Abstract
The delta subunit of RNA polymerase, RpoE, is widespread in low-G+C Gram-positive bacteria and is thought to play a role in enhancing transcriptional specificity by blocking RNA polymerase binding at weak promoter sites and stimulating RNA synthesis by accelerating core enzyme recycling. Despite the well-studied biochemical properties of RpoE, a role for this protein in vivo has not been defined in depth. In this study, we show that inactivation of rpoE in the human dental caries pathogen Streptococcus mutans causes impaired growth and loss of important virulence traits, including biofilm formation, resistance to antibiotics, and tolerance to environmental stresses. Complementation of the mutant with rpoE expressed in trans restored its phenotype to wild type. The luciferase fusion reporter showed that rpoE was highly transcribed throughout growth and that acid and hydrogen peroxide stresses repressed rpoE expression. Transcriptome profiling of wild-type and ΔrpoE cells in the exponential and early stationary phase of growth, under acid and hydrogen peroxide stress and under both stresses combined, revealed that genes involved in histidine synthesis, malolactic fermentation, biofilm formation, and antibiotic resistance were downregulated in the ΔrpoE mutant under all conditions. Moreover, the loss of RpoE resulted in dramatic changes in transport and metabolism of carbohydrates and amino acids. Interestingly, differential expression, mostly upregulation, of 330 noncoding regions was found. In conclusion, this study demonstrates that RpoE is an important global modulator of gene expression in S. mutans which is required for optimal growth and environmental adaptation.
RNA polymerase from bacteria consists of the core (α2ββ′ω) enzyme and the σ factor, the main target for gene regulation. In Gram-negative bacteria, e.g., Escherichia coli, the σ subunit is required for specific promoter recognition and transcription initiation and also for reducing unspecific binding (16). In Gram-positive bacteria, there is an additional delta (δ) subunit, RpoE, required for specificity of the enzyme. In Bacillus subtilis, promoter recognition and transcription initiation are determined by the σ subunit, but the dissociation of RNA polymerase from weak promoter sites is mediated by RpoE, which was demonstrated by biochemical studies (2, 3, 31). Sequence alignment suggests that the σA and RpoE subunits from B. subtilis may together contain the activities found in the single 70-kDa σ factor of E. coli (30, 40).
RpoE is suggested to be an abundant protein in B. subtilis; the expression of rpoE reaches a maximum value at the transition between logarithmic and stationary phases, and then the activity goes down at the stationary phase of growth (41). The rpoE mutant of B. subtilis exhibited an extended lag phase and altered morphology (41). Recent studies in Streptococcus agalactiae demonstrated a link between RpoE amount and virulence, and reduced virulence was found in the rpoE mutant (30, 55). However, no further molecular mechanisms of RpoE function in vivo with regard to these phenotypic changes have been reported.
The Gram-positive bacterium Streptococcus mutans is a major pathogen responsible for human dental caries. Its ability to form a biofilm on the tooth surface, conversion of various carbohydrates to organic acids through glycolysis, and tolerance of environmental stresses are important properties for the progression of the disease and are considered virulence traits (14, 37). In the oral environment S. mutans is exposed to rapid variations in sugar sources and concentrations and to quick changes in environmental pH and redox potential (oxidation level). All these environmental alterations require a complex and sophisticated regulation of gene expression to allow the bacterium to adapt to the changing environment and to maintain basic metabolic processes necessary for survival. The molecular mechanisms of the adaptation of S. mutans to various environmental stresses have been studied. Svensäter et al. (59) demonstrated that adaptation to acid, salt, and starvation stresses protected the cells against subsequent acid challenge at pH 3.5. The authors further analyzed the stress responses by a proteome approach and found a significant number of protein spots with altered expression under different stresses. However, these protein spots were not finally identified. It has been reported that nearly 14% of the genes in the genome were differentially expressed at acidic pH (22). Two-component systems (TCSs), comprised of a sensor kinase and a response regulator protein, are one of the primary mechanisms used by S. mutans to quickly sense and respond to environmental acid fluctuations for optimal growth (14, 22, 56, 65). S. mutans has well-adapted acid defense systems, including the F1Fo-ATPases, which pump protons out of the cell, and the malolactic fermentation (MLF) and agmatine deiminase systems, which contribute to alkalization of the cytoplasmic pH and generation of ATP as energy (37). Because it lacks catalase (the heme-containing peroxidase), which degrades hydrogen peroxide (H2O2), S. mutans depends mainly on the NADH oxidase, superoxide dismutase, and glutathione reductase activities for protection against reactive oxygen species (13, 27). Both transcriptomic (4) and phenotypic (6) studies proved that oxygen not only caused altered sugar transport activity and rate of glycolysis but also affected virulence-related traits, particularly by reducing biofilm formation.
The delta subunit RpoE of S. mutans is predicted to be a 194-amino-acid protein with a pI of 3.7 and a molecular mass of 22 kDa. The amino acid sequence alignment of S. mutans RpoE with those of other organisms shows considerable homology (see Fig. S1 in the supplemental material). The highest homology in the proteins is seen in the amino-terminal domain, which represents the core RNA polymerase binding region (40). The carboxy-terminal domain of RpoE is highly acidic because of the enriched aspartate (D) (n = 25) and glutamate (E) residues (n = 22), which is consistent with the expected function as an unstructured polyanionic polymer that mimics and displaces nucleic acids from RNA polymerase (40).
To enhance our understanding of how the changes in RNA polymerase activity might affect S. mutans biological functions on the cellular and molecular levels, we constructed an S. mutans rpoE knockout mutant and characterized its phenotype under normal and stressed conditions. The effect of these conditions on the expression of rpoE was studied using a luciferase reporter strain. We then conducted a transcriptome analysis under a range of conditions, which revealed significant and coordinated changes of gene expression in the mutant, clearly accounting for its phenotype. The data show that RpoE functions as a global modulator of gene expression in S. mutans, affecting basic virulence-related traits.
MATERIALS AND METHODS
Bacterial strains, plasmids and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown in Luria-Bertani medium (LB) (Carl-Roth, Germany) at 37°C. Streptococcus mutans UA159 strains were grown in Todd-Hewitt broth (Becton Dickinson) supplemented with 1% yeast extract (THBY) at 37°C aerobically (5% CO2 enriched) or in BM medium (39) containing 0.5% (wt/vol) glucose or sucrose under anaerobic conditions (80% N2, 10% H2, 10% CO2). For stress tolerance assays, S. mutans strains were grown in THBY supplied with 75 mM phosphate buffer. Erythromycin was included where indicated at final concentrations of 200 μg/ml for E. coli and 10 μg/ml for S. mutans, and spectinomycin was included at 50 μg/ml for E. coli and 300 μg/ml for S. mutans.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| E. coli strain Top10F′ | Strain for routine cloning work | Invitrogen |
| S. mutans strains | ||
| UA159 | Wild type; Erms Sps | ATCC 700610 |
| ΔrpoE mutant | UA159 with deletion of rpoE gene; Ermr | This study |
| ΔrpoEcomp mutant | Complementation of the rpoE mutation; Ermr Spr | This study |
| WT-luc | rpoE reporter strain carrying pFW-PrpoE-luc in wild-type background; Spr | This study |
| Plasmids | ||
| pCR2.1-TOPO | E. coli cloning vector | Invitrogen |
| pCR2.1-ΔrpoE | E. coli cloning vector containing the rpoE upstream fragment, Ermr gene, and rpoE downstream fragment | This study |
| pDL278 | E. coli-streptococcal shuttle vector; Spr | 20 |
| pDL-rpoE | pDL278 harboring the promoter region and coding sequence of rpoE; Spr | This study |
| pFW5-luc | Streptococcal suicide vector containing promoterless firefly luciferase gene; derivative of pFW5; Spr | 36 |
| pFW-PrpoE-luc | pFW5-luc harboring the promoter region of rpoE fused with luciferase gene; Spr | This study |
Ermr, erythromycin resistance; Spr, spectinomycin resistance; luc, luciferase.
Construction of an S. mutans ΔrpoE strain.
