Cell cycle coordination and regulation of bacterial chromosome segregation dynamics by polarly localized proteins
An analysis of the functions of polarity proteins and ParABS partitioning components in Caulobacter crescentus shows how a bacterial system for segregating multi-copy plasmids can be adapted to faithful unidirectional chromosome movement, and at the same time provide coupling to the cell division process.
Keywords: Caulobacter, chromosome segregation, ParA, TipN
Abstract
What regulates chromosome segregation dynamics in bacteria is largely unknown. Here, we show in Caulobacter crescentus that the polarity factor TipN regulates the directional motion and overall translocation speed of the parS/ParB partition complex by interacting with ParA at the new pole. In the absence of TipN, ParA structures can regenerate behind the partition complex, leading to stalls and back-and-forth motions of parS/ParB, reminiscent of plasmid behaviour. This extrinsic regulation of the parS/ParB/ParA system directly affects not only division site selection, but also cell growth. Other mechanisms, including the pole-organizing protein PopZ, compensate for the defect in segregation regulation in ΔtipN cells. Accordingly, synthetic lethality of PopZ and TipN is caused by severe chromosome segregation and cell division defects. Our data suggest a mechanistic framework for adapting a self-organizing oscillator to create motion suitable for chromosome segregation.
Introduction
Similar to eukaryotes, bacteria use active mechanisms to partition chromosomes equally between daughter cells. Fluorescence microscopy studies in various bacteria have shown that specific centromere-like DNA regions close to the origin of replication are translocated to opposite sides of the cell in one single event during the cell cycle. How this controlled translocation occurs at the mechanistic level is not well understood. Most of our knowledge about DNA segregation in bacteria comes from studies on plasmids, in part because they are not necessary for cell viability. Many plasmids use the Par Type 1 system for partitioning (Gerdes et al, 2010). This system has three components, here referred to as parS, ParB and ParA. Interestingly, orthologues of these components are commonly found on bacterial chromosomes (Livny et al, 2007). Although these components can have a function in the regulation of DNA replication in some bacteria (Murray and Errington, 2008), the parS/ParB/ParA system has also been shown to be involved in chromosome segregation (Gerdes et al, 2010). Plasmid and chromosome-encoded ParB proteins are known to bind to cognate, centromere-like parS DNA sequences. ParA proteins are deviant Walker-type ATPases whose weak ATPase activity is stimulated by an interaction with ParB either alone or when bound to parS (Radnedge et al, 1998; Easter and Gober, 2002; Leonard et al, 2005; Barillà et al, 2007; Bouet et al, 2007; Pratto et al, 2008). In vitro, ParA-ATP dimers bind to DNA non-specifically and in a cooperative manner, whereas ADP-bound forms do not (Leonard et al, 2005; Pratto et al, 2008; Ringgaard et al, 2009).
Recently, in vivo mechanistic insight into chromosome segregation was provided for chromosome I of Vibrio cholerae (Fogel and Waldor, 2006). Here, segregation starts at the old cell pole and occurs unidirectionally with a duplicated parSI/ParBI partition complex moving to the opposite, new pole (Fogel and Waldor, 2006). ParAI localizes in a cloud-like structure that extends from the new pole to near the parSI/ParBI focus at the old pole. Shrinking of the ParAI structure towards the new pole is accompanied with the translocation of parSI/ParBI, suggestive of a so-called ‘pulling' mechanism (Fogel and Waldor, 2006). Such a mechanism has also been invoked for plasmid partitioning. For instance, ParA of plasmid pB171 forms DNA-bound structures that perpetually extend and shrink over the nucleoid and in-between plasmids (Ebersbach et al, 2006; Ringgaard et al, 2009). These continuous cycles are correlated with back-and-forth motions of plasmids, which result in time-averaged equidistribution of plasmids along the cell length. In the case of both plasmid pB171 and V. cholerae chromosome I, it has been proposed that parS/ParB interaction with the edge of ParA nucleoprotein structures stimulates ParA-ATPase activity, leading to disassembly of ParA subunits and hence retraction of the ParA structure (Fogel and Waldor, 2006; Ringgaard et al, 2009). However, the continuous rounds of ParA assembly/disassembly producing perpetual plasmid movement and the single DNA translocation event per cell cycle of bacterial chromosomes are clearly distinct. What generates this difference is not clear.
Another important aspect of chromosome segregation that is difficult to address with plasmid models is its necessary temporal and spatial coordination with other cell cycle events. How this cell cycle coordination can occur is poorly understood. Some major advances have been achieved in Caulobacter crescentus, in which segregation of the parS/ParB partition complex is linked to cell division (Mohl et al, 2001) through MipZ, a cell division inhibitor that associates with parS/ParB partition complexes (Thanbichler and Shapiro, 2006). In C. crescentus, the DNA spreads throughout the cell, and as in V. cholerae, segregation is unidirectional (Jensen and Shapiro, 1999; Viollier et al, 2004). After replication of the parS locus at the old pole, one parS/ParB copy is rapidly translocated to the new pole (Viollier et al, 2004; Thanbichler and Shapiro, 2006; Toro et al, 2008). MipZ, which inhibits the cell division protein FtsZ, forms polar gradients through an association with ParB (Thanbichler and Shapiro, 2006). Hence, segregation of parS/ParB triggers the formation of a bipolar MipZ gradient, which has been proposed to result in the preferential assembly of the FtsZ cytokinetic ring at midcell in which the MipZ concentration is thought to be lowest (Thanbichler and Shapiro, 2006). Although MreB has been proposed to affect parS/ParB segregation (Gitai et al, 2005), data obtained from a mutagenized form of ParA suggests that ParA has a greater function in this process (Toro et al, 2008). The dynamics of ParA are, however, unknown in this organism.
Division has also been linked to cell polarity in C. crescentus through the landmark polarity factor TipN (Lam et al, 2006). C. crescentus is a highly polarized bacterium, forming a predivisional cell with a flagellum and a stalk (a thin extension of the cell body) at opposite poles. Asymmetric division yields a flagellated ‘swarmer' cell slightly shorter than its sibling, the ‘stalked' daughter cell. TipN, through its localization at the new pole, ensures proper positioning of new-pole markers such as the flagellum, whereas it has no effect on the positioning of old-pole markers (such as the stalk) or on the asymmetric localization of the daughter cell fate determinant CtrA (Huitema et al, 2006; Lam et al, 2006). Surprisingly, TipN appears to affect cell division placement, as ΔtipN cells, unlike wild type (Terrana and Newton, 1975; Quardokus and Brun, 2002), often constrict closer to the old pole, generating a reversed asymmetry in daughter cell size (Lam et al, 2006).
How the polarity factor TipN spatially affects cell division is puzzling given the distance between the site of division and the new-pole location of TipN. Here, we present evidence suggesting that TipN exerts its long-distance effect on cell division positioning by regulating ParA and parS/ParB segregation dynamics. Our study provides mechanistic insights into how rapid and directional DNA segregation can be achieved and regulated. Moreover, our findings suggest that extrinsic regulation of the parS/ParB/ParA system has a profound effect on several aspects of cell cycle coordination.
Results and discussion
TipN alters the timing and positioning of FtsZ ring formation
Our study was initially motivated by the intriguing observation that unlike wild-type cells, ΔtipN cells frequently appear to divide closer to the old pole (Lam et al, 2006). To quantify the distribution of the division defect within the ΔtipN population, we measured the division ratios of constricting cells, defined by the length between the old pole and the site of cell constriction divided by the total cell length (Supplementary Figure S1A) using a DivJ–CFP fusion as an old-pole marker (Wheeler and Shapiro, 1999). Most wild-type cells had an average division ratio of 0.537 (with a standard error of the mean of ±0.001, n=1433; Supplementary Figure S1B) that is consistent with a previous electron microscopy study (Terrana and Newton, 1975). In contrast, the majority of ΔtipN cells had a division ratio of 0.468 (±0.001, n=1766; Supplementary Figure S1B). Thus, TipN clearly affects where division occurs.
