Abstract
The assembly of SNARE proteins into a tight complex has been hypothesized to drive membrane fusion. A model of the initial fusion pore as a proteinaceous channel formed by SNARE proteins places their membrane anchors in separate membranes. This leaves the possibility of a final assembly step that brings the membrane anchors together and drives fusion pore expansion. The present study develops a model for expansion in which the final SNARE complex zipping step drives a transition from a proteinaceous fusion pore to a lipidic fusion pore. An estimate of the energy released upon merger of the helical segment of the SNARE motif with the helical membrane anchor indicates that completing the assembly of a few SNARE complexes can overcome the elastic energy that opposes lipid bilayer deformation into a narrow fusion pore. The angle between the long axes of membrane anchor and SNARE motif serves as a useful reaction coordinate for this transition. Energy was calculated as a function of this angle, incorporating contributions from membrane bending, SNARE complex assembly, membrane anchor flexing, and hydrophobic interactions. The rate of this transition was evaluated as a process of diffusion over the barrier imposed by these combined energies, and the rates estimated were consistent with experimental measurements.
Introduction
Soluble N-ethylmaleimide-sensitive factor attachment receptor (SNARE) proteins catalyze the fusion of membranes from distinct cellular compartments during membrane trafficking (1, 2). The synaptic SNARE proteins, synaptobrevin, SNAP-25, and syntaxin play essential roles in exocytosis, catalyzing the fusion of secretory vesicles with the plasma membrane. Homologous SNARE motifs within these proteins form a very tight complex comprising 4 coiled α-helices (3, 4). By forming these complexes SNARE proteins can act as a fusion machine to drive the merger of membranes (5–8). It has been noted that assembly of the SNARE complex in the fashion of a zipper will pull membranes closer together (9, 10), and some studies indicate that this assembly occurs in stages to drive sequential steps in fusion (11–14). However, there is also evidence against a pure zipping mechanism (15), and the mechanisms by which SNARE proteins deform and remodel lipid bilayers remain unclear.
Evidence has been presented suggesting that the membrane anchor of syntaxin lines the initial fusion pore of exocytosis. Mutations that change the side chain size and charge at specific sites can either increase or decrease flux through the fusion pore in a manner suggesting that these residues face the pore lumen (16, 17). Only sites along one α-helical face of the syntaxin membrane anchor have this particular effect. With one intriguing exception (18), mutations outside the membrane anchors of SNARE proteins do not alter the flux through a fusion pore (15). An independent line of investigation of the curvature elasticity of fusion pores indicated that the lipidic pore forms after the initial fusion pore (19). These results support the hypothesis that SNARE proteins catalyze membrane fusion by forming an intermediate gap junction-like channel that spans the vesicle and plasma membranes (20). The likely partner to syntaxin in forming the fusion pore through the vesicle membrane would be synaptobrevin, a SNARE protein anchored in the vesicle. In a fusion pore formed by SNARE protein membrane anchors, these helical segments must be in different membranes and cannot be part of the SNARE complex. Yet the SNARE complex can extend to include the membrane anchors (21), and deleting the membrane anchors reduces its stability (21, 22). Thus, completing the assembly of the SNARE complex can drive a late step of membrane fusion to expand the initial proteinaceous pore and initiate a transition to a pore composed of lipid.
This late step is difficult to envisage. A fusion pore must expand as exocytosis progresses, but models based on proteinaceous fusion pores do not offer a clear picture of the ensuing expansion. Fusion pores composed of lipid can expand due to the fluid nature of lipid bilayers, and theories have been developed to understand this process (23, 24). An initial proteinaceous fusion pore must undergo a conversion in its composition to allow lipid to replace protein. During this transition, lipid molecules must intercalate between the protein segments that line the fusion pore. Hypothetical mechanisms for such a process are difficult to find in the literature (3, 10).
The present study formulates a hypothesis for this transition and develops a theoretical model for fusion pore expansion in terms of graduation from protein to lipid. An analysis of the relevant energies begins with an evaluation of two key contributions, one arising from the bending of lipid bilayers and the other arising from the final SNARE complex assembly step. Completing the assembly of a small number of SNARE complexes yields sufficient energy to shift the balance in favor of a lipidic pore. This transition is opposed by a transient increase in hydrophobic energy as the hydrocarbon interiors of lipid bilayers come in contact with water. Summing this and other contributions leads to an energy as a function of membrane anchor tilt angle. This energy profile presents a barrier to fusion, and incorporating this profile into a rate theory based on diffusion over a barrier yielded rates of fusion pore dilation that can be compared to experimental measurements.
Energy Differences Between Proteinaceous and Lipidic Pores
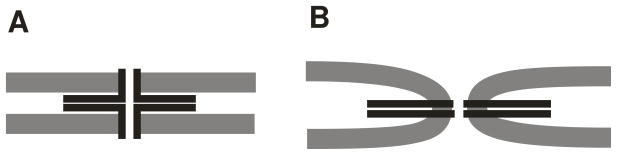
The two key structures are depicted in Fig. 1. Fig. 1A depicts a proteinaceous fusion pore lined by SNARE protein membrane anchors through the two lipid bilayers. The SNARE motifs are associated in a SNARE complex and hold the membranes together. SNARE complexes actually contain four SNARE motifs, but for the present purpose of illustration the SNARE motifs attached to the two membrane anchors are all we need. Fig. 1B shows the fusion pore after a structural transition in which the membranes have fused and the pore has become predominantly lipidic.
Fig. 1.
The dilation of a fusion pore is envisaged as a transition from a proteinaceous gap-junction-like structure (A) in which SNARE protein membrane anchors line the pore through the two membranes. Straightening out the SNARE proteins bends the membrane into a quasi-toroidal structure (B). The transition from A to B thus converts the fusion pore from protein to lipid.
Fig. 1 identifies two important structural changes, each of which embodies a major energetic contribution. First, the lipid bilayers in Fig. 1B are deformed by bending, and this bending energy opposes the transition. Second, the SNARE proteins initially have their membrane anchors and SNARE motifs at right angles. During the transition the membrane anchor rotates and is incorporated into the SNARE complex. Eliminating the right angles brings the two α-helical segments of each SNARE protein into a single longer α-helix. This final step in SNARE complex zipping releases energy. Each of these contributions to the energy difference can be estimated.
Lipid bilayer bending energy
Lipid bilayers deformed into shapes such as depicted in Fig. 1B have been studied using continuum models of membrane elasticity (25–27). However, this approach becomes inaccurate for very high curvatures such as are seen in fusion pores (28). Furthermore, the transition depicted in Fig. 1 also incurs a change in Gaussian curvature, and there is no satisfactory measurement for the Gaussian curvature modulus of a lipid bilayer. A self-consistent field theory has been developed that provides energy estimates based on a molecular description of lipid bilayers (29). This approach does not depend on the continuum approximation of classical elasticity theory, nor does it require knowledge of the Gaussian curvature modulus. In agreement with continuum models (27), the potential energy surface has a shallow minimum for a metastable pore (29), indicating that the energy variations with pore size will be small. The estimate of ~17 kT (for zero membrane tension from Fig. 8 of Katsov et al., 2004) provides a suitable starting point. Based on a comparison of interfacial tensions, Katsov et al indicated that scaling by 2.5 yields numbers that are relevant to biological membranes. This gives 42.5 kT, which will be used here for the lipid bending energy of the structure depicted in Fig. 1B.