PCR ligation mutagenesis (35) was used for gene knockout in the S. mutans wild-type chromosome. However, the rpoE knockout mutant was not obtained, presumably due to the importance of this gene. To increase the efficiency of recombination with a higher concentration of the whole DNA construct, the rpoE upstream fragment, the Ermr gene, and the rpoE downstream fragment were amplified separately using primer pairs RP1F/PR2R-AscI, ErmF-AscI/ErmR-FseI, and RP3F-FseI/RP4R (Table 2), and then these three fragments were cloned step by step into the vector pCR2.1-TOPO to construct pCR2.1-ΔrpoE. The resulting plasmid was transformed into E. coli Top10F′ cells. Positive clones were confirmed by PCR, sequencing, and restriction enzyme digestion. The insertion with the complete rpoE up-Ermr-rpoE down sequence was released from pCR2.1-ΔrpoE by HindIII (NEB) and XhoI (NEB) digestion and then was transformed into S. mutans to generate ΔrpoE mutants through double homologous recombination. The deletion of rpoE was confirmed by PCR and sequencing.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ → 3′)a | Description |
|---|---|---|
| RP1F | CGAGCTCTTCGGTAGAATCGG | rpoE upstream |
| PR2R-AscI | GGCGCGCCTTCACGAATGTCTGCATCTG | rpoE upstream |
| RP3F-FseI | GGCCGGCCATTCCAGATGAAGATTTAGAC | rpoE downstream |
| RP4R | GCTTCTGGAGCAACATCACA | rpoE downstream |
| ErmF-AscI | GGCGCGCCCCGGGCCCAAAATTTGTTTGAT | Ermr gene |
| ErmR-FseI | GGCCGGCCAGTCGGCAGCGACTCATAGAAT | Ermr gene |
| CPF-BamHI | TTCAGGATCCGATAAAAAGCAGCGTTCATA | rpoE complementation |
| CPR-HindIII | AGTCAAGCTTTTATTCTCCTTCTTCATCTTCC | rpoE complementation |
| RL5F-NcoI | TAGGCCATGGATAAAAAGCAGCGTTCATAT | rpoE reporter |
| RL5R-NcoI | CGTTCCATGGTGGTTCTCCTTATCATAATC | rpoE reporter |
| hisCF | CCCCTACTGGCATTTACAAG | QPCR, hisC |
| hisCR | CGATTGACCGCCAAAACTGA | QPCR, hisC |
| fruCF | TTGTTGCTGGTGGTGTTTTGG | QPCR, fruC |
| fruCR | GAGTCATAACGCCCATCGCA | QPCR, fruC |
| levDF | CAGTAGATGATTTTGCGGAAAC | QPCR, levD |
| levDR | GCTGTTGTCAAAGGGCTTCC | QPCR, levD |
| msmEF | GTTTGCTTTAGCGGGAACAG | QPCR, msmE |
| msmER | AATCGAATTTGGTTGGGAAAAG | QPCR, msmE |
| smu322F | TGCCTGTTAAGGTGATTATTGG | QPCR, SMU.322 |
| smu322R | ATAAAGAAGCCTAGACTTGTCC | QPCR, SMU.322 |
Restrictions sites are underlined.
Complementation of the rpoE mutation in trans.
To complement the rpoE mutation, a fragment of 1.2 kb containing the entire rpoE coding sequence plus the promoter region was amplified by PCR using primers CPF-BamHI and CPR-HindIII from the chromosomal DNA of S. mutans and inserted into the shuttle vector pDL278. The resulting plasmid, pDL-rpoE, was then transformed into the ΔrpoE mutant strain to generate the complementing ΔrpoEcomp strain. The complementation of rpoE was confirmed by PCR and sequencing.
Antibiotic resistance.
Overnight cultures of S. mutans strains were 1:10 diluted in fresh THBY and grown to an optical density at 600 nm (OD600) of ∼0.5; 0.2% of the log-phase cells were then added to fresh THBY medium which contained antibiotics at different concentrations. After 16 h of incubation at 37°C aerobically, visible growth of bacteria was determined by turbidity by eye. The antibiotics included kanamycin (100 mg/ml in H2O), tetracycline (25 mg/ml in 70% ethanol), ampicillin (100 mg/ml in H2O), and rifampin (25 mg/ml in methanol) at final concentrations of 0.1, 1, 10, 100, and 500 μg/ml. A second experiment was carried out with a smaller concentration range to determine the exact growth-inhibitory concentration.
Acid and H2O2 stress killing assays.
The ability of S. mutans strains to survive acidic and oxidative stress challenge was determined as described by Wen et al. (69) with some modifications. Briefly, overnight cultures of the wild-type, ΔrpoE, and ΔrpoEcomp strains were diluted 20-fold in fresh THBY and grown to mid-exponential phase (OD600 of ∼0.5) in THBY at pH 7.5, and two aliquots (1 ml) of each cell suspension were pelleted by centrifugation (12,000 × g, 30 s). One aliquot of the culture was used to assay the survival of “unadapted” cells by directly resuspending them in THBY with the challenge stresses pH 3.0 or pH 7.5 plus 20 mM H2O2. Another aliquot of each culture was resuspended in the same volume of THBY buffered at pH 5.0 or pH 7.5 plus 2 mM H2O2 and incubated for 2 h to induce an adaptation response. The resulting “adapted” cells were then exposed to the challenging stresses of pH 3.0 or pH 7.5 plus 20 mM H2O2. After incubation for 30 min, cells were collected by centrifugation (12,000 × g, 30 s) and serially diluted with fresh THBY. Aliquots of 100 μl of appropriate dilutions of each strain were spread in four to eight replicates onto THBY agar plates. The survival rate was expressed as the percentage of cells surviving the challenge for 30 min compared to the total number of viable cells before treatment.
Biofilm structure under CLSM.
The ability of S. mutans strains to form stable biofilms was assessed by growing the cells in 96-well flat-bottom black microtiter plates (Corning, Germany) containing THBY or BM medium supplemented with 0.5% (wt/vol) sucrose under anaerobic conditions and grown for 16 h. Precultures were grown at 37°C aerobically for 13 to 15 h to reach stationary phase. The precultures were then diluted 1:10 in fresh THBY and grown until log phase (OD600 of ∼0.5). The cells were collected by centrifugation (12,000 × g, 30 s) and resuspended in fresh medium to reach an OD600 of ∼0.01. Aliquots of 200 μl of the cell suspension were inoculated into 12 parallel wells of the microtiter plates and grown under anaerobic conditions. Several plates were prepared to quantify the biofilms at different time points. After growth, the culture medium was withdrawn, and the plates were rinsed with 0.85% NaCl to remove the planktonic and loosely bound cells. The biofilms were stained with 50 μl Live/Dead BacLight stain (L7012; Invitrogen) working solution, and the biofilm architecture was recorded using a FluoView FV1000 confocal laser scanning microscope (CLSM) (Olympus, Germany). In each experiment, the exciting laser intensity, background level, and contrast were maintained at the same level. For each sample, two replicas and at least three random fields per replica were analyzed. Using a 40× objective lens, a series of optical cross-section images were acquired at 2-μm-depth intervals from the surface, through the vertical axis of the specimen. Three-dimensional confocal images were reconstituted by the software Imaris (BitPlane AG, Switzerland). For biofilm quantification, the plates with biofilms were rinsed three times with deionized water. The biofilms were stained with 50 μl of 0.1% crystal violet and incubated at room temperature for 15 min. After rinsing two times with deionized water, the bound dye was extracted from the stained cells by adding 200 μl of 99% ethanol and shaking overnight at room temperature to facilitate full release of the dye. The biomass was quantified by measuring the absorbance of the dye solution at 620 nm in a Wallac Victor instrument (Perkin-Elmer, Germany). Each experiment was carried out at least three times.
Construction of a rpoE-luc reporter strain.
To construct a luciferase transcriptional reporter fusion to the rpoE promoter, the promoter region of rpoE (0.6 kb) was amplified by PCR using primers RL5F-NcoI and RL5R-NcoI from the chromosomal DNA of strain UA159. The promoter region was then cloned into pFW5-luc to generate the plasmid pFW-PrpoE-luc. The resulting construct was confirmed by restriction analysis, PCR, and sequencing. The confirmed plasmid was integrated into the chromosome of S. mutans via single-crossover homologous recombination to generate the reporter strain WT-luc, with the promoter region followed by the luciferase gene and the rest of the plasmid inserted before the intact rpoE promoter and coding sequence. The positive clones were confirmed by PCR, sequencing, and luciferase assay.
Analysis of rpoE gene expression using the luciferase reporter assay.
