The key function of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide
The ribosome monitors the nascent peptides and stalls in response to specific peptide sequences through ill-defined mechanism(s). This study by the Mankin laboratory identifies a posttranscriptionally modified 23S rRNA nucleotide located at the entrance of the ribosome exit tunnel together with a known nascent chain sensor nucleotide as key players in the recognition and relay of the stalling signal to the peptidyl transferase centre.
Keywords: erythromycin, nascent peptide, ribosome, rRNA, translation
Abstract
The ribosome is able to monitor the structure of the nascent peptide and can stall in response to specific peptide sequences. Such programmed stalling is used for the regulation of gene expression. The molecular mechanisms of the nascent-peptide recognition and ribosome stalling are unknown. We identified the conserved and posttranscriptionally modified 23S rRNA nucleotide m2A2503 located at the entrance of the ribosome exit tunnel as a key component of the ribosomal response mechanism. A2503 mutations abolish nascent-peptide-dependent stalling at the leader cistrons of several inducible antibiotic resistance genes and at the secM regulatory gene. Remarkably, lack of the C2 methylation of A2503 significantly function induction of expression of the ermC gene, indicating that the functional role of posttranscriptional modification is to fine-tune ribosome–nascent peptide interactions. Structural and biochemical evidence suggest that m2A2503 may act in concert with the previously identified nascent-peptide sensor, A2062, in the ribosome exit tunnel to relay the stalling signal to the peptidyl transferase centre.
Introduction
All polypeptides synthesized by the ribosome pass through the nascent-peptide exit tunnel (NPET), which originates at the peptidyl transferase centre (PTC), spans the body of the large ribosomal subunit, and opens at its opposite side. Although viewed initially as an inert hole, the tunnel is recognized now as an important functional entity of the ribosome. In particular, the tunnel has a central function in the remarkable ability of the ribosome to sense the structure of the nascent peptide and modulate the progression of protein synthesis in response to specific nascent-peptide sequences (Tenson and Ehrenberg, 2002; Jenni and Ban, 2003). Nonetheless, despite the functional significance, nascent-peptide recognition and the ensuing ribosomal response remain among the least understood fundamental properties of the ribosome.
The nascent-peptide-dependent ribosome response is often manifested in the form of translation arrest, referred to as ‘ribosome stalling,' that controls expression of a number of bacterial and eukaryotic genes (Morris and Geballe, 2000; Gong and Yanofsky, 2002; Nakatogawa and Ito, 2002; Fang et al, 2004; Chiba et al, 2009; Tanner et al, 2009). In an important class of genes regulated by this mechanism are those conferring antibiotic resistance, in particular, inducible erm genes that protect bacteria from macrolide antibiotics (Weisblum, 1995; Ramu et al, 2009). The prototype macrolide erythromycin and its derivatives bind in the ribosome exit tunnel and inhibit translation by obstructing the tunnel (Schlunzen et al, 2001; Tu et al, 2005). The methyltransferase encoded in the erm genes renders cells drug resistant by methylating 23S rRNA in the macrolide-binding site. Expression of the ermC gene (the most extensively analysed member of the erm family) is activated in the presence of erythromycin because of nascent-peptide-dependent ribosome stalling at the ninth codon of the 19-codon regulatory ORF ermCL located upstream of ermC (Figure 1A). The formation of stalled ribosome complex (SRC) triggers rearrangement of the mRNA secondary structure, resulting in activation of ermC translation (Gryczan et al, 1980; Horinouchi and Weisblum, 1980). The ribosome stalled at the ermCL ORF carries peptidyl-tRNA with the 9-amino-acid nascent-peptide MGIFSIFVI, whose C-terminal 4-amino-acid sequence, IFVI, is critical for stalling. Through a poorly understood mechanism, the presence of this sequence and the inducing antibiotic in the exit tunnel induces an allosteric change in the PTC, which leads to arrest of translation because the stalled ribosome is unable to catalyse formation of the next peptide bond (Vazquez-Laslop et al, 2008).
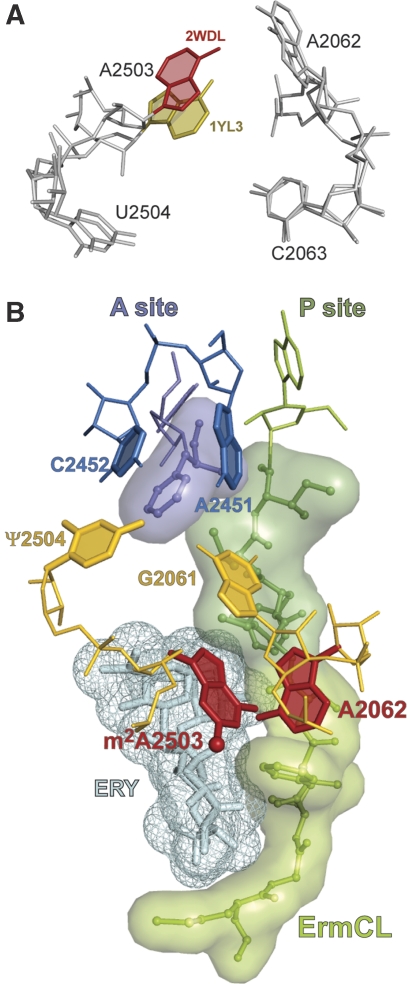
Figure 1.
Nascent peptide in the ribosome exit tunnel. (A) The structure of the inducible ermC operon where the ermC gene is preceded by a regulatory ORF ermCL. Drug- and nascent-peptide-dependent ribosome stalling at ermCL ORF changes the conformation of the mRNA intergenic region (schematically shown as a two-hairpin structure), thereby releasing translational attenuation of ermC. (B, C) Erythromycin and the ErmCL nascent peptide in the ribosome exit tunnel (viewed from the PTC down the tunnel). In the vacant tunnel (B), the nascent-peptide sensor, A2062, is free to rotate into the tunnel lumen. Binding of antibiotic (‘ERY') narrows the tunnel (C). In the constricted tunnel, the ErmCL nascent peptide drives A2062 toward the tunnel wall, where it comes into close proximity to A2503. (D) Conformational flexibility of A2062. The orientations of the A2062 base are shown for the apo structure of the Haloarcula marismortui 50S ribosomal subunit (blue) (PDB accession number 3CC2) (Blaha et al, 2008) and for the 50S subunit complexed with a transition state analog (biege) (1VQ7) (Schmeing et al, 2005). The A2503 base is coloured red. A possible hydrogen bond between A2062 and A2503 is indicated by a dashed line.