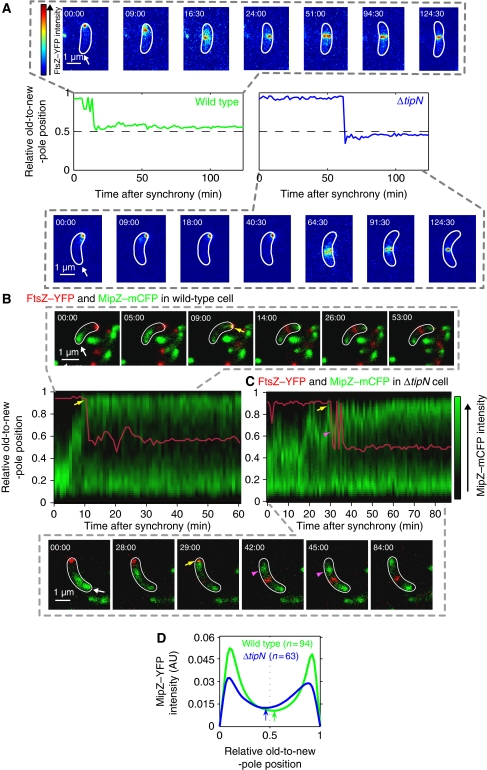
How C. crescentus selects its division site is not completely understood. MipZ is thought to restrict FtsZ polymerization to the region of lowest MipZ concentration (Thanbichler and Shapiro, 2006). As the bipolar MipZ gradients appear symmetric, it was suggested that FtsZ ring assembly may occur at midcell and that unequal growth rate between the sides flanking the FtsZ ring may cause the asymmetric division. An asymmetry in FtsZ ring positioning had been measured in dividing C. crescentus cells (Quardokus and Brun, 2002), but the assembly of the FtsZ ring occurs well before cell constriction is initiated (Kelly et al, 1998; Aaron et al, 2007). Therefore, it was conceivable that the FtsZ ring would form at midcell, but that its position would become progressively asymmetric over time because of unequal growth between sides. To test this idea, we quantified the temporal and spatial distribution of FtsZ by time-lapse microscopy starting with synchronized swarmer cell populations producing FtsZ–YFP. The relative position of FtsZ–YFP along the long cell axis in individual cells was plotted as a function of time after cell cycle synchronization (Figure 1A). In wild-type cells, FtsZ–YFP moved from the new pole to a central region and soon formed a ring (band) at a 0.536±0.003 position (n=190 cells), and this asymmetric position did not significantly change for the remainder of the cell cycle (Figure 1A; Supplementary Movie S1). Thus, it is the asymmetric location of the FtsZ ring, rather than an asymmetry in growth rate, that dictates the asymmetric division. Supporting this notion, in ΔtipN cells, the FtsZ ring stabilized on average at a 0.445±0.003 position (n=114 cells) relative to the long cell axis (Figure 1A; Supplementary Movie S2), which corresponds well to the average position of the division site of this mutant (Supplementary Figure S1B).
Figure 1.
FtsZ and MipZ dynamics in wild-type and ΔtipN cells. (A) Time-lapse microscopy of FtsZ–YFP in wild-type (MT199) and ΔtipN (CJW2563) cells after synchrony. The expression of ftsZ–yfp was induced with 0.5 mM vanillic acid 2.5 h before synchronization and imaging. Images were acquired every 1.5 min and the cells were identified using MicrobeTracker. The FtsZ–YFP signal in representative wild-type and ΔtipN cells is shown for selected time points as an overlay with the MicrobeTracker cell outline (the old pole is marked by the arrow). The graphs show the trace of the relative FtsZ–YFP position along the cell length over time. (B) Time-lapse microscopy of MipZ–mCFP and FtsZ–YFP in a wild-type background (strain CJW3455). FtsZ–YFP expression was induced with 0.5 mM vanillic acid 1 h before cell synchronization. Time-lapse results from a representative cell are shown as an overlay with MicrobeTracker cell outlines in white (the old pole is marked by the arrow.). The relative position of FtsZ–YFP (red trace) is indicated over a kymograph of the MipZ–mCFP signal profile (green) along the cell length as a function of time after synchrony. Yellow arrows show first instance of MipZ–mCFP and FtsZ–YFP colocalization at the new pole. (C) Same as (B) except in a ΔtipN background (strain CJW3612). Purple arrowheads show backwards motion of MipZ–mCFP. (D) Time-lapse recordings of MipZ–YFP in wild-type (CJW2022) and ΔtipN (CJW3366) cells after synchrony. Images were acquired every 1.5 min and the cells were identified using MicrobeTracker. Shown are profiles of the mean MipZ–YFP signal along the cell long axis of 94 wild-type cells (green) and 63 ΔtipN cells (blue) from the time point when MipZ–YFP becomes bipolar (yellow arrow in (B) and (C) to the onset of cell constriction). The arrows show the minima in MipZ–YFP intensity for each strain.
Another striking difference between wild-type and ΔtipN backgrounds was that FtsZ–YFP remained at the new pole for a longer time in ΔtipN cells, and consequently, FtsZ ring formation (represented by the stabilization of FtsZ–YFP localization at an off-centre position) was considerably delayed (Figure 1A). Under our experimental conditions, FtsZ–YFP ring formation occurred on average 11.5±0.7 min after cell synchronization in wild-type cells (n=190), whereas in ΔtipN cells (n=114), FtsZ–YFP ring formation was observed at about 42.6±1.7 min after cell synchronization. Thus, the ΔtipN mutation causes a significant delay in FtsZ ring formation (Kolmogorov Smirnov (K-S) test, P<0.0001), while having no effect on the timing of cell constriction or cell separation (see below).
TipN affects parS/ParB/MipZ segregation dynamics
MipZ regulates the timing of FtsZ ring formation during the cell cycle (Thanbichler and Shapiro, 2006), raising the possibility that the delay in FtsZ ring formation in ΔtipN cells is due to abnormal MipZ dynamics. Kymographs made from time-lapse sequences of wild-type cells producing MipZ–mCFP (Figure 1B) displayed the expected dynamics during the cell cycle (Thanbichler and Shapiro, 2006). Before DNA replication, MipZ–mCFP localized at the old pole with the single parS/ParB partitioning complex. Following initiation of DNA replication and duplication of parS, a second MipZ–YFP signal rapidly moved to the new pole through its interaction with the segregating parS/ParB complex (Thanbichler and Shapiro, 2006). When MipZ–mCFP reached the new pole, it displaced FtsZ–YFP from the new pole (Figure 1B, yellow arrow). FtsZ–YFP then moved to its off-centre position where it formed a ring. In ΔtipN cells, it took much longer for MipZ–mCFP to reach the new pole (Figure 1C), explaining the delay in FtsZ ring assembly in these mutant cells.
In addition to being slower in ΔtipN cells, the motion of MipZ–mCFP was erratic with many changes in direction. Even when MipZ–CFP reached the new pole (Figure 1C, yellow arrow), it usually temporarily moved back to the cell interior, often multiple times (Figure 1C, purple arrowheads). To examine the spatial distribution of the MipZ gradients along the cell length, we used MipZ–YFP-expressing cells and averaged their fluorescent profiles from the first time MipZ–YFP reached the new pole to the onset of cell constriction. In wild-type cells (n=94), the minimum of the MipZ–YFP signal was slightly biased towards the new pole with a mean value of 0.530±0.004 (Figure 1D, green arrow), whereas in ΔtipN cells (n=63), this minimum was shifted towards the old pole with a mean value of 0.460±0.008 (Figure 1D, blue arrow), most likely because of the common backwards motions of MipZ after FtsZ displacement from the new pole (Figure 1C, see images at time points 42 and 45 min for examples). This likely contributes to the difference in relative FtsZ–YFP ring positioning along the cell length between wild-type and ΔtipN cells.
