Abstract
The soil bacterium Arthrobacter nitroguajacolicus Rü61a contains the linear plasmid pAL1, which codes for the degradation of 2-methylquinoline. Like other linear replicons of actinomycetes, pAL1 is characterized by short terminal inverted-repeat sequences and terminal proteins (TPpAL1) covalently attached to its 5′ ends. TPpAL1, encoded by the pAL1.102 gene, interacts in vivo with the protein encoded by pAL1.101. Bioinformatic analysis of the pAL1.101 protein, which comprises 1,707 amino acids, suggested putative zinc finger and topoisomerase-primase domains and part of a superfamily 2 helicase domain in its N-terminal and central regions, respectively. Sequence motifs characteristic of the polymerization domain of family B DNA polymerases are partially conserved in a C-terminal segment. The purified recombinant protein catalyzed the deoxycytidylation of TPpAL1 in the presence of single-stranded DNA templates comprising the 3′-terminal sequence (5′-GCAGG-3′), which in pAL1 forms the terminal inverted repeat, but also at templates with 5′-(G/T)CA(GG/GC/CG)-3′ ends. Enzyme assays suggested that the protein exhibits DNA topoisomerase, DNA helicase, and DNA- and protein-primed DNA polymerase activities. The pAL1.101 protein, therefore, may act as a replicase of pAL1.
Linear plasmids have been identified in higher plants, fungi, and many bacteria. Most of these linear genetic elements are characterized by terminal inverted-repeat sequences and terminal proteins (TPs) covalently attached to their 5′ ends. The presence of TPs is a consequence of their mode of DNA replication, which in linear plasmids of plants, yeasts, and fungi is initiated at the termini by using the TP as a primer and proceeds by strand displacement. The DNA polymerases encoded by these linear elements are of the viral B type, related to those of contemporary adenoviruses and Bacillus phages (29). The mechanism of protein-primed DNA replication has been studied in detail, especially for the model of Bacillus subtilis phage φ29, which uses a monomeric B-family DNA polymerase for both the TP-primed initiation reaction and DNA elongation, resulting in continuous, full-length replication of both strands of the φ29 genome (5, 6, 7, 26, 42, 43, 44). In contrast to the linear plasmids of eukaryotes, linear chromosomes and plasmids of Streptomyces spp. replicate bidirectionally from an internal origin (9). This replication mechanism encounters the problem that discontinuous lagging-strand synthesis from RNA-primed Okazaki fragments leaves recessed 5′ ends at both telomeres when the distal RNA primers are removed. In the Streptomyces linear replicons, the TP serves as a primer for filling in these single-stranded gaps (2, 58). Both the TP and a telomere-associated protein (Tap), which is presumed to recruit the TP and position it at the telomere, are necessary for the propagation of Streptomyces replicons in their linear forms (3).
The TP and the Tap protein are highly conserved among many Streptomyces species. On the other hand, several studies have suggested a considerable diversity of streptomycetal replication systems. Replication in linear form of the plasmid pRL2 of Streptomyces sp. strain 44414, for example, requires the pRL2.3c and pRL2.4 genes, coding for a putative TP and a presumed Tap-helicase protein, respectively (59, 60). The linear chromosome of Streptomyces griseus IFO13350 also has an unusual telomere-associated protein, which has a DnaB-like helicase C-terminal domain (54). The linear plasmid SCP1 of Streptomyces coelicolor A3(2) requires the SCP1.127 gene, coding for a unique TP, and SCP1.125 for replication of its telomeres (24, 52). Linear plasmids also occur in other actinomycetes besides the streptomycetes, e.g., in several members of the rhodococci and mycobacteria, but their replicative proteins have not been studied yet.
The genus Arthrobacter has been classified in the family Micrococcaceae within the order Actinomycetales. The 113-kb conjugative plasmid pAL1, which confers on Arthrobacter nitroguajacolicus Rü61a the ability to utilize the N-heteroaromatic compound 2-methylquinoline, is so far the only described linear plasmid within this genus. The termini of pAL1 show the inverted-repeat sequence 5′-CCTGC…GCAGG-3′, and its 5′ ends are capped with proteins (34, 36). The terminal protein TPpAL1 is encoded by the pAL1.102 locus, which is located close to the “right” terminus of pAL1 (30). The adjacent gene pAL1.101 codes for a putative telomere-associated protein, which, however, is much larger than the Tap proteins of Streptomyces linear replicons. We hypothesized that the pAL1.101 protein might play a different, or more versatile, role in the replication of linear DNA than the Tap proteins (36). In this study, we show that the protein interacts with TPpAL1 in vivo. As a precondition for biochemical studies, we established a protocol for the preparation of recombinant pAL1.101 protein. The first data on its catalytic properties suggest that it exhibits DNA topoisomerase, DNA helicase, and DNA- and TP-primed DNA polymerase activities. It thus can be considered a novel replicative enzyme, which we have named REPpAL1.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The strains and plasmids used in this study are listed in Table S1 in the supplemental material. For isolation of total DNA, A. nitroguajacolicus Rü61a(pAL1) was grown in mineral salts medium (35) on 8 mM sodium benzoate at 30°C. For the preparation of crude extract supernatant for use in the deoxycytidylation assay, A. nitroguajacolicus Rü61a(pAL1) was grown in lysogeny broth (LB) (45). A. nitroguajacolicus Rü61a(pAL1, pART2-ORF102) was grown in mineral salts medium on 4 mM 4-hydroxyquinaldine in the presence of 140 μg/ml kanamycin. E. coli clones containing derivatives of pART2 or pET-22b(+) were grown in LB with 50 μg/ml kanamycin or 100 μg/ml ampicillin, respectively, at 37°C. For synthesis of maltose binding protein (MBP) and REPpAL1 fusion protein, Escherichia coli Rosetta 2(DE3)(pLysSRARE2) harboring either pET22b-malE-his6 or pET22b-ORF101 was grown in LB with ampicillin (100 μg/ml), chloramphenicol (34 μg/ml), and autoinduction solutions 5052 and M (53) at 30°C. Cells were harvested by centrifugation and stored at −80°C prior to use.
DNA techniques.
Total DNA of A. nitroguajacolicus Rü61a(pAL1) was isolated according to the method of Rainey et al. (39). Plasmid DNA was isolated with the E.Z.N.A. Plasmid Miniprep kit (peqlab, Erlangen, Germany). Gel extraction of DNA fragments from agarose gels was performed with the Perfectprep gel cleanup kit (Eppendorf, Hamburg, Germany). For cloning purposes, DNA fragments were purified with the High Pure PCR Product Purification kit (Roche Diagnostics GmbH, Mannheim, Germany). Standard protocols were used for agarose gel electrophoresis, restriction digestion, and DNA ligation (45). ORF101 of pAL1 was amplified by PCR with Phusion Hot Start High-Fidelity DNA Polymerase (Finnzymes Oy, Espoo, Finland), using total DNA of A. nitroguajacolicus Rü61a(pAL1) as the template. The primers and all oligonucleotides used are listed in Table S2 in the supplemental material. Competent E. coli and A. nitroguajacolicus Rü61a(pAL1) cells were generated as described in references 22 and 17, respectively. Plasmid inserts and flanking regions were verified by sequencing. Southern transfer of DNA from polyacrylamide gels to nylon membranes (Parablot NY plus; Macherey-Nagel, Dülmen, Germany) was done by capillary blotting. Colorimetric immunodetection of digoxigenin (DIG)-labeled DNA was performed using Anti-DIG-AP (Fab fragments from anti-digoxigenin antibody conjugated with alkaline phosphatase), nitroblue tetrazolium chloride, and 5-bromo-4-chloro-3-indolyl phosphate as described previously (40).
Protein purification.