Besides ermC, expression of several other erm genes is also controlled by drug- and nascent-peptide-dependent translation arrest (Murphy, 1985; Hue and Bechhofer, 1992; Kwon et al, 2006). The sequences of the peptides encoded in regulatory ORFs of various inducible erm genes exhibit considerable variation (reviewed in Ramu et al, 2009). Almost nothing is known about molecular mechanisms of the ribosomal stalling during translation of these diverse leader peptides; it is even unclear whether related or different ribosomal elements are engaged in response to regulatory peptides of different erm genes.
The phenomenon of gene regulation through programmed, nascent-peptide-dependent ribosome stalling expands beyond antibiotic resistance genes. In Escherichia coli, the nascent-peptide-controlled translation arrest at the secM ORF activates expression of the secA gene, whereas ribosome stalling at the tnaC ORF regulates expression of the tryptophanase operon (Gong and Yanofsky, 2002; Nakatogawa and Ito, 2002). In both cases, the details of the nascent-peptide recognition in the NPET remain elusive, although the PTC emerges as the universal destination site of the stalling signal (Ito et al, 2010).
Biochemical and structural analyses reveal the ribosome as an RNA machine. The major functional centres of the ribosome, such as decoding and PTCs, are built of rRNA (Noller, 1991; Ramakrishnan, 2002). Although the extensions of several ribosomal proteins (L4, L22, L23) reach the exit tunnel, the walls of the NPET are formed primarily of rRNA residues, suggesting that rRNA-based mechanisms should have a central function in the nascent-peptide recognition and response. This notion is further supported by the clustering of modified nucleotides around the tunnel. About one-third of the posttranscriptionally modified residues in rRNA of the bacterial large ribosomal subunit are found in the vicinity of the NPET (Chow et al, 2007). Such conspicuous congregation of modified nucleotides close to the nascent-peptide passage strongly points to their possible involvement in functional interactions with the protein being made. However, until now, only sketchy data are available about function of rRNA in the nascent-peptide-mediated modulation of translation, and no evidence revealing physiological significance of posttranscriptional modifications in the ribosome tunnel has been obtained.
Several rRNA mutations that affect activation of expression of genes controlled by secM or tnaC ORFs have been mapped (Nakatogawa and Ito, 2002; Cruz-Vera et al, 2005; Lawrence et al, 2008; Yang et al, 2009). Yet, the lack of direct evidence that rRNA mutations prevent ribosome stalling makes interpretation of the data difficult.
Importantly, it is essentially unknown how the information about the presence of the stalling peptide sequence in the tunnel is communicated to the PTC. Recent cryo-EM reconstructions of the ribosome stalled at tnaC showed proximity of the nascent peptide to certain rRNA residues in the exit tunnel and allowed proposal of several putative routes that could relay the stalling signal from the NPET to the PTC (Seidelt et al, 2009). Unfortunately, the available data do not allow differentiation between alternative pathways. Furthermore, mere proximity of a nascent peptide to a certain rRNA nucleotide in the tunnel does not necessarily reveal functional interaction.
In our previous work, we identified a conserved 23S rRNA residue, A2062, located in the PTC-proximal segment of the NPET as a key component of the ErmCL-peptide-sensing mechanism (Vazquez-Laslop et al, 2008). The mutations of A2062 prevented ermC induction by interfering with ErmCL-dependent ribosome stalling. We showed that conformational rearrangement of A2062, induced by its interaction with the ErmCL nascent peptide in the tunnel constricted by erythromycin, must be somehow relayed to the PTC in order to trigger translation arrest (Vazquez-Laslop et al, 2008). However, the mechanism of communication between the tunnel and the PTC, which is central to the nascent-peptide-mediated ribosome response, was not identified, although several putative signal-relay pathways have been proposed (Vazquez-Laslop et al, 2008; Seidelt et al, 2009; Chirkova et al, 2010).
Here, we present biochemical, genetic, and structural evidence that conserved and posttranscriptionally modified 23S rRNA residue m2A2503 has a key function in the molecular mechanism of the nascent-peptide recognition and response. We demonstrate that m2A2503 may act in concert with A2062 in relaying the signal from the exit tunnel to the peptidyl transferase active site and that this mechanism enables the ribosomal response to ErmCL and several other stalling nascent peptides. Furthermore, we show that posttranscriptional modification of A2503 is important for its functions in the nascent-peptide response mechanism, thereby presenting the first example of participation of a posttranscriptional modification in modulating functional interactions between the ribosome and the polypeptide in the exit tunnel.
Results
Our previous studies of nascent-peptide-dependent ribosome stalling at the ermCL regulatory ORF of the ermC gene revealed the critical importance of the 23S rRNA residue A2062 as a nascent-peptide sensor. It remained unclear, however, how the stalling signal, sensed by A2062, is transmitted to the PTC. A2062 is one of the most flexible nucleotides in the NPET (Fulle and Gohlke, 2009), and in various ribosomal crystallographic complexes it is seen in dramatically different conformations (Hansen et al, 2002; Blaha et al, 2008; Voorhees et al, 2009). In the vacant NPET, A2062 base can protrude into the tunnel lumen but must move closer to the tunnel wall when erythromycin and the ErmCL nascent peptide fill the tunnel (Figure 1B and C). This rearrangement places A2062 into immediate proximity to another 23S rRNA residue, A2503. The distance between the N7 atom of A2062 and the exocyclic amino group of A2503 can be as little as 2.9 Å, consistent with the formation of a hydrogen bond between these two bases (Ban et al, 2000; Blaha et al, 2008) (Figure 1D). A2503 is located in the NPET, but its immediate nucleotide neighbours reach into the active site of the PTC. Like most of the known functionally important nucleotides, A2503 is highly conserved (>99% conservation in bacteria and eukarya). Furthermore, in E. coli, A2503 is one of the few 23S rRNA residues that are posttranscriptionally modified; this underscores its potential functional significance in the ribosome. The conservation of A2503, its posttranscriptional modification, and its location within the ribosome make this rRNA residue a suitable candidate to have a central function in relaying the stalling signal from the nascent peptide in the NPET to the PTC. To test this hypothesis, we first asked whether posttranscriptional modification of A2503 in 23S rRNA affects inducible expression of the ermC gene.
