Abstract
How life emerged on this planet is one of the most important and fundamental questions of science. Although nearly all details concerning our origins have been lost in the depths of time, there is compelling evidence to suggest that the earliest life might have exploited the catalytic and self-recognition properties of RNA to survive. If an RNA based replicating system could be constructed in the laboratory, it would be much easier to understand the challenges associated with the very earliest steps in evolution and provide important insight into the establishment of the complex metabolic systems that now dominate this planet. Recent progress into the selection and characterization of ribozymes that promote nucleotide synthesis and RNA polymerization are discussed and outstanding problems in the field of RNA-mediated RNA replication are summarized.
Synthetic RNA replication systems could provide insight into how life first emerged. Recently developed ribozymes that promote nucleotide synthesis and polymerization bring us closer to this goal.
Cell division is a fundamental biological process in which genetic information is duplicated and shared between daughter cells. In extant cellular life, DNA serves as the repository of genetic information, but its replication is complicated by the daunting size and complex structural organization of modern genomes. For this reason, multiple enzymes are required to ensure faithful genomic replication in all higher life forms. Notably, simpler replicating systems such as viruses, have smaller genomes and tend to use correspondingly more error-prone replicative machinery (Kunkel and Bebenek 2000; Gago, Elena et al. 2009). Presumably, if the initial organisms on this planet also had small genomes, then the earliest genomic replication could have been a relatively simple and error-prone process compared with the complex replicative strategies of modern life.
ABC of Life: Abiotic to Biotic Chemistry, the Emergence of the RNA World
Modern biology is built up from a dense web of chemical reactions that are maintained by the catalytic reactions of hundreds of enzymes. These catalysts are all one dimensional, aperiodic polymers, built from protein or nucleic acids. Such polymers are able to adopt a diverse range of complex three-dimensional folds and by virtue of their linear sequence are simple to encode. A particular advantage of these biological polymers is that each polymer type is defined by a small set of monomers that can be polymerized by a single self-consistent chemistry. This type of polymer construction empowers evolution with a mechanism to rapidly adapt existing polymer folds to new functions by the mutation/recombination of polymer sequence. Based on the intrinsic simplicity of modern biological polymers, the earliest biological systems appear overwhelmingly likely to have been polymer based as well (Joyce 2002).
The emergence of the earliest biological replicating systems must have required considerable abiotic chemical organization. Such abiotic chemistry would not have immediately disappeared upon the emergence of life and would have provided chemical “sustenance” to nurture the increasing metabolic complexity of emergent life as it evolved. As a result if the first biological polymer can ever be reliably identified it would place an important constraint on the abiotic environments possible on the early Earth. This polymer would be the bridge between the world of abiotic chemistry and the biotic world of enzymatic reactions.
Based on this logic, the earliest replicative polymers should satisfy three primary conditions: (1) The initial polymers should have an intrinsic mechanism to facilitate their replication either abiotically or biotically. Ideally the polymer's monomer subunits should be able to be polymerized by a single uniform chemistry. (2) Abiotically, monomers should be easily synthesized and this synthesis should be compatible with abiotic polymerization. Abiotic polymerization from abiotically synthesized monomers would have presumably resulted in the synthesis of more monomer than polymer and would provide the earliest replicating systems with a source of monomers before the evolution of a biological metabolism. (3) The resulting polymers should intrinsically be endowed with the ability to promote a broad range of chemistry. In particular the metabolic synthesis of monomer units from abiotic sources should be tenable using the polymers themselves as catalysts. This monomer synthesis would serve as the basis for one of the earliest metabolic reactions associated with replication.
RNA is the simplest aperiodic polymer that biologically and chemically has been shown to satisfy the first and last of these three conditions. RNA is comprised of four distinct monomers, which can form a double-stranded RNA helix by simple and predictable pairing rules. The ability to form a regular homoduplex provides a fundamental mechanism for the templated replication of RNA strands and is used by simple RNA systems such as viruses to replicate. Equally important, RNA can fold into a variety of complex three-dimensional shapes that promote a broad range of chemical reactions (Wilson and Szostak 1999; Ellington, Chen et al. 2009). Biologically, RNA plays a fundamental role in modern life being responsible for ribosomal translation of mRNA into protein (Nissen, Hansen et al. 2000), RNase P and tRNA maturation (Pannucci, Haas et al. 1999; Marquez, Chen et al. 2006), riboswitches and gene regulation (Tucker and Breaker 2005), and as a template for the extension of telomeres by telomerase (Qiao and Cech 2008). The ribosome together with other critical biological RNA catalysts, such as RNase P, has been interpreted as a relic of an “RNA World,” where RNA and not protein was the dominant catalyst (Crick 1958; Gilbert 1986; Orgel 2004). This evidence together with the metabolic importance of RNA and the nucleotide cofactors across biology (White 1976; Benner et al. 1989), strongly suggests that RNA played an important role early in evolution and that it might have been involved in the very first autocatalytic reactions.
Abiotic Nucleotide Synthesis
Until recently a primary difficulty in declaring RNA the earliest biological polymer has been the absence of convincing evidence that activated RNA monomers can be produced by abiotic processes. This has caused some to speculate that another even simpler polymer preceded RNA in the early evolution of life (Orgel 2004; Joyce and Orgel 2006). Although it has been known for some time that the purine and pyrimidine nitrogenous bases (Robertson and Miller 1995; Zubay and Mui 2001; Hill and Orgel 2002) and ribose sugar (Ricardo et al. 2004; Gesteland et al. 2006) required to construct ribonucleosides can be synthesized separately from plausible prebiotic compounds, the creation of a glycosidic linkage has been problematic. Heating purine bases together with ribose in dehydrating conditions leads to the production of nucleosides in low yield (Fuller et al. 1972) but the synthesis of pyrimidine nucleotides has proven difficult (Orgel 2004).
In 2009, Powner et al. elegantly demonstrated that pyrimidine ribonucleotides could be formed from the same precursor molecules used to make a pyrimidine and ribose, with the exception that these molecules first be reacted to generate an intermediate, 2-aminooxazole. This clever approach bypasses the need to synthesize a glycosidic linkage and produces a reasonable yield of a pyrimidine 2′,3′-cyclic phosphate nucleotide monomer (Powner et al. 2009). It is currently unknown how purine monomers could be efficiently synthesized by abiotic mechanisms. Nevertheless, if prebiotic routes for both pyrimidine and purine nucleotides can be found, then the abiotic synthesis of RNA polymers via their condensation (Ferris et al. 1996; Monnard et al. 2003) or via the efficient polymerization of 3′-5′ cyclic nucleotides (Costanzo et al. 2009) might be sufficient to trigger the emergence of an RNA based replicating system. Such additional experimental evidence would remove the primary objections to an RNA early hypothesis and is an exciting area of current research (Ricardo and Szostak 2009).