The TPpAL1 protein with an N-terminal fusion to MBP was purified from E. coli ER2508(pLysSRARE, pMal-c2x-ORF102) as described previously (30). REPpAL1 protein was isolated with an N-terminal MBP-His7 and a C-terminal His6 fusion from E. coli Rosetta 2(DE3)(pLysSRARE2, pET22b-ORF101). Cell suspensions in 20 mM Tris-HCl buffer (pH 7.4) containing 400 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 M NDSB-201 [3-(1-pyridino)-1-propanesulfonate], 1 mM MgCl2, and 10 U/ml of benzonase were incubated for 30 min at room temperature and sonicated briefly, and crude extract supernatant was obtained by centrifugation. The crude extract (50 ml, from 20 g wet biomass) was applied to an amylose column (5 ml), which was then washed with 20 mM Tris-HCl buffer (pH 7.4) containing 400 mM NaCl and 1 mM EDTA. REPpAL1 was eluted with 20 mM maltose dissolved in the same buffer supplemented with 200 mM NDSB-201. Subsequently, the protein was subjected to size exclusion chromatography on a HiLoad 26/60 Superdex 200 column (GE Healthcare, Munich, Germany) in 20 mM Tris-HCl (pH 7.4; 400 mM NaCl, 1 mM EDTA). The pooled fractions were mixed with NDSB-201 to a concentration of 200 mM and concentrated in a U-Tube concentrator 15H-30 (Novagen).
For control experiments, MBP-His6 protein was prepared from E. coli Rosetta 2(DE3)(pLysSRARE2, pET22b-malE-His6)—i.e., from an E. coli strain that, apart from the pAL1.101 gene, is isogenic to the clone used for isolation of the REPpAL1 fusion protein—by the same amylose affinity chromatography protocol as that applied for the first step of REPpAL1 purification.
Protein concentrations were determined by the bicinchoninic acid method (48). Proteins separated in SDS-polyacrylamide gels were stained with ethyl violet and zincon (10).
In vivo formaldehyde cross-linking and identification of proteins cross-linked with TPpAL1.
Cell suspensions of A. nitroguajacolicus Rü61a(pAL1, pART2-ORF102), washed in phosphate-buffered saline (pH 7.4) and adjusted to an optical density at 600 nm (OD600) of 10, were incubated in 0%, 0.2%, and 0.5% (by volume) formaldehyde for 30 min at 22°C. The reaction was stopped by addition of glycine (0.125 M), the cells were washed in phosphate-buffered saline, resuspended in denaturing buffer (50 mM sodium phosphate buffer, pH 8.0, 300 mM NaCl, 1% [wt/vol] SDS), and disrupted by sonication. Cell extracts were incubated with Ni2+-nitrilotriacetate (NTA) agarose beads for 10 min at room temperature. Beads with adsorbed proteins were recovered by centrifugation using Spin Columns (Perfectprep; Eppendorf, Hamburg, Germany) and washed with 50 mM phosphate buffer, 300 mM NaCl, pH 8. The bound protein complexes were eluted with phosphate buffer adjusted to pH 5.5, concentrated, and separated in SDS-polyacrylamide gels (10.8% acrylamide; Coomassie stain). Protein bands that, according to immunodetection on a corresponding Western blot (anti-His5 antibody), contained covalent complexes with TPpAL1-His8 were excised from the polyacrylamide gel. In-gel tryptic digestion was performed according to the method of Stauber et al. (49). Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses were conducted as described previously (50), using an Ultimate 3000 nano-liquid chromatography system (Dionex, Sunnyvale, CA) and an LTQ Orbitrap XL (Thermo, Bremen, Germany) mass spectrometer. The measured MS/MS spectra were matched with tryptic amino acid sequences deduced from all six reading frames of pAL1 (GenBank accession no. AM286278.2) and to those deduced from the genome of Arthrobacter aurescens TC1 (CP000474) using SEQUEST as previously described (49).
DNA relaxation assay.
In all assays for catalytic activities of REPpAL1, 35 mM Tris-HCl, 72 mM KCl, 5 mM dithiothreitol, 5 mM MgCl2 (pH 8) was used as a basal buffer. For the detection of DNA relaxation activity of REPpAL1, pET-22b(+) plasmid DNA was incubated with the purified protein in the presence or absence of MgCl2. Aliquots withdrawn at appropriate time intervals were quenched in liquid N2 and subsequently separated in an agarose gel. DNA topoisomerase I from calf thymus (Takara Bio Europe); the site- and strand-specific endonuclease Nt.Bpu10I (Fermentas GmbH, St.Leon-Rot, Germany), which creates a single nick in pET-22b(+); and the restriction endonuclease HindIII to generate linearized plasmid DNA were used for control assays. The DNA forms generated were separated in agarose gels. By adding 1 μg/ml ethidium bromide to the gel buffer, it was possible to separate relaxed covalently closed circular DNA, as generated by topoisomerase I, from other DNA forms (11).
Helicase assays.
The 5′-DIG labeled 29-mer oligonucleotide DIG29basic, hybridized to either (i) the fully complementary oligomer blunt29; (ii) the 29-mer 29-3, generating 6-nucleotide 3′ overhangs; or (iii) the 29-mer 29-5, generating 6-nucleotide 5′ overhangs, was used as a DNA substrate for a strand displacement assay (for the oligonucleotide sequences, see Table S2 in the supplemental material). Hybridization of DIG29basic (33 μM) and the respective unlabeled complementary oligonucleotide (66 μM) was performed in a thermocycler using a stepwise gradient from 98°C (10 min) through 80°C (2 min), 73°C (10 min), and 70°C, 65°C, 60°C, 50°C, 40°C, and 30°C (2 min each) to 20°C. Hybridized double-stranded DNA (dsDNA) substrates, after dilution in H2O, were mixed with a 100-fold excess of unlabeled competitor DNA, which in the case of complete unwinding of the dsDNA, mediated by helicase activity of REPpAL1, outcompetes rehybridization of DIG29basic and thus releases the labeled 29-mer as single-stranded DNA (ssDNA). The respective dsDNA substrate, competitor DNA, and REPpAL1 protein (or MBP-His6 in control samples) were incubated in the presence or absence of ATP or dATP, separated in a polyacrylamide gel, and transferred by Southern blotting to a nylon membrane. Colorimetric detection of DIG-labeled fragments was performed with anti-DIG-AP antibodies (Fab fragments conjugated with alkaline phosphatase; Roche), p-nitrotetrazolium blue, and 5-bromo-4-chloro-3-indolyl phosphate. Helicase activity was also measured in a continuous fluorometric assay, which was based on the displacement of the fluorescent dye DAPI (4′,6-diamidino-2-phenylindole) from dsDNA upon DNA unwinding (16). pUC18 DNA, linearized with HindIII, PstI, or SmaI, was used as a substrate. To test 5′-capped dsDNA as a potential substrate, an ∼3-kb stretch of DNA was amplified using pET-22b(+) as a template and 5′-biotinylated primers (see Table S2 in the supplemental material), and the amplicon was purified from an agarose gel. Fluorescence measurements were performed in a Jasco FP-6500 spectrofluorimeter, using excitation and emission wavelengths of 345 and 467 nm and bandwidths of 1 and 10 nm for the excitation and emission splits, respectively. The value for 100% unwinding was obtained by subtracting the fluorescence of an equimolar amount of ssDNA (dsDNA substrate denatured at 98°C and quenched in liquid N2) (FssDNA) from the initial fluorescence of dsDNA (FdsDNA). These values were determined for each set of reaction conditions and each DNA substrate tested. The observed fluorescence change (Fobs) divided by (FdsDNA − FssDNA) indicates the extent of unwinding. Initial unwinding rates were estimated from the initial slopes of the kinetic traces. The percentage of total DNA unwound was multiplied by the concentration of base pairs in the reaction mixture, and the total concentration of unwound base pairs was divided by the time required for complete unwinding to estimate the apparent rate (16, 41).
Deoxycytidylation of TPpAL1.
The template specificity of REPpAL1-catalyzed deoxycytidylation of TPpAL1 was analyzed in an in vitro assay as described previously (30).
DNA polymerase assays.
Protein-primed DNA amplification by REPpAL1 was analyzed using the 285-bp dsDNA template l285 (see Table S2 in the supplemental material) with purified MBP-TPpAL1 as a primer. The product was treated with proteinase K prior to agarose gel electrophoresis. The ability of REPpAL1 to elongate a DNA primer-template hybrid was tested with the left50/left13t hybrid molecule (see Table S2 in the supplemental material). Annealing of the two oligomers was performed in a thermocycler using a stepwise gradient from 98°C (2 min) through 65°C (2 min), 55°C (10 min), and 50°C, 40°C, and 30°C (2 min each) to 10°C. The hybrid DNA was incubated with dATP, dGTP, dTTP, dithiothreitol, [α-32P]dCTP, and purified REPpAL1 protein; controls were performed with MBP-His6 instead of REPpAL1. The reaction products were analyzed in a 15% polyacrylamide gel.