SNARE complex zipping energy
The final step of SNARE complex zipping suggests two possible driving forces. First, contacts can form between the two membrane anchors and these contacts stabilize a SNARE complex (4, 21). This contribution would include hydrophobic contacts arising from the predominantly hydrophobic side-chains exposed to water in the proteinaceous fusion pore (17). These side chains will move from a polar to a hydrophobic environment during the transition. It should also be noted that stabilizing contacts between adjacent membrane anchors within the proteinaceous pore will be lost during the transition. These contacts would stabilize the initial state, but the magnitude of this contribution is difficult to assess.
A second potential contribution to zipping arises from the merger of the two helical segments of each SNARE protein. The cooperative nature of the helix-coil transition polypeptides dictates an energetic cost of juxtaposing segments in different states. This cooperativity reflects the ease of extending an already formed helix versus the difficulty of nucleating a new helix in a coil background. For the SNARE protein configuration illustrated in Fig. 1A, placing a short unstructured linker between two α-helical segments creates energetically costly borders. Straightening out the linker and merging these two helices to create the structure depicted in Fig. 1B eliminates these borders and lowers the free energy.
The free energy change of merging two helical segments can be expressed in a straightforward manner within the framework of the theory of the helix-coil transition (30). This theory assigns statistical weights to each residue depending on its identity and on whether or not the adjacent residue is in the same state. Thus, elongating a helix by one residue multiplies the statistical weight of a configuration by the elongation parameter s. Nucleating a helix multiplies the statistical weight of a configuration by a product of the elongation parameter and the nucleation parameter, sσ. For a coiled segment with a neutral intrinsic preference between helix and coil, the change in free energy for one SNARE protein to undergo the transition is simply kT ln(σ). (Note that Fig. 1A shows only a right angle but not the actual linker. Although one SNARE protein contains two helix-coil boundaries on each side of the linker, the statistical weight only contains one factor of σ. This follows the original Zimm-Bragg formulation and enables us to use parameter values based on fits of experimental data to this theory.)
Values for σ obtained by fitting Zimm-Bragg theory to helix melting data range from 10−5 to 2.1×10−2 (31). Several amino acids in the synaptobrevin and syntaxin linkers have σ values of 10−5, and most of the amino acids have low σ values. Indeed, the linker sequences could reflect adaptations to maximize the energy cost of breaking an α-helix, so for the present study σ will be taken at the low end of the range. Taking the elongation parameter values (31) for the 10 amino acid linker sequences and multiplying them together gives 0.82 for syntaxin and 0.78 for synaptobrevin. Thus, for these linker sequences elongation parameters are not expected to make a significant contribution to helix completion in these two proteins. Elongation factors will therefore be ignored. Taking σ = 10−5 gives a favorable free energy change of 11.5 kT per SNARE protein.
The formation of contacts between the SNARE protein membrane anchors will be addressed later, but for now we can use the helix-completion free energy just estimated to assess its effectiveness in driving the transition illustrated in Fig. 1. Each SNARE complex has two membrane-spanning SNARE proteins, so two SNARE complexes can provide enough energy (46 kT) to overcome the 42.5 kT of membrane bending energy required to create a lipidic fusion pore. Based on conductance measurements, 5–8 SNARE complexes have been proposed to form a fusion pore (17). With 6 there will be 12 helix-completing SNARE proteins releasing a total free energy of 138 kT. This is more than enough to overcome the energy cost of bending membrane.
The Energy Barrier for the Transition
For SNARE complex zipping to be effective in driving the transition we need a viable pathway between the two states shown in Fig. 1, which does not incur too high an energy cost. As the SNARE proteins straighten out, the proximal parts of the membrane anchors will spread out and expose lipid acyl chains to water (Fig. 2). This creates unfavorable contact between water and hydrophobic surfaces, and raises the free energy substantially. This section addresses the issue of the hydrophobic energy barrier of the transition. First a simple approach was used and found to give a prohibitively high energy. Then an approach incorporating two more realistic features was tested and found to reduce the height of the energy barrier to levels that make this zipping mechanism tenable.
Fig. 2.
A. An intermediate state of fusion pore dilation illustrates a structure during the transition from the proteinaceous state (Fig. 1A) to the lipidic state (Fig. 1B). Tilting of the membrane anchors exposes triangular domains of lipid hydrocarbon to water. θ0 represents the angle of tilt of the membrane anchor and b is the thickness of the hydrocarbon region of the bilayer. Eq. 2 gives the area of one of those sections in terms of b, θ0, and the number of membrane anchors. B. The structure pictured in A is modified to incorporate 1) membrane anchor bending and 2) water exclusion from the junction of the two bilayer interiors as they approach one another. The sections of hydrocarbon exposed to water are no longer triangular but are now bounded by curves and no longer confined to a plane. A circle in the left corner with radius w probes solvent accessible surface to determine the area from which water is excluded. θ0 and θb denote the angles between the bilayer normal and membrane anchors at the bottom and top of the membrane, respectively.
Hydrophobic Interactions During Pore Expansion
The exposure of the hydrocarbon bilayer interior to water as the membrane anchors tilt is illustrated in Fig. 2A. The hydrocarbon-water contact zones appear as triangular sections between adjacent membrane anchors. Hydrophobic energy (Eh) is generally taken as proportional to the area of a contact region.
| (1) |
A is the area of a hydrocarbon-water interface and β is an empirical factor determined experimentally. For aliphatic hydrocarbons β has been estimated from plots of free energy versus water accessible surface area to be in the range of 20–33 cal/Å2, and β = 25 cal/Å2 has been suggested as a generic value (32). Carbon-carbon double bonds reduce this factor to below 20 cal/Å2 (33) and biological membranes have cholesterol and acyl chains with some double bonds so for the present analysis β will be taken as 22 cal/Å2 (0.0374 kT/Å2).
It is important to note that values of β based on interfacial tension measurements of macroscopic interfaces are approximately 3 times greater than the above values based on thermodynamic measurements (34). This discrepancy has been attributed to various factors such as surface roughness, curvature, and differences between microscopic and macroscopic scales (35–37). The larger value of β from interfacial tension measurements has been used previously to estimate energy costs of creating hydrocarbon-water contact in hypothesized fusion intermediates, and on this basis it was argued that very large energies would make the formation of structures with substantial hydrophobic-water contacts prohibitive (38). However, it is not clear which parameter should be used in such calculations. The lower values have been used effectively to account for the stability of proteins (39, 40) and micelles (41). Furthermore, a computer simulation has generated similar low values for the association of α-helices (42). The present analysis will proceed using the lower thermodynamic estimate of β.
