and PF10_0355) had highly positive values of Tajima's D and Fu and Li's F indices, indicative of balancing selection (these positive values, respectively, indicate fewer rare alleles and fewer singletons than expected under neutrality) (table 1). The PF10_0348 gene had previously been studied in a Gambian population in which similar indices were obtained, but the positive result for the other two genes is unprecedented. The remaining three genes analyzed showed negative values of these indices.
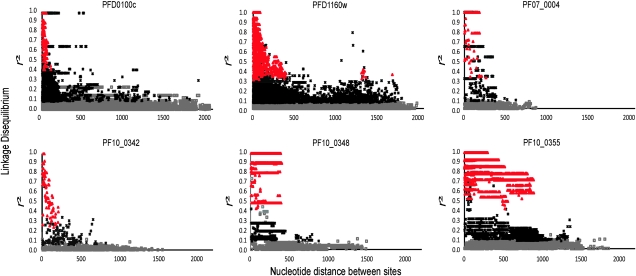
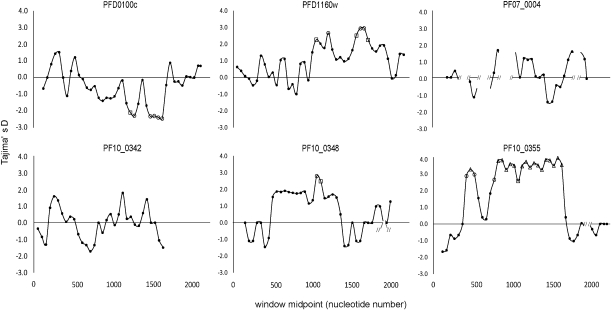
There is evidence of extensive recombination in all these genes within the population, as LD indices are maximal between sites that are closely situated together, and significant values are mostly among sites separated by <0.5 kb (fig. 2). The PF10_0355 gene has a marked dimorphic structure throughout much of its sequence (supplementary fig. S2, Supplementary Material online), so there are more extended strong LD values in this gene (fig. 2). A high recombination rate indicates that signatures of selection on one part of a gene are not likely to be reflected in the pattern of nucleotide polymorphism throughout the gene, so sliding window analyses were performed for Tajima's D (fig. 3) and Fu and Li's F (supplementary fig. S3, Supplementary Material online). These show exceptionally high values of both indices for aligned nucleotides 1000–1200 and 1500–1900 in PFD1160w and aligned nucleotides 500–1200 in PF10_0348, with PF10_0355 having very high values throughout most of its dimorphic sequence and lower values only near the 5′- and 3′-ends of the alignment. The PFD1160w gene additionally shows an unusual signature of strong LD between a limited number of sites widely separated (∼1500 bp apart) in the gene, indicating that there may be epistatic interactions between polymorphisms in different parts of the protein. The other three genes did not show any sliding windows of significantly elevated Tajima's D or Fu and Li's F values, consistent with their lack of overall departure from neutrality. In contrast, two windows of significantly negative values for PFD0100c are likely to reflect directional selection on polymorphisms in that gene.
FIG. 2.
LD in each of the six gene loci studied in the Kenyan population. The r2 values for all pairwise tests between polymorphic sites are shown. Red and black symbols indicate values that are statistically significant (red points remain significant after Bonferroni correction for the multiple tests), whereas open gray symbols indicate nonsignificant values.
FIG. 3.
Sliding window analysis of Tajima's D index along the aligned sequences of all alleles for each of the six genes studied in the Kenyan population. The x axis shows the midpoint of contiguous windows of 100 bp with step size of 50 bp for the portions of the genes sequenced (nucleotide positions in each gene are given as those in the allele of the reference genome strain 3D7): PFD0100c exon 1 positions 61–2124; PFD1160w exon 1 positions 1–2205; PF07_0004 exon 2 positions 124–2781 (positions on x axis condensed as repeat sequences were deleted); PF10_0342 positions 4–1683; PF10_0348 positions 97–2028; and PF10_0355 positions 70 – 2241. Symbols for points indicating windows with significant departures from zero: P < 0.05, open square; P < 0.01, open circle; P < 0.001, open triangle.