TOWARD A REPLICATING SYSTEM: NUCLEOTIDE SYNTHESIS BY RNA
Prebiotic sources of nucleotides would have been quickly used by a rapidly growing population of ribo-organisms and would have presented a bottleneck to early evolution. To flourish, early RNA based life would have needed to develop the ability to synthesize nucleotide monomers from some more abundant supply of prebiotic material. Based on the chemistry of nucleotide synthesis in modern metabolism it would be satisfying if these abundant prebiotic compounds included ribose and the nucleotide bases (Joyce 1989; Robertson and Miller 1995; Orgel 1998). If so a solid abiotic chemical framework would exist for the evolution of RNA-mediated nucleotide synthesis reactions that resemble those found in modern metabolism.
Pyrimidine nucleotides and purine nucleotides (via salvage pathways) are synthesized by the formation of a glycosidic linkage using phosphoribosyl 1-pyrophosphate (PRPP) and the appropriate nucleobase. Given the fundamental importance of this chemistry to modern metabolism and its potential compatibility with abiotic supplies of ribose and nucleobases (Lau and Unrau 2009), the ability of RNA to mediate such chemistry serves as a potential bridge between the abiotic and biotic RNA World of nucleotide synthesis. However, the chemistry of glycosidic bond formation presents a number of hurdles for RNA catalysts. Relative to protein, RNA has relatively few functional groups and the nucleotide monomers from which an RNA catalyst must be built are considerably bulkier. Further, the chemistry of nucleotide synthesis is strongly influenced by nucleobase composition. Pyrimidine nucleotides are thermodynamically and kinetically much more difficult to form than purine nucleotides (Lau et al. 2004) making nucleotide synthesis by RNA an interesting enzymatic challenge.
One way to evaluate the ability of RNA to mediate such chemistries is to artificially select and evolve ribozymes in the laboratory using in vitro selection (Ellington and Szostak 1990; Robertson and Joyce 1990; Tuerk and Gold 1990). We have used this approach to isolate both pyrimidine nucleotide and purine nucleotide synthase ribozymes that are able to promote the formation of a glycosidic linkage between a base (4-thiouracil and 6-thioguanine, respectively) and tethered PRPP (Unrau and Bartel 1998; Lau et al. 2004). As expected based on the difficulty of pyrimidine glycosidic bond formation, purine nucleotide synthases were much more abundant in sequence space and were 50–100 times more efficient than their pyrimidine synthase counterparts. Interestingly pyrimidine nucleotide synthase ribozymes use charge stabilization to promote glycosidic bond formation (Unrau and Bartel 2003) presumably by the precise positioning of the ribozyme's phosphodiester backbone (Dinner et al. 2001). Recently we made the interesting discovery that a ribozyme selected for its ability to mediate chemistry between ribose and 6-thioguanosine could also promote purine nucleotide synthesis when the ribose substrate was substituted with PRPP (Lau and Unrau 2009). This promiscuous ribozyme suggests that metabolically relevant ribozymes making use of a small metabolite such as ribose could have easily evolved to promote nucleotide synthesis with PRPP. Together with our purine and pyrimidine nucleotide synthases, this in vitro evidence strongly suggests that RNA folds able to promote nucleotide synthesis would not have been difficult to discover early in the evolution of a RNA World.
Satisfyingly, both purine and pyrimidine nucleotide synthase ribozymes were able to robustly discriminate between even quite closely related nucleobase substrates. The family A pyrimidine nucleotide synthase showed a marked preference for 4-thiouracil over any other pyrimidine tested (including uracil), whereas the purine nucleotide synthases reacted two to three orders of magnitude slower when 6-thiopurine was substituted for 6-thioguanine (Unrau and Bartel 1998; Lau et al. 2004). This ability to accurately distinguish small substrates is a hallmark of the metabolic enzymes and provides encouraging evidence that in an early RNA World metabolically relevant ribozymes would have been able to efficiently distinguish and hence regulate the synthesis of important small molecule metabolites essential to life.
RNA REPLICATION: RNA POLYMERASES AND LIGASES
A RNA-mediated metabolism could only have been sustained if the RNA catalysts required to sustain metabolism were produced or repaired faster than they degraded. Although RNA repair is relatively uncommon in modern metabolism (Chan, Zhou et al. 2009), there exist numerous examples of RNA modification and editing that make use of short guide RNAs to promote chemistry at specific sites within a target RNA (Kiss 2002; Madison-Antenucci, Grams et al. 2002; Matera, Terns et al. 2007). Guide RNAs, which generally speaking are RNAs having the reverse complement of some target sequence, could have been very useful early in evolution and might have directed RNA ligase ribozymes to specifically ligate together short RNA fragments in a sequence specific fashion. This would allow the construction of larger and more complex metabolically useful ribozymes from simpler RNA elements. If in turn these RNA fragments and guide sequences were transcribed from short RNA “genomic elements” this would not only allow the replication of a relatively complex, but fragmented RNA genome, it would only require two replicative activities: RNA ligation and a RNA polymerase capable of transcribing and replicating the short RNA genomic elements. As the phosphodiester chemistry of RNA ligation and polymerization are related (Fig. 1) their emergence from an abiotic system could be evolutionarily linked and provide a simple route to biological replication.
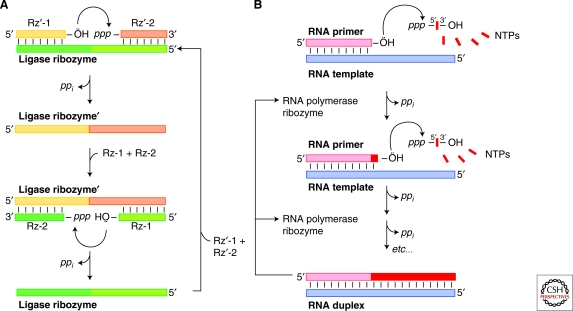
Figure 1.
RNA-catalyzed polymerization and cross-replication by ligation. (A) Cartoon schematic of cross-replication of RNA ligase ribozymes: A ligase ribozyme (colored in two shades of green) catalyzes the ligation of two orange oligonucleotides (Rz′-1 and Rz′-2) to generate a ligase ribozyme that catalyzes the ligation of two green oligonucleotides (Rz-1 and Rz-2) to regenerate the first ligase ribozyme. (Adapted from Lincoln and Joyce 2009.) (B) Cartoon schematic scheme of ribozyme-catalyzed RNA polymerization. The complete extension of an RNA primer (pink) according to the sequence of a template (blue) by an RNA polymerase produces an RNA duplex.