Samples from each time point were collected and treated with chloramphenicol to reduce the intracellular enzymatic activity, and the OD600 was measured. Following treatment with chloramphenicol (50 μg/ml) and short cooling on ice, the samples were washed and resuspended in the same volume of 20 mM Tricine buffer (pH 7.8). One hundred microliters of each sample was mixed with 3× assay buffer (75 mM Tricine, 15 mM MgSO4, 1.5 mM dithiothreitol [DTT], 900 μM ATP, 3 mg/ml bovine serum albumin [BSA], 2% wt/vol d-glucose, pH 7.8) and incubated at room temperature for 20 min before injection of 100 μl d-luciferin (120 μM dissolved in 20 mM Tricine, pH 7.8). d-Luciferin (Carl-Roth, Germany) stock solution (1 mg/ml) was dissolved in 20 mM Tricine, aliquoted, and stored at −70°C. Luminescence was recorded with a Wallac Victor2 instrument and normalized against the OD600 to calculate the relative light units (RLU). All measurements were done in duplicate in at least two independent experiments.
Microarray design.
A customized whole-genome microarray containing all open reading frames (ORFs) as well as all noncoding regions of Streptococcus mutans UA159 (gb|AE014133.1) was used in this study. For microarray design, the eArray platform by Agilent was utilized (Agilent Technologies, Santa Clara, CA). For each ORF and intergenic region, three antisense probes (60 bp in length) were designed, and each probe was in duplicate on the array. The positive and negative controls from Agilent and the spike-in control for GFPmut2 and RFPExpress were included.
Sample preparation for microarray analysis.
Overnight cultures of wild-type and ΔrpoE S. mutans were diluted 1:20 in fresh THBY medium buffered with 75 mM phosphate at pH 7.5 and grown at 37°C aerobically to an OD600 of ∼0.5. An aliquot of each log-phase cell culture was withdrawn as a control. At the same time point, the other part of the log-phase cells was collected (12,000 × g, 30 s) and incubated in buffered THBY as follows: (i) pH 7.5, (ii) pH 5.0, (iii) pH 7.5 plus 2 mM H2O2, and (iv) pH 5.0 plus 2 mM H2O2. After 2 h of treatment, samples were collected (12,000 × g, 30 s). For each sample, two biological replicas were collected. Two volumes of RNA protect (Qiagen) were immediately added to 1 volume of each sample and then mixed by vortexing for 5 s and incubated at room temperature for 5 min. Pellets were collected by centrifugation (5,000 × g, 5 min) and were stored at −70°C. For RNA isolation, the cell pellets were resuspended and lysed by incubation in Tris buffer (10 mM Tris and 1 mM EDTA, pH 8.0) containing 2.5 mg/ml lysozyme (Sigma, Germany) and 50 U/ml mutanolysin (Sigma) at room temperature for 45 min, followed by vortexing for 3 min in the presence of 50 μg sterile, acid-washed glass beads (diameter, 106 μm; Sigma) (60). RNA extraction was performed using the RNeasy mini kit (Qiagen). The integrity and quality of RNA were assessed by electrophoresis on a denaturing formaldehyde agarose gel. Traces of genomic DNA were removed by RNase-free DNase I (Qiagen) digestion (13.5 U, room temperature, 45 min), and the RNA was purified using the RNeasy mini kit (Qiagen). The concentration of RNA was measured with a Nanodrop 1000 spectrophotometer (Peqlab, Germany). The genomic DNA contamination was checked by using RNA directly as a template for PCR. Furthermore, the RNA quality was checked with a Bioanalyzer (Agilent, Germany) before labeling.
Microarray analysis.
For wild-type and mutant strains, two biological replicas and two technical replicas for dye swabs were used in a microarray study. RNA samples were labeled either with Cy3 or Cy5 using the ULS fluorescent labeling kit (Kreatech, Germany). The degree of labeling was calculated by the following formula: percent labeling = (340 × pmol dye × 100%)/(ng nucleic acid × 1,000). Nine hundred nanograms of each Cy3- and Cy5-labeled RNA was fragmented and hybridized to the microarray at 65°C for 17 h using the Agilent hybridization chamber according to the manufacturer's instructions. The arrays were scanned using the Agilent DNA microarray scanner, and the raw data were extracted using Agilent Feature Extraction software (v9.5). The data were processed by using Bioconductor packages written in R language (http://www.r-project.org/). The Linear Models for Microarray Analysis (LIMMA) package (70) was used for background correction, Lowess normalization of the two channels of one array, quantile normalization between different arrays, and identification of differentially expressed genes. Genes with a log2 fold change of >1.0 and a P value of <0.05 were selected for gene expression pattern discovery using clustering analysis from Genesis (58). KegArray (Kanehisa Laboratories; http://www.genome.jp/download/) was used for integrated analysis of gene expression profiles together with KEGG pathways. The genes were categorized into functional groups according to functional categories file NC_004350.ptt in the S. mutans clusters of orthologous groups (COGs) of proteins, which was downloaded from ftp://ftp.ncbi.nih.gov/genomes/Bacteria/Streptococcus_mutans.
QPCR.
The reverse transcription reaction, performed on the same RNA samples which were used for microarray experiments, was carried out using 1.5 μg random hexamers and 200 U SuperScript III reverse transcriptase for 2-μg RNA samples in total volume of 30 μl (Invitrogen). The genomic DNA contamination was checked by use of a control without SuperScript III reverse transcriptase. Oligonucleotides primers for quantitative PCR (QPCR) were designed using Primer3 (http://frodo.wi.mit.edu/primer3/). For each set of primer pairs, a standard amplification curve was plotted (critical threshold cycle against log of concentration), and only those primer pairs with slopes of about −3 were considered reliable. Quantitative PCR was performed using the LightCycler 480 system (Roche, Germany), and the reaction mixtures were prepared using Quantitect SYBR green PCR kit (Qiagen), including a nontemplate control. The expression of the hypothetical protein (SMU.322) was constant in the wild-type and ΔrpoE mutant strains under all tested conditions in the microarray results, and thus this gene was chosen as a reference gene for quantification. Changes in the level of gene expression were calculated using the ΔΔCT method (51). All steps were performed according to the manufacturer's protocols. All measurements were done in triplicate reactions.
Microarray data accession number.
Microarray data have been deposited at NCBI-GEO (accession no. GSE22333).
RESULTS AND DISCUSSION
The ΔrpoE mutant has a growth defect.
To determine the function of the RpoE protein in S. mutans, a deletion of rpoE was created by replacing the coding sequence with an erythromycin antibiotic resistance cassette through homologous recombination. The rpoE mutation was complemented in trans with plasmid pDL-rpoE, which contained the promoter region and the entire rpoE coding sequence.
The ΔrpoE strain showed an extended lag phase and slower growth, and it failed to reach the same final optical density as the parent strain (see Fig. S2 in the supplemental material). Moreover, the ΔrpoE strain had a strong tendency to clump when entering the late exponential phase and produced flocs which sedimented to the bottom of the culture vessel. Complementation of rpoE restored the growth phenotype of the wild-type strain with respect to growth rate, lack of clumping, and final OD600; however, the lag phase of the complemented strain was still longer than that of the wild type.
Interestingly, the ΔrpoE mutant had a cell morphology similar to that of the parent strain as determined using scanning electron microscopy (see Fig. S3 in the supplemental material). Moreover, transmission electron microscopy analysis found no obvious defect in cell division or the cell membrane of the ΔrpoE strain (data not shown). These observations indicate that the growth defect of the ΔrpoE mutant was due to other genetic and metabolic changes.
RpoE deficiency reduces resistance to antibiotics.
The ability of S. mutans to resist antibiotics, including kanamycin, tetracycline, rifampin, and ampicillin, was tested. Compared to the wild-type strain, the ΔrpoE mutant failed to grow at 1 μg/ml tetracycline and 100 μg/ml kanamycin. Thus, lower concentrations from 0.1 to 1 μg/ml tetracycline or 10 to 100 μg/ml kanamycin were tested in a second experiment. The data show that the ΔrpoE mutant was more sensitive to these two antibiotics and could not grow at above 0.6 μg/ml tetracycline or 80 μg/ml kanamycin (see Table ST1 in the supplemental material). The reduced tolerance of the mutant strain to both kanamycin and tetracycline, which interfere with translation by targeting the 30S ribosomal subunit of the ribosome (49, 62), indicated a deficiency in ribosome function. For the antibiotic rifampin, which inhibits transcription by binding to the β subunit of the RNA polymerase (21), and the β-lactam antibiotic ampicillin, which disrupts the cross-linking of peptidoglycan chains in cell wall synthesis (63), no obvious difference was found between the wild-type and ΔrpoE strains (data not shown).