Indigenous posttranscriptional modification of A2503 modulates the ribosomal response to the nascent peptide
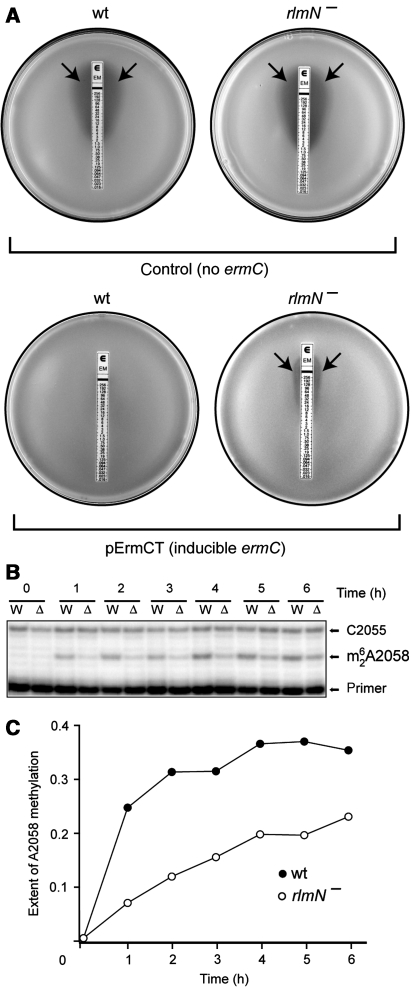
A2503 in 23S rRNA of E. coli and other bacteria is posttranscriptionally methylated at the C2 atom because of the action of RlmN methyltransferase (Toh et al, 2008). The lack of this modification has little effect upon cell growth in rich media, although rlmN− cells slowly lose to wild type in a cogrowth competition. To test the effect of A2503 modification upon erythromycin- and nascent-peptide-dependent induction of ermC, the rlmN gene was deleted from the chromosome of the antibiotic-hypersensitive acrB− E. coli strain. The lack of A2503 posttranscriptional modification did not influence interaction of erythromycin with its ribosomal target: ribosomes isolated from rlmN+ and rlmN− strains bound erythromycin with the same affinity (KD, 0.18±0.03 μM) (see Supplementary Information for experimental details). In agreement with this observation, the untransformed rlmN+ and rlmN− strains showed comparable sensitivity to erythromycin in the broth dilution assay (MIC 1.5–2 μg/ml, Supplementary Table S1). When a constitutively expressed ermC gene was introduced into these two strains on a pErmCTP plasmid, the erythromycin MIC increased to 2 mg/ml but, again, remained the same for both strains. This demonstrates that the lack of A2503 modification does not affect ermC translation or binding of the drug to the ribosome.
The rlmN+ and rlmN− strains were then transformed with the plasmid pErmCT, encoding the inducible ermC operon. Strikingly, in this case, a four-fold higher concentration of erythromycin in the liquid media was required to inhibit growth of rlmN+ compared with the rlmN− strain (Supplementary Table S1), indicating that rlmN+ cells more readily activate expression of the inducible ermC. This observation was further confirmed by the E-test, in which cells are exposed to a gradient of erythromycin concentrations on agar plates. Expression of the inducible ermC gene made the rlmN+ cells resistant to a notably higher concentration of erythromycin as compared with the rlmN− strain (Figure 2A). As this effect was restricted exclusively to the inducible ermC, these data suggest that it is the induction of the ermC expression rather than its translation per se that is affected by the lack of posttranscriptional modification at A2503.
Figure 2.
The effects of indigenous posttranscriptional modification of A2503 upon ermC induction. (A) The E-test analysis of erythromycin resistance of the E. coli cells that either do not carry the ermC gene (control) or carry a plasmid pErmCT with inducible ermC. The rlmN+ cells (wt) or rlmN− cells (lacking posttranscriptional modification of A2503) were plated onto agar plates and overlaid with an E-strip containing a gradient of erythromycin concentration. Inhibition of cell growth is manifested as a clear zone around the strip (arrows). (B) Primer-extension analysis of the induction of 23S rRNA modification by ErmC upon exposure of cells to erythromycin. Exponential wild-type (W) or rlmN− (Δ) cells carrying inducible ermC were induced with 32 μg/ml erythromycin, and the extent of ErmC-catalysed A2058 dimethylation in 23S rRNA at specified time points was analysed by primer extension. In the presence of ddGTP terminator, reverse transcriptase stops at the dimethylated A2058 but advances to C2055 when A2058 remains unmodified. (C) Quantification of the intensities of the primer-extension bands on the gel shown in (B), representing the fraction of the modified 23S rRNA.
We further verified this conclusion by following the kinetics of ermC induction. ErmC catalyses dimethylation of A2058 in 23S rRNA, and the presence of m62A2058 can be monitored by primer extension. After addition of a subinhibitory concentration of erythromycin (32 μg/ml) to early exponential cells carrying the inducible ermC operon, the level of A2058 dimethylation increased significantly more rapidly in the rlmN+ cells compared with the rlmN− mutant, indicating a more rapid activation of ermC expression (Figure 2B and C). This effect was paralleled by a faster resumption of growth of the rlmN+ versus the rlmN− cells upon exposure to erythromycin (Supplementary Figure S1). As induction of ermC expression by erythromycin is controlled by nascent-peptide-dependent ribosome stalling, these data argue that posttranscriptional modification of A2503 to m2A by RlmN methyltransferase affects the ability of the ribosome to respond to the stalling signal encoded in the ErmCL nascent peptide.