As MipZ is associated with the parS/ParB complex, we examined the segregation dynamics of this complex (Thanbichler and Shapiro, 2006). Without TipN, translocation of the parS/CFP–ParB complex from one pole to the other took longer with an average speed of 0.038±0.001 μm/min in ΔtipN cells (n=79) compared with 0.109±0.006 μm/min in wild-type cells (n=94). Translocation was also more erratic, displaying many directional changes (Supplementary Figure S2A). The origin of replication had a similarly erratic motion in ΔtipN cells (Supplementary Figure S2B), consistent with its proximity to the parS locus. DAPI staining revealed no major defect in overall DNA organization at a gross level (Supplementary Figure S2C).
The MipZ segregation and FtsZ localization defects in ΔtipN cells were complemented by expression of plasmid-encoded TipN (Supplementary Figure S2D). Our data thus argue that TipN affects the timing and positioning of FtsZ ring formation by affecting the segregation of the MipZ-associated parS/ParB partition complex.
TipN regulates ParA localization dynamics
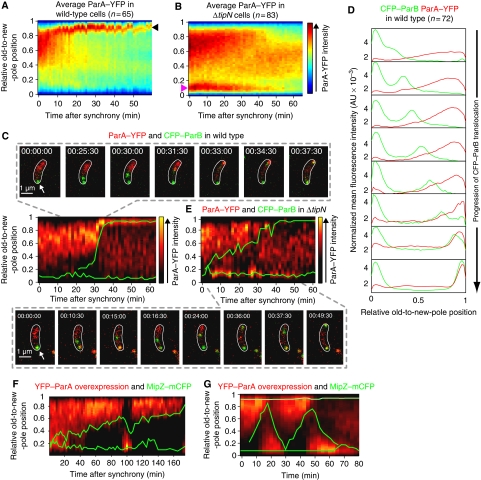
How can TipN, a protein that localizes at the new pole (Huitema et al, 2006; Lam et al, 2006), affect parS/ParB segregation? To address this question, we examined the spatial and temporal localization of ParA. In C. crescentus, parA is part of an operon that includes gidA, gidB, parA and parB. We generated strains in which the chromosomal parA open reading frame was substituted with a parA–yfp fusion while maintaining the native promoter, the operon structure and the upstream parS locus. Maintaining endogenous expression was important to determine the correct localization pattern as expression from heterologous promoters can lead to abnormal localization pattern as shown for the ParA homologue Soj in Bacillus subtilis (Murray and Errington, 2008). Our ParA–YFP fusion was functional (Supplementary data; Supplementary Figure S3A–C). Time-lapse microscopy and kymograph analysis of the average ParA–YFP signal over time in synchronized wild-type cells (n=65) revealed a dynamic and consistent pattern (Figure 2A). During the beginning of the cell cycle, ParA–YFP formed a ‘cloud' that extended from the new pole towards the old pole. About 10–20 min into the cell cycle, the ParA–YFP cloud rapidly condensed into a focus at the new pole (Figure 2A, arrowhead) and this new-pole accumulation persisted for the rest of the cell cycle. The ParA–YFP signal was constant during the cell cycle (Supplementary Figure S3D), consistent with western blot analysis of ParA levels (Mohl and Gober, 1997). Thus, the rapid condensation of the ParA–YFP cloud observed in wild-type cells is caused by a change in protein localization as opposed to degradation. The level of ParA–YFP signal was similar in the ΔtipN background (Supplementary Figure S3D), but its localization pattern exhibited two important differences. First, the retraction of the ParA–YFP cloud was slow (Figure 2B) and the protein accumulated at the old pole (Figure 2B, pink arrowhead). Second, unlike in wild-type cells, the ParA–YFP signal failed to accumulate at the new pole in ΔtipN cells (Figure 2B).
Figure 2.
ParA dynamics in wild-type, ΔtipN and parA-overexpressing cells. (A) Kymograph of the average ParA–YFP signal intensity along the cell length as a function of time after synchrony. Wild-type cells (CJW3010) were imaged every 1.5 min by time-lapse microscopy and analysed using MicrobeTracker. (B) Same as (A) except that the average spatial distribution of ParA–YFP over time was obtained from imaging ΔtipN cells (CJW3011). (C) Time-lapse microscopy of ParA–YFP and CFP–ParB in a wild-type background (strain CJW3367). The results from a representative cell are shown as an overlay between the relative position of CFP–ParB (green trace) and a kymographic representation of the ParA–YFP signal profile (red) along the cell length as a function of time after synchrony. CFP–ParB synthesis was induced with 0.03% xylose for 1 h before synchronization and imaging. (D) Profiles of the mean CFP–ParB and ParA–YFP intensity profiles of wild-type cells (n=72) from time-lapse sequences described in (C). (E) Same as (C) except in a ΔtipN background (strain CJW3376). (F) Time-lapse microscopy of YFP–ParA and MipZ–mCFP in cells slightly overproducing YFP–ParA (strain CJW3373). YFP–ParA overproduction was induced with 0.03% xylose for 2 h before cell synchronization and imaging. The results from a representative cell are shown as an overlay between the relative position of MipZ–mCFP (green trace) and a kymographic representation of the ParA–YFP signal profile (red) along the cell length as a function of time after synchrony. (G) Same as (F) except that the overproduction of YFP–ParA was induced with 10 times more xylose (0.3%) for 2 h before cell synchronization and imaging.
To understand how ParA dynamics may affect parS/ParB segregation, we covisualized ParA–YFP and CFP–ParB during the cell cycle. Kymograph analysis of time-lapse sequences from synchronized cell populations revealed that the condensation of the ParA–YFP cloud towards the new pole was temporally and spatially correlated with CFP–ParB segregation (Figure 2C; Supplementary Movie S3). When the duplicated CFP–ParB focus (Figure 2C, green trace) reached the ParA–YFP signal, the ParA–YFP cloud rapidly retracted to form a focus at the new pole (Figure 2C, red, 31:30 to 37:30 min; Supplementary Movie S3). Averaging the ParA–YFP and CFP–ParB signals from multiple cells showed that the translocating CFP–ParB focus follows in the wake of a retracting ParA–YFP gradient (Figure 2D).
In ΔtipN cells, rapid shifts in ParA–YFP localization between poles correlated with back-and-forth movements and stalls of the segregating CFP–ParB focus (Figure 2E; Supplementary Movie S4), as if the back-and-forth motion of ParA was attracting the partitioning complex in opposite directions. Transient localizations of ParA–YFP behind the translocating CFP–ParB focus typically preceded the direction reversal of CFP–ParB towards the old pole (Figure 2E; Supplementary Movie S4). These phenotypes were complemented by producing TipN in trans (Supplementary Figure S3G). Altogether, the data suggest that the localization defect of ParA–YFP in ΔtipN cells causes the slower, erratic translocation of the parS/ParB complex.
Further evidence that a change in ParA dynamics affects parS/ParB/MipZ segregation came from examining DNA segregation when ParA was slightly overproduced. Overexpression of a yfp–parA fusion from the xylose-inducible promoter (Pxyl) from a second locus had considerable effects on several aspects of ParA dynamics and localization (Figure 2F). First, YFP–ParA tended to form larger cloud structures in swarmer cells, often expanding all the way to the old pole. Second, the condensation of the YFP–ParA cloud was slower, accompanied with a correspondingly slower segregation of the parS/ParB/MipZ–mCFP complex. Third, ParA–YFP occasionally exhibited oscillations between new and old poles, which were associated with the reversal of parS/ParB/MipZ–mCFP movement (Figure 2F, 90–110 min).