RpoE is required for stress tolerance.
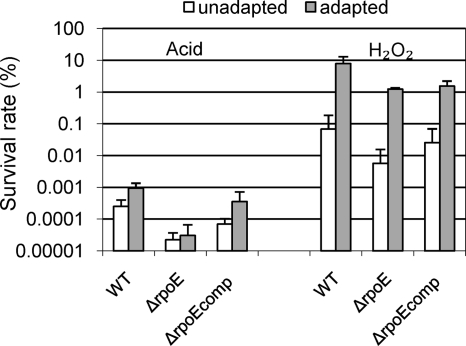
The ability of the rpoE deletion strain of S. mutans to tolerate environmental stresses, particularly acid and H2O2 stresses, was determined. Cells in the exponential phase of growth were split into two parts; one was directly subjected to the killing stress without adaptation, while the other was treated for 2 h with sublethal adaptive stress before killing stress. As shown in Fig. 1, the preadaptation did protect both wild-type and mutant strains from the more severe stress, as indicated by the higher survival rate of “adapted” cells. However, the survival rate of the ΔrpoE mutant was about 10 times lower than that of the parent strain, both with and without preadaptation. Genetic complementation could only partially restore the tolerance of the wild-type strain. One possible reason for this might be the multiple copies (∼20 to 30) of the plasmid carrying the rpoE gene in the complementation strain, which could cause global metabolic changes in the cells.
FIG. 1.
Effect of preadaptation to killing stress on the survival of S. mutans. The assay for “unadapted” cells was carried out by directly transferring mid-exponential-phase cells to challenge stresses at pH 3.0 (left) or 20 mM H2O2 (right) for 30 min. The assay for “adapted” cells was carried out by incubating mid-exponential-phase cells at pH 5.0 (left) or 2 mM H2O2 (right) for 2 h to induce an adaptation response before exposing them to the challenging stresses. Survival was determined by plating four to six replicates on THBY agar plates. The results are expressed as the percentage of the cells surviving the challenge for 30 min in comparison to the total viable cells before treatment. Data are the averages and standard deviations from three independent experiments.
Inactivation of rpoE causes alterations in biofilm structure.
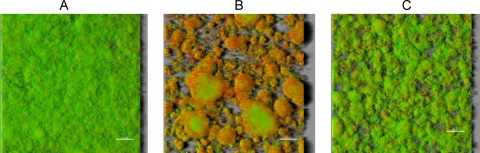
The capacity of the rpoE deletion strain of S. mutans to form biofilms in the presence of 0.5% sucrose in both rich medium (THBYS) and minimal medium (BMS) was determined. Notably, the ΔrpoE strain showed a different biofilm architecture, especially in BMS medium (Fig. 2). The biofilm of the ΔrpoE mutant was nonhomogeneous and comprised of large cell clusters. It appeared remarkably denser than the wild-type biofilm in distinct areas. Moreover, we observed strong staining of the mutant biofilm with propidium iodide (red color), indicating damaged membranes. Biomass quantification using crystal violet staining showed that the ΔrpoE mutant formed about 10% more biofilm than the wild type in THBYS medium (see Fig. S4 in the supplemental material), but there was no obvious change in biomass in BMS medium.
FIG. 2.
Structures of S. mutans biofilms. S. mutans wild-type (A), ΔrpoE (B), and ΔrpoEcomp (C) strains were grown in a 96-well microtiter plate in BMS medium for 16 h under anaerobic condition. The biofilms were stained with Live/Dead BacLight viability stain, and images of biofilms at various depths were taken by confocal laser scanning microscopy (CLSM). Three-dimensional images were reconstituted with the software Imaris. The shadows at the right sides of pictures indicate the biofilm height.
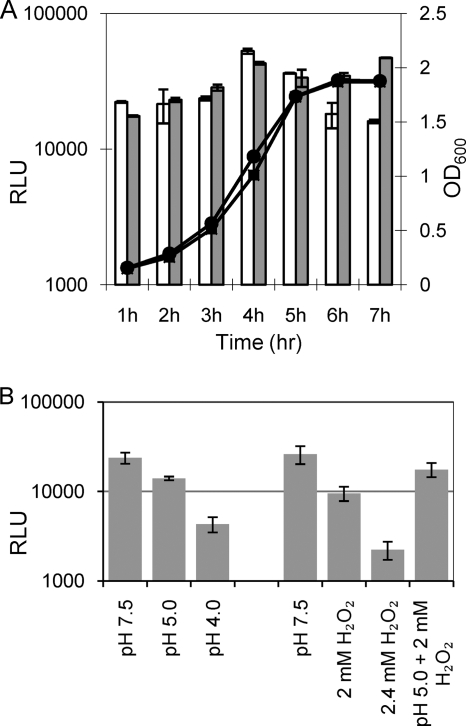
Expression of rpoE during growth and under stress.
A reporter strain, WP-luc, containing the promoter region of rpoE fused to a promoterless luciferase gene was constructed to characterize rpoE expression. The promoter of rpoE showed high activity, reaching a maximum at the late logarithmic phase of growth (Fig. 3 A). Its expression declined in the stationary phase, but only in unbuffered medium. In buffered medium, rpoE expression remained constant at the maximum value in the stationary phase (Fig. 3A). In a previous study (41), a decline in rpoE expression in the stationary growth phase was reported. Since these experiments were performed in unbuffered medium, our data suggest that low pH rather than the growth phase may have caused the decline in rpoE expression. Indeed, the pH of the S. mutans culture in the stationary phase was below 5.0 in the medium without buffering, while in a buffered medium it remained at 6.5. Furthermore, we showed that the rpoE promoter activity was repressed under acid stress. A lower promoter activity was found at pH 5.0, and it went down quickly when the pH dropped to 4.0 (Fig. 3B). The rpoE promoter activity was also very sensitive to H2O2 stress, and it went down as the concentration increased from 2.0 to 2.4 mM. Interestingly, the repression of the rpoE promoter by H2O2 was relieved when weak acid and H2O2 stress were combined.
FIG. 3.
Expression of the rpoE gene during growth and under stress. Luciferase activity was measured in the reporter strain WP-luc carrying a rpoE::luc fusion in the wild-type background. Relative light units were calculated by normalizing luminescence against optical density, indicated at the right axis (OD600). (A) Expression of the rpoE gene during growth. White bars, luciferase expression in unbuffered THBY medium; gray bars, luciferase expression in phosphate-buffered THBY medium. At least three independent experiments were carried out, and duplicate samples were measured each time. Representative results are shown. above. •, growth (OD600) of the reporter strain in unbuffered THBY; ▪, growth in buffered THBY. (B) Expression of the rpoE gene under stress. The luciferase activity was measured in log-phase WP-luc after a 2-h treatment with acid stress, H2O2 stress, or both acid and H2O2 stresses combined. Means and standard deviations are shown for at least two independent experiments; each experiment contained four replicas.
Transcriptome profiling of the ΔrpoE mutant.
The ΔrpoE mutant showed less resistance to low pH and H2O2, and the adaptation conditions (pH 5.0 or 2 mM H2O2) were shown to provide protection against stronger stresses; therefore, they were selected for microarray studies to further investigate the role of RpoE in stress adaptation. The combination of both stresses was also studied, since the reporter strain analysis (Fig. 3B) had shown that both stresses together had a less inhibitory effect on rpoE expression than H2O2 stress alone. Furthermore, cells from the logarithmic and early stationary phases of growth were studied to analyze the role of RpoE under normal growth conditions.
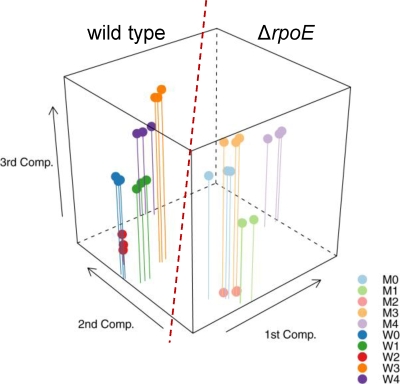
The raw data extracted from the Agilent microarray images were processed by using the Bioconductor software packages. A principal-component analysis was performed on the expression data from all samples to highlight their variability. As shown in Fig. 4, biological replicates grouped closely together. The data sets of the wild-type and ΔrpoE mutant strains were clearly separated by the first two components, explaining 48% of total variability. The data sets of combined acid and H2O2 stresses were distant from all other samples when only the first two components were plotted; major differences between the log and early stationary phases and acid or H2O2 stress became visible when the third component was also plotted. In sum, these three components explain 61.1% of the variability in gene expression between the samples. The Linear Models for Microarray Analysis (LIMMA) package was used for the identification of differentially expressed genes. Comparison of the transcription profiles of the ΔrpoE mutant and the wild type revealed a total of 550 genes that were differentially expressed (log2 fold change of ≥1.0; P < 0.05) when all experimental conditions were combined (see Fig. S5 in the supplemental material).