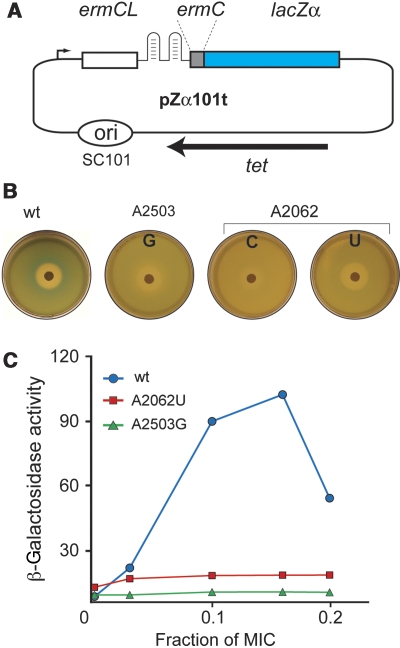
Mutation of A2503 abolishes expression of the ermC-based reporter
Having discovered that the lack of posttranscriptional modification of A2503 interferes with the nascent-peptide recognition, we then tested what effect a more dramatic change in the structure of A2503, more specifically the base mutation, would have upon the ribosomal response to the stalling nascent peptide. Out of the three possible mutations of A2503, only the G mutant was able to support cell growth in the absence of wild-type ribosomes. This mutation was expressed in the engineered SQK15 E. coli strain that lacks chromosomal rrn alleles and in which rRNA is expressed from a plasmid-borne rRNA operon (Asai et al, 1999). In addition, the SQK15 strain is capable of α-complementation, which allows for the use of the reporter pZa101tet where ribosome stalling at the ermCL ORF activates expression of the β-galactosidase α-peptide (Bailey et al, 2008) (Figure 3A). In X-Gal- and IPTG-containing plates, SQK15 cells with wild-type ribosomes formed a bright blue halo around the erythromycin disk, showing drug-induced activation of the reporter expression. In contrast, the A2503G mutant showed no blue halo, indicating that the rRNA mutation interfered with the induction mechanism (Figure 3B). This qualitative observation was verified by measuring the level of β-galactosidase activity in lysates of cells with wild-type or mutant ribosomes exposed to inducing concentrations of the antibiotic: the mutation of A2503, similar to the A2062 mutations, completely suppressed drug- and nascent-peptide-dependent induction of the ermC-based lacZ reporter (Figure 3C). Mutations of several other tested nucleotides in the vicinity of A2062, including residues U2586, U2609, and 750 loop implicated by previous studies in ribosomal response to SecM or TnaC stalling peptides, had little if any effect upon the reporter expression (Supplementary Figure S2 and Table S2). Thus, A2503 emerged as a major player in ribosomal response to the ErmCL nascent peptide.
Figure 3.
The effect of the A2503 and A2062 mutations upon induction of the ermC-based reporter. (A) The structure of the ermC-based pZα101t reporter. A portion of the lacZ gene encoding the β-galactosidase α-peptide is fused to the first two codons of ermC. Ribosome stalling at the ermCL ORF induces expression of the ermC-lacZα fusion. (B) Drug-diffusion induction of the pZα101t reporter in E. coli cells expressing wild-type or mutant ribosomes. The plates contain a lawn of SQK15/pZα101t cells grown on the surface of LB agar plates supplemented with ampicillin, tetracycline, IPTG, and X-gal. Expression of the reporter is induced by diffusion of erythromycin from the paper disk containing 0.5 mg of the drug. (C) β-Galactosidase activity (nmol/min/mg) in the wild-type and mutant SQK15 cells containing the pZα101t reporter.
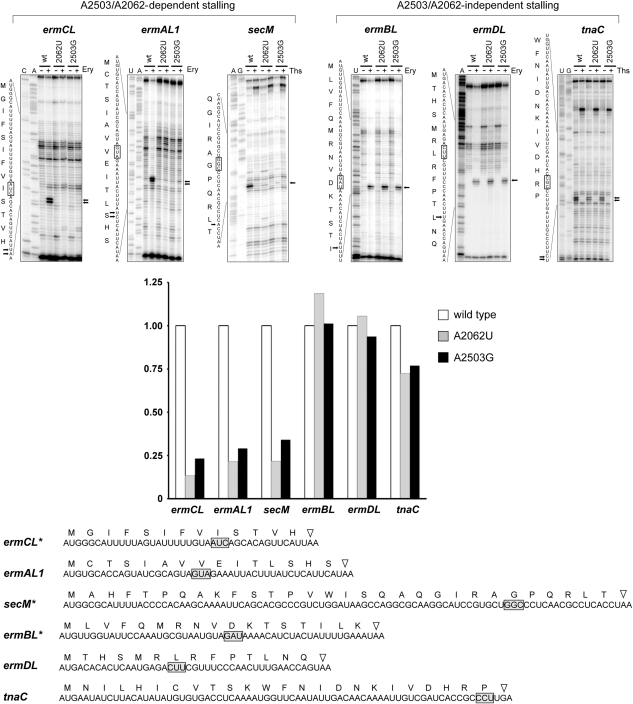
Mutations at A2503 completely prevent ribosome stalling at the ermCL leader ORF
Nascent-peptide-dependent induction of a plasmid-borne reporter in vivo is a complex process. Hypothetically, the rRNA mutations may modulate various aspects of the reporter expression. Therefore, it was important to test whether the mutation at A2503 specifically affects the nascent-peptide-dependent ribosome stalling. Having verified that the isolated wild-type and mutant ribosomes were active in in vitro protein synthesis (Supplementary Figure S3) and readily able to bind erythromycin as can be judged from the MIC values (Supplementary Table S2), we tested their capacity to form SRC at the ermCL ORF by toeprinting assay (Hartz et al, 1988; Muto et al, 2006; Vazquez-Laslop et al, 2008). The ermCL template was translated by wild-type or mutant ribosomes in the absence or presence of erythromycin, and formation of the stalled complex was detected by extension of a primer annealed to mRNA downstream of the stalling site (Figure 4). In agreement with our previous findings (Vazquez-Laslop et al, 2008), wild-type ribosomes readily formed SRC at the ninth (Ile) codon of the ermCL gene in the presence of erythromycin as revealed by the appearance of a characteristic toeprint band on the primer-extension gel. In contrast, neither the A2503G mutant ribosomes nor (as we previously determined) the A2062U ribosomes were able to form the stalled complex. As erythromycin was used at a concentration sufficient to saturate binding to wild-type and mutant ribosomes, the most plausible explanation of this result is that A2503 as well as A2062 are directly involved in the formation of SRC in response to the presence of erythromycin and the ErmCL stalling peptide in the tunnel.
Figure 4.
Toeprinting analysis of the effects of rRNA mutations on nascent-peptide-dependent ribosome stalling. The regulatory ORF templates were translated in a cell-free translation system under either the nonstalling or stalling conditions, and formation of the stalled complex was monitored by primer extension. Ribosome stalling signals on the gels and corresponding sequences are indicated by arrows. The codon located in the P site of the stalled ribosome is boxed. Addition of erythromycin (Ery) is necessary to cause ribosome stalling during translation of the leader erm templates (ermCL, ermAL1, ermBL, and ermDL). Thiostrepton (Ths), an inhibitor of translation, was added to the indicated reactions directed by the secM and tnaC templates to demonstrate that appearance of toeprint signals on these cistrons depends on their active translation. Sequencing lanes are marked. Quantification of the normalized relative intensity of the stalling signal band is shown in the bar graph. The complete nucleotide sequences of the ORFs used in cell-free translation and the amino-acid sequences of the encoded peptides are shown below the gels. Stop codons are indicated by triangles. The ORFs marked with asterisks have been modified from the original wild-type versions by truncating the 5′ end (secM) or 3′ end (ermCL and ermBL).