When the SNARE protein membrane anchors tilt by an angle θ0, a straightforward geometric calculation yields the hydrophobic contact area for one of the triangles in Fig. 2A between two adjacent membrane anchors of a pore formed by N chains as
| (2) |
b is the length of an edge of the membrane anchor that is exposed to the lipid bilayer hydrocarbon interior prior to tilting (Fig. 2A), and it is assumed that this length remains constant. Note that Eq. 2 is the area between the membrane anchors. The hydrocarbon-water contact areas contributed by the membrane anchors themselves are not included because this contact exists prior to tilting and will not increase.
b will be taken as the thickness of the hydrocarbon interior of a fluid lipid bilayer. In fluid, fully-hydrated lipid bilayers composed of dioleolylphosphatidylcholine the carbonyls of the acyl chains of opposing monolayers are separated by 29 Å (43). The hydrophobic region of each chain begins one carbon-carbon bond deeper in the membrane, reducing the thickness of the hydrophobic region by ~3 Å. Furthermore, the value of 29 Å was for phospholipids containing 18-carbon chains and biological membranes have a roughly equal proportion of 16-carbon chains. With an average of 17 carbons per acyl chain the thickness of the hydrophobic region will be reduced by ~ 2 Å (44), leading to a value of b = 24 Å for use here.
For a pore formed by 6 SNARE complexes there will be a total of 12 triangular sections of water-hydrocarbon contact. For these 12 sections Eqs. 1 and 2 gives 112 kT when the membrane anchors are fully horizontal. But at this stage fusion will be complete and the opposing hydrocarbon sections from the two membranes will merge to eliminate the water-hydrocarbon interface. Even tilting half way to θ0 = π/4 will raise the energy to 84 kT for N=6 and 90 kT for N=8. This energy is relatively insensitive to further increases in N because of a cancellation of two opposing factors. As the number of sections between adjacent membrane anchors increases, the area per section decreases. Thus, adding more SNARE complexes can provide more energy, and ultimately enough of them could overcome the hydrophobic energy barrier. Given the shallow energy minima of fusion pores (27, 29), changes in membrane bending energy that would accompany the formation of a larger lipidic pore will have little impact compared to the 23 kT per SNARE complex increase in driving force. A proteinaceous fusion pore thus effectively focuses the energy derived from the assembly of multiple SNARE complexes.
Expansion of a Flexible Pore
The above calculations were based on the assumption that the membrane anchors and lipid hydrocarbon interior are rigid. In fact, the membrane anchors can bend and this will reduce the area of hydrocarbon-water contact. Furthermore, the lipids from the opposing bilayers can collapse together to close out narrow regions so that as θ0 increases the adjacent zones of hydrocarbon will merge. Fig. 2B illustrates these additional features as a modification of the rigid model shown in Fig. 2A.
To address membrane anchor bending in response to the torque applied through the linker, we will introduce θb for the angle at the upper end of a bilayer, and continue to use θ0 for the angle at the distal end (Fig. 2B). Thus, when the anchor is bent, θ0 ≠ θb, and the difference gives the degree of bending. θb will be used later in this work as a reaction coordinate to follow the extent of the transition.
The theory of elasticity gives the energy of bending a rod as (45)
| (3) |
where s is the coordinate that follows the contour of the long axis of the rod, R(s) is the radius of curvature at s, and κ is the coefficient of flexural rigidity. Equilibrium molecular dynamics has been used to estimate values of κ for α-helices composed of various amino acids, and except for polypeptides composed of glycine, κ values were composition independent and about 1000 kT Å (46). For a 30 Å α-helix, uniform bending to 0.24 radians costs 1 kT. This much bending should reduce the hydrocarbon-water contact area in Fig. 2B and thus reduce the energy barrier of membrane anchor tilting.
Next, consider the effect of reduced water accessibility in the narrow gap between the proximal zones of hydrocarbon of the two bilayers (Fig. 2B). As the membrane anchors and adjacent hydrocarbon-water surfaces rotate the opposing lipid bilayers will approach one another. The surfaces will collapse together when the gap closes down to the size of a molecule of water. In a classical treatment of this problem, the water accessible surface area of a protein was evaluated based on contact area with a water molecule-sized spherical probe (32). Fig. 2B shows this probe at the junction between the bilayers. The region excluding water extends away from the junction to a distance of
| (4) |
This length delimits the zone between the hydrocarbon regions of the two fusing bilayers from which a sphere of radius w is excluded. For nonpolar surfaces the appropriate value is w = 1.6 Å (32).
This same effect will apply to the mostly hydrophobic side chains of the SNARE protein membrane anchors. As θb increases the proximal ends of the membrane anchors will merge and eliminate energetically costly contacts between these side chains and water. Taking each membrane anchor as a cylinder with a radius of 3.5 Å, the surface area per cylinder exposed to the pore lumen will be 3.5π(1–2/N)L, where L is the cylinder length and N is the number of SNARE complexes. The length of the stretch of membrane anchor with water excluded from its surface is then δ from Eq. 4. Multiplying the area of this stretch by β gives
| (5) |
This can be viewed as the hydrophobic membrane anchor contribution to the energy of the final zipping step.
This calculation of hydrophobic energy treats the interaction essentially as a contact force. The distance dependence of the hydrophobic interaction is not well understood but extending the range of this force will stabilize the intermediate structures with tilted membrane anchors. Two lipid bilayers interact by long range attractive forces between the hydrocarbon interior and short range repulsive forces between the head groups. As the membrane anchors tilt the attractions of the exposed hydrocarbon bilayer interiors will no longer be directly opposed by the head group repulsions. This attraction can drive membrane anchor tilting and draw the hydrocarbon regions together. The strongest part of the attraction between hydrophobic surfaces appears at a separation of ~2–10 Å (37). This study also showed that there was no energy barrier to the association of hydrophobic surfaces. Computer simulations of the association of poly-leucine α-helices also indicated that the attraction has a range of nearly 10 Å, with no barrier obstructing association (although poly-alanine had a someone shorter range of attraction and a low barrier) (42). Incorporating a distance dependence into the hydrophobic energy will add considerably to the stabilization of the tilted intermediate state of the fusion pore. The simplest way to incorporate the range of the hydrophobic interaction into the present analysis is to increase w. Here w = 3.2 Å will be tested and compared with results obtained with 1.6 Å.