FIG. 4.
Principal-component analysis of microarray samples. The first three components, explaining 61.1% of total sample variability in gene expression, are plotted. The distance between samples on the plot is proportional to the variance between these samples by expression of genes clustered in the first three components. The samples of the ΔrpoE mutant are shown in a lighter color. W, wild type; M, ΔrpoE; 0, before treatment, log-phase cells; 1, early-stationary-phase cells; 2, acid-stressed cells; 3, H2O2-stressed cells; 4, acid- and H2O2-stressed cells.
Noncoding regions.
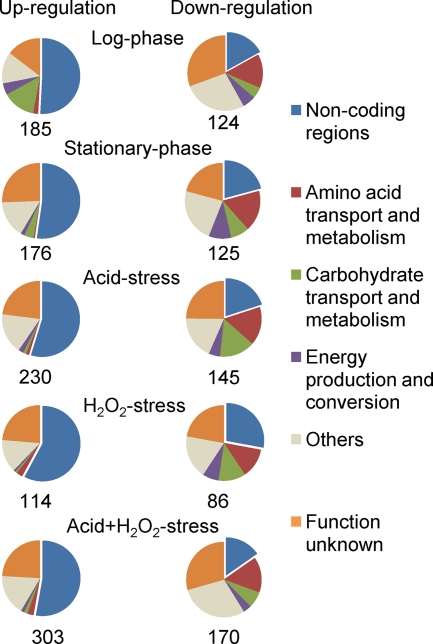
A significant number of noncoding regions were found to be differentially expressed in the mutant. Under all experimental conditions, more than 50% of the total number of upregulated transcripts were noncoding regions (Fig. 5). Among the downregulated transcripts, noncoding regions represented about 20 to 30%. Since rpoE is postulated to reduce the specificity of binding of the RNA polymerase to DNA, lack of RpoE might offer the opportunity for transcription of DNA with weak promoters, especially since the majority of differentially expressed noncoding regions were upregulated. Thus, the global changes in gene expression observed in the rpoE mutant could indirectly be related to the massive changes in expression of intergenic regions, some of which might have regulatory functions.
FIG. 5.
Differentially expressed coding and noncoding regions in the ΔrpoE mutant compared to the wild type under five experimental conditions. For each condition, the percentages of up- and downregulated coding regions and noncoding regions are shown in pie charts. Coding regions are further split into the various functional categories. The total number of differentially expressed genes is indicated below each pie chart.
It has been shown that small regulatory RNAs (sRNAs) have a broad effect on gene regulation in bacteria, and the majority of them are located inside the intergenic regions (38, 67). In streptococci, sRNAs involved in the regulation of virulence factors have been reported (54). The fasX and pel sRNAs in S. pyogenes were shown to be growth phase dependent and regulated the expression of surface proteins and exotoxins (33, 43). The rivX sRNA had been shown to be an effector of the virulence regulator CovR and Mga in S. pyogenes (52). In S. pneumoniae, five highly similar sRNAs (ccnABCDE) that are under the direct control of CiaR of the CiaRH system were identified. Among them, ccnDE had been shown to be involved in stationary-phase autolysis (25), while ccnA expression reversed some phenotypes of a ΔciaR mutant (66).
Our microarray showed that 21 noncoding regions had a differential transcription level in the mutant (log2 fold change of ≥1.0; P < 0.05) under all tested conditions, indicating a common mechanism dependent on RpoE function rather than experimental conditions. Sixteen of these 21 noncoding regions were kept for further analysis because they were present on the list of predicted small RNAs in S. mutans (http://www.oralgen.lanl.gov/oralgen/bacteria/analysis/srna_result/oralgen_srna_result_old/NC_004350/NC_004350.html). The expression patterns of all the coding genes and noncoding regions were analyzed by clustering using Genesis. Seven of these 16 noncoding regions had expression patterns similar to those of their up- or downstream coding genes, and thus they were considered cotranscribed regions and were ignored in further analyses. Interestingly, the remaining nine noncoding regions were all upregulated in the mutant, independent of adjacent genes, suggesting that they might represent regulatory RNAs (see Table ST2 in the supplemental material). Despite the developing knowledge of the roles of small regulatory RNA (sRNA) in bacteria, the computational prediction of sRNAs remains a challenge due to the lack of conventional calculation modeling (11). The prediction of sRNAs relies mainly on comparative genome analysis, in combination with thermodynamic stability or termination signals (11). Thus, the TransTermHP web service (http://transterm.cbcb.umd.edu/query.php) was used for detection of Rho-independent transcription terminators, and three of these nine transcripts were found to contain terminators. The first and second noncoding regions (smu-r1 and smu-r2) contained putative promoters (see Fig. S6 and S7 in the supplemental material) predicted by the SoftBerry web service (SoftBerry Inc., Mount Kisco, NY), while the third one had too short a sequence length (114 bp in total, but with only 45 bp before the putative terminator) to find a putative promoter. The secondary structures of smu-r1and smu-r2 (see Fig. S6 and S7 in the supplemental material) were predicted using the RNAfold web service (24). The potential targets of smu-r1and smu-r2 were predicted using the TargetRNA web service (64; http://snowwhite.wellesley.edu/targetRNA/index.html) and sRNATarget (75; http://ccb.bmi.ac.cn/starpecker/). The overlap from these two prediction services was considered a potential mRNA target (see Fig. S6 and S7 in the supplemental material). However, experimental confirmation of these two sRNAs and their interaction with target mRNAs will be needed for further conclusions.
Global changes caused by the lack of the rpoE gene.
Knockout of the rpoE gene resulted in significantly reduced expression of 24 genes (log2 fold change of ≥1.0; P < 0.05) under all tested conditions. Table 3 lists these genes together with their fold change values. Several genes that had a strongly correlated function are also presented in the table despite a slightly lower cutoff value. Interestingly, the majority of these genes were downregulated, suggesting a common regulatory mechanism independent of growth phase, pH, and H2O2 stress. Their physiological roles will be discussed below.
TABLE 3.