Cross Catalytic Ligation Strategies
An ingenious continuous molecular evolution approach, developed by Kim and Joyce (Kim and Joyce 2004) and further optimized by Lincoln and Joyce (Lincoln and Joyce 2009) has made possible the study of cross-catalytic systems built entirely of RNA. In this ribozyme-based system an RNA ligase ribozyme hybridizes to two oligonucleotide substrates and specifically catalyzes their ligation (Fig. 1A). The ligated RNA results in a second ligase ribozyme that in turn can hybridize to two other oligonucleotide substrates catalyzing in turn the production of the first ribozyme. This cross-catalytic system was found to promote the continuous exponential doubling of RNA. A similar system was developed and optimized by the Lehman laboratory, which divided the Azoarcus Group I ribozyme into four parts and made use of its intrinsic ligation capability to autocatalytically reassemble the ribozyme (Hayden and Lehman 2006; Draper et al. 2008). These fascinating systems all make use of guide sequences built into the RNA catalyzing ligation. If in the future these systems can be engineered to use generic guide RNAs supplied in trans these systems would have tremendous in vitro evolutionary potential and make possible the synthesis of arbitrary RNA components from a set of short RNAs and their corresponding guide sequences.
Although these systems have elegantly shown the potential for RNA to replicate without the addition of other components and provide insight into how a complex multicomponent RNA world might function, these experiments do not address how the RNA fragments being ligated together are themselves produced. Returning to our initial argument, if the emergence of life rapidly consumed abiotically generated RNA fragments it might be expected that early in evolution ligation strategies could have evolved hand in hand with ribozyme polymerases able to transcribe short nucleotide sequences that ultimately become substrates for ligation. In this model, as the RNA World became more established and complex, improvements in RNA polymerization would make possible the gradual concatenation of RNA genomic elements into longer and longer elements making the importance of RNA ligation less and less important as overall genome size grew.
Polymerization from Short RNA Genomic Elements
Although nature abounds with replicative strategies, nucleic acid based replication always involves the synthesis of sense and antisense strands following the canonical base pairing rules of nucleic acid. The fundamental symmetry of this copying process—where sense stand is copied to antisense to sense ad infinitum—is always broken in biological systems with the synthesis of one stand being elevated over the other so as to allow gene expression. This is most clear in the highly evolved organisms where dsDNA is the genetic material and mRNA is transcribed, but it is also true for the simplest homopolymeric systems, such as the RNA based plant viroids, where more plus strand is expressed during rolling circle replication than the minus. Notably in the case of the plant viroids this asymmetry does not depend on translation but only on the functional aspects of the expressed RNAs (Soll et al. 2001). In an early RNA world such asymmetry would have been equally important for the stable expression of RNA based metabolic enzymes. If such “transcriptional” asymmetry could be combined with the replication of short RNA genomic elements, then together with the ligation strategies just discussed a self-consistent system of RNA replication could be constructed that simultaneously allows genomic replication and expression of metabolic ribozymes.
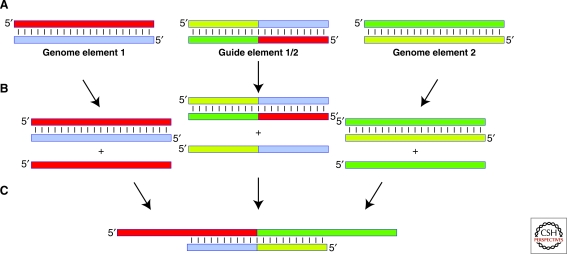
We favor a replicative model whereby the earliest genetic components were short dsRNAs. Although not fundamental for replication, dsRNA has a number of interesting virtues. First and in contrast to single-strand or folded RNAs the RNA duplex has a uniform and predictable double helix that makes it easily recognizable as a genetic component by replicative enzymes. Second, it is intrinsically difficult to copy single-stranded RNA without creating a RNA duplex. Thus generation of a reverse complement strand from a single-stranded RNA genome requires some structural mechanism to avoid the genome becoming double-stranded by default. Although such strategies are possible and are used by degenerate RNA systems such as RNA-X (Konarska and Sharp 1989) it is unclear how such strategies could be generalized to arbitrary sequence (Bartel 1999). A RNA polymerase ribozyme able to differentially transcribe from both strands of a short RNA duplex could however produce double-stranded genomic copies and an excess of single-stranded RNA that can be ligated by subsequent steps into functional RNA components (Fig. 2).
Figure 2.
Genomic elements, their asymmetrical transcription resulting in genomic replication and synthesis of functional RNAs by ligation. (A) Short double-stranded elements define the total RNA genome together with guide sequences important for the later synthesis of full length functional RNAs. (B) Asymmetrical transcription from either strand of the duplex genomic element results in the synthesis of multiple genomic element copies together with an excess of one strand. This asymmetry could be generated by having one end of the genomic element fray with a higher propensity than the other. (C) Excess single-stranded transcripts can in turn hybridize to guide element sequences making them substrates for ligase enzymes and allowing the synthesis of long highly functional RNAs.
RNA POLYMERIZATION: EVOLVING RNA LIGASES
The advent of in vitro selection provided a mechanism to directly select template directed RNA ligase ribozymes. RNA ligation, which involves the nucleophilic attack of a 3′-hydroxyl from one substrate onto the 5′-triphosphate of a second, is identical to the chemistry required for RNA polymerization (Fig. 1). Initial efforts in the 1990s led to the isolation and structural characterization of the highly efficient Class I ligase ribozyme (Bartel and Szostak 1993; Ekland and Bartel 1995; Ekland et al. 1995). Different classes of ligase ribozymes were subsequently isolated by in vitro selection, including the hc ribozyme and its variants isolated from the Tetrahymena group 1 intron (Jaeger, Wright et al. 1999; Yoshioka, Ikawa et al. 2004) and smaller ligases such as the L1 ligase (Robertson and Ellington 1999; Robertson and Scott 2007). Unfortunately, these ribozymes were all much slower than the class I ligase implying that highly efficient ligases might be rare in sequence space. Recently however, the DSL ligase (Ikawa, Tsuda et al. 2004; Voytek and Joyce 2007) was evolved to rival the catalytic efficiency of the class I ligase. This second example of an efficient RNA ligase ribozyme strongly suggests that a diverse range of ligase ribozymes could have been available early in evolution. Further characterization of these new ligases should generalize our understanding of RNA mediated phosphodiester bond formation and by virtue of their identical chemistry provide insight into the function of RNA polymerases.