To calculate the hydrocarbon-water contact area between two adjacent, bent membrane anchors, note that at any given position, s, along the membrane anchor, the increment in θ is dθ = ds/R(s). Integrating then gives
| (6) |
An increment, ds, has a projection on the horizontal plane of dx = sin(θ(s))ds. Using the above expression for θ(s) and integrating dx gives
| (7) |
The distance between two adjacent membrane anchors at s is then
| (8) |
The increment in area of an infinitesimally thin trapezoidal section between two adjacent membrane anchors is
| (9) |
With Eq. 8 and its derivative, the area can be expressed as an integral of Eq. 9. Multiplying by β gives the hydrophobic contribution to the fusion pore expansion energy.
| (10) |
Note that this integral over s ends at b-δ (δ from Eq. 4) to account for the area around the top of the membrane anchor from which water is taken as excluded.
Combining Eqs. 3, 5, and 10 provides an expression for the energy of fusion pore expansion as the membrane anchors tilt.
| (11) |
If we set θb = θ(b) then R(s) can be found as the function that minimizes this energy. The double integral form of Eq. 10 makes applying the standard methods of variational calculus very difficult. As an alternative approach 1/R(s) is expanded in a power series.
| (12) |
Using Eq. 6 to express θ(s), and taking Eq. 12 to 2nd order yields
| (13) |
Using Eq. 4 then gives the membrane anchor bending contribution as
| (14) |
And Eq. 10 gives the hydrophobic contribution as
| (15) |
With these explicit expressions for Eκ and Eh, one can minimize E of Eq. 11 to determineθ0 and the coefficients of the power series for 1/R(s) in Eq. 12, subject to the constraint that θb is fixed according to Eq. 13 as
| (16) |
This constrained minimization was carried out with MathCad v. 14, varying A, B, C, and θ0. Adding cubic or quartic terms to Eq. 12 failed to reduce the minimum energy significantly; variations were on the order of 0.1%, indicating that the power series expansion generates a quantitative solution to this minimization problem. The constraint θ0>0 was imposed because negative values lead to unphysical situations. The double integral in Eq. 15 prolonged the minimization, and the process was dramatically shortened by expanding sin(θ0 + the power series) as the sine of a sum of two angles and replacing the sine and cosine of the power series with first and second order Taylor expansions, respectively. This approximation is reasonable because the power series, which represents the total bend in the membrane anchors, never exceeds 1/R ~ 0.01 Å−1. In instances where the minimization was performed with the double integral expression, the effect of this approximation was found to be on the order of 0.1%.
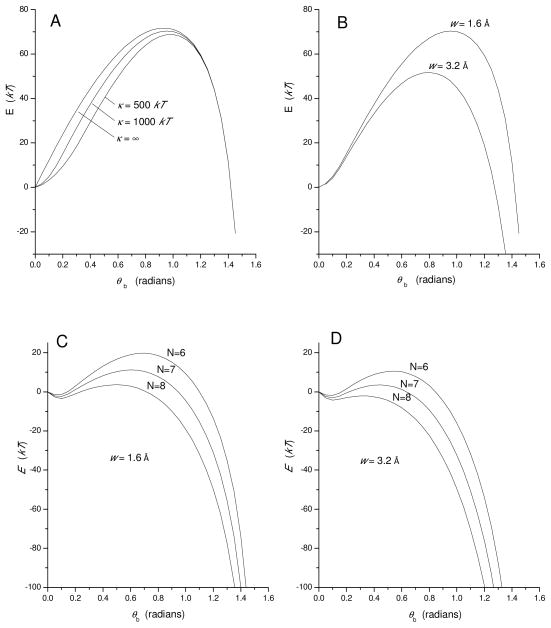
The energy barriers obtained by this method are illustrated in Fig. 3. Fig. 3A shows plots of energy versus tilt angle for w=1.6 Å, and κ=500 kT, 1000 kT, and ∞. Introducing flexibility to the membrane anchors had a relatively small effect on the energy barrier, reducing the peak and moving it to the right. However, when summed with a large driving force, shifting the peak to the right will also reduce the height of the energy barrier. Keeping κ=1000 kT and comparing w=1.6 Å and 3.2 Å shows that varying the range of the hydrophobic interaction has a much greater impact on the energy barrier (Fig. 3B). How these barriers influence the rate of the fusion pore dilation transition will be considered next.
Fig. 3.
Energy profiles obtained by minimizing Eκ + Eh + Es (Eq. 11) for fusion pore expansion, plotted versus tilt angle θb (see Fig. 2B). A and B present the sum of hydrophobic energy and α-helix bending energy for N=6 SNARE complexes. Varying the flexural rigidity, κ, of the α-helix from 500 to 1000 kT and making it absolutely stiff (κ=∞) have a moderate influence on the energy barrier (A). Varying the parameter representing the range of the hydrophobic interaction, w, from 1.6 to 3.2 Å has a much stronger influence on the energy barrier (B). C and D present this minimized contribution from Eq. 11 added to a linear interpolation of the helix completion energy plus the membrane bending energy. The profiles were calculated for N = 6, 7, and 8 SNARE complexes, with w = 1.6 Å (C) and 3.2 Å (D).
The Rate of Fusion Pore Expansion
A full reaction coordinate energy
To calculate a complete energy profile along the reaction coordinate, the various contributions evaluated above were summed. The helix completion energy arising from N SNARE complexes was taken as 2N ln(σ) with σ = 10−5 and the elastic energy of a lipidic pore was taken as 42.5 kT, as described above. These two energies were added and multiplied by θb/(π/2, so that as the tilting progresses from 0 to π/2, the driving force makes a contribution to the energy that is linear in θb. The energy from minimizing Eq. 11 was then added to this linear function to give complete energy profiles for N=6, 7, and 8. Figs. 3C and 3D show the energy barriers obtained with w=1.6 Å and 3.2 Å, respectively. These plots show that for many conditions, fusion pores encounter reasonably low energy barriers during a transition driven by tilting the membrane anchors. Thus, allowing the membrane anchors to bend and the bilayers to coalesce during the zipping process leads to a significant reduction in the energy barrier to fusion pore expansion. Increasing the number of SNARE complexes also lowers the barrier.
The Rate of Barrier Crossing
The energy profiles in Figs. 3C and 3D will now be used to estimate the rate of the transition. For a process governed by diffusion over a barrier the rate, α, can be calculated as (47) (see Ch. 7 of (48))
| (17) |
Three of the parameters in this equation, E†, φ0, and φ†, can be obtained from the complete energy plots in Figs. 3C and 3D. E† is the energy difference between the initial energy minimum and the peak of the energy barrier. φ0 and φ† are the quadratic coefficients for the energy as a function of θb around initial minimum and around the peak of the barrier, respectively. They were estimated from quadratic fits for the five points around the initial dip and the peak of each plot in Figs. 3C and 3D.
f, the coefficient of friction, remains to be determined. It depends on the flow of the surrounding medium as the transition progresses. As the SNARE membrane anchors tilt and zip, lipid flows through the fusion pore, encountering resistance from shearing forces and from friction between the two lipid monolayers as they slide past one another; water-lipid shear makes a negligible contribution (49). The lipid flow as θb increases closely resembles the flux of lipid through a fusion pore driven by a tension difference between the two membranes (compare Figs. 4A and 4B). The hydrodynamics of the lipid flux illustrated in Fig 4A was analyzed by Chizmadzhev et al. (49), who derived an analytical expression for the velocity as a function of the tension gradient. Their tension-velocity representation will be used here to develop a representation relevant to the present case where energy varies with θb.