RESULTS
REPpAL1 is a multidomain protein.
REPpAL1, the product of the pAL1.101 gene, comprises 1,707 amino acids (aa) and has a predicted molecular mass of ∼186.8 kDa. In its C-terminal region, it shows sequence similarity to part of the Tap proteins of Streptomycetes, e.g., 29% identity of aa 1446 to 1663 of REPpAL1 to aa 415 to 678 of TapL of Streptomyces lividans. Closer homologs of REPpAL1 occur in the genus Rhodococcus. Several gene products of comparable size, encoded by rhodococcal linear plasmids, exhibit >30% overall identity to REPpAL1. Examples are the pROB01-00090 protein of Rhodococcus opacus B4 and the pREL1_00080 protein of Rhodococcus erythropolis strain PR4, which have been annotated as putative telomere-binding proteins, and the protein encoded by the RHA1_ro10009 locus of pRHL2 of Rhodococcus jostii RHA1.
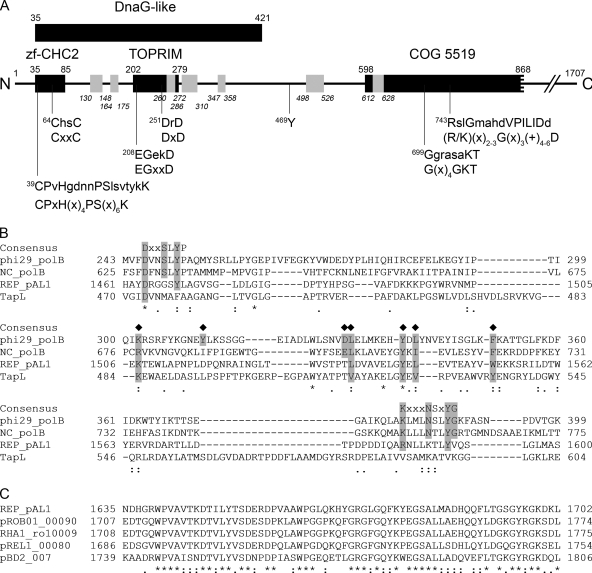
A search for conserved domains in REPpAL1 revealed similarity of its N-terminal region to DnaG-type proteins; this region includes fully conserved CHC2-type zinc finger and topoisomerase-primase (Toprim) domains (Fig. 1 A). The consensus of a “zinc ribbon” (19), as well as the EGXXD and DXD motifs, which in Toprim-containing enzymes are involved in coordination of Mg2+ and catalysis (1, 47), are conserved in REPpAL1, as well as in its rhodococcal orthologs. Residue Y469 of REPpAL1 is also strictly conserved among the related rhodococcal proteins. A centrally located region (aa 598 to 868), which contains Walker A- and Walker B-like motifs potentially involved in nucleotide binding, aligns with part of the COG5519 domain of superfamily 2 helicases (Fig. 1A). Besides the Walker A and B boxes (56), which are conserved in all helicases, residues 809MGITGS814 of REPpAL1 might correspond to motif III (+X+TGS, where + is hydrophobic and X is any amino acid) of helicase superfamily 2 (20); however, due to the substantial divergence of motif sequences, it is difficult to predict helicase motifs solely from an amino acid sequence (8). Further downstream in the REPpAL1 sequence, a CXXC(X)13-14HXXC motif at positions 1155 to 1175 that is also conserved in the rhodococcal homologs might form a binding site for divalent metal ions.
FIG. 1.
Bioinformatic analysis of the REPpAL1 protein. (A) Hypothetical domain architecture and predicted sequence motifs of the N-terminal region of the REPpAL1 protein. The amino acid sequence of REPpAL1 (the gene product of pAL1.101; accession no. CAL09956) is represented by a line. The black bars, with residue numbers above each bar, indicate predicted conserved domains, namely, a DnaG-like domain (COG 0358; E value, 0.005), a CHC2 zinc finger subdomain (smart00400; E value, 0.001), and a Toprim subdomain (cd01029; E value, 3e-06), located within the DnaG region. A central region matched the N-terminal half of COG 5519, representing a superfamily II helicase and derivatives (E value, 3e-05). Searches for conserved domains against the CDD database were performed using the CD Search tool at NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), with the low-complexity filter inactivated and an E value threshold of 0.01. Putative conserved amino acid motifs are indicated below the bars (x, any amino acid; +, hydrophobic amino acid); the lower lines indicate the consensus sequence of the respective motif (for references, see the text), and the associated upper lines show the amino acid sequence of REPpAL1, with residues matching the consensus in uppercase. Residue Y469, which is conserved in rhodococcal homologs of REPpAL1, is also indicated. The short bars in gray, with residue numbering in italics, correspond to tryptic peptides of REPpAL1 identified in the cross-linked TPpAL1-REPpAL1 complex. For details, see the text. (B) Alignment of segments of phage φ29 DNA polymerase (phi29_polB; GenBank accession no. ACE96023), type B DNA polymerase of Neurospora crassa (NC_polB; CAA39046), REPpAL1 (REP_pAL1; CAL09956), and telomere-associated protein of S. lividans (TapL; AAO73842). The consensus sequence of motifs A and B of φ29 DNA polymerase, which is conserved among different nucleic-acid-synthesizing enzymes (7), is indicated above the sequences. Residues shaded in gray that are marked with a diamond above the sequence may represent conserved residues of the TPR-1 region, which is specific to protein-priming DNA polymerases (14, 15, 27, 37). (C) Alignment of a region rich in acidic residues, which is highly conserved among REPpAL1 (aa 1635 to 1702) and rhodococcal homologs. pROB01_00090 (BAH55508), putative telomere-binding protein of plasmid pROB01 of R. opacus B4; RHA1_ro10009 (ABH00202), possible transcriptional regulator of plasmid pRHL2 of R. jostii RHA1; pREL1_00080 (BAE45951), putative telomere-binding protein of plasmid pREL1 of R. erythropolis PR4; pBD2_007 (AAP73892), putative regulatory protein of plasmid pBD2 of R. erythropolis BD2. In panels B and C, residues conserved throughout are marked with asterisks, while residues marked with colons and dots are conserved and semiconserved substitutions, respectively.
There was no significant similarity to known conserved domains in the large C-terminal region of REPpAL1, and it was not feasible to predict putative function from sequence analysis. However, alignments tentatively suggested partial conservation of motifs A (DX2SLYP) and B (KX3NSXYG), which are characteristic of the polymerization domain of family B DNA polymerases (Fig. 1B). Residues of these motifs contribute to the polymerization active site and are involved in binding of metal ions (Asp of motif A) and binding of deoxynucleoside triphosphates (dNTPs) (Tyr residues of motifs A and B) (7). Notably, motif A- and motif B-like sequences seem to be absent in TapL of Streptomyces (Fig. 1B). A region reminiscent of the so-called TPR-1 sequence ([R/K][X]6-10[Y/W/F][X]12-16[D/E][L/I/W][X]6-8[Y/W/F]X[L/I/V/F][X]7-14[F/W/Y]), located between motifs A and B of protein-priming DNA polymerases, might be conserved in both REPpAL1 (1507K[X]26L[X]7YEL[X]7W1552) and TapL (Fig. 1B). In phage φ29 DNA polymerase, TPR-1 was demonstrated to be important for positioning of the protein primer and for transition between protein-primed and DNA-primed modes of replication (14, 15, 27, 37). A sequence corresponding to motif C of DNA polymerases (YXDTDS), which via its Asp residues contributes to metal ion binding and catalysis (7), was not obvious in REPpAL1. However, in a C-terminal region that is highly conserved among REPpAL1 and its rhodococcal homologs, a number of acidic residues are conserved (Fig. 1C), some of which might be involved in the two-metal-ion mechanism of DNA polymerases (51). Despite only distant relatedness to known proteins, sequence analysis suggested that REPpAL1 might act as a multifunctional enzyme in DNA replication; however, the functional significance of individual domains and of conserved amino acid residues remains to be investigated.