TipN interacts with ParA at the new pole
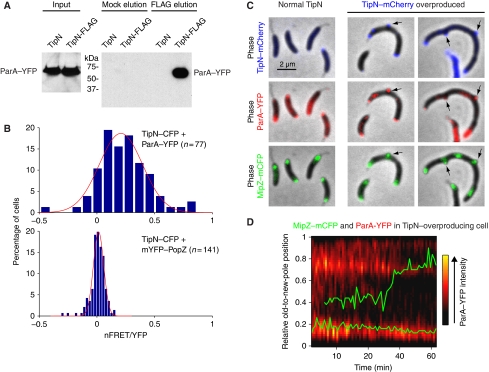
In the absence of TipN, ParA clouds failed to condense at the new pole and new ParA structures frequently formed at the opposite end behind the partitioning complex, which was correlated with stalls or backwards movement of this complex (Figure 2E). As TipN specifically localizes at the new pole (Huitema et al, 2006; Lam et al, 2006), we hypothesized that TipN may interact with ParA at that location. Consistent with this hypothesis, a TipN-FLAG fusion (produced from the native chromosomal tipN promoter in place of wild-type TipN) was able to pull down ParA–YFP from C. crescentus cells using the anti-FLAG antibody, whereas no ParA–YFP was pulled down in the control strain producing untagged TipN (Figure 3A). We confirmed that ParA and TipN interact inside cells using fluorescence resonance energy transfer (FRET) microscopy (Verveer et al, 2006). After correcting for the bleed through from strains expressing TipN–CFP and ParA–YFP alone (Supplementary Figure S4A), we obtained a mean apparent energy transfer efficiency (nFRET/YFP) of 0.21±0.03 (n=77) between TipN–CFP and ParA–YFP (Figure 3B), indicative of an interaction. Consistent with this, TipN–CFP and mYFP-PopZ, which colocalize at the new pole, but do not physically interact (Bowman et al, 2010), displayed a mean nFRET/YFP of about zero (0.014±0.005; n=141; Figure 3B; we obtained similar results when PopZ was fused to YFP and produced from its native chromosomal locus in place of PopZ; data not shown). The difference in distributions of nFRET/YFP values for TipN/ParA and TipN/PopZ fusions were highly significant (K-S test, P<0.00001).
Figure 3.
Interaction between ParA and TipN. (A) Western blotting of pull-down eluates from lysates of cells carrying ParA–YFP and TipN-FLAG (TipN-FLAG; strain CJW3359) and lysates of control cells carrying ParA–YFP and untagged TipN (TipN; strain CJW3010). Elution was carried out in the presence (FLAG) or absence (Mock) of FLAG peptide. (B) Distributions of nFRET/YFP values for TipN–CFP and ParA–YFP (strain CJW3406) and for TipN–CFP and mYFP-PopZ (strain CJW3614) at the new pole. mYFP-PopZ was induced with 0.03% xylose for 2 h before imaging. The red line is the Gaussian fit of the distributions (see Supplementary data). (C) Overlays of phase-contrast images with fluorescent images of MipZ–mCFP, ParA–YFP and TipN–mCherry localization in CJW3407 cells in which TipN–mCherry overproduction was either uninduced (normal TipN) or induced with 0.5 mM vanillic acid for 6 h (TipN–mCherry overproduced). (D) Time-lapse sequence of MipZ–mCFP (green) and ParA–YFP (red) signals in CJW3408 cells during TipN overproduction, which was induced with 0.25 mM vanillic acid for 3 h before imaging on agarose pads containing vanillic acid.
Overproduction of TipN can cause aberrant accumulation of TipN at the old pole and occasionally along lateral sides of the cell (Lam et al, 2006). ParA–YFP and partition complexes (visualized with MipZ–mCFP) were recruited to these abnormal TipN–mCherry localization sites (Figure 3C, arrows). Aberrant old-pole localization of ParA–YFP resulted in a slow and erratic segregation of the MipZ–mCFP-labelled partition complex characterized by frequent direction reversals and stalls (Figure 3D), similarly to what happens in ΔtipN cells (Figure 2E). Thus, collectively, our findings suggest that TipN is required for ParA recruitment to the new pole during condensation of the ParA structure, which in turn enables rapid and unidirectional segregation of the partition complex.
Extrinsic regulation of the parS/ParB/ParA system
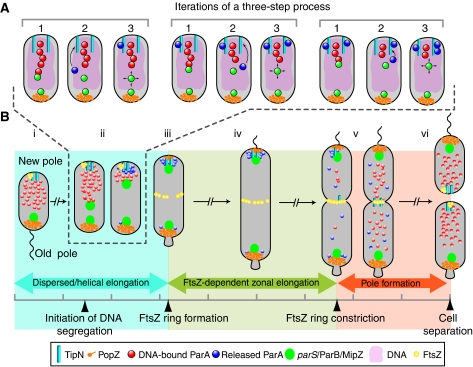
We envision a model in which the ParA cloud observed before DNA replication is primarily made of DNA-bound ParA-ATP dimers forming nucleoprotein structures. The weak intrinsic ATPase activity of ParA (Easter and Gober, 2002) would maintain this steady-state until replication is initiated. The replication process itself (Lemon and Grossman, 2001; Dworkin and Losick, 2002) and/or entropy would presumably push duplicated parS/ParB complexes apart and help one of them come in contact with the edge of the nucleoprotein structure. This interaction would lead to ParB-dependent stimulation of ParA-ATPase activity and release of ParA from the nucleoprotein structure into the cytoplasm, resulting in the shrinkage of the ParA-ATP structure (see model in Figure 7A). Space-constrained Brownian motion of chromosomal loci results in some space exploration (Elmore et al, 2005; Ebersbach et al, 2008), which presumably would allow the parS/ParB complex to interact again with the edge of the slightly smaller DNA-bound ParA-ATP structure. Repetition of this process would cause the parS/ParB complex to move closer and closer to the new pole in the wake of a ‘shrinking' ParA-ATP nucleoprotein structure. This proposed mechanism shares similarities with what has been suggested for the segregation of V. cholerae chromosome I (Fogel and Waldor, 2006) and Escherichia coli plasmid pB171 (Ringgaard et al, 2009), but how can a similar mechanism explain both the single, directional motion of the chromosome and the back-and-forth motions of the plasmid? Our data suggest that segregation of chromosomal parS/ParB in C. crescentus depends on a regulatory mechanism that prevents the released ParA molecules from reforming a ParA-ATP nucleoprotein structure behind the translocating parS/ParB complex to avoid backwards motions. We propose that TipN is an important element of this preventive mechanism.
Figure 7.
Proposed models for chromosome segregation and cell cycle coordination in C. crescentus. (A) Model for parS/ParB segregation. In wild-type cells, after initiation of DNA replication, one of the duplicated parS/ParB partition complexes comes in contact with the leading edge of a DNA-bound ParA-ATP structure (red circles) for which ParB has a strong binding affinity (step 1). (Only a small portion of the ParA structure is shown for simplicity.) ParB then stimulates ParA-ATPase activity, resulting in dissociation from the nucleoprotein filament and release of ParA molecules (blue circles), presumably ParA monomers, into the cytoplasm (step 2). This shortens the DNA-bound ParA-ATP structure and the released ParA is sequestered by TipN at the new pole to prevent regeneration of DNA-associated ParA-ATP dimers (step 3). Presumably through Brownian motion, the segregating parS/ParB reaches the new leading edge of the DNA-bound ParA-ATP structure, thereby getting closer to the new pole. Successive rounds of this cycle result in net parS/ParB translocation to the new pole. See text for more details. (B) Model for cell cycle coordination. Before DNA replication and segregation are initiated, the parS/ParB/MipZ complex is attached at the old pole by the PopZ matrix, promoting the establishment of a DNA-bound ParA-ATP structure biased towards the new pole (that is, away from the destabilizing activity of ParB at the old pole) (i). During segregation of the partition complex (described in panel A), the TipN-mediated condensation of the ParA cloud structure ensures rapid and unidirectional translocation of the partition complex (ii). Rapid segregation, which affects the timing of FtsZ ring formation, ensures an early switch between a dispersed/helical mode of growth and a primarily zonal, FtsZ ring-dependent mode of growth. Unidirectional translocation, on the other hand, affects the proper positioning of the FtsZ ring along the cell length, which dictates where the cell will divide (iii). TipN and PopZ functions maintain ParA and the partitioning complexes at the cell poles (iv) such that only a single round of segregation occurs per cell cycle. Near the end of the cell cycle, TipN delocalizes from the new cell pole, contributing to the release of ParA (v). This event, coupled with cytokinesis, which creates new cell poles free of PopZ and ParB, resets the cycle and allows for the reestablishment of the DNA-bound ParA-ATP structure in the daughter cells (vi).
