Abstract
Summary: Computational methods in molecular biology will increasingly depend on standards-based annotations that describe biological experiments in an unambiguous manner. Annotare is a software tool that enables biologists to easily annotate their high-throughput experiments, biomaterials and data in a standards-compliant way that facilitates meaningful search and analysis.
Availability and Implementation: Annotare is available from http://code.google.com/p/annotare/ under the terms of the open-source MIT License (http://www.opensource.org/licenses/mit-license.php). It has been tested on both Mac and Windows.
Contact: rshankar@stanford.edu
1 INTRODUCTION
Meta-data describing high-throughput investigations enable unambiguous interpretation of experiments, experiment reproducibility and meaningful searching and analysis of the resulting data. The microarray community has developed MAGE-TAB (Rayner, 2006), an annotation format for microarray data. MAGE-TAB allows laboratories to manage, exchange and publish well-annotated biomedical data using a spreadsheet-based paradigm. Several public repositories and analysis tools for microarray data such as ArrayExpress (Parkinson, 2009), Stanford Microarray Database (SMD) (Hubble, 2009), MeV (Saeed, 2006), Bioconductor (Gentleman, 2004) and caArray (Klemm et al., 2010) support microarray data submissions with MAGE-TAB annotations, and open source tools are available for conversion of legacy formats into MAGE-TAB (Rayner, 2009).
In order to improve the volume, quality and granularity of annotations, there is a compelling need for software that enables biologists to easily annotate such data. We describe Annotare, a tool that facilitates annotation of gene expression data in MAGE-TAB format. Annotare is available under the terms of the MIT License at http://code.google.com/p/annotare/.
2 SOFTWARE COMPONENTS
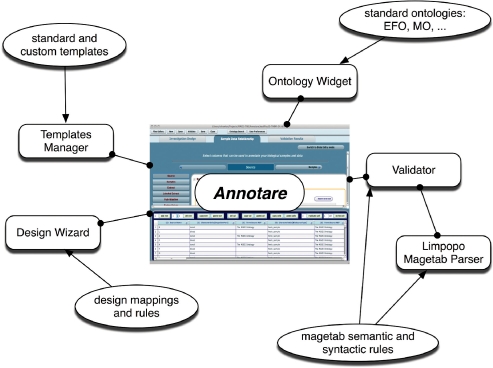
Annotare is a stand-alone desktop application that features (i) a set of intuitive editor forms to create and modify annotations; (ii) support for easy incorporation of terms from biomedical ontologies; (iii) standard templates for common experiment types; (iv) a design wizard to help create a new document; and (v) a validator that checks for syntactic and semantic violations (Fig. 1).
Fig. 1.
Annotare software components. Rectangles represent the various components and the ovals represent the resources that these components consume.
The front-end graphical user interface (GUI) uses Adobe AIR. This enables Annotare to run on multiple operating systems, and also sets the stage for future work to translate the desktop version to the web (see Section 3). Backend modules are built using Java, and the data communication between Adobe AIR and Java modules is supported by the Merapi messaging technology. Annotare has been tested on Windows XP2 and Mac OS (10.5 or greater).
2.1 Annotations editor
Annotare has a set of easy-to-use GUIs to view and modify annotations for an experiment. Using the forms, users can record details such as author's contact information, experimental design, protocols used, publications and relationships between biological materials used and data produced.
The GUI hides the syntactic complexity of MAGE-TAB as much as possible. A spreadsheet edit-and-view paradigm allows annotation of the relationships between biomaterials and data. A column designer complements the spreadsheet functionality by grouping relevant MAGE-TAB column options together, facilitating the addition or deletion of columns, while obviating the need to know the correct column ordering.
2.2 Ontology support
The most challenging part of creating MAGE-TAB annotations can be using the correct terms from appropriate biomedical ontologies to describe an experiment in an unambiguous fashion. Examples of information that use controlled vocabularies include experimental design, experimental factor types, protocol types and sample characteristics. To support use of controlled vocabularies, Annotare includes the Experiment Factor Ontology (Malone et al., 2010). Annotare exploits an ontology auto-complete function. Annotare also supports an ontology widget that is enabled with ontology look-up services of the NCBO Bioportal (http://bioportal.bioontology.org/). The widget allows users to search for and use appropriate terms from many ontologies, such as the MGED Ontology (Whetzel, 2006).