REPpAL1 interacts in vivo with the terminal protein TPpAL1.
To address the question of whether REPpAL1, together with the terminal protein TPpAL1, is part of a telomeric complex, octahistidine-tagged TPpAL1 was synthesized in the homologous host, and interacting proteins were captured by in vivo cross-linking. From cells of A. nitroguajacolicus Rü61a(pAL1, pART2-ORF102) treated with formaldehyde, the covalent complex of TPpAL1-His8 was prepared by denaturing metal chelate affinity chromatography and gel electrophoresis and subjected to trypsin digestion, and peptides were identified by LC-MS/MS analysis (see Table S3 in the supplemental material). The complex from cells treated with 0.5% formaldehyde contained fragments of TPpAL1, as well as peptides that were assigned to proteins which probably were highly expressed in the cytoplasm, such as superoxide dismutase, encoded by pAL1.014, and a predicted GroEL protein. In the tryptic digest of the protein complex purified from cells soaked in 0.2% formaldehyde, fragments of TPpAL1 (aa 34 to 49, 145 to 168, 169 to 181, and 182 to 192) and of the N-terminal region of REPpAL1 (Fig. 1A) exclusively were identified, suggesting specific interaction of these proteins in vivo.
Preparation of REPpAL1 fusion protein.
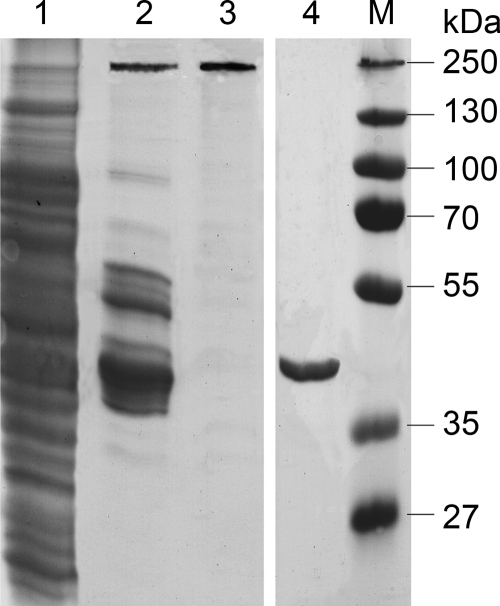
The pAL1.101 gene was expressed in E. coli cotransformed with a plasmid carrying tRNA genes for codons rarely used in E. coli (see Table S1 in the supplemental material). Since recombinant REPpAL1 proteins fused to short affinity tags were present in the insoluble fraction of the cell extract (data not shown), REPpAL1 was synthesized with an N-terminal fusion to MBP, which conferred in vitro solubility. The fusion protein (also termed REPpAL1) was prepared by affinity chromatography, followed by size exclusion chromatography (Fig. 2). Since proteins from E. coli active on DNA might contaminate preparations of REPpAL1 (even if the protein appeared electrophoretically homogeneous), we prepared MBP-His6 from E. coli Rosetta 2(DE3)(pLysSRARE2, pET22b-malE-His6)—i.e., from an E. coli strain that, apart from the pAL1.101 gene, is isogenic to the clone used for isolation of the REPpAL1 fusion protein—by the same amylose affinity chromatography protocol applied for the first step of REPpAL1 purification (Fig. 2, lane 4). This MBP-His6 preparation should have contained the same (or rather, more) E. coli contaminants as the REPpAL1 preparations (which were subjected to a second chromatographic step after amylose affinity chromatography). MBP-His6 was used as a control in all assays for catalytic functions of REPpAL1.
FIG. 2.
Preparation of REPpAL1 fusion protein. Proteins from each preparation step were separated in a denaturing (SDS) polyacrylamide gel (10.8%; stained with ethyl violet/zincon). Lane 1, crude extract supernatant of E. coli Rosetta 2(DE3)(pLysSRARE2, pET22b-ORF101); lane 2, pool after amylose affinity chromatography containing the REPpAL1 fusion protein at 231 kDa and additional amylose binding proteins; lane 3, REPpAL1 fusion protein carrying an N-terminal MBP-His7 and C-terminal His6 tag after separation of the amylose pool by size exclusion chromatography (proteins of equivalent electrophoretic quality were used for all functional studies); lane 4, MBP-His6 prepared from E. coli Rosetta 2(DE3)(pLysSRARE2, pET22b-malE-His6), i.e., from the same genetic background as the REPpAL1 fusion protein, by amylose affinity chromatography; lane M, marker proteins.
DNA relaxation activity of REPpAL1.
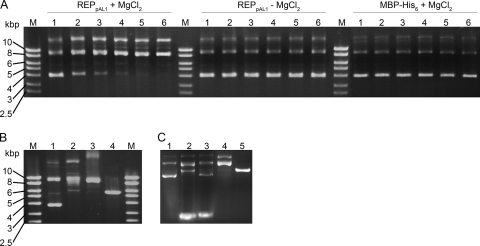
The purified REPpAL1 fusion protein catalyzed the relaxation of negatively supercoiled circular DNA. The activity was ATP independent but required Mg2+ ions (Fig. 3 A). Such a cofactor requirement, and the prediction of a Toprim domain (see above), may classify REPpAL1 as a type IA topoisomerase (47). In control experiments, treatment of plasmid DNA with topoisomerase I from calf thymus resulted in the formation of a DNA band with mobility similar to that observed in the REPpAL1-catalyzed reaction; under the conditions used, partially relaxed DNA was also visible (Fig. 3B, lane 2). However, nicked circular DNA, formed by the site-specific endonuclease Nt.Bpu10I, which is active on a single strand, showed electrophoretic mobility similar to that of the products of the topoisomerase reaction (Fig. 3B, lane 3). In order to distinguish between relaxed covalently closed circular DNA and nicked circular DNA, ethidium bromide was included in the agarose gel (Fig. 3C). Binding of the intercalating agent generates positive supercoiling in the covalently closed DNA, which significantly increases its electrophoretic mobility (11). Incubation of the plasmid DNA with an excess of topoisomerase I indeed yielded a DNA form which in the presence of ethidium bromide migrated very fast and thus is proposed to represent the covalently closed circular form (Fig. 3C, lane 3). Intercalators cannot overwind nicked DNA, which, irrespective of the presence of ethidium bromide, migrates above the linear form (Fig. 3C, lanes 4 and 5). The major DNA species formed in the REPpAL1-catalyzed reaction (Fig. 3C, lane 2) showed the same fast electrophoretic mobility as the product of the topoisomerase reaction, suggesting that REPpAL1 exhibits topoisomerase activity.
FIG. 3.