Template-Directed RNA Polymerization: Evolving the Class I Ligase
Surprisingly, Ekland and Bartel showed that a variant of the class I ligase ribozyme could be simply engineered to extend a primer by six nucleotide incorporations after a four day incubation (Ekland and Bartel 1996). This template-mediated extension required that the primer itself be hybridized to the ligase ribozyme. In vitro selection was used to overcome this problem and led to the isolation of the Round 18 ribozyme (Johnston, Unrau et al. 2001). This RNA polymerase ribozyme contains a 76-nucleotide accessory domain selected from a random sequence pool that was appended onto the 3′ end of the class I ligase ribozyme (now called the ligase core). The Round 18 ribozyme was capable of using nucleotide triphosphates in a template-dependent manner to extend 14 nucleotides of a trans RNA primer-template in 24 hours (i.e., see Fig. 1B). The engineering of the Round 18 ribozyme from the class I ligase directly demonstrates that polymerization can be evolved from RNA ligation and has yet to be shown for any other RNA ligase family.
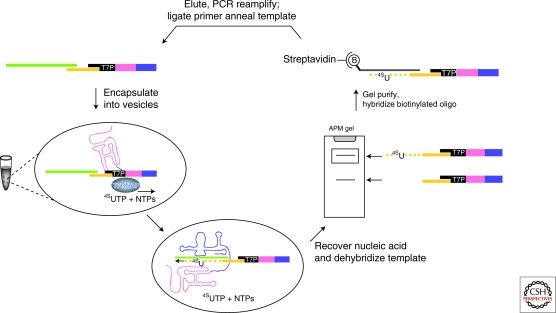
The selection of the Round 18 ribozyme required that the RNA primer being extended by the polymerase be covalently attached to the ribozyme so as to maintain a correlation between RNA function (polymerization) and phenotype (RNA sequence). To overcome this limitation and allow a true trans selection for RNA polymerization, we used a compartmentalized approach (Fig. 3) to select for an improved variant of the Round 18 ribozyme called B6.61. This polymerase is able to extend a trans primer-template duplex by 20 nucleotides and has improved fidelity relative to the Round 18 ribozyme (Zaher and Unrau 2007). In the selection for B6.61, a diverse DNA pool containing approximately 9 ×1014 variants of the Round 18 ribozyme was ligated to an RNA primer-template complex. These tagged DNA genomes were encapsulated into water-in-oil vesicles (Tawfik and Griffiths 1998; Miller et al. 2006) where they were transcribed by T7 RNA polymerase. By this strategy RNA phenotypes were confined together with their DNA genotype. If an expressed RNA could extend the primer-template, enrichment of the corresponding DNA genome was made possible. By mimicking natural selection, the in vitro encapsulated selection of B6.61 allowed for a true trans correlation between the genotype and phenotype. Similar approaches might in future allow the isolation of RNA systems that work cooperatively in order to survive within a common compartment and allow the development of artificially evolving RNA systems (Bartel and Unrau 1999).
Figure 3.
In vitro encapsulated selection scheme for trans acting RNA polymerase ribozymes. A DNA pool was generated that contained a T7 promoter allowing transcription of mutagenized Round 18 ribozyme sequence library. After ligating a RNA primer (orange)–template (green) complex to the DNA pool, genomes were encapsulated and RNA transcribed by T7 RNAP. Active RNA polymerase ribozymes extend the RNA primer tethered to their DNA genome and in the process incorporate 4-thiouridine residues in the growing strand. This allows selection of functional genomes using thiol-sensitive mercury gels and hybridization-based capture using biotinylated oligonucleotides. The captured DNA was then PCR amplified and used in a further round of selection (Zaher and Unrau 2007 and reprinted here with permission by the author).
The Evolutionary Power of Constructing Modular RNA Polymerases
Protein polymerases are modular enzymes that have clearly delimited functional folds (Werner 2007). This is seen most clearly in the well studied DNA dependent RNA polymerases where initiation requires the static recognition of a dsDNA promoter element (by the σ-factors in bacterial RNAPs) and where elongation requires a structural rearrangement of the polymerase so as to produce a DNA–RNA heteroduplex that moves within the DNA transcription bubble to ensure processive elongation (Yin and Steitz 2002; Mooney et al. 2005). Although the B6.61 polymerase cannot rival the complexity of such beautiful machines, it does compare with the much less processive, primer dependent DNA repair enzymes such as Taq polymerase. Interestingly, experiments with this commonly used polymerase have found that appending a nonspecific DNA binding protein to the enzyme can dramatically increase its processivity (Wang et al. 2004). It might therefore be possible to enhance the ability of the B6.61 polymerase to extend long templates by adding additional RNA domains that enhance the ability of the polymerase to nonspecifically bind RNA duplex. Such modularity is extensively found in large biological RNAs such as the ribosome where the functions of peptide bond formation and mRNA decoding are apportioned between the large and small subunits. Similarly, the RNase P RNA can be dissected into two major submotifs, one responsible for substrate recognition and the other catalysis (Krasilnikov et al. 2004). Presumably the evolution of such modularity would have driven the early emergence of new function and is an important element in understanding complex molecular machines.
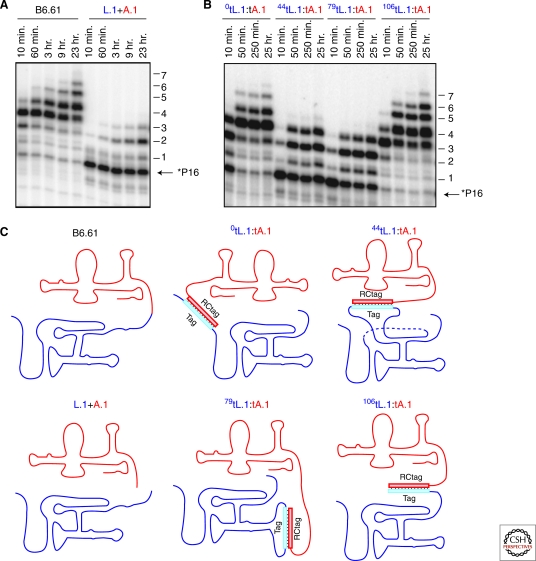
The laboratory evolution of the B6.61 ribozyme models this incremental expansion in enzyme functionality and was recently explored by characterizing the two domains of the polymerase. The ligase core and accessory domain of B6.61 are modular domains that fold independently, yet act cooperatively, to extend a trans primer-template substrate (Cheng et al., unpubl). Both domains fold well in trans and do not, based on chemical probing, appear to change their folds when transcribed separately. Notably nucleotide incorporation was completely abolished by removal of the accessory domain, but as shown in Figure 4A, adding the ligase and accessory domain in trans resulted in polymerization (last 5 lanes), albeit ∼100-fold slower than unimolecular construct (first 5 lanes). Remarkably the two domains can be joined together by a range of hybridization sites some of which retain nearly wild-type levels of activity (Fig. 4 B,C). Orientation appears crucial based on the existence of a specific cross-linking pattern between the two domains that suggest that weak tertiary interactions are responsible for the correct positioning of the two domains with respect to each other. Consistent with the three-dimensional structure of the class I ligase ribozyme (Bergman et al. 2004; Shechner et al. 2009), the 5′ and 3′ tethering sites are close together and on the same side of the ligase core as the active site, whereas tethering the accessory domain to the two internal loops of the ligase core that are distal to the active site lowers polymerization activity. This is consistent with the accessory domain being positioned over the class I ligase active site so at to constrain the primer–template and presumably nucleotide triphosphates in the vicinity of the enzyme active site. Most interestingly, a ten-fold increase in polymerization rate was observed by tethering the primer-template to the polymerase. As this increase was dependent on the accessory domain being present, improvements in the accessory domain or the selection of new complementary functional modules might be expected to further improve RNA polymerase.