A2503 acts in concert with A2062 in nascent-peptide-dependent ribosome stalling
Mutations of rRNA residues m2A2503 and A2062, located close to each other in the NPET, abolish ribosome stalling at the ermCL ORF. Do these residues act as independent sensors of the peptide or do they operate as a part of a common sensory/signal transduction pathway? In the former case, importance of each of these nucleotides in the ribosomal response to different stalling peptides might be peptide specific. In the latter scenario, the effect of mutations at one position should always parallel the effect of mutations at the other residue. To distinguish between these possibilities, we studied the effects of mutations at A2062 and A2503 upon SRC formation at a variety of regulatory ORFs.
The macrolide resistance genes ermA, ermB, and ermD are preceded by regulatory ORFs ermAL1, ermBL, and ermDL, respectively (reviewed in Ramu et al, 2009), and genetic evidence implicates drug- and nascent-peptide-dependent ribosome stalling in the mechanism of induction (Horinouchi and Weisblum, 1980; Murphy, 1985; Kwak et al, 1991; Hue and Bechhofer, 1992; Kwon et al, 2006; Min et al, 2008). The amino-acid sequences of the peptides encoded in these regulatory ORFs show considerable variation compared with ErmCL (Figure 4). In the cell-free translation reaction driven by wild-type ribosomes, erythromycin-dependent SRC formation could be clearly detected at the Val8 codon of ermAL1, the Leu7 codon of ermBL, and the Asp10 codon of ermDL (Figure 4). When wild-type ribosomes were substituted with the mutant ones, we observed that the A2062U or A2503G mutations completely abolished formation of the stalled complex at the ermAL1 ORF. Strikingly, however, neither the mutation at A2062 nor the mutation at A2503 had any effect upon stalling at the two other tested ORFs, ermBL or ermDL (Figure 4).
We further investigated how the A2062U and A2503G mutations affect drug-independent ribosome stalling. Specifically, we tested whether these mutations influence the ability of the ribosome to respond to the SecM or TnaC stalling nascent peptides. In agreement with previous findings, toeprinting analysis showed formation of SRC at the Pro24 codon of the tnaC ORF; as expected, SRC formation was stimulated by 5 mM tryptophan, showing that the properties of the in vitro-formed SRC are similar to those of the stalled complex formed in vivo (Gong and Yanofsky, 2002). Remarkably, the SRC at the 24th codon of tnaC formed irrespective of whether tnaC ORF was translated by wild-type or mutant (A2062U or A2503G) ribosomes (Figure 4). In contrast, mutations A2062U and A2503G completely abolished ribosome stalling at the Gly165 codon of secM (Figure 4), the known stalling site in the secM gene (Muto et al, 2006).
As the mutations at A2062 and A2503 exert a strictly parallel effect upon the translation arrest directed by different stalling peptides, these results argue that both residues may act as components of the same sensory/signalling pathway in the ribosome.
Discussion
Our experiments identified one of the key components of the molecular mechanism used by the ribosome to halt translation in response to specific nascent peptides. We showed that universally conserved and posttranscriptionally modified 23S rRNA nucleotide A2503 may act in conjunction with another conserved residue, A2062, to sense the presence of specific nascent-peptide sequences in the ribosome exit tunnel and relay the translation arrest signal to the PTC.
Function of A2503 in the ribosomal response to the stalling nascent peptide
A2503 is essentially invariant in bacterial and eukaryotic cytoplasmic ribosomes. Yet, despite the extreme degree of its evolutionary conservation, which points to its functional significance, the function of this residue in the ribosome remained elusive. A2503 is located at the wall of the NPET close to the PTC active site. Although this rRNA resides outside the catalytic centre, alterations in the A2503 structure directly influence the properties of the PTC. Indeed, an extra methyl group added to the C8 position of this already naturally modified nucleotide by Cfr methyltransferase renders cells resistant to an array of antibiotics that bind in the peptidyl transferase A site (Kehrenberg et al, 2005; Smith and Mankin, 2008; Giessing et al, 2009). The structural and functional link of A2503 to the PTC active site is likely used in a signal-relay mechanism that operates between the NPET and PTC.
A2503 belongs to an exquisite group of 25 naturally posttranscriptionally modified residues in the 2904-nucleotide E. coli 23S rRNA. Of these 25 nucleotides, 14 are chemically altered by methylation. Of these 14, the only rRNA nucleotide methylated at a carbon centre is m2A2503. Addition of a methyl group to sp2-hybridized C2 carbon of the adenine base is a challenging reaction. It is catalysed by the methyltransferase RlmN, the unique enzyme (besides its close relative and a likely descendant, Cfr) that methylates RNA using the radical-SAM mechanism (Toh et al, 2008; Giessing et al, 2009; Yan et al, 2010). Both the exotic chemistry required for the C2 methylation of A2503 and the functional rarity of the corresponding enzyme hint that nature has compelling reasons to tinker with the chemical structure of this conserved rRNA residue in the NPET.
Although abolishing the C2 methylation at A2503 causes only a minor growth defect (Toh et al, 2008), cells with unmodified A2503 activate expression of the inducible ermC gene at a much slower rate compared with the wild-type control (Figure 3B and C) and achieve a lower level of erythromycin resistance. As neither affinity of erythromycin to the ribosome nor translation of the constitutive ermC is affected, ribosomes lacking the modification at A2503 must be impaired in recognition and response to the stalling signal encoded in the ErmCL peptide. We believe that this result represents the first demonstration of the possible function of a posttranscriptional rRNA modification in interactions between the ribosome and the nascent peptide. A similar functional mission could be envisioned for the modifications of other rRNA residues in the NPET.
The C2 methylation does not dramatically alter chemical properties of the adenine base but is expected to distort syn–anti equilibrium of the adenine base in favour of the anti conformation. Notably, although in most reported ribosomal crystallographic structures adenine 2503 is in syn configuration, in some 70S ribosome complexes it appears in anti conformation (Petry et al, 2005; Jenner et al, 2007) (Figure 5A). This indicates that both orientations of the base are generally allowed in the context of the ribosome structure. It is conceivable that under special circumstances, for example, when a specific nascent peptide is present in the NPET, the C2 methylation may facilitate rotation of the A2503 base into an anti conformation (see below).
Figure 5.