Relaxation of supercoiled plasmid DNA by REPpAL1. (A) Agarose gel (1%, ethidium bromide stained) of plasmid DNA treated with REPpAL1. pET-22b(+) DNA (22.6 nM) was incubated at 30°C in 35 mM Tris-HCl, 72 mM KCl, 5 mM dithiothreitol (pH 8) with REPpAL1 (1.06 μM) in the presence and in the absence of 5 mM MgCl2. Control assay mixtures contained 2.4 μM MBP-His6 instead of REPpAL1. Aliquots of the reaction mixtures were quenched by immersion in liquid nitrogen directly (time zero; lanes 1) and 20, 40, 60, 80, and 100 min (lanes 2 to 6) after the reaction was started. The prominent lower and middle bands represent supercoiled and relaxed circular DNA monomers, respectively (cf. panel B), whereas the upper band may represent relaxed dimers or multimers. Estimation of the supercoiled DNA in the samples treated with REPpAL1 plus MgCl2, performed with the ImageJ program (v1.38j; open source; NIH, Bethesda, MD) after calibration with marker bands, indicated amounts of ∼63 ng, 29 ng, 13 ng, 5 ng, 2 ng, and 1 ng in lanes 1, 2, 3, 4, 5, and 6, respectively. (B) Agarose gel (1%, ethidium bromide stained) of pET-22b(+) DNA. All assay mixtures contained 22.6 nM DNA. Lane 1, plasmid as isolated; lane 2, after treatment with DNA topoisomerase I from calf thymus (5 U, 37°C, 20 h); lane 3, after treatment with the site- and strand-specific endonuclease (“nickase”) Nt.Bpu10I (1 U, 37°C, 2 h); lane 4, after linearization with restriction endonuclease HindIII (1 U, 37°C, 2 h). The same buffer (with 5 mM MgCl2) as in panel A was used for all control experiments. The diffuse DNA band with low mobility in lane 3 may represent nicked multimers, DNA still associated with enzyme, or both. (C) DNA forms of pET-22b(+), separated in a 1% agarose gel containing 1 μg/ml ethidium bromide. The concentration of plasmid DNA in all assays was 22.6 nM. Lane 1, plasmid DNA as isolated; lane 2, after treatment with 1.06 μM REPpAL1 for 2 h at 37°C; lane 3, after incubation with an excess of DNA topoisomerase I from calf thymus (10 U, 37°C, 20 h); lane 4, after treatment with the nickase Nt.Bpu10I (1 U, 37°C, 2 h); lane 5, after linearization with HindIII (1 U, 37°C, 2 h). The same buffer (with 5 mM MgCl2) as in panel A was used for all experiments. After incubation, the samples were supplemented with SDS (1%). Electrophoresis was performed at 2 V/cm for 15 min, paused for 5 min, and continued at 6 V/cm for 60 min.
DNA helicase activity of REPpAL1.
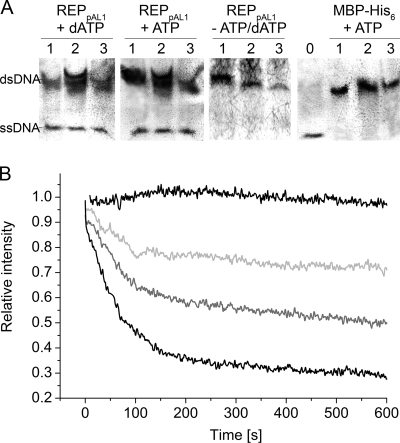
The capability of REPpAL1 to catalyze unwinding of double-stranded DNA was detected in a strand displacement assay, using oligomeric duplex DNA substrates consisting of a 5′-labeled and an unlabeled strand. REPpAL1 in the presence of ATP or dATP indeed mediated release of the labeled strand from the duplex DNA (Fig. 4 A). Another assay, based on measuring liberation of the fluorescent dye DAPI from linearized dsDNA upon DNA unwinding, was performed with HindIII-linearized plasmid DNA as a substrate. Figure 4B shows kinetic traces of the unwinding of this linear dsDNA substrate at different concentrations of the REPpAL1 protein. Assuming that the fluorescence intensity of DAPI in the presence of the respective dsDNA and in the presence of the heat-denatured DNA substrate represented 0% and 100% unwinding of DNA, respectively, the estimated initial rate of REPpAL1-catalyzed unwinding of dsDNA was in the range of 105 bp/s per enzyme monomer. Unwinding of 5′-biotinylated fully complementary dsDNA, which was tested as a model for 5′-capped linear dsDNA, occurred at a similar apparent initial rate. Such estimated unwinding rates are higher than those reported for other DNA helicases. The RecBCD enzyme, for example, showed a maximum rate of between 1,000 and 1,500 bp/s (4, 12). However, since several replicative DNA helicases have been described as much more effective when complexed with the polymerase or with accessory proteins of the replisome (13, 28, 55), it is tempting to speculate that the high apparent unwinding rate of REPpAL1 in vitro might be due to the presence of modulating domains on the same polypeptide.
FIG. 4.
DNA helicase activity of REPpAL1. (A) Immunodetection of DIG-labeled DNA oligomers after separation in a 15% polyacrylamide gel and Southern blotting. REPpAL1 catalyzed the displacement of DIG-labeled ssDNA from oligomeric DNA duplex substrates consisting of the 5′-DIG-labeled oligomer DIG29basic and complementary unlabeled strands. The duplex DNA substrates had 6-nucleotide 3′ overhangs (DIG29basic plus 29-3; lanes 1), 6-nucleotide 5′ overhangs (DIG29basic plus 29-5; lanes 2), and blunt ends (DIG29basic plus blunt29; lanes 3). For oligonucleotide sequences, see Table S2 in the supplemental material. The assays contained 10 nM duplex DNA substrate, a 100-fold excess of unlabeled competitor ssDNA, 2 mM ATP or dATP, and 0.14 μM REPpAL1 or 10 μM MBP-His6 (as the negative control) in 35 mM Tris-HCl, 72 mM KCl, 5 mM dithiothreitol, 5 mM MgCl2 (pH 8). All assay mixtures were incubated for 2 h at 30°C. The lane marked 0 shows the electrophoretic mobility of the 5′-DIG-labeled single-stranded 29-mer DIG29basic. (B) Fluorescence quenching upon unwinding of dsDNA as a function of the REPpAL1 concentration. The kinetic traces of fluorescence decrease are shown for assays containing 2 pM of MBP-His6 instead of REPpAL1 (control; upper black line) and 0.5 pM (light gray), 1 pM (dark gray), and 2 pM (lower black line) of REPpAL1 fusion protein. The dsDNA substrate (HindIII-linearized pUC18 DNA; 1 nM), DAPI (4 μM), and 5 mM ATP in 35 mM Tris-HCl, 72 mM KCl, 5 mM dithiothreitol, 5 mM MgCl2 (pH 8) was equilibrated for 1 h in the dark before the reaction was started by the addition of REPpAL1 protein. The DAPI fluorescences of dsDNA and ssDNA substrates were defined as relative intensities 1 and 0, respectively.
Many DNA helicases require a single-stranded tail as a loading site (31, 46), whereas REPpAL1 appears to also be active toward blunt-ended, 5′-capped dsDNA. Such activity is consistent with a possible role in initiating replication of linear plasmid DNA from its telomeres.
Template-specific deoxycytidylation of TPpAL1 by REPpAL1 and TPpAL1- and DNA-primed polymerase activity.
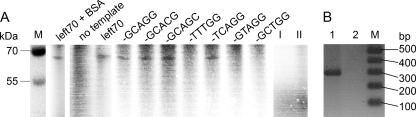
To investigate whether TPpAL1 can act as a priming protein for DNA replication, we previously developed an in vitro deoxynucleotidylation assay that contained the ssDNA template left70 (see Table S2 in the supplemental material), representing the 3′-terminal 70 nucleotides of the “left” end of pAL1; MBP-TPpAL1; crude extract of A. nitroguajacolicus Rü61a; the REPpAL1 fusion protein; ATP; and different [α-32P]dNTPs in Mg2+-containing buffer. Specific deoxycytidylation of TPpAL1 was detected in the presence of the ssDNA template, tentatively suggesting that the terminal or subterminal guanosine nucleotide at the 3′ end of pAL1 might serve as a template for the nucleotide incorporation reaction (30). When the Arthrobacter cell extract in the assay was replaced by bovine serum albumin (BSA), incorporation of [32P]dCMP into TPpAL1 occurred at similar intensity (Fig. 5 A, compare lanes left70 + BSA and left70), indicating that REPpAL1 is sufficient to catalyze the reaction.
FIG. 5.