Figure 4.
RNA polymerase ribozyme modularity: The B6.61 Ligase core and accessory domain in a range of equimolar (0.5 µm) contexts. (A) Comparison of polymerization activity of trans bimolecular constructs (L.1 + A.1) with that of B6.61. (B) Polymerization activity assay of different assemblies of the two hybridized trans bimolecular constructs as shown in panel C. (C) Cartoon schematic of the four bimolecular assemblies.
FUTURE RESEARCH DIRECTIONS: STRAND DISPLACEMENT AND INITIATION
The B6.61 polymerase fills in a primer-template to produce a RNA duplex. Currently, this dsRNA product is not a substrate for the polymerase, making it impossible to construct a replicating system of the sort sketched in Figure 2. If, however, this polymerase could be evolved to recognize and transcribe a RNA duplex, then a self-consistent RNA system could be constructed that requires only nucleotide triphosphates to replicate. Two critical challenges remain to be achieved: First and most problematic, a mechanism must be developed to allow the initiation of polymerization from a duplex RNA. Second a mechanism to expose the template strand of the duplex must be engineered. These two functionalities would allow the polymerase ribozyme to transcribe one strand of the duplex, resulting in a total of three RNA strands, one of which is by necessity must be single-stranded (Fig. 2). Because this ssRNA can hybridize to an antisense strand produced by transcription in the opposite direction a simple and effective mechanism for copying genomic elements would be built into this form of transcription. Asymmetric transcription from any particular genomic element would therefore not only provide a mechanism to copy the RNA genome but would naturally provide a source of ssRNA that could be ligated together to form larger more complex functional RNAs (Fig. 2).
RdRP Initiation: A Protein Point of View
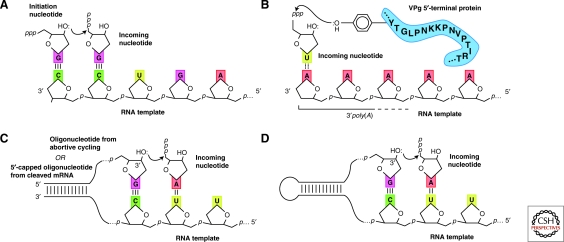
Biologically, there are two main types (Ng et al. 2008) of initiation: primer-independent (de novo synthesis) and primer-dependent. As shown in Figure 5A, de novo synthesis begins with the base pairing of an initiation nucleotide triphosphate (often this is GTP, Ng et al. 2008) to the 3′ end of the RNA template. Because this interaction is not sufficient to stabilize the single base pair thus formed, other interactions such as base stacking of aromatic protein residues with the initiation nucleotide are typically employed (Butcher et al. 2001). The extension of this first nucleotide by additional templated nucleotides can then trigger a rearrangement of protein structure that allows the formation of a stable elongation complex via an abortive cycling process (Ng et al. 2008). Primer-dependent mechanisms of initiation use a hydroxyl from a nearby source for nucleophilic attack on the triphosphate of the incoming nucleotide. Figure 5B–D depict the mechanism employed by three viral RdRPs in which a free hydroxyl is derived from: (1) a nearby protein residue (Paul et al. 1998), (2) a short oligonucleotide originating either from abortive cycling (McClure 1985) or from a cleaved mRNA (Hagen et al. 1995), or (3) by folding back a RNA template so as to produce a hairpin and a free 3′-hydroxyl that can be extended by a suitable polymerase (Laurila et al. 2002; Laurila et al. 2005).
Figure 5.
Comparison of initiation mechanisms. (A) Primer-independent (de novo) initiation B, C, and D—Primer-dependent initiation strategies: (B) “Borrowing” a hydroxyl from a nearby protein residue (C) Use of a short oligonucleotide from abortive cycling in de novo initiation or from a cleaved mRNA; (D) Template folds back to form a stable hairpin that is then extended. Adapted from (Paul et al. 1998; van Dijk et al. 2004 and reprinted with permission from The Journal of General Virology ©2004; Ng et al. 2008 and reprinted with express permission from the authors).
In all of these examples the template strand must first be exposed so that a short primer can either be synthesized (primer-independent) or provided by an external source (primer dependent). While transcribing from a RNA duplex it would be essential that the duplex be first melted by some form of helicase activity. There are several ways to design a solution to this problem using RNA, although none appear particularly straightforward. The simplest approach might be to make use of the intrinsic properties of RNA to pry open the end of the duplex. This could be achieved by first making a temporary and reversible anchor to the 3′ terminus of the template strand that would ultimately orient the template strand correctly with respect to the polymerase's active site. This ligation might be expected to mildly destabilize the terminal duplex and could be dramatically enhanced by appending a binding motif that specifically recognizes either the 5′ triphosphate on the RNA duplex or equally a 5′ cap (Huang and Yarus 1997; Zaher et al. 2006). Correctly positioned this combination of a covalent anchor on the 3′ terminus of the template strand together with tight binding to the 5′ terminus would serve to unwind the RNA duplex in a fashion sufficient to allow either primer independent or dependent initiation. After several nucleotide incorporations the polymerase structure would change, induced by forces acting through the site of RNA ligation. This change in geometry would then mediate changes in the ligation site causing RNA cleavage to be favored instead of ligation and triggering the release of the template strand from the polymerase (potentially the polymerase could use a cyclic phosphate to mediate this interaction in analogy with the reversible ligation and cleavage mediated by the hammerhead ribozyme). This same structural change would hopefully distort the geometry of the 5′ binding motif, releasing the emerging single strand and allowing a functional elongation complex to form.
Solving the Strand Displacement Problem
The initiation just described would not lead to a transcriptional complex that is directly analogous to a biological transcriptional complex. DNA dependent RNAPs form a stable transcription bubble complex (Yin and Steitz 2004; Mooney et al. 2005; Borukhov and Nudler 2008) that makes use of a RNA exit tunnel to precisely position a RNA–DNA heteroduplex with respect to the polymerase active site. A transcription bubble where the two downstream strands of DNA are reunited upstream of the RNA–DNA heteroduplex is only required for transcription of mixed polymer types and makes DNA dependent RNA transcription more complex. If RNA is being transcribed from a RNA duplex simpler topological nucleic acid structures can be used and the freshly synthesized strand need not be displaced, but instead can remain hybridized to the template strand. This change results in the original nontemplate strand being displaced and makes elongation simpler as a consequence.