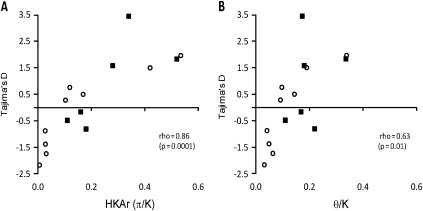
These three genes with highest values of Tajima's D (and Fu and Li's F) also had highest values of the polymorphism-versus-divergence HKA ratio (π/K), a correlation among the indices similar to that seen for a set of genes previously studied in a Gambian population (fig. 4A). This supports the suggestion that the HKA ratio (which requires fewer allelic sequences to derive an accurate estimate than Tajima's D) may be a useful screen for genes under balancing selection when population-based data are limited. As both these indices involve the pairwise nucleotide diversity parameter π as a numerator, despite being based otherwise on very different data, they are not fully independent statistically as some correlation may be due to variance in π among loci. However, by substituting Watterson's θ as the nucleotide diversity parameter (based on numbers of polymorphic sites and independent of allele frequencies) in place of π, in a modified polymorphism-versus-divergence ratio (θ/K), this can be tested for correlation with Tajima's D. A positive correlation between these indices would not be expected under neutrality, as θ occurs as a negative term toward calculating Tajima's D = (π − θ)/SD (π − θ) (Tajima 1989c). This correlation is significantly positive for the polymorphic genes studied (fig. 4B), which reflects independently concordant signals of selection from the frequency-based and polymorphism-versus-divergence indices.
FIG. 4.
Scatterplot of Tajima's D index and two diversity-versus-divergence ratios for six genes studied here in the Kenyan population (black square points) and nine genes studied previously in a Gambian population (open circle points). (A) Tajima's D correlated with the π/K ratio (the conventional HKA ratio) and (B) Tajima's D correlated with the θ/K ratio (a form of HKA ratio that is independent of allele frequencies). One of the genes (PF10_0348) had been studied in both populations (with similar results as indicated by nearly overlapping points furthest right on each scatterplot), so data for this gene were only included from one of the populations (it made no difference which) in the correlation analyses.
Discussion
It remains vital to identify important targets of immunity among the many proteins expressed by malaria parasites, apart from the relatively small set that have been studied already as vaccine candidates. This study identifies three genes to be under strong balancing selection (the surf4.2 gene PFD1160w, the msp3/6-like dblmsp gene PF10_0348, and the msp3/6-like PF10_0355), out of six highly polymorphic genes subjected to allele frequency–based analyses on a Kenyan population and tests of diversity versus divergence from P. reichenowi. One of these (PF10_0348) was previously identified as the only gene under strong balancing selection after analysis of 26 merozoite-stage genes that were not chosen on the basis of prior polymorphism data. As expected, focusing analysis on genes with high nucleotide diversity yields a higher hit rate of genes with signatures of balancing selection. The potential extra benefit of using a polymorphism-versus-divergence ratio in such a screen should encourage more sequencing of related species such as P. reichenowi (Jeffares et al. 2007; Krief et al. 2010; Prugnolle et al. 2010).
Population genomic studies on P. falciparum should identify further loci under balancing selection, and these should ideally be based on large samples from each population, such as the single endemic village community in Kenya studied here. Admixture or pooling of samples between two divergent populations could lead to elevated Tajima's D values generally (Tajima 1989a), and this can be reduced by sampling individual well-defined populations that are not derived by secondary contact between two separate populations. In contrast, a historical population expansion would tend to lead to reduced Tajima's D values (Tajima 1989b), and this appears to be a feature of P. falciparum populations in Africa (Joy et al. 2003; Verra et al. 2006). Importantly, inferences of nonneutrality should be based on comparisons across a set of loci, as we have demonstrated here and that should be realized fully in genome scale population analyses (Nielsen 2001).