Genes showing similar differential expression in the ΔrpoE mutant compared to the wild-type strain under five experimental conditions
| Category and locus tag | Gene symbol | Gene product description | Log2 fold change (ΔrpoE/wild type)a under the following condition: |
||||
|---|---|---|---|---|---|---|---|
| Log-phase growth | Stationary-phase growth | Acid stress | H2O2 stress | Acid + H2O2 stress | |||
| RNA polymerase | |||||||
| SMU.96 | rpoE | DNA-directed RNA polymerase subunit delta | −8.56 | −7.62 | −7.22 | −7.30 | −6.85 |
| Malolactic fermentation | |||||||
| SMU.137 | mleS | Malate dehydrogenase | −2.88 | −3.45 | −3.52 | −2.18 | −3.62 |
| SMU.138 | mleP | Putative malate permease | −2.45 | −2.82 | −2.88 | −1.60 | −2.93 |
| SMU.139 | oxdC | Oxalate decarboxylase | −2.60 | −3.06 | −2.89 | −1.85 | −3.37 |
| SMU.140 | gshR | Putative glutathione reductase | −2.46 | −2.91 | −2.72 | −1.61 | −2.85 |
| SMU.141 | Hypothetical protein | −2.31 | −2.72 | −2.47 | −1.35 | −2.21 | |
| Histidine synthesis | |||||||
| SMU.1260c | Hypothetical protein | −1.43 | −1.24 | −0.95 | −0.96 | −1.29 | |
| SMU.1261c | Putative phosphoribosyl-ATP pyrophosphohydrolase | −1.48 | −1.20 | −1.02 | −0.90 | −1.34 | |
| SMU.1262c | Hypothetical protein | −1.55 | −1.28 | −1.08 | −0.90 | −1.80 | |
| SMU.1263 | hisI | Putative phosphoribosyl-ATP pyrophosphatase/phosphoribosyl-AMP cyclohydrolase | −1.78 | −1.47 | −1.55 | −0.97 | −2.11 |
| SMU.1264 | hisF | Imidazole glycerol phosphate synthase subunit HisF | −1.82 | −1.46 | −1.57 | −0.84 | −2.07 |
| SMU.1265 | hisA | 1-(5-Phosphoribosyl)-5-[(5-phosphoribosylamino) methylideneamino] imidazole-4-carboxamide isomerase | −1.95 | −1.59 | −1.70 | −1.07 | −2.28 |
| SMU.1266 | hisH | Imidazole glycerol phosphate synthase subunit HisH | −2.16 | −1.83 | −1.83 | −1.22 | −2.36 |
| SMU.1267c | Hypothetical protein | −2.20 | −1.92 | −1.87 | −1.24 | −2.36 | |
| SMU.1268 | hisB | Imidazoleglycerol-phosphate dehydratase | −2.22 | −1.94 | −1.93 | −1.17 | −2.67 |
| SMU.1269 | serB | Putative phosphoserine phosphatase | −2.24 | −1.91 | −1.97 | −1.12 | −2.56 |
| SMU.1270 | hisD | Histidinol dehydrogenase | −2.15 | −1.99 | −1.86 | −1.04 | −2.39 |
| SMU.1271 | hisG | ATP phosphoribosyltransferase catalytic subunit | −2.15 | −2.14 | −1.82 | −1.07 | −2.26 |
| SMU.1272 | hisZ | Putative histidyl-tRNA synthetase | −1.96 | −2.05 | −1.59 | −0.95 | −2.09 |
| SMU.1273 | hisC | Histidinol-phosphate aminotransferase | −2.01 | −2.24 | −1.69 | −1.07 | −2.27 |
| Biofilm, adherence, virulence | |||||||
| SMU.1004 | gtfB | Glucosyltransferase I | −1.18 | −0.90 | −0.59 | −1.17 | −2.81 |
| SMU.1005 | gtfC | Glucosyltransferase Si | −1.39 | −1.45 | −1.32 | −1.33 | −1.89 |
| SMU.1396 | gbpC | Glucan-binding protein C | −2.71 | −1.79 | −1.27 | −1.28 | −3.06 |
| SMU.1397c | irvA | Transcriptional regulator | −1.40 | −1.40 | −1.26 | −1.05 | −1.38 |
| SMU.610 | spaP | Cell surface antigen SpaP | −1.92 | −0.99 | −0.59 | −1.41 | −1.85 |
| Antibiotic resistance | |||||||
| SMU.440 | Hypothetical protein | −1.16 | −1.23 | −1.02 | −0.35 | −1.22 | |
| SMU.441 | Putative transcriptional regulator | −1.22 | −1.24 | −1.07 | −0.49 | −1.19 | |
| SMU.442 | Hypothetical protein | −1.12 | −1.25 | −1.00 | −0.50 | −1.10 | |
| Other | |||||||
| SMU.1861c | Hypothetical protein | −1.20 | −1.35 | −2.21 | −1.82 | −1.79 | |
| SMU.108 | Hypothetical S-adenosylmethionine-dependent methyltransferases | 1.69 | 2.54 | 2.15 | 1.19 | 2.33 | |
| SMU.109 | Lantibiotic-related antibiotic efflux permease | 1.17 | 1.10 | 1.25 | 1.10 | 1.16 | |
| SMU.1480 | Hypothetical protein | 1.02 | 1.32 | 1.32 | 1.30 | 1.19 | |
| SMU.1642c | Hypothetical protein | 1.25 | 1.42 | 1.71 | 1.49 | 1.83 | |
| SMU.1643c | Hypothetical protein | 1.15 | 1.41 | 1.57 | 1.44 | 1.94 | |
| SMU.643 | Putative esterase | 1.08 | 1.35 | 1.11 | 1.03 | 1.44 | |
Genes that were below the cutoff of 1.0 (log2 fold change) under some conditions but had a strong correlation with other genes according to gene functional annotation or genomic location are included in the table.
(i) MLF.
One of the significant changes in the mutant was the downregulation of genes for malolactic fermentation (MLF) (SMU.137 to SMU.141). MLF protects the organism against acid stress by transforming l-malate to the weaker acid l-lactic acid and carbon dioxide. Products of this fermentation raise the cytoplasmic pH and furthermore generate ATP as an energy source for the extrusion of protons necessary for keeping the intracellular pH more alkaline than the extracellular pH (57). Therefore, the strong reduction of these genes resulted in decreased acid tolerance, which was consistent with the phenotypic results in this study (Fig. 1). The expression of MLF genes is induced by low pH, by the substrate l-malate, and to a lesser extent by the positive regulator MleR (SMU.135) (36). However, despite the strong repression of the MLF structural genes in the ΔrpoE strain, the regulator gene mleR did not show a corresponding change. Furthermore, these genes were downregulated in the ΔrpoE strain under every tested condition, regardless of growth phase or type of stress, indicating that there should exist another regulatory mechanism directly related to the role of RpoE during transcription. Moreover, the overlap of the responses to acidic and oxidative stress is confirmed by the reduction of the putative glutathione reductase GshR in the same operon. GshR is an important antioxidant that reduces glutathione disulfide to the sulfhydryl form. Downregulation of gshR might decrease the cellular oxidative protection, and this might partly explain the sensitivity of the ΔrpoE mutant to H2O2.
(ii) Histidine metabolism.
All the genes for the histidine synthesis pathway (SMU.1260c to SMU.1273) were downregulated in the ΔrpoE mutant. The pathway for histidine is interconnected with the purine synthetic pathway. The by-product of histidine biosynthesis, 5′-phosphoribosyl-4-carboxamide-5-aminoimidazole (AICAR), is an intermediate of purine biosynthesis that is rapidly recycled to ATP, which in turn is an important precursor of histidine biosynthesis (48). Since RNA polymerase is essential for purine and pyrimidine synthesis, the loss of RpoE may decrease purine synthesis and further affect histidine synthesis.
Based on base composition analyses and BLAST taxonomy data, the histidine synthesis gene cluster has been suggested as a potential genomic island in the genome of S. mutans (http://www.oralgen.lanl.gov/). By comparative genomic analyses, Maruyama et al. (45) showed that genes for histidine metabolism are conserved among oral streptococci, and they speculated that the biogenesis of histidine may have physiological importance for the survival of streptococci in the oral environment. The buffering capacity of the cytoplasm depends on the concentrations of proteins inside the cytoplasm and is one of the important pH homeostasis mechanisms (12). Histidine has an ionizable side chain with a pKa near neutrality (47) and thus proteins containing histidine residues buffer effectively near neutral pH.
(iii) Biofilm, adherence, and virulence.
Sucrose, as a common dietary disaccharide, is of particular interest because it is an important substrate for extracellular production of water-insoluble glucan which is mediated by the membrane-bound glucosyltransferases (GtfB and GtfC). Glucan is important for adherence of S. mutans to the tooth surface and for forming the extracellular polysaccharide matrix of the biofilm (34). The cell surface glucan-binding protein (GbpC) has been shown to be involved in biofilm formation and architecture (42). In the ΔrpoE strain, these genes were all downregulated, which may be the reason for the observed changes in its biofilm structure. Moreover, the expression of the cell wall-associated adhesin P1 (SapP), which mediates the sucrose-independent adherence by binding to salivary pellicles formed on the tooth surface (5), was also depressed in the ΔrpoE strain. Furthermore, the transcriptional regulator gene irvA, located immediately adjacent to gbpC, promotes biofilm formation via induction of gbpC and sapP expression (77) and was repressed in the ΔrpoE strain. Since the adhesion, glucan-producing, and glucan-binding enzymes are considered conserved virulence factors of S. mutans, different virulence of the ΔrpoE strain might be expected.
(iv) Resistance to antibiotics.
Although the transcription levels of the 30S ribosomal proteins S1 RpsA (SMU.1200c) and S2 RpsB (SMU.2032) were similar in the microarray data, a lower transcription level of an operon related to polyketide antibiotic resistance, SMU.440-SMU.441 (46), was observed in the ΔrpoE mutant under all conditions. The hypothetical protein SMU.440 is conserved in some dental pathogenic bacteria and is associated with polyketide-like antibiotic resistance. The adjacent protein, SMU.441, belongs to the MarR protein family of transcriptional regulators, which is involved in controlling multiple antibiotic resistances (46). Moreover, the adjacent SMU.442 showed a similar expression pattern. Since proteins with related functions are often clustered into the same operon in bacteria, SMU.442 might also be involved in antibiotic resistance. Hence, the sensitivity of the ΔrpoE mutant to the polyketide antibiotic tetracycline might in part be due to the repression of SMU.440 to SMU.442.