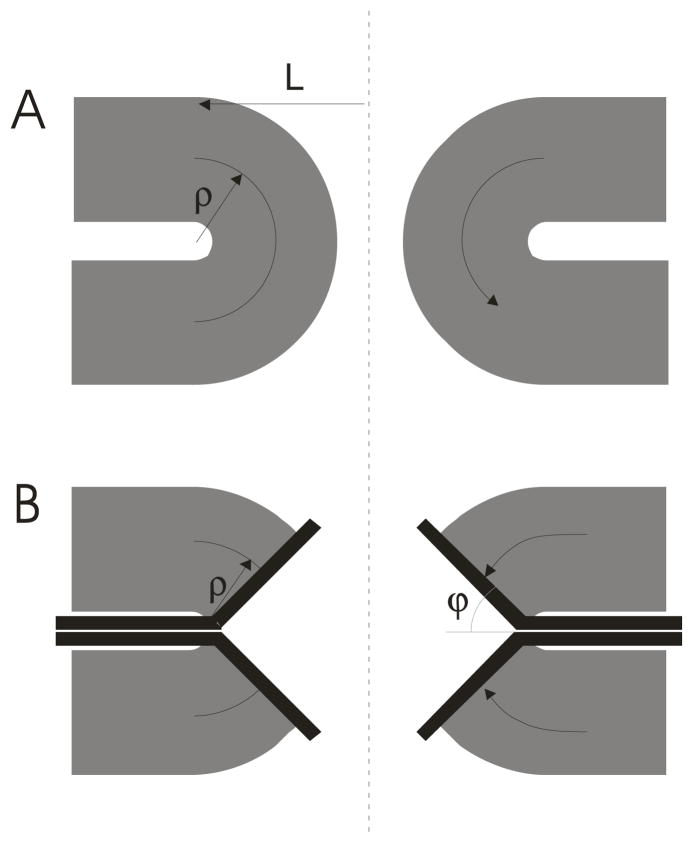
Fig. 4.
A. A toroidal fusion pore composed of lipid. When the two fusing membranes are under different tensions lipid flows along the curved arrow on the right. ρ represents the radius of the semicircle that defines the toroidal pore. L represents the distance from the pore axis (vertical dashed line) to the outer rim of the pore (49). B. The flow of lipid in a fusion pore during the transition follows a segment of the trajectory taken by lipid in a toroidal pore in A.
Lipid velocity v(ϕ) along a contour at the bilayer midplane in Fig. 4A is related to the rate of change of θb as
| (18) |
ρ is the radius of curvature of the contour at the leading edge of the growing pore; for a toroid this is ρ in Fig. 4A. ϕ is the angle between the membrane anchors and the horizontal plane (Fig. 4B), so θb = π/2 − ϕ. (Use of the variable ϕ in this analysis follows Chizmadzhev et al. (49).) At the fusion pore-planar membrane junction (where ϕ = π/2) the velocity is taken as v0. For a toroidal pore these two velocities are related by
| (19) |
where L is the distance from the vertical axis of the pore to its rim (again where ϕ = π/2; Fig. 4B). (Separate expressions were used previously for the inner and outer monolayers of the pore (49), but for the present analysis that distinction is unnecessary). Chizmadzhev et al. (49) showed that v0 is related to the tension difference between the two membranes, Δσ, by the expression
| (20) |
ηs and ηr are coefficients of shear and relative viscosity, respectively, and will both be taken here as η = 10−6 g/s (49). g=ρ/L. I(g) and J(g) are ‘form factors’ that reflect the geometry of the fusion pore. These form factors were expressed as integrals evaluated over a complete toroidal fusion pore in the interval −π/2 to π/2. In the present analysis the pore is only partially formed (Fig. 4B), so form factors were determined by integrating from −π/2 to −ϕ and ϕ to π/2. Combining Eqs. 18–20 gives a relation between the tension difference and the rate of change of θb.
| (21) |
The rate of change of energy was found to be (49)
| (22) |
In a brief time interval the change in energy for a small displacement δu is
| (23) |
From Eq. 19 we obtain
| (24) |
These displacements can be expressed in terms of changes in θb with the aid of Eq. 18.
| (25) |
Combining Eqs. 23 and 25 gives
| (26) |
Eliminating Δσ from Eqs. 21 and 26 gives
| (27) |
From this expression the coefficient of friction emerges as
| (28) |
f in Eq. 28 was evaluated here taking ρ = 3 nm (the closest distance between two bilayers determined by hydration forces (50)), L = 5.5 nm (ρ+half the bilayer thickness+ fusion pore radius of ~0.5 nm (17)), and ϕ=π/2−θb at the peaks in Fig. 3. f was found to range from 1.81 × 10−4 to 1.85 × 10−4 erg s. For a short range hydrophobic interaction (w = 1.6 Å) the rate constants obtained from Eq. 17 were α =7.8 × 10−5 s−1, 0.20 s−1, and 145 s−1 for N=6, 7, and 8, respectively. For w=3.2 Å the rate constants were 1.01 s−1, 390 s−1, and 2520 s−1, respectively. Although it is generally recognized that exocytosis takes place with millisecond kinetics, most kinetic studies have not determined the rate of the transition examined here. However, amperometry experiments in PC12 cells have examined the kinetics of fusion pore transitions in detail and determined the rate of the transition of a fusion pore to a dilating state at saturating [Ca2+] to be 700 sec−1 (51), and this falls in the faster end of the range of values calculated here.
DISCUSSION
This study analyzed the energetics of a hypothetical mechanism for the dilation of a proteinaceous fusion pore involving conversion to lipid. This mechanism posits a final step of SNARE complex assembly in which zipping and helix completion drive membrane anchors tilting. This mechanism allows zipping to perform mechanical work, deforming the membrane and prying open the fusion pore. This mechanism offers a parallel with a hypothetical mechanism of muscle contraction in which a helix-coil transition within myosin has been proposed to generate contractile force (52). Here a structural transition in the SNARE proteins drives the dilation of a protein-lined fusion pore, exerting a bending force on lipid bilayers so that two membranes can fuse. This model was used to derive a range of rates for the transition that includes an experimental measurement without adjusting free parameters. Thus, this coupling of SNARE protein zipping to fusion pore dilation provides a plausible mechanism for a key step in biological fusion during exocytosis.