Template specificity of REPpAL1-catalyzed deoxycytidylation of TPpAL1 (A) and TPpAL1-primed amplification of dsDNA (B). (A) Deoxynucleotidylation assay mixtures contained 1.0 μM purified MBP-TPpAL1 protein, 0.1 μM REPpAL1 fusion protein, 1 mM ATP, 1 μM DNA template, [α-32P]dCTP (0.33 μM; 111 TBq/mmol; PerkinElmer, Rodgau-Jügesheim, Germany), and either 0.05 mg/ml BSA (lane left70 + BSA) or crude extract (soluble proteins) of A. nitroguajacolicus Rü61a(pAL1) (0.33 mg protein/ml; all other lanes) in 35 mM Tris-HCl, 72 mM KCl, 5 mM dithiothreitol, 5 mM MgCl2 (pH 8). The samples were incubated for 16 h at 30°C. After treatment with 10 U DNase I at 30°C for 1 h, samples were separated in a 10% SDS-polyacrylamide gel, and radiolabeled proteins were detected with a phosphorimager (PharosFX Plus; Bio-Rad Laboratories). The template left70 represents the 3′-terminal 70 nucleotides of the left end of pAL1 with the terminal sequence GCAGG-3′, which is conserved at both 3′ ends of pAL1. Other templates (20-mer ssDNA) (see Table S2 in the supplemental material) showed variations of the 3′-terminal sequence, as indicated above the lanes. In lanes I and II, MBP-TPpAL1 and REPpAL1, respectively, were replaced by equimolar concentrations of MBP-His6 protein; the control assays contained left70 DNA template. (B) Lane 1, dsDNA (285-bp l285 [see Table S2 in the supplemental material]; 0.3 μM), MBP-TPpAL1 (65 nM) as a protein primer, ATP (2 mM), and dNTPs (200 μM each) were incubated with REPpAL1 fusion protein (0.1 μM) at 30°C in 35 mM Tris-HCl, 72 mM KCl, 5 mM dithiothreitol, 5 mM MgCl2 (pH 8) for 24 h. A control assay was performed in the same way, except that REPpAL1 was replaced by MBP-His6 protein (lane 2). Samples were treated with 0.5 mg/ml proteinase K prior to electrophoresis in a 2% agarose gel (ethidium bromide stain). Estimation of the amount of DNA in the 285-bp band, performed with the ImageJ program (v1.38j; open source; NIH, Bethesda, MD) after calibration with marker bands, indicated about 51 ng dsDNA in lane 1.
Deoxycytidylation of TPpAL1 at both the left70 template and the 20-mer 5′-CAGTTCGCATCTATTGCAGG-3′—which, aside from the terminal inverted-repeat sequence GCAGG-3′, consists of random nucleotides—suggests that distal regions of the 3′ end are not essential for the reaction (Fig. 5A, lane -GCAGG). To address the requirement for a conserved terminal sequence, the 3′ end of the 20-mer was varied. The REPpAL1-catalyzed deoxycytidylation of TPpAL1 required conservation of the third and fourth nucleotides (CA) of the template sequence, whereas a G→T transversion of the distal nucleotide of the 3′-terminal inverted-repeat sequence did not interfere with the reaction. Surprisingly, incorporation of dCMP into TPpAL1 occurred at several templates having GG-3′, GC-3′, and CG-3′ ends, suggesting that under the assay conditions used, deoxycytidylation of TPpAL1 by REPpAL1 does not show strict preference for the 3′-terminal or the subterminal guanosine nucleotide of the template (Fig. 5A). In the case of subterminal initiation, replication at the pAL1 telomere in vivo should involve a “sliding-back” mechanism, as shown for protein-primed replication of phage genomes, to maintain the integrity of the DNA ends (25, 32, 33). Subterminal initiation followed by “sliding back” for transition from protein-primed initiation to DNA elongation was proposed to be a general feature of protein-primed replication systems (7, 33).
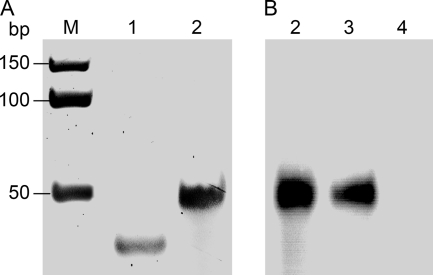
Isothermal amplification of a 285-bp dsDNA template using TPpAL1 as a protein primer resulted in production of a distinct, full-length amplification product (Fig. 5B), suggesting that REPpAL1 can act as a combined helicase and protein-primed DNA polymerase. To determine whether REPpAL1 also catalyzes DNA-primed elongation reactions, a primer extension assay was carried out using a 13-nucleotide DNA primer hybridized to a 50-mer DNA template. As shown in Fig. 6, lane 3, the primer was extended up to the full length of the template, resulting in a 50-bp dsDNA product.
FIG. 6.
DNA primer elongation by REPpAL1. Shown are a polyacrylamide gel (15%; ethidium bromide stained) (A) and an autoradiograph of the polyacrylamide gel (B), both detected with a phosphorimager (PharosFX Plus; Bio-Rad Laboratories). Lane 1 shows the DNA substrate, i.e., the 13-mer primer (left13t) hybridized to the 50-mer template (left50). For oligonucleotide sequences, see Table S2 in the supplemental material. Elongation assay mixtures contained the unlabeled DNA hybrid left50/left13t (1 μM) (lane 1); 66 nM (each) dATP, dGTP, dTTP, and [α-32P]-dCTP; and either GoTaq DNA polymerase (A and B, lanes 2), REPpAL1 fusion protein (88 nM) (B, lane 3), or MBP-His6 protein (800 nM) (lane 4) in 35 mM Tris-HCl, 72 mM KCl, 5 mM dithiothreitol, 5 mM MgCl2 (pH 8). The GoTaq-catalyzed elongation reaction shown in the ethidium bromide-stained gel (A, lane 2) was performed with 5 U of DNA polymerase, whereas for the autoradiographic detection (B, lane 2), only 0.1 U of GoTaq DNA polymerase was used in the assay, in order to achieve a signal intensity similar to that of the REPpAL1-catalyzed elongation reaction (lane 3). The assay mixtures were incubated for 1 h at 22°C. Lane M, dsDNA marker.
DISCUSSION
With a combination of DNA topoisomerase, DNA helicase, and protein-primed, as well as DNA-primed, DNA polymerase activities, REPpAL1 is a novel type of replicative enzyme. It is obvious that more detailed biochemical studies must be performed to quantitatively characterize its function. Nevertheless, the present data show that the protein in principle provides all the catalytic activities necessary for protein-primed synthesis of ssDNA at a parental duplex DNA. However, template-dependent REPpAL1-catalyzed deoxynucleotidylation of TPpAL1, as well as amplification of dsDNA with TPpAL1 as a primer, required very long incubation times (Fig. 5), suggesting that the protein-priming step proceeded very inefficiently. Moreover, our attempts to amplify long (several-kbp) DNA templates, using REPpAL1 as the only replicative enzyme and either TPpAL1 or DNA oligomers as the primer, have failed (data not shown), indicating low processivity of the DNA polymerase under the conditions used. Even though we cannot exclude the possibility that the MBP tags fused to both TPpAL1 and REPpAL1 impede the deoxynucleotidylation and polymerization steps, it is conceivable that in the in vivo situation, accessory protein factors contribute to efficient protein priming and improve the processivity of the DNA polymerase. Accessory proteins are common components of bacterial replisomes and of replicative complexes of phage genomes (21). Remarkably, in vitro replication of full-length (19,285-bp) TP-linked φ29 DNA by the strand displacement mechanism could be accomplished with only the presence of TP and φ29 DNA polymerase (5), but highly efficient DNA amplification also required the dsDNA-binding phage protein p6, which recognizes the φ29 DNA ends and forms a nucleoprotein complex, and the ssDNA-binding (SSB) protein p5 (6). Protein-protein- and protein-DNA interaction studies will be required to identify all the components of a possible replicative protein complex on pAL1.
Another key question concerns the mode of replication of pAL1 in vivo. REPpAL1, presumably in complex with accessory proteins, might catalyze protein-primed replication of full-length pAL1-DNA initiated at the telomeres. Alternatively, if replication of pAL1 is initiated from an internal origin, REPpAL1 might be part of a telomere complex that binds to 3′ overhangs of replication intermediates, unwinds secondary structures of the overhangs, and fills in the recessed 5′ ends.
The DNA region representing an origin of replication may be predicted by analysis of the DNA strand compositional asymmetry. In most bidirectionally replicated genomes, origins of replication are represented by a minimum in the cumulative GC skew diagram (18). For pAL1, the cumulative GC skew plot exhibits three local minima at about 32 kbp, 58 to 61 kbp, and 73 kbp; however, the global minimum actually localizes at the “right” end of pAL1 (see Fig. S1 in the supplemental material).