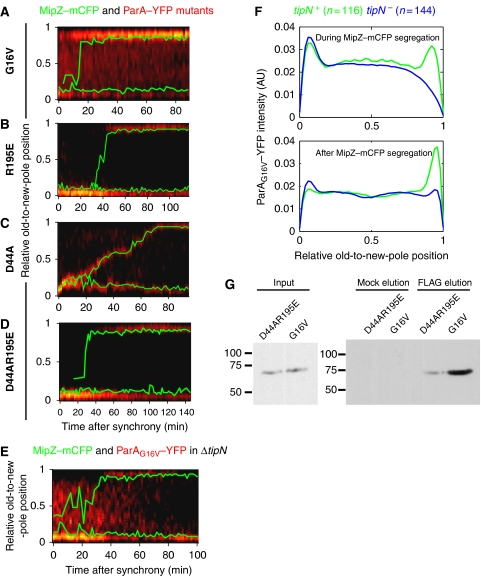
To evaluate our partitioning model, we generated point mutations in ParA that are known to interfere with the ATPase cycle and localization of Thermus thermophilis and B. subtilis ParA homologues (Soj) (Leonard et al, 2005; Hester and Lutkenhaus, 2007; Murray and Errington, 2008), which are closely related to C. crescentus ParA (see Supplementary data). Accordingly, a G16V mutant (G12V in B. subtilis Soj) is expected to be ATP-binding proficient, but DNA-binding defective. A R195E mutant (R189E in B. subtilis Soj) should be proficient for ATP-binding, dimerization and ATP hydrolysis, but defective for DNA binding (Hester and Lutkenhaus, 2007). On the other hand, a D44A mutant (D40A in B. subtilis Soj) should bind ATP, dimerize and bind DNA, but should be defective in ATP hydrolysis (Leonard et al, 2005; Murray and Errington, 2008). As ParA is essential for viability in C. crescentus (Mohl and Gober, 1997), a YFP fusion of each ParA mutant was synthesized from Pxyl at the chromosomal xylX locus in an otherwise wild-type background. All three ParA–YFP mutants, which were produced as stable, full-length proteins (Supplementary Figure S3E and F), exhibited distinct localization pattern during the cell cycle (Figure 4A–D).
Figure 4.
Mutational analysis of ParA. (A–D) Kymographs of a specific ParA–YFP mutant (in red) in single cells. The traces of MipZ–mCFP are shown in green. The synthesis of each ParA mutant was induced for 1.5 h with 0.3% xylose before synchronization and imaging. (A) ParAG16V–YFP; strain CJW3337. (B) ParAR195E–YFP; strain CJW3369. (C) ParAD44A–YFP; strain CJW3346. (D) ParAD44A R195E–YFP; strain CJW3507. (E) ParAG16V–YFP in a ΔtipN cell (strain CJW3338) as in (A–D). (F) Mean integrated intensity of ParAG16V–YFP signal in wild-type (CJW3337) and ΔtipN (CJW3338) cells during MipZ–mCFP segregation (top) and after MipZ–mCFP reaches the new pole until the onset of cell constriction (bottom). (G) Western blotting of pull-down eluates from lysates of cells carrying TipN-FLAG and ParAD44AR195E–YFP or ParAG16V–YFP (strains CJW3538 or CJW3537). Elution was carried out in the presence (FLAG) or absence (Mock) of FLAG peptide.
ParAG16V–YFP accumulated at the old and new poles before MipZ–mCFP reached the new pole and shifted to a strong, predominantly new-pole localization upon the completion of MipZ–mCFP segregation (Figure 4A). We observed no colocalization between ParAG16V–YFP and the parS/ParB/MipZ–mCFP complex during its translocation (Figure 4A).
The ParAR195E–YFP mutant displayed a strong polar localization, initially at the old pole and then at both poles after the partition complex reached the new pole (Figure 4B). It also failed to colocalize with the segregating MipZ–mCFP focus.
Conversely, the ParAD44A–YFP mutant always colocalized with MipZ–mCFP even during its segregation (Figure 4C), indicating interaction with ParB, in agreement with the B. subtilis Soj mutagenesis study (Murray and Errington, 2008). This association had a dominant-negative effect on the rate of segregation (Figure 4C). ParA–YFP carrying a double R195E and D44A mutation failed to colocalize with the partition complex during segregation (Figure 4D), similarly to the single R195E mutant (Figure 4B) and in contrast to the D44A single mutant (Figure 4C). All three mutants are expected to bind ATP and to dimerize (Leonard et al, 2005; Hester and Lutkenhaus, 2007; Murray and Errington, 2008). The only expected difference is that D44A should be able to bind DNA, unlike the other two mutants. This result suggests that ParB most strongly interacts with the DNA-bound ParA-ATP structure, in accord with our model. Consistent with this notion, we showed by FRET microscopy that CFP–ParB and ParAD44A–YFP interact as they exhibited a mean nFRET/YFP of 0.31± 0.03 (Supplementary Figure S5A). Conversely, colocalized CFP–ParB and DivJ–YFP, which are not expected to interact and served here as a negative control, had a mean nFRET/YFP ratio close to zero (−0.008±0.002; n=30; Supplementary Figure S5A; Supplementary data). Furthermore, in accord with our DNA-binding assumptions, the assumed DNA-binding proficient C. crescentus ParA–YFP and ParAD44A–YFP colocalized with the nucleoid in a heterologous E. coli system, whereas the proposed DNA-binding defective G16V and R195E mutants displayed a diffuse cytoplasmic localization (Supplementary Figure S5B).
Interestingly, after taking into consideration the segregation delay in the ΔtipN mutant, only the G16V mutant, and not the R195E and D44A single or double mutants, displayed a distinct localization pattern in the absence of TipN (Figure 4E). When TipN was present, ParAG16V–YFP localization was bipolar during segregation of the MipZ–mCFP-labelled partition complex and switched to a primarily new-pole localization after completion of MipZ–mCFP translocation (Figure 4A). In the absence of TipN, ParAG16V–YFP accumulated at the old pole throughout most of the cell cycle and only displayed a weak and intermittent new-pole accumulation after the completion of MipZ–mCFP segregation (Figure 4E).
Averaging the distribution of ParAG16V–YFP signal along the cell length during and after MipZ–mCFP segregation (Figure 4F) revealed specific differences in ParAG16V–YFP localization between wild-type and ΔtipN backgrounds. During MipZ–mCFP segregation, ParAG16V–YFP showed a clear accumulation at the new pole in wild-type cells (n=107), but not in ΔtipN cells (n=144; Figure 4F). After the completion of MipZ–mCFP segregation (i.e. after the partition complex reached the new pole), ParAG16V–YFP accumulated almost exclusively at the new pole in wild-type cells, whereas this strong new-pole accumulation was virtually absent in ΔtipN cells (Figure 4F). As the G16V mutant is unable to bind DNA (Supplementary Figure S5B) and hence fails to form nucleoprotein filaments, its TipN-dependent localization at the new pole supports a model in which TipN recruits the cytoplasmic ParA molecules released from the DNA-bound structure during segregation of parS/ParB. A corresponding mutation to G16V in the closely related T. thermophilis has been proposed to block the dimerization step (Leonard et al, 2005), suggesting the possibility that TipN might have a higher affinity for a monomeric form of ParA. Localization data (Figure 4E–F; Supplementary Figure S6A) and pull-down assays (Figure 4G) are consistent with the G16V mutant having a higher affinity for TipN than the D44A R195E double mutant, supportive of a conformational difference between these mutants.