2.3 Standard templates
A researcher should not have to start from a blank slate in order to annotate experiments. Annotare provides a set of standard templates, covering common species and experimental designs (i.e. a time series). Users can select templates that best match experiments and get pre-formatted MAGE-TAB that can then be completed with experiment-specific data. Custom templates can also be created and saved for use in future experiments.
2.4 Design wizard
In addition to experiment templates, Annotare has a design wizard that helps users create a MAGE-TAB. The wizard takes the user through a series of questions eliciting information about the experimental design, the number of channels, the labels used for each channel, and platform and protocol information. Based on the user's answers, the wizard generates partial annotations that the user can then complete using the editor. In the process of generating annotations, the wizard taps into an internally stored knowledge base of rules and mappings that connect various experiment designs, species, technology vendors, array designs and protocols.
2.5 Validator
The MAGE-TAB specification imposes a set of syntactic and semantic rules on the layout and content of MAGE-TAB documents. Users can invoke Annotare's validator component at any time in order to check if a document complies with these rules. The validator flags any violations as errors, warnings or missing data. It employs the Limpopo Parser, a library for MAGE-TAB parsing and validation, developed by ArrayExpress.
3 DISCUSSION
Annotare is a collaborative open-source software development effort involving many institutions. The tool is freely available from Annotare's project web site http://code.google.com/p/annotare/. Updates and improvements are planned in response to current usability studies. A web-based version of Annotare is also under development. Not only will a web-based version be able to take advantage of finding key ontology terms or publications via the internet, but it will be configurable so that it can be directly connected to a software package or database. Both ArrayExpress and SMD will provide access to the web-based Annotare tool to construct and view high-throughput experimental annotations.
In addition to the web-based version of Annotare, future work will provide support for the MAGE-TAB version 1.1 as well as RNA-seq data. In particular, Annotare will be extended to allow researchers to annotate their RNA-seq or ChIP-seq experiments to satisfy the MINSEQE data sharing requirements for high-throughput sequence data (A.Brazma et al., submitted for publication).
ACKNOWLEDGEMENTS
The authors acknowledge the MGED community, particularly Dr J. Klemm, for critical input and evaluation of this software package.
Funding: US National Institutes of Health (the National Human Genome Research Institute, the National Institute of Biomedical Imaging and Bioengineering and the National Cancer Institute) [grant number P41 HG003619]. EC Funding SLING contract number (to E.H. 226073), and EMBL funding (to T.B. and H.P.).
Conflict of Interest: none declared.
REFERENCES
- Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubble J, et al. Implementation of GenePattern within the Stanford Microarray Database. Nucleic Acids Res. 2009;37:D898–D901. doi: 10.1093/nar/gkn786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm J, et al. The caBIG® Life Sciences Distribution. In: Ochs MF, et al., editors. Biomedical Informatics for Cancer Research. 1st. New York, NY, USA: Springer; 2010. pp. 253–266. [Google Scholar]
- Malone J, et al. Modeling sample variables with an experimental factor ontology. Bioinformatics. 2010;26:1112–1118. doi: 10.1093/bioinformatics/btq099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson H, et al. ArrayExpress update–from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37:D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner TF, et al. A simple spreadsheet-based, MIAME-supportive format for microarray data: MAGE-TAB. Bioinformatics. 2006;7:489. doi: 10.1186/1471-2105-7-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner TF, et al. MAGETabulator, a suite of tools to support the microarray data format MAGE-TAB. Bioinformatics. 2009;25:279–280. doi: 10.1093/bioinformatics/btn617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Whetzel PL, et al. The MGED Ontology: a resource for semantics-based description of microarray experiments. Bioinformatics. 2006;22:866–873. doi: 10.1093/bioinformatics/btl005. [DOI] [PubMed] [Google Scholar]