In a number of studies on actinomycetal linear plasmids, their replication regions containing the internal origin were identified by cloning DNA fragments of the linear plasmid into a vector that was unable to replicate in actinomycetes. Inserts comprising the replication region conferred on the recombinant plasmid the ability to replicate autonomously in circular form in the actinomycete (for examples, see references 23, 38, and 57). In an analogous approach, we inserted fragments of pAL1 into a pUC18-based vector containing the cmx gene, encoding a chloramphenicol efflux protein, as a selectable marker. Restriction fragments of total pAL1 DNA, as well as PCR-generated fragments in sizes of 5 to 10 kbp, which cover overlapping internal parts of pAL1, including the regions which in the cumulative GC-skew diagram shows local minima, were used to generate plasmid libraries. However, transformation of a pAL1-deficient mutant of A. nitroguajacolicus Rü61a failed to yield plasmid-containing clones; the occasional chloramphenicol-resistant transformants all contained ectopic insertions of plasmid DNA into the genome (data not shown). Since our present attempts to isolate an internal region required for replication failed, either the relevant fragment was not part of the libraries tested or loci necessary for replication are dispersed on pAL1. Alternatively, replication of pAL1 might indeed be initiated at the telomere. More detailed studies will be required to elucidate the mode of replication of the linear Arthrobacter plasmid pAL1.
Supplementary Material
Acknowledgments
We thank R. Brandsch (University of Freiburg, Freiburg, Germany) for kindly providing the vector pART2, A. Steinbüchel (Münster, Germany) for access to the phosphorimager, Katja Parschat and Heiko Niewerth for construction of pART2-ORF102, and Katja Parschat for experiments aimed at the identification of the replication region of pAL1 and for discussions and helpful comments on the manuscript. We also thank Almut Kappius for excellent technical assistance.
This work was supported by grants FE 383/11-1 and 11-2 from the Deutsche Forschungsgemeinschaft (DFG) to S.F. and by a 6-month research grant from the University of Münster to S.K.
Footnotes
Published ahead of print on 30 July 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aravind, L., D. D. Leipe, and E. V. Koonin. 1998. Toprim: a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 26:4205-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bao, K., and S. N. Cohen. 2001. Terminal proteins essential for the replication of linear plasmids and chromosomes in Streptomyces. Genes Dev. 15:1518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bao, K., and S. N. Cohen. 2003. Recruitment of terminal protein to the ends of Streptomyces linear plasmids and chromosomes by a novel telomere-binding protein essential for linear DNA replication. Genes Dev. 17:774-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianco, P. R., L. R. Brewer, M. Corzett, R. Balhorn, Y. Yeh, S. C. Kowalczykowski, and R. J. Baskin. 2001. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature 409:374-378. [DOI] [PubMed] [Google Scholar]
- 5.Blanco, L., A. Bernad, J. M. Lázaro, G. Martín, C. Garmendia, and M. Salas. 1989. Highly efficient DNA synthesis by the phage φ29 DNA polymerase. Symmetrical mode of DNA replication. J. Biol. Chem. 264:8935-8940. [PubMed] [Google Scholar]
- 6.Blanco, L., J. M. Lázaro, M. De Vega, A. Bonnin, and M. Salas. 1994. Terminal protein-primed DNA amplification. Proc. Natl. Acad. Sci. U. S. A. 91:12198-12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanco, L., and M. Salas. 1996. Relating structure to function in φ29 DNA polymerase. J. Biol. Chem. 271:8509-8512. [DOI] [PubMed] [Google Scholar]
- 8.Caruthers, J. M., and D. B. McKay. 2002. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12:123-133. [DOI] [PubMed] [Google Scholar]
- 9.Chang, P.-C., and S. N. Cohen. 1994. Bidirectional replication from an internal origin in a linear Streptomyces plasmid. Science 265:952-954. [DOI] [PubMed] [Google Scholar]
- 10.Choi, J. K., K. H. Tak, L. T. Jin, S. Y. Hwang, T. I. Kwon, and G. S. Yoo. 2002. Background-free fast protein staining in sodium dodecyl sulfate polyacrylamide gel using counterion dyes, zincon and ethyl violet. Electrophoresis 23:4053-4059. [DOI] [PubMed] [Google Scholar]
- 11.Dexheimer, T. S., and Y. Pommier. 2008. DNA cleavage assay for the identification of topoisomerase I inhibitors. Nat. Protoc. 3:1736-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dillingham, M. S., M. R. Webb, and S. C. Kowalczykowski. 2005. Bipolar DNA translocation contributes to highly processive DNA unwinding by RecBCD enzyme. J. Biol. Chem. 280:37069-37077. [DOI] [PubMed] [Google Scholar]
- 13.Dong, F., S. E. Weitzel, and P. H. von Hippel. 1996. A coupled complex of T4 DNA replication helicase (gp41) and polymerase (gp43) can perform rapid and processive DNA strand-displacement synthesis. Proc. Natl. Acad. Sci. U. S. A. 93:14445-14461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dufour, E., J. Méndez, J. M. Lázaro, M. de Vega, L. Blanco, and M. Salas. 2000. An aspartic acid residue in TPR-1, a specific region of protein-priming DNA polymerases, is required for the functional interaction with primer terminal protein. J. Biol. Chem. 304:289-300. [DOI] [PubMed] [Google Scholar]
- 15.Dufour, E., I. Rodríguez, J. M. Lázaro, M. de Vega, and M. Salas. 2003. A conserved insertion in protein-primed DNA polymerases is involved in primer terminus stabilisation. J. Mol. Biol. 331:781-794. [DOI] [PubMed] [Google Scholar]
- 16.Eggleston, A. K., N. A. Rahim, and S. C. Kowalczykowski. 1996. A helicase assay based on the displacement of fluorescent, nucleic-acid binding ligands. Nucleic Acids Res. 24:1179-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gartemann, K. H., and R. Eichenlaub. 2001. Isolation and characterization of IS1409, an insertion element of 4-chlorobenzoate-degrading Arthrobacter sp. strain TM1, and development of a system for transposon mutagenesis. J. Bacteriol. 183:3729-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grigoriev, A. 1998. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 26:2286-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grishin, N. V. 2000. C-terminal domains of Escherichia coli topoisomerase I belong to the zinc-ribbon superfamily. J. Mol. Biol. 299:1165-1177. [DOI] [PubMed] [Google Scholar]
- 20.Hall, M. C., and S. W. Matson. 1999. Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol. 34:867-877. [DOI] [PubMed] [Google Scholar]
- 21.Hamdan, S. M., and C. C. Richardson. 2009. Motors, switches, and contacts in the replisome. Annu. Rev. Biochem. 78:205-243. [DOI] [PubMed] [Google Scholar]
- 22.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 23.Hiratsu, K., S. Mochizuki, and H. Kinashi. 2000. Cloning and analysis of the replication origin and the telomeres of the large linear plasmid pSLA2-L in Streptomyces rochei. Mol. Gen. Genet. 263:1015-1021. [DOI] [PubMed] [Google Scholar]
- 24.Huang, C.-H., H.-H. Tsai, Y.-G. Tsay, Y.-N. Chien, S.-L. Wang, M.-Y. Cheng, C.-H. Ke, and C. W. Chen. 2007. The telomere system of the Streptomyces linear plasmid SCP1 represents a novel class. Mol. Microbiol. 63:1710-1718. [DOI] [PubMed] [Google Scholar]
- 25.Illana, B., L. Blanco, and M. Salas. 1996. Functional characterization of the genes coding for the terminal protein and DNA polymerase from bacteriophage GA-1. Evidence for a sliding-back mechanism during protein-primed GA-1 DNA replication. J. Mol. Biol. 264:453-464. [DOI] [PubMed] [Google Scholar]
- 26.Kamtekar, S., A. J. Berman, J. Wang, J. M. Lázaro, M. de Vega, L. Blanco, M. Salas, and T. A. Steitz. 2004. Insights into strand displacement and processivity from the crystal structure of the protein-primed DNA polymerase of bacteriophage φ29. Mol. Cell 16:609-618. [DOI] [PubMed] [Google Scholar]
- 27.Kamtekar, S., J. Méndez, J. M. Lázaro, M. de Vega, L. Blanco, and M. Salas. 2006. The φ29 DNA polymerase:protein-primer structure suggests a model for the initiation to elongation transition. EMBO J. 25:1335-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, S., H. G. Dallmann, C. S. McHenry, and K. J. Marians. 1996. Coupling of a replicative polymerase and helicase: a τ-DnaB interaction mediates rapid replication fork movement. Cell 84:643-650. [DOI] [PubMed] [Google Scholar]
- 29.Klassen, R., and F. Meinhardt. 2007. Linear protein-primed replicating plasmids in eukaryotic microbes, p. 187-226. In F. Meinhardt and R. Klassen (ed.), Microbial linear plasmids. Microbiology monograph 7. Springer, Heidelberg, Germany.