Plasmid-encoded parS/ParB/ParA systems are thought to be self-organizing and to act without host factors (Gerdes et al, 2010). Without extrinsic interference, the dynamics of these systems lead to back-and-forth motion of plasmids over short times and hence equidistribution when time averaged. These dynamics are analogous to the oscillatory behaviour of the related, self-organizing Min system in E. coli. However, our findings imply that these intrinsic, self-perpetuating dynamics are inconsistent with chromosome segregation, in which duplicated origins are partitioned to opposite poles in one single, rapid event during the cell cycle. We propose that in C. crescentus, this distinct single-event partitioning process is achieved through the regulatory action of polarly localized proteins such as TipN. As before DNA replication, ΔtipN cells form normal ParA cloud structures, TipN is likely not involved in nucleation or stabilization of the ParA structure at the new pole. Instead, collectively, our data suggest a model in which TipN interrupts the ParB-stimulated ParA-ATPase cycle by trapping cytoplasmic ParA intermediates at the new pole after their release from the nucleoprotein structure during segregation. This step would prevent reassembly of ParA into nucleoprotein structures behind the migrating parS/ParB, ensuring its rapid translocation in one constant direction.
Existence of partially compensatory mechanisms
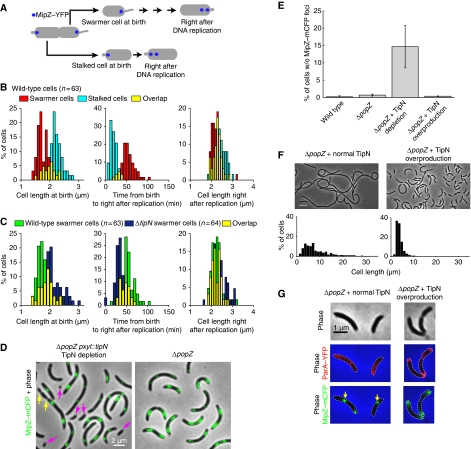
A ΔtipN mutation caused a significant increase in cell length within the cell population (Supplementary Figure S1B; K-S test, P<0.00001), but remained viable. We found that two processes partially compensate for the loss of TipN function. One of them is mediated by a cell size-dependent sensing mechanism. In wild-type cells, asymmetric division leads to swarmer daughter cells that are shorter and that replicate and segregate parS/ParB/MipZ–mCFP later than their stalked siblings (Figure 5A and B). Quantitative analysis of time-lapse sequences showed that swarmer daughter cells initiate DNA replication at similar cell length as stalked daughter cells (Figure 5B). This suggests that cells have to reach a certain size before initiating DNA replication, explaining the delay in swarmer progeny. By affecting division placement and growth (see below), the ΔtipN mutation generated swarmer cells considerably longer than wild type (Figure 5C). However, these longer ΔtipN swarmer cells initiated DNA replication and segregation sooner than wild-type swarmer cells (Figure 5C). In other words, ΔtipN swarmer cells display a shorter G1 phase, thereby considerably reducing the extra period of growth during G1 and partially compensating for the cell length increase at birth. In the absence of such cell size-sensing mechanism, the ΔtipN mutation would cause swarmer cells to become ever longer with each successive division.
Figure 5.
Mechanisms compensating for a loss of TipN function. (A) Schematic showing the different stages plotted in (B) and (C). (B, C) Synchronized predivisional cells of wild-type (CJW2022) and ΔtipN (CJW3366) strains carrying mipZ–yfp were grown on agarose pads containing M2G and imaged every 1.5 min. The time right after DNA replication in the swarmer and stalked progeny was determined by the appearance of a second MipZ–YFP focus. Cell identification and image analysis were performed using MicrobeTracker. (B) Distributions of (1) the length of wild-type swarmer and stalked cells at birth, (2) times between birth and right after DNA replication and (3) cell lengths right after DNA replication. (C) Distribution of (1) cell lengths at birth, (2) times between birth and right after DNA replication and (3) cell length right after DNA replication, starting with wild-type or ΔtipN swarmer cells. (D) Overlays of phase contrast and MipZ–mCFP images of ΔpopZ cells carrying mipZ–mcfp (strain CJW3599) and ΔpopZ cells carrying mipZ–mcfp and depleted of TipN (growth without xylose for 17h; strain CJW3543). Purple arrows show minicells lacking MipZ–mCFP signal and yellow arrows show the creation of these cells. (E) Percentage of the cell population lacking MipZ–mCFP from wild-type cells (CB15N), ΔpopZ cells (CJW3599) and ΔpopZ cells (CJW3543) either depleted of TipN (growth without xylose for 17 h) or overproducing TipN (growth in 0.3% xylose). Shown is the mean±s.d. from three independent experiments. (F) Top, phase-contrast images of ΔpopZ cells (CJW3512) either producing TipN–mYFP at a normal level (growth in PYE with 0.2% glucose) or overproducing TipN–mYFP (growth in PYE with 0.3% xylose for 17 h). Scale bar, 5 μm. Bottom, distribution of cell lengths under each condition. (G) Phase-contrast and overlays of ParA–YFP (red) and MipZ–mCFP (green) in a ΔpopZ background carrying tipN under Pxyl on a low-copy-number plasmid (strain CJW3043). Overexpression of tipN was either uninduced with 0.2% glucose or induced with 0.3% xylose for 5 h.
The second compensatory mechanism is mediated by PopZ, a multifunctional protein that assembles into a matrix at the cell poles (Bowman et al, 2008; Ebersbach et al, 2008). PopZ anchors parS/ParB at the pole after segregation through an interaction with ParB (Bowman et al, 2008; Ebersbach et al, 2008). In our model, TipN ensures speedy and directional segregation and without this regulation, the parS/ParB complex displays erratic, back-and-forth motions over the DNA. However, the erratic motion in the ΔtipN background stops when parS/ParB gets captured by PopZ at the pole; from then on, parS/ParB can no longer respond to the aberrant ParA cloud inside the cells (Figure 2B and E) because it is attached to the PopZ matrix. A similar effect could be observed when ParA–YFP was overproduced. These conditions occasionally resulted in ParA oscillations, as reported above (Figure 2F). As large overproduction of ParA causes cell filamentation (Mohl and Gober, 1997), the elongated ParA–YFP-overproducing cells often displayed more than two partition complexes, with the unattached complexes—but not the polarly localized, PopZ-attached complexes—moving back and forth synchronously with ParA oscillations (Figure 2G). Overproduction of ParA may presumably saturate TipN-binding sites, resulting in oscillatory behaviour of unattached parS/ParB, as observed for plasmids.
In addition to this parS/ParB anchoring function, PopZ appears involved in the polar maintenance of ParA. Mutant analysis suggests that PopZ has an affinity for non-DNA-binding forms of ParA (Supplementary Figure S6; Supplementary data). This property, together with the polar attachment of parS/ParB, minimizes the ΔtipN defects. Such synergistic function between PopZ and TipN is substantiated by the synthetic lethal phenotype of a double ΔpopZ ΔtipN mutation (Ebersbach et al, 2008). Depletion of TipN in ΔpopZ cells growing in minimal M2G medium caused a severe increase in cell division events producing cells lacking a partition complex (Figure 5D and E), suggesting that the synthetic lethal phenotype is due to a severe chromosome segregation defect.
ΔpopZ cells were elongated in both minimal M2G and rich peptone-yeast extract (PYE) media, but cell filamentation was particularly severe in PYE medium (Supplementary Figure S6E). Overexpression of tipN, however, was able to suppress this cell filamentation phenotype, even in PYE medium (Figure 5F), with the mean cell length being reduced from 9.0±0.4 μm (n=305) in the absence of TipN overproduction to 3.83±0.03 μm (n=3463) when TipN was overproduced. Suppression of cell filamentation was correlated with a rescue of the defects in ParA localization and parS/ParB/MipZ–mCFP polar attachment (Figure 5G). These data are consistent with TipN and PopZ acting synergistically in the regulation of chromosome segregation in C. crescentus. The existence of compensatory mechanisms attests to the importance of this regulation.