Immunological analyses of allelic protein products of each of the three hits from the current study can now be prioritized. Initial studies on the proteins expressed by these genes support the idea that they could be targets of immunity. The SURFIN4.2 protein accumulates in the parasitophorous vacuole (the compartment between the intraerythrocytic parasite and the erythrocyte cytoplasm) and appears to be transported to the infected erythrocyte membrane as well as being associated with released merozoites. High concentrations of rabbit antibodies raised to SURFIN4.2 had an inhibitory effect on erythrocyte invasion, suggesting that the protein may be accessible to inhibitory antibodies in vivo (Winter et al. 2005). The DBLMSP is specifically located on the merozoite surface, and a recombinant protein fragment incorporating the DBL domain showed binding to human erythrocyte surface that was neuraminidase and trypsin sensitive and that could be inhibited by murine antibodies raised to the protein (Wickramarachchi et al. 2009). Although expression of PF10_0355 transcript is not narrowly stage specific, one report indicates that antibodies raised to a recombinant protein fragment recognize the merozoite surface (Singh et al. 2009), and other data indicate it is present on a minority of merozoites within a laboratory cloned line (Knuepfer E and Holder AA, personal communication). This raises the important question of whether these proteins are variantly expressed in abundance or cellular location within or on the parasite surface, as well as being structurally polymorphic among allelic forms.
Although each of these genes occurs at a single locus, it is possible that some polymorphisms are derived from more complex evolutionary history than would classically occur at a single-copy gene. Gene copy number variation has been identified to be widespread in the P. falciparum genome, not only in subtelomeric regions where it is concentrated but also at many loci on different chromosomes (Kidgell et al. 2006; Ribacke et al. 2007; Cheeseman et al. 2009; Mackinnon et al. 2009). For example, the dblmsp gene (PF10_0348) has a paralogous copy in 3 of 14 laboratory isolates studied (Tetteh et al. 2009), and one of the polymorphic genes without a selective signature in this study, the surfin4.1 gene (PFD0100c), has six copies in laboratory isolate FCR3 and one copy in other isolates (Mphande et al. 2008).
Importantly for future studies, we show that the allele frequency–based Tajima's D and the polymorphism-versus-divergence ratio (π/K or the allele frequency–independent Watterson's θ/K) are significantly positively correlated, indicating that signatures of selection are discernable from each type of index with some concordance. Thus, 2D tests based on allele frequency distribution spectra and ratios of polymorphism to interspecific divergence can be recommended (Innan 2006; Zhai et al. 2009), for large-scale genome-wide screens to identify further signatures of balancing selection on malaria parasites.
Supplementary Material
Supplementary tables S1 and S2 and figures S1–S3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We are grateful to Tabitha Mwangi for leading the epidemiological study that provided the population sample of Plasmodium falciparum parasites, Brett Lowe for encouragement of this investigation and coordination of laboratory management, Alan Thomas and Clemens Kocken for provision of Plasmodium reichenowi DNA, and all colleagues who have discussed ideas on this investigation.
This work is published with the permission of the director of the Kenya Medical Research Institute (KEMRI). This work was supported by Wellcome Trust (074695/Z/04/B).
References
- Andres AM, Hubisz MJ, Indap A, et al. (12 co-authors) Targets of balancing selection in the human genome. Mol Biol Evol. 2009;26:2755–2764. doi: 10.1093/molbev/msp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurrecoechea C, Brestelli J, Brunk BP, et al. (25 co-authors) PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–D543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum J, Thomas AW, Conway DJ. Evidence for diversifying selection on erythrocyte-binding antigens of Plasmodium falciparum and P. vivax. Genetics. 2003;163:1327–1336. doi: 10.1093/genetics/163.4.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1:85–100. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubb KL, Bovee D, Buckley D, et al. (12 co-authors) Scan of human genome reveals no new loci under ancient balancing selection. Genetics. 2006;173:2165–2177. doi: 10.1534/genetics.106.055715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth D. Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2006;2:e64. doi: 10.1371/journal.pgen.0020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IH, Gomez-Escobar N, Carret CK, Ivens A, Stewart LB, Tetteh KK, Conway DJ. Gene copy number variation throughout the Plasmodium falciparum genome. BMC Genomics. 2009;10:353. doi: 10.1186/1471-2164-10-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheesman S, Tanabe K, Sawai H, O'Mahony E, Carter R. Strain-specific immunity may drive adaptive polymorphism in the merozoite surface protein 1 of the rodent malaria parasite Plasmodium chabaudi. Infect Genet Evol. 2009;9:248–255. doi: 10.1016/j.meegid.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Conway DJ. Natural selection on polymorphic malaria antigens and the search for a vaccine. Parasitol Today. 1997;13:26–29. doi: 10.1016/s0169-4758(96)10077-6. [DOI] [PubMed] [Google Scholar]
- Conway DJ, Cavanagh DR, Tanabe K, et al. (12 co-authors) A principal target of human immunity to malaria identified by molecular population genetic and immunological analyses. Nat Med. 2000;6:689–692. doi: 10.1038/76272. [DOI] [PubMed] [Google Scholar]
- Conway DJ, Polley SD. Measuring immune selection. Parasitology. 2002;125:S3–S16. doi: 10.1017/s0031182002002214. [DOI] [PubMed] [Google Scholar]
- Cortes A, Mellombo M, Mueller I, Benet A, Reeder JC, Anders RF. Geographical structure of diversity and differences between symptomatic and asymptomatic infections for Plasmodium falciparum vaccine candidate AMA1. Infect Immun. 2003;71:1416–1426. doi: 10.1128/IAI.71.3.1416-1426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MU, Hartl DL. Plasmodium falciparum: worldwide sequence diversity and evolution of the malaria vaccine candidate merozoite surface protein-2 (MSP-2) Exp Parasitol. 2007;115:32–40. doi: 10.1016/j.exppara.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Fu Y-X, Li W-H. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayton K, Gaur D, Liu A, et al. (15 co-authors) Erythrocyte binding protein PfRH5 polymorphisms determine species-specific pathways of Plasmodium falciparum invasion. Cell Host Microbe. 2008;4:40–51. doi: 10.1016/j.chom.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A. Linkage disequilibrium in finite populations. Theor Appl Genet. 1968;38:226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kreitman R, Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H. Modified Hudson-Kreitman-Aguade test and two-dimensional evaluation of neutrality tests. Genetics. 2006;173:1725–1733. doi: 10.1534/genetics.106.056242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffares DC, Pain A, Berry A, et al. (15 co-authors) Genome variation and evolution of the malaria parasite Plasmodium falciparum. Nat Genet. 2007;39:120–125. doi: 10.1038/ng1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy DA, Feng X, Mu J, et al. (12 co-authors) Early origin and recent expansion of Plasmodium falciparum. Science. 2003;300:318–321. doi: 10.1126/science.1081449. [DOI] [PubMed] [Google Scholar]
- Kidgell C, Volkman SK, Daily J, et al. (14 co-authors) A systematic map of genetic variation in Plasmodium falciparum. PLoS Pathog. 2006;2:e57. doi: 10.1371/journal.ppat.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krief S, Escalante AA, Pacheco MA, et al. (18 co-authors) On the diversity of malaria parasites in African apes and the origin of Plasmodium falciparum from Bonobos. PLoS Pathog. 2010;6:e1000765. doi: 10.1371/journal.ppat.1000765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llinas M, Bozdech Z, Wong ED, Adai AT, DeRisi JL. Comparative whole genome transcriptome analysis of three Plasmodium falciparum strains. Nucleic Acids Res. 2006;34:1166–1173. doi: 10.1093/nar/gkj517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinnon MJ, Li J, Mok S, Kortok MM, Marsh K, Preiser PR, Bozdech Z. Comparative transcriptional and genomic analysis of Plasmodium falciparum field isolates. PLoS Pathog. 2009;5:e1000644. doi: 10.1371/journal.ppat.1000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli A, Cheesman S, Hunt P, Culleton R, Raza A, Mackinnon M, Carter R. A genetic approach to the de novo identification of targets of strain-specific immunity in malaria parasites. Proc Natl Acad Sci U S A. 2005;102:814–819. doi: 10.1073/pnas.0405097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Mphande FA, Ribacke U, Kaneko O, Kironde F, Winter G, Wahlgren M. SURFIN4.1, a schizont-merozoite associated protein in the SURFIN family of Plasmodium falciparum. Malar J. 2008;7:116. doi: 10.1186/1475-2875-7-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J, Awadalla P, Duan J, McGee KM, Keebler J, Seydel K, McVean GA, Su XZ. Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome. Nat Genet. 2007;39:126–130. doi: 10.1038/ng1924. [DOI] [PubMed] [Google Scholar]