Significant changes in the ΔrpoE strain during growth.
In addition to the changes described above, which occurred under all tested conditions, RpoE deficiency profoundly affected cellular physiology during growth, and it had a major impact on the expression of genes for amino acid transport and metabolism, carbohydrate transport and metabolism, and energy production and conversion (Fig. 6; see Table ST3 in the supplemental material).
FIG. 6.
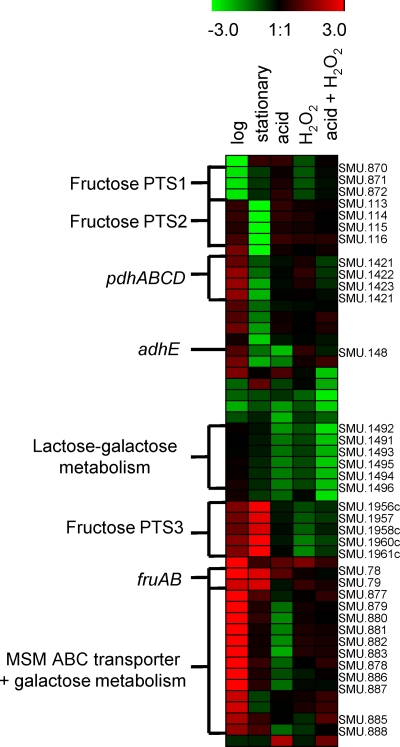
Hierarchical clustering of differentially expressed genes involved in carbohydrate transport and metabolism and in energy production and conversion. Gene expression ratios are calculated as log2 fold changes, comparing the ΔrpoE mutant to the wild-type strain. Red, upregulated genes; green, downregulated genes.
(i) Carbohydrate transport, carbohydrate metabolism, energy production, and energy conversion.
Carbohydrate metabolism by fermentation is the principal source of energy production for S. mutans, which possesses an incomplete tricarboxylic acid (TCA) cycle (7). Furthermore, the ability of S. mutans to catabolize a variety of carbohydrates plays an important role in pathogenesis, since its acidic end products, e.g., lactic acid, formate, and acetate, reduce the extracellular pH significantly and induce caries. The major uptake mechanism for sugars in S. mutans depends on the phosphoenolpyruvate-phosphotransferase system (PTS), which mediates the internalization and phosphorylation of various sugars. It consists of two nonspecific energy coupling components, enzymes EI and EII and Hpr protein, and various substrate-specific multiprotein permeases (23).
The rpoE mutation caused dramatic changes in carbohydrate transport and metabolism (Fig. 6). The transcription of the PTS1 operon (SMU.871-SMU.872), the primary PTS for fructose uptake (68), was reduced in the ΔrpoE mutant at the logarithmic phase of growth. Of note, the putative transcriptional repressor of the fructose operon, SMU.870, had the same expression pattern as SMU.871 and SMU.872, indicating that the SMU.870 protein may function as a positive regulator. Thus, the ΔrpoE mutant failed to have this PTS; instead, an alternative sugar uptake system, the multiple-sugar-binding (MSM) ABC transporter genes, including the downstream operon (SMU.877 to SMU.888), was highly induced. The MSM transporter is responsible for the uptake of a wide range of sugars that are structurally related to raffinose, including disaccharides (melibiose and sucrose) and monosaccharides (glucose, fructose, and galactose) (8, 61). However, no obvious change in the transcription of the putative positive regulator of the MSM operon (SMU.876) was found. Hence, there might be other regulatory mechanisms of MSM-mediated sugar transport and metabolism.
Wen et al. showed that genes in the fructose-specific PTS2 operon (SMU.113 to SMU.116) were constitutively expressed at a basic level (68), but no further report on the regulation of this PTS was found. Our microarray results showed clear induction of this operon at the early stationary phase in the wild type, while deletion of rpoE abolished such induction. However, the transcription level of the fructose/mannose-specific PTS3 Lev (SMU.1956c to SMU.1961c) increased in the ΔrpoE mutant. Moreover, upregulation of fructan hydrolase genes fruA (SMU.78) and fruB (SMU.79) was observed. The expression of PTS3 Lev and FruA is repressed by fructose-specific PTS1 and PTS2 through carbon catabolite repression, which allows bacteria to metabolize the readily used sugars instead of nonpreferred ones (72, 73, 74). Thus, the inhibitory effect of PTS3 Lev was relieved because of reduced repressor PTS1 and PTS2 in the ΔrpoE mutant. However, such changes might hamper efficient energy production, since the downregulation of energy generation genes (pdhABCD and adhE) was observed in the early stationary phase. Since the optimization of carbohydrate utilization is crucial for oral streptococci, the inefficient sugar metabolism may be part of the reason for the growth deficiency and reduced stress tolerance of the ΔrpoE mutant.
(ii) Amino acid transport.
Genes involved in amino acid transport and metabolism were significantly changed, the majority of which were downregulated in the ΔrpoE mutant. Two osmoprotectant uptake ABC transporters, OpuA (SMU.1062 to SMU.1063) and OpuC (SMU.2116 to SMU.2119), were downregulated in the ΔrpoE mutant. Osmoprotectant molecules are soluble under physiological conditions and bear no net charge at neutral pH. Their accumulation in the cytosol is important for maintaining cell turgor and for stabilization of proteins, which is crucial for osmotic stress adaptation of bacteria (29). Thus, the reduced level of osmoprotectant ABC transporters in the ΔrpoE mutant could be a disadvantage for its survival in the fluctuating oral environment.
(iii) Iron transport.
Interestingly, the putative ferrichrome transporter genes (SMU.995 to SMU.998) were induced in the ΔrpoE strain under every condition, except to a smaller degree under oxidative stress. Sufficient levels of iron are necessary for bacterial survival and growth, but the accumulation of iron can lead to the production of toxic oxygen radicals via Fenton chemistry (17, 76). Thus, its intracellular transport must be tightly controlled, and such induction might cause toxicity to the ΔrpoE strain.
Deficiency of the rpoE mutant in stress responses.
In addition to the changes of the ΔrpoE strain during growth, the loss of RpoE also caused deficiency in stress defense compared to the wild-type strain. As an example, the lactose-galactose pathway, encoded by the lac operon (SMU.1491 to SMU.1496), which generates the important substrate glucose 1-phosphate for glycolysis (1), was downregulated in the ΔrpoE mutant compared to the wild type under all stress conditions (Fig. 6; see Fig. S8 in the supplemental material). The high-affinity manganese and iron transporter operon sloABCR (SMU.182 to SMU.186) was induced in the wild type under acid or H2O2 stress; however, the ΔrpoE strain did not have induction under H2O2 stress. The metal ions manganese and iron are cofactors for superoxide dismutase, the enzyme necessary for oxidative defense (44, 53). Recent studies suggest a role of the SloABC transporter and its regulator SloR for the regulation of virulence in S. mutans, including adherence, biofilm formation, genetic competence, metal ion homeostasis, oxidative stress tolerance, and antibiotic resistance (9, 50, 53).
Stress as an additional modulator of the transcriptome profile of S. mutans.
The wild-type and mutant strains had similar responses to specific stresses, which revealed that stresses act as additional modulators of gene regulation in S. mutans, and some of these responses are not dependent on RpoE. Gene expression changes corresponding to either acid or H2O2 stress could be found under the combined-stress condition. The copper-transporting ATPase CopA (SMU.426), transcriptional regulator CopY (SMU.424), and chaperone CopZ (SMU.427), for instance, were upregulated under acidic and combined-stress conditions in both the wild-type and mutant strains (see Fig. S8 in the supplemental material). Interestingly, under H2O2 and combined-stress conditions, the strong induction of SMU.191c to SMU.217c was found in both strains. Within SMU.191c to SMU.217c, five genes (SMU.191c, SMU.198c, SMU.201c, SMU.207c, and SMU.208c) encode putative transposases and integrase, while SMU.194c encodes a bacteriophage P2-associated protein and SMU.196c encodes an immunogenic secreted protein. Transposases and integrases promote the movement of DNA segments to new locations, through a “cut-and-paste” mechanism, without a requirement for sequence homology (26). Transposase genes are the most abundant genes in both sequenced genomes and environmental metagenomes (10). Their mobile nature on the one hand may cause inactivation and mutation of structural genes in bacterial genome; however, on the other hand, they diversify and enrich the genomes and offer a selective advantage to the bacteria (26, 28). H2O2 has been reported to induce transposition activity in Burkholderia cenocepacia (19) and to increase transcription of transposases in Porphyromonas gingivalis (18). The increased transposase activity under oxidative stress was expected to contribute to the genomic rearrangement and diversity for better survival under environmental stress.