Thermodynamically the free energy of the states sampled by polymerization should be nearly equivalent at any particular extension:
| 1 |
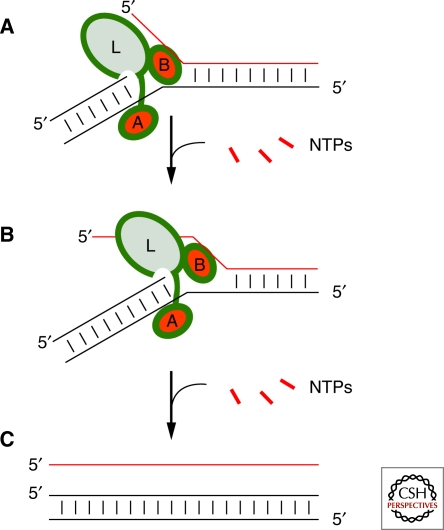
where n and m are the nth and mth nucleotide extension, RNA–RNA represents the interactions formed in the substrate strands, and RNA-polymerase the interactions made specifically by the polymerase with the substrate. As the formation of a base pair in the downstream product helix is on average compensated by the disruption of an upstream base pair, ΔG°RNA-RNA should remain small except for pathological sequences. Ideally ΔG°RNA-polymerase can be made small as well by ensuring that the polymerase is only topologically associated with substrate RNA strands and does not make any specific contacts with the replicating nucleic acid. This can be achieved by building RNA domains onto the polymerase that are able to surround but not bind to either the upstream or downstream helixes. This clamp would provide a mechanism to ensure that the polymerase always stays associated with its template making it much more processive (Fig. 6, Accessory domain). Equally important the emerging single-strand must be prevented from hybridizing to the template strand of the duplex immediately upstream of the site of nucleotide incorporation. A helix placed transversely between the template strand and the strand being displaced by polymerization might achieve this aim provided it was rigidly anchored to the duplex sliding clamp (Fig. 6, third domain). Failure to achieve strand displacement would prevent polymerization and is essential to success. Similarly, improving the stability of the B6.61 accessory domain and ligase core by further engineering and in vitro selection could well result in a suitable duplex sliding clamp. This clamp could then be selected to function with the new strand displacement motif by selecting for polymerization from artificially prepared mimics of a replication complex (Fig. 7).
Figure 6.
Evolving a processive strand-displacing RNA polymerase ribozyme from the B6.61 RNA polymerase. (A) After formation of an initiation complex, which exposes the template strand, a short RNA primer sequence is either synthesized de novo or allowed to hybridize. (B) Elongation requires that nucleotides immediately upstream of the site of nucleotide incorporation are single-stranded. If interactions between the ligase core (L) and the accessory domain (Circled A) can be improved so that the freshly synthesized strand together with template are held robustly and nonspecifically the polymerase should become much more processive. If in addition these two domains can be rigidly connected to a third domain (Circled B) that ensures dehybridization of the incoming duplex, transcription from dsRNA as well as ssRNA templates would be possible. This elongation complex should remain invariant in shape as it slides along the template. (C) Transcription ends with the release of one strand from the original duplex RNA and the duplex RNA being regenerated with a freshly synthesized RNA strand.
Figure 7.
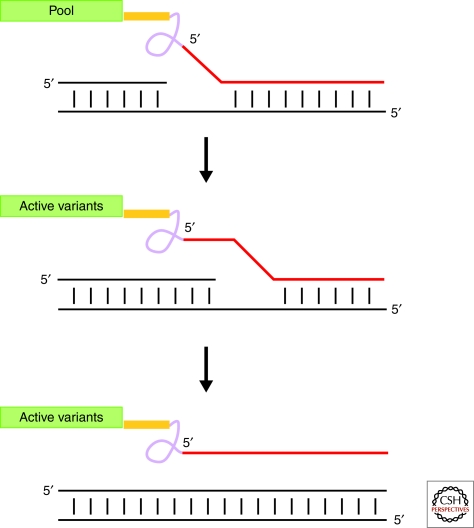
Selecting for strand displacement in an RNA polymerase ribozyme. A pool of potential strand displacing RNA polymerase ribozymes is tethered by a long flexible linker to a RNA (red strand) that is partially complementary to the template strand (bottom black strand). The unhybridized nucleotides in the red strand are not able to hybridize to the template strand resulting in the formation of an elongation complex mimic. Transcription by the tethered polymerase that is able to displace the red strand results in the polymerase disassociating from the extending primer template duplex (black strands). Freed polymerases can then be differentially recovered and amplified.
Selecting polymerases for strand displacement should be the first priority for improving RNA dependent polymerase ribozymes. Such an enzyme even if it lacks the ability to initiate could still allow a number of very interesting experiments. For example, with the appropriate primers such a polymerase could mediate rolling circle replication of an RNA genome. If this circular genome contained a hammerhead ribozyme motif together with the polymerase ribozyme sequence, then primed rolling circle transcription of both plus and minus strands would allow the creation of a simple autocatalytic system requiring only primer and nucleotides to replicate. This would make possible the artificial continuous evolution of the RNA polymerase itself and with suitable selection pressure might rapidly lead to the evolution of initiation strategies that allow the controlled expression of other potentially metabolically relevant RNAs.
CONCLUSIONS
Based on a number of independent lines of laboratory research RNA appears well suited to have served as the first replicative polymer on this planet. More importantly the catalytic and genetic properties intrinsic to RNA might in the near future make possible the first artificial forms of life. Such systems would allow the exploration of many important and outstanding questions concerning the robustness and early evolution of life and provide critical insight into the difficulty of evolving life elsewhere in the universe.