It is significant that the energetics of this transition reflects a balance between very large opposing forces. This makes the transition very sensitive to small changes in key parameters. Modest changes in the membrane bending rigidity, α-helix flexural rigidity, the coefficient relating hydrophobic contact area to energy, the range of the hydrophobic interaction, and the energy of the final SNARE complex zipping step can make the energy barrier much larger or much smaller. Other energetic modes not considered in this model, such as sagging of the bilayer surface or other elastic deformations of the lipids around the exposed hydrocarbon region, as well as tilting of hydrocarbon chains, might allow the system to reduce the energy barrier below that estimated here. On the other hand, breaking of contacts between adjacent membrane anchors would increase the barrier.
While balancing large opposing forces makes it more difficult to make quantitative theoretical predictions, this makes the underlying biological process more sensitive to conditions. This is precisely a feature one expects to find in a biological system that produces large and rapid structural changes in response to a biological signal. Triggering membrane fusion on millisecond time scales requires large forces that can be released in response to ligand binding. The sensitivity of the helix-coil transition to capping interactions makes helix completion an ideal mechanism for the transduction of such signals. This would enable a protein to clamp the fusion apparatus in an inactive state by intermolecular capping through hydrogen bonds with backbone carbonyls and amides of the linker region. Withdrawal of that protein would disrupt the capping and trigger fusion. Conversely, intramolecular capping within the linker sequence could be relieved by another protein forming hydrogen bonds with a capping side chain in the linker region. The Ca2+ sensor synaptotagmin I inhibits SNARE protein mediated fusion of liposomes in the absence of Ca2+ (53), and this could result from helix capping. Ca2+ could thus initiate fusion pore dilation by binding to synaptotagmin I to disrupt this interaction and reduce the stability of the helix-coil junction. The helix-coil transition is also sensitive to environment, so that the dipping of the linkers into the membrane (54, 55) could also favor helix completion.
The high stability of the heterotrimeric SNARE complex has been invoked as a source for the large energy required to drive fusion (5, 10). The present theory offers a more detailed picture of how this energy could be utilized. For helix completion to deform the membranes as proposed here, a rigid base is required to transmit force to the membrane anchor. In order for helix completion to tilt the membrane anchor and pry the fusion pore to open wider, the cytoplasmic portion most be held firmly in place. Incorporating the SNARE motifs of the proteins anchored in different membranes into one complex prevents these parts of the protein from rotating relative to the membrane during helix completion. The stability of the SNARE complex thus provides a means of transferring the force generated during zipping to the lipid bilayer rather than generating an unproductive rotation of the SNARE motif into an orientation perpendicular to the plane of the membrane.
This model also suggests a function for the formation of fusion pores from multiple SNARE protein membrane anchors. Combining multiple SNARE complexes to form a proteinaceous fusion pore can focus the energy and provide the driving force necessary to overcome the hydrophobic energy barrier that obstructs the transition to a lipidic pore. Experimental results support the involvement of multiple SNARE complexes in exocytosis. A cooperativity analysis has suggested that at least 3 SNARE complexes are necessary for fusion (56). The conductance of a fusion pore suggests a channel formed by 5–8 SNARE complexes (17, 57). Bilayer fusion in vitro requires ~8 SNARE complexes (58). An analysis of SNARE protein cleavage by chlostridial toxins suggests that fusion may require as many as 15 SNARE complexes (59).
Studies of liposomes have shown that replacing the membrane anchor with a lipid link reduces the fusogenicity of SNARE proteins (60). The present model suggests that experiments should focus on the linkers as pivotal regions of SNARE proteins. Manipulating the linkers between the SNARE motif and membrane anchor can reduce fusion, but the results do not provide a clear test of this model (61). Lengthening the linker slows down fusion, possibly because the greater entropy of a longer coil reduces the energy of helix completion. Insertion of a proline into the linker slowed fusion in some experiments but had no effect in other experiments (61). However, while proline breaks α-helices, it increases the stiffness of the polypeptide so that the impact on the transmission of force to the membrane anchor cannot be assessed. Mutations in the linker region of syntaxin slow fusion pore dilation during Ca2+-triggered exocytosis (15, 62). Mutations in the linker region of synaptobrevin increased fusion pore lifetime by 40%, although these effects were not statistically significant (63). It is hoped that the present work will motivate more critical experiments to test the role of the linker motif in SNARE-mediated membrane fusion.
Models for membrane fusion have generally focused on the two alternative mechanisms of 1) lipid stalk formation (64) and 2) formation of a gap junction-like proteinaceous pore (20). Experiments have suggested that lipid bilayers can fuse by the stalk mechanism (65), and this mechanism can account for the rate of viral fusion (66). One particularly attractive feature of the lipid stalk model is that fusion can proceed through a sequence of intermediates while minimizing hydrocarbon-water contact. But when fusion is driven by a protein complex capable of releasing a substantial quantity of energy, transient exposure of the lipid hydrocarbon to water becomes a possibility. This may be the most reliable way to induce fusion rapidly in response to a specific biological signal.
Acknowledgments
I thank Fred Cohen, Ed Chapman, Jim Weisshaar, Qiang Cui, and Tom Record for helpful discussions and comments. This work was funded by NIH grant NS44057.