Molecular mechanism of the translation arrest induced by the presence of antibiotic and stalling nascent peptide in the ribosome exit tunnel. (A) Possible syn–anti rotation of the A2503 base. The orientation of A2503 in different crystallographic complexes of T. thermophilus 70S ribosome (PDB accession numbers 2WDL and 1YL3 for the syn and anti conformations, respectively) (Jenner et al, 2005; Voorhees et al, 2009). (B) Molecular interactions leading to programmed ribosome stalling. The 9-amino-acid ErmCL nascent peptide was modelled in the exit tunnel of the T. thermophilus 70S ribosome (pdb accession number 2WDL) according to Tu et al (2005) and energy minimized. Erythromycin was docked by aligning the T. thermophilus structure, with the structure of H. marismortui 50S subunit complexed with erythromycin (1YI2; Tu et al, 2005). In the ribosome stalled at the ermCL ORF, the 9-amino-acid ErmCL nascent peptide (green, sticks and semitransparent surface) esterifies tRNA bound in the ribosomal P site (only the 3′ adenosine of the peptidyl-tRNA is shown). The presence of erythromycin (cyan, sticks and mesh) in the tunnel constrains the peptide placement, compelling the critical C-terminal sequence of the peptide (dark green) to come into direct contact with the A2062 base (red) and forcing it into a conformation where it would clash with m2A2503 (red). Reorientation of m2A2503, possibly facilitated by posttranscriptional methylation at C2 (ball), is then communicated to the PTC active site. Change in the position of G2061 and/or U2504 (pseudouridine, Ψ, in E. coli) (both shown in gold) will likely change the opening of the A2451/C2452 (blue) A-site cavity and prevent proper accommodation of the aminoacyl moiety of the A-site aminoacyl-tRNA (purple).
One of the important aspects of our findings is the strikingly parallel effect of A2503 and A2062 mutations upon the ribosomal ability to respond to a regulatory peptide. If a mutation at one of the rRNA residues prevents stalling at a specific ORF (ermCL, ermAL1, secM), then mutation at the other position would also abolish such stalling; if, on the other hand, translation arrest is insensitive to a mutation at either A2503 or A2062, then altering the second nucleotide also has no effect upon stalling (ermBL, ermDL, tnaC). These observations, as well as close proximity of the two residues in the ribosome tertiary structure, argue that both nucleotides could be part of the same nascent-peptide response mechanism and may operate in a concerted manner, like a single sensory/communication module.
The location of A2062 and m2A2503—in the NPET close to the PTC—is suitable for these rRNA residues to serve the function of nascent-peptide sentinels. We envision the following mode of operation of the nascent-peptide sensory/response mechanism based on engagement of these nucleotides. Although A2062 is directly exposed in the tunnel, m2A2503 is less accessible and additionally shielded by antibiotic binding. Therefore, although A2062 has the function of primary sensor of the nascent peptide, m2A2503 likely relays the stalling signal to the PTC. In the absence of a macrolide antibiotic, the tunnel's aperture is sufficiently wide for the unfolded nascent peptide to avoid steric clash with A2062 even when the adenine base projects into the lumen of the tunnel (Figure 1B). Binding of a macrolide antibiotic narrows the tunnel (Figure 1C) and leaves such a small space for the peptide that its close encounter with A2062 becomes unavoidable. (In the case of SecM, an unusually contorted conformation of the nascent peptide (Woolhead et al, 2006; Yap and Bernstein, 2009) in the absence of a drug bound in the tunnel may cause a similar effect.) Steric clash with the nascent-peptide chain would force the A2062 base to rotate closer to the wall. Sequence-specific contacts with the critical C-terminal amino acids of the peptide likely coerce A2062 to adopt a conformation that not only brings it in proximity to m2A2503 but also leads to displacement of the m2A2503 base, possibly involving a syn–anti transition facilitated by the presence of a posttranscriptionally added C2-methyl group.
The nascent-peptide-dependent translation arrest results from failure of the PTC to catalyse peptide bond formation (or, in the case of tnaC, peptide release) (Gong and Yanofsky, 2002; Muto et al, 2006; Vazquez-Laslop et al, 2008). There are several scenarios that could explain how the stalling signal relayed through m2A2503 to the PTC active site can inhibit catalysis (Figure 5B). G2061 is an important component of the PTC catalytic centre. It is hydrogen bonded to A2451, a key constituent of the A-site amino-acid-binding pocket (Nissen et al, 2000; Schmeing et al, 2005) and may also directly interact with the aminoacyl moiety of the A-site-bound aminoacyl-tRNA (Bogdanov, 2003). Even a small shift in the position of G2061 is likely to result in disruption of the PTC activities (Bayfield et al, 2001). In the active conformation, the base of G2061 is arranged parallel to syn adenine 2503. Changing the precise placement of m2A2503 (e.g. rotating its base from syn to trans orientation) and additionally rearranging the G2061 neighbour A2062 would displace G2061, resulting in inactive conformation of the PTC active site.
Another, but not necessarily alternative, scenario involves the immediate neighbour of m2A2503, U2504 (which in E. coli is posttranscriptionally converted to pseudouridine). The exact position of this residue was proposed to control the opening of the cavity formed by the splayed out bases of A2451 and C2452 where the side chain of the A-site amino acid binds (Gurel et al, 2009). It is easy to envision how reorientation of m2A2503 can displace U2504 and prevent the A-site cavity from accommodating the incoming aminoacyl-tRNA.
The ribosome uses different mechanisms to recognize different stalling peptides
The critical sequences of all of the presently characterized stalling peptides are located at the peptide's C-terminus. It means that recognition of the translation arrest signal encoded in all these peptides takes place in the ‘upper chamber' of the NPET, proximal to the PTC. It was tempting to think that the same universal mechanism accounts for the ribosomal response to the variety of stalling peptides. Therefore, having found that the A2503 and A2062 mutations completely abolished ribosome stalling at the ermCL regulatory ORF (Figure 4), we anticipated that the mutations of these residues would have a similarly profound effect upon ribosomal response to all the nascent peptides that directly programmed translation arrest. Indeed, erythromycin-dependent stalling at the ermAL1 ORF and drug-independent stalling at the secM ORF were abolished by either A2503 or A2062 mutations. Unexpectedly, however, erythromycin-dependent stalling at two other erm regulatory ORFs, ermBL and ermDL, as well as translation arrest at the tnaC ORF were not affected (Figure 4). This finding provides the first unequivocal evidence that different classes of stalling peptides engage different sensors in the tunnel and possibly activate principally different stalling mechanisms. In this regard, it is worth noting that the putative signal-relay mechanism that operates through m2A2503 and A2062 is generally compatible with one of the hypothetical pathways (‘R3') proposed to propagate the stalling signal resulting from the presence of TnaC peptide in the ribosome tunnel (Seidelt et al, 2009). As neither A2503 nor A2062 mutations have any significant effect upon stalling at the tnaC ORF (Figure 4), our results indicate that this route does not have a major function in sensing TnaC and argue that alternative routes (possibly routes R1 or R2 proposed by Seidelt et al (2009) are more important for the ribosomal response to the TnaC peptide. Investigation of the mechanism of the ribosomal response to TnaC, ErmBL, and ErmDL peptides may illuminate both similarities and idiosyncrasies of different ways the ribosome deals with the functional signals encoded in the nascent peptides.