- 30.Kolkenbrock, S., and S. Fetzner. 2010. Identification and in vitro deoxynucleotidylation of the terminal protein of the linear plasmid pAL1 of Arthrobacter nitroguajacolicus Rü61a. FEMS Microbiol. Lett. 304:169-176. [DOI] [PubMed] [Google Scholar]
- 31.Lohman, T. M., and K. P. Bjornson. 1996. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 65:169-214. [DOI] [PubMed] [Google Scholar]
- 32.Martín, A. C., L. Blanco, P. García, and M. Salas. 1996. In vitro initiation of pneumococcal phage Cp-1 DNA replication occurs at the third 3′ nucleotide of the linear template: a stepwise sliding-back mechanism. J. Mol. Biol. 260:369-377. [DOI] [PubMed] [Google Scholar]
- 33.Méndez, J., L. Blanco, J. A. Esteban, A. Bernad, and M. Salas. 1992. Initiation of φ29 DNA replication occurs at the second 3′-nucleotide of the linear template: a sliding-back mechanism for protein-primed DNA replication. Proc. Natl. Acad. Sci. U. S. A. 89:9579-9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Overhage, J., S. Sielker, S. Homburg, K. Parschat, and S. Fetzner. 2005. Identification of large linear plasmids in Arthrobacter spp. encoding the degradation of quinaldine to anthranilate. Microbiology 151:491-500. [DOI] [PubMed] [Google Scholar]
- 35.Parschat, K., B. Hauer, R. Kappl, R. Kraft, J. Hüttermann, and S. Fetzner. 2003. Gene cluster of Arthrobacter ilicis Rü61a involved in the degradation of quinaldine to anthranilate. J. Biol. Chem. 278:27483-27494. [DOI] [PubMed] [Google Scholar]
- 36.Parschat, K., J. Overhage, A. W. Strittmatter, A. Henne, G. Gottschalk, and S. Fetzner. 2007. Complete nucleotide sequence of the 113-kilobase linear catabolic plasmid pAL1 of Arthrobacter nitroguajacolicus Rü61a and transcriptional analysis of genes involved in quinaldine degradation. J. Bacteriol. 189:3855-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pérez-Arnaiz, P., E. Longás, L. Villar, J. M. Lázaro, M. Salas, and M. de Vega. 2007. Involvement of phage φ29 DNA polymerase and terminal protein subdomains in conferring specificity during initiation of protein-primed DNA replication. Nucleic Acids Res. 35:7061-7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Picardeau, M., C. Le Dantec, and V. Vincent. 2000. Analysis of the internal replication region of a mycobacterial linear plasmid. Microbiology 146:305-313. [DOI] [PubMed] [Google Scholar]
- 39.Rainey, F. A., N. Ward-Rainey, R. M. Kroppenstedt, and E. Stackebrandt. 1996. The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: proposal of Nocardiopsaceae fam. nov. Int. J. Syst. Bacteriol. 46:1088-1092. [DOI] [PubMed] [Google Scholar]
- 40.Roche Molecular Biochemicals. 1995. The DIG system user's guide for filter hybridization. Boehringer Mannheim GmbH, Mannheim, Germany.
- 41.Roman, L. J., and S. C. Kowalczykowski. 1989. Characterization of the helicase activity of the Escherichia coli RecBCD enzyme using a novel helicase assay. Biochemistry 28:2863-2873. [DOI] [PubMed] [Google Scholar]
- 42.Salas, M. 1991. Protein-priming of DNA replication. Annu. Rev. Biochem. 60:39-71. [DOI] [PubMed] [Google Scholar]
- 43.Salas, M., L. Blanco, J. M. Lázaro, and M. de Vega. 2008. The bacteriophage φ29 DNA polymerase. IUBMB Life 60:82-85. [DOI] [PubMed] [Google Scholar]
- 44.Salas, M., R. Freire, M. S. Soengas, J. A. Esteban, J. Méndez, A. Bravo, M. Serrano, M. A. Blasco, J. M. Lázaro, L. Blanco, C. Gutiérrez, and J. M. Hermoso. 1995. Protein-nucleic acid interactions in bacteriophage φ29 DNA replication. FEMS Microbiol. Rev. 17:73-82. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 46.Singleton, M. R., and D. B. Wigley. 2002. Modularity and specialization in superfamily 1 and 2 helicases. J. Bacteriol. 184:1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sissi, C., and M. Palumbo. 2009. Effects of magnesium and related divalent metal ions in topoisomerase structure and function. Nucleic Acids Res. 37:702-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 49.Stauber, E. J., A. Busch, B. Naumann, A. Svatos, and M. Hippler. 2009. Proteotypic profiling of LHCI from Chlamydomonas reinhardtii provides new insights into structure and function of the complex. Proteomics 9:398-408. [DOI] [PubMed] [Google Scholar]
- 50.Stauber, E. J., A. Fink, C. Markert, O. Kruse, U. Johanningmeier, and M. Hippler. 2003. Proteomics of Chlamydomonas reinhardtii light-harvesting proteins. Eukaryot. Cell 2:978-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steitz, T. A. 1999. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 274:17395-17398. [DOI] [PubMed] [Google Scholar]
- 52.Stoll, A., M. Redenbach, and J. Cullum. 2007. Identification of essential genes for linear replication of an SCP1 composite plasmid. FEMS Microbiol. Lett. 270:146-154. [DOI] [PubMed] [Google Scholar]
- 53.Studier, F. W. 2005. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 41:207-234. [DOI] [PubMed] [Google Scholar]
- 54.Suzuki, H., K. Marushima, Y. Ohnishi, and S. Horinouchi. 2008. A novel pair of terminal protein and telomere-associated protein for replication of the linear chromosome of Streptomyces griseus IFO13350. Biosci. Biotechnol. Biochem. 72:2973-2980. [DOI] [PubMed] [Google Scholar]
- 55.Trego, K. S., and D. S. Parris. 2003. Functional interaction between the herpes simplex virus type 1 polymerase processivity factor and origin-binding proteins: Enhancement of UL9 helicase activity. J. Virol. 77:12646-12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warren, R., W. W. L. Hsiao, H. Kudo, M. Myhre, M. Dosanjh, A. Petrescu, H. Kobayashi, S. Shimizu, K. Miyauchi, E. Masai, G. Yang, J. M. Stott, J. E. Schein, H. Shin, J. Khattra, D. Smailus, Y. S. Butterfield, A. Siddiqui, R. Holt, M. A. Marra, S. J. M. Jones, W. W. Mohn, F. S. L. Brinkman, M. Fukuda, J. Davies, and L. D. Eltis. 2004. Functional characterization of a catabolic plasmid from polychlorinated-biphenyl-degrading Rhodococcus sp. strain RHA1. J. Bacteriol. 186:7783-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang, C.-C., Y.-H. Chen, H.-H. Tsai, C.-H. Huang, T.-W. Huang, and C. W. Chen. 2006. In vitro deoxynucleotidylation of the terminal protein of Streptomyces linear chromosomes. Appl. Environ. Microbiol. 72:7959-7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, R., Y. Yang, P. Fang, C. Jiang, L. Xu, Y. Zhu, M. Shen, H. Xia, J. Zhao, T. Chen, and Z. Qin. 2006. Diversity of telomere palindromic sequences and replication genes among Streptomyces linear plasmids. Appl. Environ. Microbiol. 72:5728-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, R., H. Xia, P. Guo, and Z. Qin. 2009. Variation in the replication loci of Streptomyces linear plasmids. FEMS Microbiol. Lett. 290:209-216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.