References
- 1.Rothman JE. Intracellular membrane fusion. Adv Second Messenger Phosphoprotein Res. 1994;29:81–96. doi: 10.1016/s1040-7952(06)80008-x. [DOI] [PubMed] [Google Scholar]
- 2.Ferro-Novick S, Jahn R. Vesicle fusion from yeast to man. Nature. 1994;370:191–193. doi: 10.1038/370191a0. [DOI] [PubMed] [Google Scholar]
- 3.Sutton RB, Fassauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 4.Poirier MA, Xiao W, Macosko JC, Chan C, Shin YK, Bennett MK. The synaptic SNARE complex is a parallel four-stranded helical bundle. Nat Struct Biol. 1998;5:765–769. doi: 10.1038/1799. [DOI] [PubMed] [Google Scholar]
- 5.Fasshauer D. Structural insights into the SNARE mechanism. Biochim Biophys Acta. 2003;1641:87–97. doi: 10.1016/s0167-4889(03)00090-9. [DOI] [PubMed] [Google Scholar]
- 6.Brunger AT. Structure and function of SNARE and SNARE-interacting proteins. Q Rev Biophys. 2005;38:1–47. doi: 10.1017/S0033583505004051. [DOI] [PubMed] [Google Scholar]
- 7.Jackson MB, Chapman ER. Fusion pores and fusion machines in Ca2+-triggered exocytosis. Annu Rev Biophys Biomol Struct. 2006;35:135–160. doi: 10.1146/annurev.biophys.35.040405.101958. [DOI] [PubMed] [Google Scholar]
- 8.Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 9.Hanson PI, Heuser JE, Jahn R. Neurotransmitter release - four years of SNARE complexes. Curr Opin Neurobiol. 1997;7:310–315. doi: 10.1016/s0959-4388(97)80057-8. [DOI] [PubMed] [Google Scholar]
- 10.Harbury PA. Springs and zippers: coiled coils in SNARE-mediated membrane fusion. Structure. 1998;6:1487–1491. doi: 10.1016/s0969-2126(98)00147-6. [DOI] [PubMed] [Google Scholar]
- 11.Hua SY, Charlton MP. Activity-dependent changes in partial VAMP complexes during neurotransmitter release. Nat Neurosci. 1999;2:1078–1083. doi: 10.1038/16005. [DOI] [PubMed] [Google Scholar]
- 12.Xu T, Rammner B, Margittai M, Artalejo AR, Neher E, Jahn R. Inhibition of SNARE complex assembly differentially affects kinetic components of exocytosis. Cell. 1999;99:713–722. doi: 10.1016/s0092-8674(00)81669-4. [DOI] [PubMed] [Google Scholar]
- 13.Melia TJ, Weber T, McNew JA, Fisher LE, Johnston RJ, Parlati F, Mahal LK, Sollner TH, Rothman JE. Regulation of membrane fusion by the membrane-proximal coil of the t-SNARE during zippering of SNAREpins. J Cell Biol. 2002;158:929–940. doi: 10.1083/jcb.200112081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sorensen JB, Wiederhold K, Muller EM, Milosevic I, Nagy G, de Groot BL, Grubmuller H, Fasshauer D. Sequential N- to C-terminal SNARE complex assembly drives priming and fusion of secretory vesicles. Embo J. 2006;25:955–966. doi: 10.1038/sj.emboj.7601003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han X, Jackson MB. Structural transitions in the synaptic SNARE complex during Ca2+-triggered exocytosis. J Cell Biol. 2006;172:281–293. doi: 10.1083/jcb.200510012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han X, Jackson MB. Electrostatic interactions between the syntaxin membrane anchor and neurotransmitter passing through the fusion pore. Biophys J. 2005;88:L20–22. doi: 10.1529/biophysj.104.056739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han X, Wang CT, Bai J, Chapman ER, Jackson MB. Transmembrane segments of syntaxin line the fusion pore of Ca2+-triggered exocytosis. Science. 2004;304:289–292. doi: 10.1126/science.1095801. [DOI] [PubMed] [Google Scholar]
- 18.Fang Q, Berberian K, Gong LW, Hafez I, Sorensen JB, Lindau M. The role of the C terminus of the SNARE protein SNAP-25 in fusion pore opening and a model for fusion pore mechanics. Proc Natl Acad Sci U S A. 2008;105:15388–15392. doi: 10.1073/pnas.0805377105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z, Jackson MB. Membrane Bending Energy and Fusion Pore Kinetics in Ca2+-Triggered Exocytosis. Biophys J. 2010 doi: 10.1016/j.bpj.2010.02.043. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindau M, Almers W. Structure and function of fusion pores in exocytosis and ectoplasmic membrane fusion. Curr Opin Cell Biol. 1995;7:509–517. doi: 10.1016/0955-0674(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 21.Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poirier MA, Hao JC, Malkus PN, Chan C, Moore MF, King DS, Bennett MK. Protease resistance of syntaxin.SNAP-25.VAMP complexes. Implications for assembly and structure. J Biol Chem. 1998;273:11370–11377. doi: 10.1074/jbc.273.18.11370. [DOI] [PubMed] [Google Scholar]
- 23.Chizmadzhev YA, Cohen FS, Shcherbakov A, Zimmerberg J. Membrane mechanics can account for fusion pore dilation in stages. Biophys J. 1995;69:2489–2500. doi: 10.1016/S0006-3495(95)80119-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chizmadzhev YA, Kuzmin PI, Kumenko DA, Zimmerberg J, Cohen FS. Dynamics of fusion pores connecting membranes of different tensions. Biophys J. 2000;78:2241–2256. doi: 10.1016/S0006-3495(00)76771-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozlovsky Y, Kozlov MM. Stalk model of membrane fusion: solution of energy crisis. Biophys J. 2002;82:882–895. doi: 10.1016/S0006-3495(02)75450-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markin VS, Albanesi JP. Membrane fusion: stalk model revisited. Biophys J. 2002;82:693–712. doi: 10.1016/S0006-3495(02)75432-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson MB. Minimum Membrane Bending Energies of Fusion Pores. J Membr Biol. 2009;231:101–115. doi: 10.1007/s00232-009-9209-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 29.Katsov K, Muller M, Schick M. Field theoretic study of bilayer membrane fusion. I. Hemifusion mechanism. Biophys J. 2004;87:3277–3290. doi: 10.1529/biophysj.103.038943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zimm BH, Bragg JK. Theory of the phase transition between helix and random coil in polypeptide chains. Journal of Chemical Physics. 1959;31:526–535. [Google Scholar]
- 31.Wojcik J, Altmann K-H, Scheraga HA. Helix-coil stability contants for the naturally occurring amino acids in water. XXIV. Half-cystine parameters from random poly9hydroxybutylglutamine-co-Smethylthio-L-cysteine. Biopolymers. 1990;30:121–134. doi: 10.1002/bip.360300112. [DOI] [PubMed] [Google Scholar]
- 32.Richards FM. Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- 33.Reynolds JA, Gilbert DB, Tanford C. Empirical Correlation Between Hydrophobic Free Energy and Aqueous Cavity Surface Area. Proc Natl Acad Sci U S A. 1974;71:2925–2927. doi: 10.1073/pnas.71.8.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanford C. Interfacial free energy and the hydrophobic effect. Proc Natl Acad Sci U S A. 1979;76:4175–4176. doi: 10.1073/pnas.76.9.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dill KA, Truskett TM, Vlachy V, Hribar-Lee B. Modeling water, the hydrophobic effect, and ion solvation. Annu Rev Biophys Biomol Struct. 2005;34:173–199. doi: 10.1146/annurev.biophys.34.040204.144517. [DOI] [PubMed] [Google Scholar]