Footnotes
Editors: David Deamer and Jack W. Szostak
Additional Perspectives on The Origins of Life available at www.cshperspectives.org
REFERENCES
- Bartel DP, Szostak JW 1993. Isolation of new ribozymes from a large pool of random sequences. Science 261: 1411–1418 [DOI] [PubMed] [Google Scholar]
- Bartel DP, Unrau PJ 1999. Constructing an RNA world. Trends Cell Biol 9: M9–M13 [PubMed] [Google Scholar]
- Benner SA, Ellington AD, Tauer A 1989. Modern metabolism as a palimpsest of the RNA world. Proc Natl Acad Sci 86: 7054–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman NH, Lau NC et al. 2004. The three-dimensional architecture of the class I ligase ribozyme. RNA 10: 176–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borukhov S, Nudler EC 2008. RNA polymerase: The vehicle of transcription. Trends Microbiol 16: 126–134 [DOI] [PubMed] [Google Scholar]
- Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI 2001. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410: 235–240 [DOI] [PubMed] [Google Scholar]
- Chan CM, Zhou C, Huang RH 2009. Reconstituting bacterial RNA repair and modification in vitro. Science 326: 247. [DOI] [PubMed] [Google Scholar]
- Costanzo G, Pino S, Cicirielli F, Di Mauro E 2009. Generation of long RNA chains in water. J Biol Chem 284: 33206–33216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FH 1958. On protein synthesis. Symp Soc Exp Biol 12: 138–163 [PubMed] [Google Scholar]
- Dinner AR, Blackburn GM, Karplus M 2001. Uracil-DNA glycosylase acts by substrate autocatalysis. Nature 413: 752–755 [DOI] [PubMed] [Google Scholar]
- Draper WE, Hayden EJ, Lehman N 2008. Mechanisms of covalent self-assembly of the Azoarcus ribozyme from four fragment oligonucleotides. Nucleic Acids Res 36: 520–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekland EH, Bartel DP 1995. The secondary structure and sequence optimization of an RNA ligase ribozyme. Nucleic Acids Res 23: 3231–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekland EH, Bartel DP 1996. RNA-catalysed RNA polymerization using nucleoside triphosphates. Nature 382: 373–376 [DOI] [PubMed] [Google Scholar]
- Ekland EH, Szostak JW, Bartel DP 1995. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science 269: 364–370 [DOI] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW 1990. In vitro selection of RNA molecules that bind specific ligands. Nature 346: 818–822 [DOI] [PubMed] [Google Scholar]
- Ellington AD, Chen X, Robertson M, Syrett A 2009. Evolutionary origins and directed evolution of RNA. Int J Biochem Cell Biol 41: 254–265 [DOI] [PubMed] [Google Scholar]
- Ferris JP, Hill AR Jr, Lui R, Orgel LE 1996. Synthesis of long prebiotic oligomers on mineral surfaces. Nature 381: 59–61 [DOI] [PubMed] [Google Scholar]
- Fuller WD, Sanchez RA, Orgel LE 1972. Studies in prebiotic synthesis. VI. Synthesis of purine nucleosides. J Mol Biol 67: 25–33 [DOI] [PubMed] [Google Scholar]
- Gago S, Elena SF, Flores R, Sanjuan R 2009. Extremely high mutation rate of a hammerhead viroid. Science 323: 1308. [DOI] [PubMed] [Google Scholar]
- Gesteland RF, Cech T, Atkins JF 2006. The RNA World: The nature of modern RNA suggests a prebiotic RNA world. Cold Spring Harbor Laboratory Press, NY [Google Scholar]
- Gilbert SD 1986. The RNA world. Nature 319: 618 [Google Scholar]
- Hagen M, Tiley L, Chung TD, Krystal M 1995. The role of template-primer interactions in cleavage and initiation by the influenza virus polymerase. J Gen Virol 76: 603–611 [DOI] [PubMed] [Google Scholar]
- Hayden EJ, Lehman N 2006. Self-assembly of a group I intron from inactive oligonucleotide fragments. Chem Biol 13: 909–918 [DOI] [PubMed] [Google Scholar]
- Hayden EJ, von Kiedrowski G, Lehman N 2008. Systems chemistry on ribozyme self-construction: Evidence for anabolic autocatalysis in a recombination network. Angew Chem Int Ed Engl 47: 8424–8428 [DOI] [PubMed] [Google Scholar]
- Hill A, Orgel LE 2002. Synthesis of adenine from HCN tetramer and ammonium formate. Orig Life Evol Biosph 32: 99–102 [DOI] [PubMed] [Google Scholar]
- Huang F, Yarus M 1997. Versatile 5′ phosphoryl coupling of small and large molecules to an RNA. Proc Natl Acad Sci 94: 8965–8969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa Y, Tsuda K, Matsumura S, Inoue T 2004. De novo synthesis and development of an RNA enzyme. Proc Natl Acad Sci 101: 13750–13755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger L, Wright MC, Joyce GF 1999. A complex ligase ribozyme evolved in vitro from a group I ribozyme domain. Proc Natl Acad Sci 96: 14712–14717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston WK, Unrau PJ, Lawrence MS, Glasner ME, Bartel DP 2001. RNA-catalyzed RNA polymerization: Accurate and general RNA-templated primer extension. Science 292: 1319–1325 [DOI] [PubMed] [Google Scholar]
- Joyce GF 1989. RNA evolution and the origins of life. Nature 338: 217–224 [DOI] [PubMed] [Google Scholar]
- Joyce GF 2002. The antiquity of RNA-based evolution. Nature 418: 214–221 [DOI] [PubMed] [Google Scholar]
- Joyce GF, Orgel LE 2006. Progress towards understanding the origin of the RNA world. In RNA World, 3rd ed Cold Spring Harber Laboratory Press, Cold Spring Harber, NY [Google Scholar]
- Kim DE, Joyce GF 2004. Cross-catalytic replication of an RNA ligase ribozyme. Chem Biol 11: 1505–1512 [DOI] [PubMed] [Google Scholar]
- Kiss T 2002. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148 [DOI] [PubMed] [Google Scholar]
- Konarska MM, Sharp PA 1989. Replication of RNA by the DNA-dependent RNA polymerase of phage T7. Cell 57: 423–431 [DOI] [PubMed] [Google Scholar]
- Krasilnikov AS, Xiao Y, Pan T, Mondragon A 2004. Basis for structural diversity in homologous RNAs. Science 306: 104–107 [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Bebenek K 2000. DNA replication fidelity. Annu Rev Biochem 69: 497–529 [DOI] [PubMed] [Google Scholar]
- Lau MW, Unrau PJ 2009. A promiscuous ribozyme promotes nucleotide synthesis in addition to ribose chemistry. Chem Biol 16: 815–825 [DOI] [PubMed] [Google Scholar]
- Lau MW, Cadieux KE, Unrau PJ 2004. Isolation of fast purine nucleotide synthase ribozymes. J Am Chem Soc 126: 15686–15693 [DOI] [PubMed] [Google Scholar]
- Laurila MR, Makeyev EV, Bamford DH 2002. Bacteriophage φ6 RNA-dependent RNA polymerase: Molecular details of initiating nucleic acid synthesis without primer. J Biol Chem 277: 17117–17124 [DOI] [PubMed] [Google Scholar]