- Mwangi TW, Fegan G, Williams TN, Kinyanjui SM, Snow RW, Marsh K. Evidence for over-dispersion in the distribution of clinical malaria episodes in children. PLoS One. 2008;3:e2196. doi: 10.1371/journal.pone.0002196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R. Statistical tests of selective neutrality in the age of genomics. Heredity. 2001;86:641–647. doi: 10.1046/j.1365-2540.2001.00895.x. [DOI] [PubMed] [Google Scholar]
- Pattaradilokrat S, Cheesman SJ, Carter R. Linkage group selection: towards identifying genes controlling strain specific protective immunity in malaria. PLoS One. 2007;2:e857. doi: 10.1371/journal.pone.0000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley SD, Chokejindachai W, Conway DJ. Allele frequency based analyses robustly identify sites under balancing selection in a malaria vaccine candidate antigen. Genetics. 2003;165:555–561. doi: 10.1093/genetics/165.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley SD, Conway DJ. Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics. 2001;158:1505–1512. doi: 10.1093/genetics/158.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley SD, Tetteh KK, Lloyd JM, Akpogheneta OJ, Greenwood BM, Bojang KA, Conway DJ. Plasmodium falciparum merozoite surface protein 3 is a target of allele-specific immunity and alleles are maintained by natural selection. J Infect Dis. 2007;195:279–287. doi: 10.1086/509806. [DOI] [PubMed] [Google Scholar]
- Prugnolle F, Durand P, Neel C, et al. (13 co-authors) African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. Proc Natl Acad Sci U S A. 2010;107:1458–1463. doi: 10.1073/pnas.0914440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribacke U, Mok BW, Wirta V, Normark J, Lundeberg J, Kironde F, Egwang TG, Nilsson P, Wahlgren M. Genome wide gene amplifications and deletions in Plasmodium falciparum. Mol Biochem Parasitol. 2007;155:33–44. doi: 10.1016/j.molbiopara.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Rozas J. DNA sequence polymorphism analysis using DnaSP. Methods Mol Biol. 2009;537:337–350. doi: 10.1007/978-1-59745-251-9_17. [DOI] [PubMed] [Google Scholar]
- Singh S, Soe S, Weisman S, Barnwell JW, Perignon JL, Druilhe P. A conserved multi-gene family induces cross-reactive antibodies effective in defense against Plasmodium falciparum. PLoS One. 2009;4:e5410. doi: 10.1371/journal.pone.0005410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. DNA polymorphism in a subdivided population: the expected number of segregating sites in the two-subpopulation model. Genetics. 1989a;123:229–240. doi: 10.1093/genetics/123.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. The effect of change in population size on DNA polymorphism. Genetics. 1989b;123:597–601. doi: 10.1093/genetics/123.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989c;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetteh KK, Stewart LB, Ochola LI, Amambua-Ngwa A, Thomas AW, Marsh K, Weedall GD, Conway DJ. Prospective identification of malaria parasite genes under balancing selection. PLoS One. 2009;4:e5568. doi: 10.1371/journal.pone.0005568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verra F, Chokejindachai W, Weedall GD, Polley SD, Mwangi TW, Marsh K, Conway DJ. Contrasting signatures of selection on the Plasmodium falciparum erythrocyte binding antigen gene family. Mol Biochem Parasitol. 2006;149:182–190. doi: 10.1016/j.molbiopara.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Volkman SK, Sabeti PC, DeCaprio D, et al. (28 co-authors) A genome-wide map of diversity in Plasmodium falciparum. Nat Genet. 2007;39:113–119. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- Weedall GD, Preston BM, Thomas AW, Sutherland CJ, Conway DJ. Differential evidence of natural selection on two leading sporozoite stage malaria vaccine candidate antigens. Int J Parasitol. 2007;37:77–85. doi: 10.1016/j.ijpara.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Wickramarachchi T, Cabrera AL, Sinha D, et al. (11 co-authors) A novel Plasmodium falciparum erythrocyte binding protein associated with the merozoite surface, PfDBLMSP. Int J Parasitol. 2009;39:763–773. doi: 10.1016/j.ijpara.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Winter G, Kawai S, Haeggstrom M, Kaneko O, von Euler A, Kawazu S, Palm D, Fernandez V, Wahlgren M. SURFIN is a polymorphic antigen expressed on Plasmodium falciparum merozoites and infected erythrocytes. J Exp Med. 2005;201:1853–1863. doi: 10.1084/jem.20041392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai W, Nielsen R, Slatkin M. An investigation of the statistical power of neutrality tests based on comparative and population genetic data. Mol Biol Evol. 2009;26:273–283. doi: 10.1093/molbev/msn231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.