Effects of regulation of the parS/ParB/ParA system on cell cycle coordination
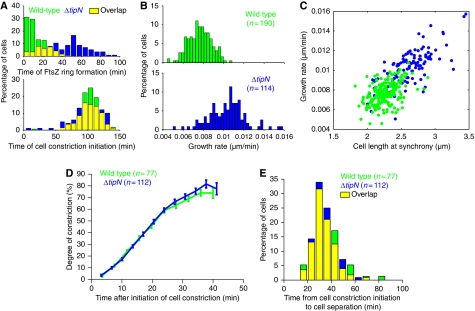
Although the requirement of DNA segregation for division has been previously documented in C. crescentus (Mohl et al, 2001; Figge et al, 2003; Thanbichler and Shapiro, 2006), we found that parS/ParB segregation does not control the timing of cell constriction as wild-type and ΔtipN cells constricted at similar times (K-S test, P>0.2; Figure 6A), despite the significant delay in FtsZ ring formation in ΔtipN cells (K-S test, P<0.0001; Figure 6A). Thus, the timing of FtsZ ring formation does not dictate that of cell constriction initiation, and divisome maturation (i.e. recruitment of other cell division proteins) can occur rapidly and does not require early FtsZ ring formation. Although the positioning of the division plane along the cell length is abnormal (Figure 1A), the rate of cell constriction and the timing of cell separation appeared normal in ΔtipN cells (Figure 6D and E). This argues that regulation of MipZ-associated parS/ParB/ParA translocation influences where cell division occurs along the long cell axis (through the positioning of the FtsZ ring), but not when or how fast the cell constricts and separates.
Figure 6.
Effects of ΔtipN mutation on the temporal and spatial regulation of cell division and cell growth. (A) Top, distributions of the time from synchrony to the appearance of the FtsZ–YFP ring. Bottom, distributions of the time from synchrony to the appearance of cell constriction. These distributions were obtained from time-lapse sequences of wild-type (MT199; n=190) and ΔtipN (CJW2563; n=114) cells as described in Figure 1A. (B) Distributions of rates of cell-length elongation during the time-lapse sequences described in Figure 1A. (C) Rates of cell-length growth were plotted as a function of the cell length at time=0 min in Figure 1A. (D) Mean degree of cell constriction ±s.e.m. of wild-type (CB15N) and ΔtipN (CJW1407) cells after the time of cell constriction initiation. Following synchronization, cells were imaged every 1.5 min by time-lapse microscopy and analysed by MicrobeTracker. (E) Distributions of time gaps between initiation of cell constriction initiation and cell separation for cell populations in (D).
In addition to mispositioning the division plane, ΔtipN cells elongated significantly faster than wild-type cells (Figure 6B; 10.3±0.2 nm/min for ΔtipN (n=114) versus 7.7±0.1 nm/min for wild type (n=190); K-S test, P<0.00001) despite similar cell cycle durations. A possible source for this difference may come from the observation that C. crescentus uses two modes of cell elongation: a dispersed/helical mode of growth along the entire cell length (Figge et al, 2004; Aaron et al, 2007) before FtsZ ring formation and a largely FtsZ ring-dependent, zonal mode of growth after FtsZ ring assembly (Aaron et al, 2007). The delay in FtsZ ring formation in ΔtipN cells increases the duration of dispersed/helical growth compared with that in wild-type cells, suggesting that the dispersed/helical growth is faster than the zonal one. In addition, as ΔtipN swarmer cells are on average longer than wild-type cells (Figure 5C), they may have more available sites for new cell wall insertion, providing an explanation for the greater rate of cell elongation. Consistent with a dependence of growth rate on cell size, the length of swarmer cells (i.e. after synchrony) was correlated with the rate of growth for both wild-type and ΔtipN cells (Figure 6C).
Taken together, our data suggest that by affecting the timing of FtsZ ring formation, TipN-dependent regulation of parS/ParB translocation dynamics does not affect the timing of cell constriction initiation, but instead affects the time and hence relative contribution of dispersed/helical growth versus largely FtsZ-dependent, zonal growth (Aaron et al, 2007). In addition, our findings argue that the erratic, less-directional translocation of parS/ParB/MipZ in the ΔtipN mutant affects the positioning of the FtsZ ring and cell constriction along the cell length (Figure 1). Extrinsic regulation of parS/ParB/MipZ translocation would thus control cell growth in time and cell division in space.
After division, the segregation system needs to be reset in the daughter cells. TipN delocalization from the new pole in the late predivisional stage likely contributes to this event (Supplementary Figure S4B; see Supplementary data). This delocalization presumably lowers the local concentration of TipN and releases ParA, allowing them to reform a DNA-bound ParA-ATP structure. This structure preferentially forms away from ParB destabilizing activity, which is restricted at the old pole of each daughter cell after cytokinesis through attachment to PopZ. These proposed TipN- and PopZ-dependent mechanisms would ensure that translocation of the partition complex occurs efficiently in only one direction and only once per cell cycle, as summarized in Figure 7B.
Adapting the parS/ParB/ParA system for chromosome segregation
The distribution of TipN and PopZ among α-proteobacteria suggests conserved function in this large, medically and agriculturally important class of bacteria. However, the conservation of chromosomal parS/ParB/ParA systems is even broader and spans across bacterial phyla (Livny et al, 2007), suggesting a widespread function in chromosome segregation. Our findings argue that these parS/ParB/ParA partitioning systems may require extrinsic regulation to prevent back-and-forth motions, which are unsuitable for chromosome segregation. Our work suggests a mechanistic framework by which this regulation can be achieved and integrated with the cell cycle through polarly localized factors.
Materials and methods
Strains, plasmids and growth conditions
C. crescentus was grown at 30°C in PYE or M2G supplemented (Ely, 1991) with kanamycin (5 μg/ml), oxytetracycline (1 μg/ml), spectinomycin/streptomycin (30/5 μg/ml) or gentamycin (5 μg/ml) when appropriate. Vanillic acid (0.5 mM or 0.25) or xylose (0.3 or 0.03%) was added to the medium to induce the Pvan or Pxyl promoters, respectively. Exponentially growing cultures were used for all experiments. Plasmids were mobilized from E. coli strain S17-1 into C. crescentus by conjugation (Ely, 1991) or electroporation. Plasmids and strains are listed in Supplementary Table S1 and their mode of construction is provided in the Supplementary data. Synchronized populations of C. crescentus swarmer cells were obtained using a technique described previously (Evinger and Agabian, 1977) with some modifications (Angelastro et al, 2010).
Microscopy and image analysis
Cells were imaged at room temperature (RT, ∼22°C) or at 31°C using an objective heater on either a Nikon Eclipse ti-U fitted with a 100X phase-contrast objective and an Andor ixonEM+ 888 camera or a Nikon E1000 microscope fitted with a 100X phase-contrast objective and a Hamamatsu Orca-ER LCD camera. Cells were immobilized on 1% agarose pads with M2 buffer or M2G medium containing vanillic acid or xylose when appropriate. Images were taken with NIS-Elements (Nikon) or Metamorph software (MDS Analytical Technologies).
FRET microscopy and image analysis were performed as described in the Supplementary data.
Pull-down experiments
Two liters of CJW3359, CJW3010, CJW3538 and CJW3537 cell cultures (OD660 <0.6) were harvested and washed with 50 ml of buffer A (40 mM HEPES (pH 7.4), 150 mM KCl in 20% glycerol). Cells were pelleted again and resuspended in 36 ml of buffer A. Cross-linking was carried out for 30 min with 2 mM dithiobis(succinimidylpropionate) at RT. The reaction was quenched by the addition of Tris/HCl (pH 8.0) to a final concentration of 0.5 M and incubated at RT for 15 min. The cells were pelleted and resuspended in 25 ml of buffer A containing a protease inhibitor cocktail (Roche). Following cell lysis by French Press, the crude lysate was clarified by centrifugation and the pellet was resuspended in 20 ml of buffer B (buffer A plus 0.1% Triton-X-100 and protease inhibitor cocktail) and incubated at 4°C for 3 h. The membrane protein extract was then clarified by ultracentrifugation (30 000 g, 15 min) before 0.5 ml ANTI-FLAG® M2 Affinity Gel (Sigma) was added. Following overnight incubation at 4°C, unbound proteins were removed by passing the mixture over a column. The Affinity Gel was washed on the column with a total of 36 ml of buffer B in 2 ml batches. The last 1.5 ml of the wash was used for the mock elution. Then, TipN-FLAG was eluted with 1.5 ml buffer B supplemented with 100 μM 3 × FLAG peptides. The elutions (including the mock elution) were concentrated by trichloroacetic acid/deoxycholic acid precipitation and resuspended in 12.5 μl of 2 × loading buffer plus 12.5 μl 1 M Tris (pH 8). The samples (10 μl) were separated on an SDS–PAGE gel.