- Laurila MR, Salgado PS, Stuart DI, Grimes JM, Bamford DH 2005. Back-priming mode of φ6 RNA-dependent RNA polymerase. J Gen Virol 86: 521–526 [DOI] [PubMed] [Google Scholar]
- Lincoln TA, Joyce GF 2009. Self-sustained replication of an RNA enzyme. Science 323: 1229–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison-Antenucci S, Grams J, Hajduk SL 2002. Editing machines: the complexities of trypanosome RNA editing. Cell 108: 435–438 [DOI] [PubMed] [Google Scholar]
- Marquez SM, Chen JL, Evans D, Pace NR 2006. Structure and function of eukaryotic Ribonuclease P RNA. Mol Cell 24: 445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP 2007. Non-coding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol 8: 209–220 [DOI] [PubMed] [Google Scholar]
- McClure WR 1985. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem 54: 171–204 [DOI] [PubMed] [Google Scholar]
- Miller OJ, Bernath K, Agresti JJ, Amitai G, Kelly BT, Mastrobattista E, Taly V, Magdassi S, Tawfik DS, Griffiths AD 2006. Directed evolution by in vitro compartmentalization. Nat Methods 3: 561–570 [DOI] [PubMed] [Google Scholar]
- Monnard PA, Kanavarioti A, Deamer DW 2003. Eutectic phase polymerization of activated ribonucleotide mixtures yields quasi-equimolar incorporation of purine and pyrimidine nucleobases. J Am Chem Soc 125: 13734–13740 [DOI] [PubMed] [Google Scholar]
- Mooney RA, Darst SA, Landick R 2005. Sigma and RNA polymerase: an on-again, off-again relationship? Mol Cell 20: 335–345 [DOI] [PubMed] [Google Scholar]
- Ng KK, Arnold JJ, Cameron CE 2008. Structure-function relationships among RNA-dependent RNA polymerases. Curr Top Microbiol Immunol 320: 137–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930 [DOI] [PubMed] [Google Scholar]
- Orgel LE 1998. The origin of life—a review of facts and speculations. Trends Biochem Sci 23: 491–495 [DOI] [PubMed] [Google Scholar]
- Orgel LE 2004. Prebiotic chemistry and the origin of the RNA world. Crit Rev Biochem Mol Biol 39: 99–123 [DOI] [PubMed] [Google Scholar]
- Pannucci JA, Haas ES, Hall TA, Harris JK, Brown JW 1999. RNase P RNAs from some Archaea are catalytically active. Proc Natl Acad Sci 96: 7803–7808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AV, van Boom JH et al. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393: 280–284 [DOI] [PubMed] [Google Scholar]
- Powner MW, Gerland B et al. 2009. Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature 459: 239–242 [DOI] [PubMed] [Google Scholar]
- Qiao F, Cech TR 2008. Triple-helix structure in telomerase RNA contributes to catalysis. Nat Struct Mol Biol 15: 634–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricardo A, Szostak JW 2009. Origin of life on earth. Sci Am 301: 54–61 [DOI] [PubMed] [Google Scholar]
- Ricardo A, Carrigan MA, Olcott AN, Benner SA 2004. Borate minerals stabilize ribose. Science 303: 196. [DOI] [PubMed] [Google Scholar]
- Robertson MP, Ellington AD 1999. In vitro selection of an allosteric ribozyme that transduces analytes to amplicons. Nat Biotechnol 17: 62–66 [DOI] [PubMed] [Google Scholar]
- Robertson DL, Joyce GF 1990. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 344: 467–468 [DOI] [PubMed] [Google Scholar]
- Robertson MP, Miller SL 1995. An efficient prebiotic synthesis of cytosine and uracil. Nature 375: 772–774 [DOI] [PubMed] [Google Scholar]
- Robertson MP, Scott WG 2007. The structural basis of ribozyme-catalyzed RNA assembly. Science 315: 1549–1553 [DOI] [PubMed] [Google Scholar]
- Shechner DM, Grant RA et al. 2009. Crystal structure of the catalytic core of an RNA-polymerase ribozyme. Science 326: 1271–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll D, Nishimura S, Moore PB 2001. RNA. Pergamon Press [Google Scholar]
- Tawfik DS, Griffiths AD 1998. Man-made cell-like compartments for molecular evolution. Nat Biotechnol 16: 652–656 [DOI] [PubMed] [Google Scholar]
- Tucker BJ, Breaker RR 2005. Riboswitches as versatile gene control elements. Curr Opin Struct Biol 15: 342–348 [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gold LC 1990. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505–510 [DOI] [PubMed] [Google Scholar]
- Unrau PJ, Bartel DP 1998. RNA-catalysed nucleotide synthesis. Nature 395: 260–263 [DOI] [PubMed] [Google Scholar]
- Unrau PJ, Bartel DP 2003. An oxocarbenium-ion intermediate of a ribozyme reaction indicated by kinetic isotope effects. Proc Natl Acad Sci 100: 15393–15397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk AA, Makeyev EV, Bamford DH 2004. Initiation of viral RNA-dependent RNA polymerization. J Gen Virol 85: 1077–1093 [DOI] [PubMed] [Google Scholar]
- Voytek SB, Joyce GF 2007. Emergence of a fast-reacting ribozyme that is capable of undergoing continuous evolution. Proc Natl Acad Sci 104: 15288–15293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Prosen DE, Mei L, Sullivan JC, Finney M, Vander Horn PB 2004. A novel strategy to engineer DNA polymerases for enhanced processivity and improved performance in vitro. Nucleic Acids Res 32: 1197–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner F 2007. Structure and function of archaeal RNA polymerases. Mol Microbiol 65: 1395–1404 [DOI] [PubMed] [Google Scholar]
- White HB III 1976. Coenzymes as fossils of an earlier metabolic state. J Mol Evol 7: 101–104 [DOI] [PubMed] [Google Scholar]
- Wilson DS, Szostak JW 1999. In vitro selection of functional nucleic acids. Annu Rev Biochem 68: 611–647 [DOI] [PubMed] [Google Scholar]
- Yin YW, Steitz TA 2002. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science 298: 1387–1395 [DOI] [PubMed] [Google Scholar]
- Yin YW, Steitz TA 2004. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell 116: 393–404 [DOI] [PubMed] [Google Scholar]
- Yoshioka W, Ikawa Y, Jaeger L, Inoue T 2004. A ligase ribozyme obtained from a structured pool. Nucleic Acids Symp Ser (Oxf) 48: 209–210 [DOI] [PubMed] [Google Scholar]
- Zaher HS, Unrau PJ 2007. Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA 13: 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Watkins RA, Unrau PJ 2006. Two independently selected capping ribozymes share similar substrate requirements. RNA 12: 1949–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubay G, Mui T 2001. Prebiotic synthesis of nucleotides. Orig Life Evol Biosph 31: 87–102 [DOI] [PubMed] [Google Scholar]
- Bartel DP 1999. Re-creating an RNA replicase. In RNA World, 2nd ed Cold Spring Harber Laboratory Press, Cold Spring Harber, NY [Google Scholar]