Materials and methods
Time course of induction of ermC-dependent 23S rRNA methylation
The acrB− version of the E. coli strain JM109 and the rlmN− derivative were transformed with the pErmCT plasmid carrying inducible ermC operon under control of the Ptac promoter. The overnight cultures, grown in LB/ampicillin media, were diluted 200-fold in fresh LB media containing 100 μg/ml ampicillin and 0.1 mM IPTG and grown to O.D.600=0.1, at which point the expression of ermC was induced by addition of 32 μg/ml erythromycin. Cells were incubated with shaking at 37°C. Aliquots of the cultures (0.5 A600) were withdrawn at specified time points, cells were spun down, and total cellular RNA was extracted using the RNeasy kit (Qiagen). The extent of dimethylation of A2058 in 23S rRNA by ErmC methyltransferase was determined by primer extension, using the primer L2063 (Supplementary Table S3) and ddGTP terminator. The amount of radioactivity in the bands in gels was quantified using phosphorimager and plotted.
Construction of the reporter system for monitoring antibiotic-dependent ribosome stalling
The E. coli strain SQK15 lacking chromosomal rrn alleles and capable of α-complementation was prepared from the strain SQ171 (Asai et al, 1999) through recombineering (Datsenko and Wanner, 2000). The details are described in Supplementary Information.
The pErmZα reporter plasmid (Bailey et al, 2008) was modified to carry tetracycline resistance marker and pSC101 origin of replication (Figure 3A). The resulting plasmid, pErmZ101t, was introduced in the SQK15 strain. Inducible expression of the lacZα reporter gene in the pErmZ101t plasmid was tested using the disk-diffusion assay as described (Bailey et al, 2008).
The β-galactosidase activity was quantified using the standard assay adapted to a 96-well format.
An antibiotic-hypersensitive acrB− variant of the E. coli strain JM109 was constructed by recombineering using DNA from the Keio collection strain JW0451 (Baba et al, 2006) as a PCR template for preparation of the donor DNA. The rlmN gene was then inactivated with a similar approach using JW2501 strain from the same collection as a source of DNA.
Other procedures
Site-directed mutagenesis of 23S rRNA, MIC determination, studying of antibiotic binding to the ribosomes, and toeprinting analysis were carried out as previously described (Xiong et al, 2005; Vazquez-Laslop et al, 2008). Experimental details can be found in Supplementary Information.
Supplementary Material
Acknowledgments
We thank C Squires for providing the strain SQ171 and advice on its use, B Llano-Sotelo for help with analysis of the binding data, D Mulhearn for helping to model the nascent peptide, V Ramakrishnan for providing pdb coordinates of ribosome complexes prior to publication, and E Westhof for advice on the properties of modified adenines. This work was supported by the grant MCB-0824739 from National Science Foundation, USA.
Footnotes
The authors declare that they have no conflict of interest.
References
- Asai T, Zaporojets D, Squires C, Squires CL (1999) An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc Natl Acad Sci USA 96: 1971–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey M, Chettiath T, Mankin AS (2008) Induction of ermC expression by ‘non-inducing' antibiotics. Antimicrob Agents Chemother 52: 866–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2000) The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289: 905–920 [DOI] [PubMed] [Google Scholar]
- Bayfield MA, Dahlberg AE, Schulmeister U, Dorner S, Barta A (2001) A conformational change in the ribosomal peptidyl transferase center upon active/inactive transition. Proc Natl Acad Sci USA 98: 10096–10101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaha G, Gurel G, Schroeder SJ, Moore PB, Steitz TA (2008) Mutations outside the anisomycin-binding site can make ribosomes drug-resistant. J Mol Biol 379: 505–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov AA (2003) Some structural aspects of the peptidyltransferase reaction. Mol Biol 37: 436–439 [PubMed] [Google Scholar]
- Chiba S, Lamsa A, Pogliano K (2009) A ribosome-nascent chain sensor of membrane protein biogenesis in Bacillus subtilis. EMBO J 28: 3461–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirkova A, Erlacher MD, Clementi N, Zywicki M, Aigner M, Polacek N (2010) The role of the universally conserved A2450-C2063 base pair in the ribosomal peptidyl transferase center. Nucl Acids Res 38: (advance online publication, 7 April 2010; doi:10.1093/nar/gkq213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CS, Lamichhane TN, Mahto SK (2007) Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. Chem Biol 2: 610–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Vera LR, Rajagopal S, Squires C, Yanofsky C (2005) Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol Cell 19: 333–343 [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang P, Spevak CC, Wu C, Sachs MS (2004) A nascent polypeptide domain that can regulate translation elongation. Proc Natl Acad Sci USA 101: 4059–4064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulle S, Gohlke H (2009) Statics of the ribosomal exit tunnel: implications for cotranslational peptide folding, elongation regulation, and antibiotics binding. J Mol Biol 387: 502–517 [DOI] [PubMed] [Google Scholar]
- Giessing AM, Jensen SS, Rasmussen A, Hansen LH, Gondela A, Long K, Vester B, Kirpekar F (2009) Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria. RNA 15: 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F, Yanofsky C (2002) Instruction of translating ribosome by nascent peptide. Science 297: 1864–1867 [DOI] [PubMed] [Google Scholar]
- Gryczan TJ, Grandi G, Hahn J, Grandi R, Dubnau D (1980) Conformational alteration of mRNA structure and the posttranscriptional regulation of erythromycin-induced drug resistance. Nucl Acids Res 8: 6081–6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurel G, Blaha G, Moore PB, Steitz TA (2009) U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome. J Mol Biol 389: 146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JL, Schmeing TM, Moore PB, Steitz TA (2002) Structural insights into peptide bond formation. Proc Natl Acad Sci USA 99: 11670–11675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartz D, McPheeters DS, Traut R, Gold L (1988) Extension inhibition analysis of translation initiation complexes. Methods Enzymol 164: 419–425 [DOI] [PubMed] [Google Scholar]