Growth curve measurements
Cultures (three for each condition) in exponential phase of growth were diluted to final OD660 of 0.01. Two hundred fifty microliters were used to inoculate 96-well plates in triplicates (9 wells per strain condition). Readings were taken every 5 min with continuous shaking at 30°C.
Western blots
Western blots were performed using antibodies at the following concentrations: GFP and derivatives (YFP and YFP), antibody JL-8 (Sigma) at 1:1000; ParB, anti-ParB at 1:5000; MreB, anti-MreB at 1:10 000.
Supplementary Material
Acknowledgments
We are grateful to M Thanbichler and L Shapiro for strains, J Gober for antibodies, O Sliusarenko and T Emonet for help with MATLAB, G Jentsch and S Poggio for helpful discussions and the members of the Jacobs-Wagner laboratory for valuable discussions and critical reading of an earlier version of the paper. This work was supported by the National Institutes of Health (GM065835 to CJ-W). WS was partially supported by a predoctoral fellowship (1F31GM083627). HCL was supported by the Edward L Tatum Fellowship from Yale University. CJ-W is a Howard Hughes Medical Institute investigator.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C (2007) The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol 64: 938–952 [DOI] [PubMed] [Google Scholar]
- Angelastro PS, Sliusarenko O, Jacobs-Wagner C (2010) Polar localization of the CckA histidine kinase and cell cycle periodicity of the essential master regulator CtrA in Caulobacter crescentus. J Bacteriol 192: 539–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barillà D, Carmelo E, Hayes F (2007) The tail of the ParG DNA segregation protein remodels ParF polymers and enhances ATP hydrolysis via an arginine finger-like motif. Proc Natl Acad Sci USA 104: 1811–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouet J-Y, Ah-Seng Y, Benmeradi N, Lane D (2007) Polymerization of SopA partition ATPase: regulation by DNA binding and SopB. Mol Microbiol 63: 468–481 [DOI] [PubMed] [Google Scholar]
- Bowman GR, Comolli LR, Gaietta GM, Fero M, Hong S-H, Jones Y, Lee JH, Downing KH, Ellisman MH, McAdams HH, Shapiro L (2010) Caulobacter PopZ forms a polar subdomain dictating sequential changes in pole composition and function. Mol Microbiol 76: 173–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, Moerner WE, Earnest T, Shapiro L (2008) A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134: 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin J, Losick R (2002) Does RNA polymerase help drive chromosome segregation in bacteria? Proc Natl Acad Sci USA 99: 14089–14094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easter J, Gober JW (2002) ParB-stimulated nucleotide exchange regulates a switch in functionally distinct ParA activities. Mol Cell 10: 427–434 [DOI] [PubMed] [Google Scholar]
- Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C (2008) A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell 134: 956–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G, Ringgaard S, Møller-Jensen J, Wang Q, Sherratt DJ, Gerdes K (2006) Regular cellular distribution of plasmids by oscillating and filament-forming ParA ATPase of plasmid pB171. Mol Microbiol 61: 1428–1442 [DOI] [PubMed] [Google Scholar]
- Elmore S, Müller M, Vischer N, Odijk T, Woldringh CL (2005) Single-particle tracking of oriC-GFP fluorescent spots during chromosome segregation in Escherichia coli. J Struct Biol 151: 275–287 [DOI] [PubMed] [Google Scholar]
- Ely B (1991) Genetics of Caulobacter crescentus. Meth Enzymol 204: 372–384 [DOI] [PubMed] [Google Scholar]
- Evinger M, Agabian N (1977) Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol 132: 294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figge RM, Divakaruni AV, Gober JW (2004) MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol 51: 1321–1332 [DOI] [PubMed] [Google Scholar]
- Figge RM, Easter J, Gober JW (2003) Productive interaction between the chromosome partitioning proteins, ParA and ParB, is required for the progression of the cell cycle in Caulobacter crescentus. Mol Microbiol 47: 1225–1237 [DOI] [PubMed] [Google Scholar]
- Fogel MA, Waldor MK (2006) A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev 20: 3269–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Howard M, Szardenings F (2010) Pushing and pulling in prokaryotic DNA segregation. Cell 141: 927–942 [DOI] [PubMed] [Google Scholar]
- Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L (2005) MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120: 329–341 [DOI] [PubMed] [Google Scholar]
- Hester CM, Lutkenhaus J (2007) Soj (ParA) DNA binding is mediated by conserved arginines and is essential for plasmid segregation. Proc Natl Acad Sci USA 104: 20326–20331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH (2006) Bacterial birth scar proteins mark future flagellum assembly site. Cell 124: 1025–1037 [DOI] [PubMed] [Google Scholar]
- Jensen RB, Shapiro L (1999) The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc Natl Acad Sci USA 96: 10661–10666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AJ, Sackett MJ, Din N, Quardokus E, Brun YV (1998) Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev 12: 880–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H, Schofield WB, Jacobs-Wagner C (2006) A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell 124: 1011–1023 [DOI] [PubMed] [Google Scholar]
- Lemon KP, Grossman AD (2001) The extrusion-capture model for chromosome partitioning in bacteria. Genes Dev 15: 2031–2041 [DOI] [PubMed] [Google Scholar]
- Leonard TA, Butler PJ, Löwe J (2005) Bacterial chromosome segregation: structure and DNA binding of the Soj dimer—a conserved biological switch. EMBO J 24: 270–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J, Yamaichi Y, Waldor MK (2007) Distribution of centromere-like parS sites in bacteria: insights from comparative genomics. J Bacteriol 189: 8693–8703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohl DA, Easter J, Gober JW (2001) The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus. Mol Microbiol 42: 741–755 [DOI] [PubMed] [Google Scholar]
- Mohl DA, Gober JW (1997) Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell 88: 675–684 [DOI] [PubMed] [Google Scholar]
- Murray H, Errington J (2008) Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell 135: 74–84 [DOI] [PubMed] [Google Scholar]
- Pratto F, Cicek A, Weihofen WA, Lurz R, Saenger W, Alonso JC (2008) Streptococcus pyogenes pSM19035 requires dynamic assembly of ATP-bound ParA and ParB on parS DNA during plasmid segregation. Nucleic Acids Res 36: 3676–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quardokus EM, Brun YV (2002) DNA replication initiation is required for mid-cell positioning of FtsZ rings in Caulobacter crescentus. Mol Microbiol 45: 605–616 [DOI] [PubMed] [Google Scholar]
- Radnedge L, Youngren B, Davis M, Austin S (1998) Probing the structure of complex macromolecular interactions by homolog specificity scanning: the P1 and P7 plasmid partition systems. EMBO J 17: 6076–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringgaard S, van Zon J, Howard M, Gerdes K (2009) Movement and equipositioning of plasmids by ParA filament disassembly. Proc Natl Acad Sci USA 106: 19369–19374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrana B, Newton A (1975) Pattern of unequal cell division and development in Caulobacter crescentus. Dev Biol 44: 380–385 [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L (2006) MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126: 147–162 [DOI] [PubMed] [Google Scholar]
- Toro E, Hong S-H, McAdams HH, Shapiro L (2008) Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc Natl Acad Sci USA 105: 15435–15440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verveer PJ, Rocks O, Harpur AG, Bastiaens PI (2006) Measuring FRET by sensitized emission. Cold Spring Harb Protoc 2006: 4597. [DOI] [PubMed] [Google Scholar]
- Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L (2004) Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci USA 101: 9257–9262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler RT, Shapiro L (1999) Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol Cell 4: 683–694 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.