- 36.Sharp KA, Nicholls A, Fine RF, Honig B. Reconciling the magnitude of the microscopic and macroscopic hydrophobic effects. Science. 1991;252:106–109. doi: 10.1126/science.2011744. [DOI] [PubMed] [Google Scholar]
- 37.Meyer EE, Rosenberg KJ, Israelachvili J. Recent progress in understanding hydrophobic interactions. Proc Natl Acad Sci U S A. 2006;103:15739–15746. doi: 10.1073/pnas.0606422103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chernomordik LV, Kozlov MM. Protein-lipid interplay in fusion and fission of biological membranes. Annu Rev Biochem. 2003;72:175–207. doi: 10.1146/annurev.biochem.72.121801.161504. [DOI] [PubMed] [Google Scholar]
- 39.Baldwin RL. Energetics of protein folding. J Mol Biol. 2007;371:283–301. doi: 10.1016/j.jmb.2007.05.078. [DOI] [PubMed] [Google Scholar]
- 40.Rose GD, Geselowitz AR, Lesser GJ, Lee RH, Zehfus MH. Hydrophobicity of amino acid residues in globular proteins. Science. 1985;229:834–838. doi: 10.1126/science.4023714. [DOI] [PubMed] [Google Scholar]
- 41.Tanford C. The Hydrophobic Effect. John Wiley and Sons; New York: 1980. [Google Scholar]
- 42.MacCallum JL, Moghaddam MS, Chan HS, Tieleman DP. Hydrophobic association of alpha-helices, steric dewetting, and enthalpic barriers to protein folding. Proc Natl Acad Sci U S A. 2007;104:6206–6210. doi: 10.1073/pnas.0605859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hristova K, White SH. Determination of the hydrocarbon core structure of fluid dioleoylphosphocholine (DOPC) bilayers by x-ray diffraction using specific bromination of the double-bonds: effect of hydration. Biophys J. 1998;74:2419–2433. doi: 10.1016/S0006-3495(98)77950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis BA, Engelman DM. Lipid bilayer thickness varies linearly with acyl chain length in fluid phosphatidylcholine vesicles. J Mol Biol. 1983;166:211–217. doi: 10.1016/s0022-2836(83)80007-2. [DOI] [PubMed] [Google Scholar]
- 45.Boal D. Mechanics of the Cell. Cambridge University Press; Cambridge: 2002. [Google Scholar]
- 46.Choe S, Sun SX. The elasticity of alpha-helices. J Chem Phys. 2005;122:244912. doi: 10.1063/1.1940048. [DOI] [PubMed] [Google Scholar]
- 47.Kramers HA. Brownian motion in a field of force. Physica. 1940;7:284–304. [Google Scholar]
- 48.Jackson MB. Molecular and Cellular Biophysics. Cambridge University Press; Cambridge: 2006. [Google Scholar]
- 49.Chizmadzhev YA, Kumenko DA, Kuzmin PI, Chernomordik LV, Zimmerberg J, Cohen FS. Lipid flow through fusion pores connecting membranes of different tensions. Biophys J. 1999;76:2951–2965. doi: 10.1016/S0006-3495(99)77450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parsegian VA, Fuller N, Rand RP. Measured work of deformation and repulsion of lecithin bilayers. Proc Natl Acad Sci U S A. 1979;76:2750–2754. doi: 10.1073/pnas.76.6.2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang CT, Bai J, Chang PY, Chapman ER, Jackson MB. Synaptotagmin-Ca2+ triggers two sequential steps in regulated exocytosis in rat PC12 cells: fusion pore opening and fusion pore dilation. J Physiol. 2006;570:295–307. doi: 10.1113/jphysiol.2005.097378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harrington WF, Rodgers ME, Davis JS. Functional aspects of the myosin rod in muscle contraction. In: Squire JW, editor. Molecular Mechanisms in Muscular Contraction. Macmillan Press; London: 1990. pp. 241–257. [Google Scholar]
- 53.Chicka MC, Hui E, Liu H, Chapman ER. Synaptotagmin arrests the SNARE complex before triggering fast, efficient membrane fusion in response to Ca2+ Nat Struct Mol Biol. 2008;15:827–835. doi: 10.1038/nsmb.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kweon DH, Kim CS, Shin YK. The membrane-dipped neuronal SNARE complex: a site-directed spin labeling electron paramagnetic resonance study. Biochemistry. 2002;41:9264–9268. doi: 10.1021/bi025934+. [DOI] [PubMed] [Google Scholar]
- 55.Kweon DH, Kim CS, Shin YK. Insertion of the membrane-proximal region of the neuronal SNARE coiled coil into the membrane. J Biol Chem. 2003;278:12367–12373. doi: 10.1074/jbc.M211123200. [DOI] [PubMed] [Google Scholar]
- 56.Hua Y, Scheller RH. Three SNARE complexes cooperate to mediate membrane fusion. Proceedings of the National Academy of Sciences. 2001;98:8065–8070. doi: 10.1073/pnas.131214798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Z, Zhang Z, Jackson MB. Synaptotagmin IV modulation of vesicle size and fusion pores in PC12 cells. Biophys J. 2010 doi: 10.1016/j.bpj.2009.11.024. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Domanska MK, Kiessling V, Stein A, Fasshauer D, Tamm LK. Single vesicle millisecond fusion kinetics reveals number of SNARE complexes optimal for fast SNARE-mediated membrane fusion. J Biol Chem. 2009;284:32158–32166. doi: 10.1074/jbc.M109.047381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Montecucco C, Schiavo G, Pantano S. SNARE complexes and neuroexocytosis: how many, how close? Trends Biochem Sci. 2005;30:367–372. doi: 10.1016/j.tibs.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 60.McNew JA, Weber T, Parlati F, Johnston RJ, Melia TJ, Sollner TH, Rothman JE. Close is not enough: SNARE-dependent membrane fusion requires an active mechanism that transduces force to membrane anchors. J Cell Biol. 2000;150:105–117. doi: 10.1083/jcb.150.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McNew JA, Weber T, Engelman DM, Sollner TH, Rothman JE. The length of the flexible SNAREpin juxtamembrane region is a critical determinant of SNARE-dependent fusion. Mol Cell. 1999;4:415–421. doi: 10.1016/s1097-2765(00)80343-3. [DOI] [PubMed] [Google Scholar]
- 62.Lam AD, Tryoen-Toth P, Tsai B, Vitale N, Stuenkel EL. SNARE-catalyzed Fusion Events Are Regulated by Syntaxin1A-Lipid Interactions. Mol Biol Cell. 2008;19:485–497. doi: 10.1091/mbc.E07-02-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kesavan J, Borisovska M, Bruns D. v-SNARE actions during Ca(2+)-triggered exocytosis. Cell. 2007;131:351–363. doi: 10.1016/j.cell.2007.09.025. [DOI] [PubMed] [Google Scholar]
- 64.Kozlov MM, Leikin SL, Chernomordik LV, Markin VS, Chizmadzhev YA. Stalk mechanism of vesicle fusion. Intermixing of aqueous contents. Eur Biophys J. 1989;17:121–129. doi: 10.1007/BF00254765. [DOI] [PubMed] [Google Scholar]
- 65.Lee J, Lentz BR. Evolution of lipidic structures during model membrane fusion and the relation of this process to cell membrane fusion. Biochemistry. 1997;36:6251–6259. doi: 10.1021/bi970404c. [DOI] [PubMed] [Google Scholar]
- 66.Kuzmin PI, Zimmerberg J, Chizmadzhev YA, Cohen FS. A quantitative model for membrane fusion based on low-energy intermediates. Proc Natl Acad Sci U S A. 2001;98:7235–7240. doi: 10.1073/pnas.121191898. [DOI] [PMC free article] [PubMed] [Google Scholar]