- Horinouchi S, Weisblum B (1980) Posttranscriptional modification of mRNA conformation: mechanism that regulates erythromycin-induced resistance. Proc Natl Acad Sci USA 77: 7079–7083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hue KK, Bechhofer DH (1992) Regulation of the macrolide-lincosamide-streptogramin B resistance gene ermD. J Bacteriol 174: 5860–5868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Chiba S, Pogliano K (2010) Divergent stalling sequences sense and control cellular physiology. Biochem Biophys Res Commun 393: 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner L, Rees B, Yusupov M, Yusupova G (2007) Messenger RNA conformations in the ribosomal E site revealed by X-ray crystallography. EMBO Rep 8: 846–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner L, Romby P, Rees B, Schulze-Briese C, Springer M, Ehresmann C, Ehresmann B, Moras D, Yusupova G, Yusupov M (2005) Translational operator of mRNA on the ribosome: how repressor proteins exclude ribosome binding. Science 308: 120–123 [DOI] [PubMed] [Google Scholar]
- Jenni S, Ban N (2003) The chemistry of protein synthesis and voyage through the ribosomal tunnel. Curr Opin Struct Biol 13: 212–219 [DOI] [PubMed] [Google Scholar]
- Kehrenberg C, Schwarz S, Jacobsen L, Hansen LH, Vester B (2005) A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: methylation of 23S ribosomal RNA at A2503. Mol Microbiol 57: 1064–1073 [DOI] [PubMed] [Google Scholar]
- Kwak JH, Choi EC, Weisblum B (1991) Transcriptional attenuation control of ermK, a macrolide-lincosamide-streptogramin B resistance determinant from Bacillus licheniformis. J Bacteriol 173: 4725–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon AR, Min YH, Yoon EJ, Kim JA, Shim MJ, Choi EC (2006) ErmK leader peptide: amino acid sequence critical for induction by erythromycin. Arch Pharm Res 29: 1154–1157 [DOI] [PubMed] [Google Scholar]
- Lawrence M, Lindahl L, Zengel JM (2008) Effects on translation pausing of alterations in protein and RNA components of the ribosome exit tunnel. J Bacteriol 190: 5862–5869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min YH, Kwon AR, Yoon EJ, Shim MJ, Choi EC (2008) Translational attenuation and mRNA stabilization as mechanisms of erm(B) induction by erythromycin. Antimicrob Agents Chemother 52: 1782–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DR, Geballe AP (2000) Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol 20: 8635–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E (1985) Nucleotide sequence of ermA, a macrolide-lincosamide-streptogramin B determinant in Staphylococcus aureus. J Bacteriol 162: 633–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto H, Nakatogawa H, Ito K (2006) Genetically encoded but nonpolypeptide prolyl-tRNA functions in the A site for SecM-mediated ribosomal stall. Mol Cell 22: 545–552 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Ito K (2002) The ribosomal exit tunnel functions as a discriminating gate. Cell 108: 629–636 [DOI] [PubMed] [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA (2000) The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930 [DOI] [PubMed] [Google Scholar]
- Noller HF (1991) Ribosomal RNA and translation. Annu Rev Biochem 60: 191–227 [DOI] [PubMed] [Google Scholar]
- Petry S, Brodersen DE, Murphy FV 4th, Dunham CM, Selmer M, Tarry MJ, Kelley AC, Ramakrishnan V (2005) Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell 123: 1255–1266 [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V (2002) Ribosome structure and the mechanism of translation. Cell 108: 557–572 [DOI] [PubMed] [Google Scholar]
- Ramu H, Mankin A, Vazquez-Laslop N (2009) Programmed drug-dependent ribosome stalling. Mol Microbiol 71: 811–824 [DOI] [PubMed] [Google Scholar]
- Schlunzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F (2001) Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413: 814–821 [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Huang KS, Strobel SA, Steitz TA (2005) An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature 438: 520–524 [DOI] [PubMed] [Google Scholar]
- Seidelt B, Innis CA, Wilson DN, Gartmann M, Armache JP, Villa E, Trabuco LG, Becker T, Mielke T, Schulten K, Steitz TA, Beckmann R (2009) Structural insight into nascent polypeptide chain-mediated translational stalling. Science 326: 1412–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LK, Mankin AS (2008) Transcriptional and translational control of the mlr operon, which confers resistance to seven classes of protein synthesis inhibitors. Antimicrob Agents Chemother 52: 1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner DR, Cariello DA, Woolstenhulme CJ, Broadbent MA, Buskirk AR (2009) Genetic identification of nascent peptides that induce ribosome stalling. J Biol Chem 284: 34809–34818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenson T, Ehrenberg M (2002) Regulatory nascent peptides in the ribosomal tunnel. Cell 108: 591–594 [DOI] [PubMed] [Google Scholar]
- Toh SM, Xiong L, Bae T, Mankin AS (2008) The methyltransferase YfgB/RlmN is responsible for modification of adenosine 2503 in 23S rRNA. RNA 14: 98–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu D, Blaha G, Moore PB, Steitz TA (2005) Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121: 257–270 [DOI] [PubMed] [Google Scholar]
- Vazquez-Laslop N, Thum C, Mankin AS (2008) Molecular mechanism of drug-dependent ribosome stalling. Mol Cell 30: 190–202 [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V (2009) Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol 16: 528–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum B (1995) Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob Agents Chemother 39: 797–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhead CA, Johnson AE, Bernstein HD (2006) Translation arrest requires two-way communication between a nascent polypeptide and the ribosome. Mol Cell 22: 587–598 [DOI] [PubMed] [Google Scholar]
- Xiong L, Korkhin Y, Mankin AS (2005) Binding site of the bridged macrolides in the Escherichia coli ribosome. Antimicrob Agents Chemother 49: 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F, Lamarre JM, Rohrich R, Wiesner J, Jomaa H, Mankin AS, Fujimori DG (2010) RlmN and Cfr are radical SAM enzymes involved in methylation of ribosomal RNA. J Am Chem Soc 132: 3953–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Cruz-Vera LR, Yanofsky C (2009) 23S rRNA nucleotides in the peptidyl transferase center are essential for tryptophanase operon induction. J Bacteriol 191: 3445–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap MN, Bernstein HD (2009) The plasticity of a translation arrest motif yields insights into nascent polypeptide recognition inside the ribosome tunnel. Mol Cell 34: 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.