Interestingly, the combination of two stresses triggered new responses. The pyruvate dehydrogenase complex PdhABCD (SMU.1421 to SMY.1424), which oxidizes pyruvate to acetyl coenzyme A (acetyl-CoA) and CO2 (15, 32), was strongly induced when acid and H2O2 stresses were combined, while it was only weakly affected by single stress. Accordingly, the alcohol dehydrogenase AdhCD (SMU.129 to SMY.130), which catabolizes the conversion of acetyl-CoA to alcohol (71), was derepressed under combined-stress conditions. These data suggest that S. mutans has sophisticated regulatory mechanisms, instead of simple combination, to protect itself against different stress sources.
Confirmation of gene expression by quantitative PCR.
The differential expression of four genes was confirmed by quantitative PCR in all samples using the same RNA sources as used for the microarray analysis (see Table ST4 in the supplemental material). The first gene in the histidine metabolism operon, hisC, was downregulated in the ΔrpoE mutant under every tested condition, similar to the microarray results. More pronounced induction of the sugar transporter genes levD and msmE was found in the log phase and early stationary phase of growth by quantitative PCR in comparison to the microarray data. The reduction of the expression of the transporter gene fruC at the early stationary phase in the mutant strain was also confirmed with both methods.
Summary of RpoE function in S. mutans.
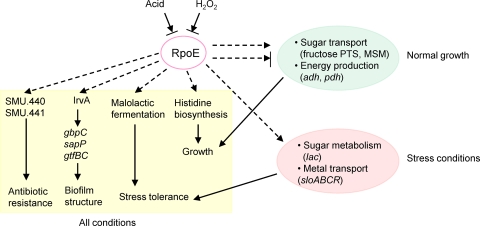
The phenotypic and transcriptomic results allow us to develop a concept of the global role of RpoE in S. mutans, likely caused by its modification of the transcriptional specificity of the RNA polymerase. As shown in Fig. 7, rpoE transcription was inhibited by acid and H2O2. The lack of RpoE interfered with various biological functions, including growth, acid tolerance, biofilm formation, and resistance to antibiotics under all experimental conditions. Under normal growth conditions, RpoE had a significant effect on sugar transport and energy production, both of which affect cell growth. Under stress conditions, rpoE knockout caused deficiency in sugar metabolism and metal transport, resulting in reduced growth and survival of the mutant. Thus, RpoE acts as a global modulator of gene expression. It is critical for optimal growth and quick adaptation to changing environmental conditions and potentially increases the virulence of S. mutans.
FIG. 7.
Model of RpoE function in S. mutans. The yellow shadow shows functions influenced by RpoE under all conditions, including growth, acid tolerance, biofilm structure, and antibiotic resistance. The green part shows the effect of RpoE on sugar transport and energy production under normal growth conditions, while the red part shows the role of RpoE in sugar metabolism and metal transport under stress conditions.
Supplementary Material
Acknowledgments
We thank Mathias Müsken for introduction to CLSM. We gratefully acknowledge Robbert Geffers for microarray technical support and Michael Reck for the microarray design.
X. L. Xue was supported by a CSC-Helmholtz Joint Fellowship.
Footnotes
Published ahead of print on 30 July 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abranches, J., Y. Y. Chen, and R. A. Burne. 2004. Galactose metabolism by Streptococcus mutans. Appl. Environ. Microbiol. 70:6047-6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achberger, E. C., and H. R. Whiteley. 1981. The role of the delta peptide of the Bacillus subtilis RNA polymerase in promoter selection. J. Biol. Chem. 256:7424-7432. [PubMed] [Google Scholar]
- 3.Achberger, E. C., M. D. Hilton, and H. R. Whiteley. 1982. The effect of the delta subunit on the interaction of Bacillus subtilis RNA polymerase with bases in a SP82 early gene promoter. Nucleic Acids Res. 10:2893-2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn, S. J., and R. A. Burne. 2007. Effects of oxygen on biofilm formation and the AtlA autolysin of Streptococcus mutans. J. Bacteriol. 189:6293-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahn, S. J., S. J. Ahn, Z. T. Wen, L. J. Brady, and R. A. Burne. 2008. Characteristics of biofilm formation by Streptococcus mutans in the presence of saliva. Infect. Immun. 76:4259-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahn, S. J., S. J. Ahn, C. M. Browngardt, and R. A. Burne. 2009. Changes in biochemical and phenotypic properties of Streptococcus mutans during growth with aeration. Appl. Environ. Microbiol. 75:2517-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci.U. S. A. 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ajdic, D., and V. T. Pham. 2007. Global transcriptional analysis of Streptococcus mutans sugar transporters using microarrays. J. Bacteriol. 189:5049-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arirachakaran, P., E. Benjavongkulchai, S. Luengpailin, D. Ajdic, and J. A. Banas. 2007. Manganese affects Streptococcus mutans virulence gene expression. Caries Res. 41:503-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aziz, R. K., M. Breitbart, and R. A. Edwards. 2010. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 38:4207-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Backofen, R., and W. R. Hess. 2010. Computational prediction of sRNAs and their targets in bacteria. RNA Biol. 7:33-42. [DOI] [PubMed] [Google Scholar]
- 12.Baker-Austin, C., and M. Dopson. 2007. Life in acid: pH homeostasis in acidophiles. Trends Microbiol. 15:165-171. [DOI] [PubMed] [Google Scholar]
- 13.Baldeck, J. D., and R. E. Marquis. 2008. Targets for hydrogen-peroxide-induced damage to suspension and biofilm cells of Streptococcus mutans. Can. J. Microbiol. 54:868-875. [DOI] [PubMed] [Google Scholar]
- 14.Biswas, I., L. Drake, D. Erkina, and S. Biswas. 2008. Involvement of sensor kinases in the stress tolerance response of Streptococcus mutans. J. Bacteriol. 190:68-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlsson, J., U. Kujala, and M. B. Edlund. 1985. Pyruvate dehydrogenase activity in Streptococcus mutans. Infect. Immun. 49:674-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chamberlin, M. J. 1974. The selectivity of transcription. Annu. Rev. Biochem. 43:721-775. [DOI] [PubMed] [Google Scholar]
- 17.Clancy, A., J. W. Loar, C. D. Speziali, M. Oberg, D. E. Heinrichs, and C. E. Rubens. 2006. Evidence for siderophore-dependent iron acquisition in group B streptococcus. Mol. Microbiol. 59:707-721. [DOI] [PubMed] [Google Scholar]
- 18.Diaz, P. I., N. Slakeski, E. C. Reynolds, R. Morona, A. H. Rogers, and P. E. Kolenbrander. 2006. Role of oxyR in the oral anaerobe Porphyromonas gingivalis. J. Bacteriol. 188:2454-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drevinek, P., A. Baldwin, L. Lindenburg, L. T. Joshi, A. Marchbank, S. Vosahlikova, C. G. Dowson, and E. Mahenthiralingam. 2010. Oxidative stress of Burkholderia cenocepacia induces insertion sequence-mediated genomic rearrangements that interfere with macrorestriction-based genotyping. J. Clin. Microbiol. 48:34-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunny, G. M., L. N. Lee, and D. J. LeBlanc. 1991. Improved electroporation and cloning vector system for gram-positive bacteria. Appl. Environ. Microbiol. 57:1194-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Floss, H. G., and T. W. Yu. 2005. Rifamycin—mode of action, resistance, and biosynthesis. Chem. Rev. 105:621-632. [DOI] [PubMed] [Google Scholar]
- 22.Gong, Y., X. L. Tian, T. Sutherland, G. Sisson, J. Mai, J. Ling, and Y. H. Li. 2009. Global transcriptional analysis of acid-inducible genes in Streptococcus mutans: multiple two-component systems involved in acid adaptation. Microbiology 155:3322-3332. [DOI] [PubMed] [Google Scholar]
- 23.Gorke, B., and J. Stulke. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6:613-624. [DOI] [PubMed] [Google Scholar]
- 24.Gruber, A. R., R. Lorenz, S. H. Bernhart, R. Neubock, and I. L. Hofacker. 2008. The Vienna RNA websuite. Nucleic Acids Res. 36:W70-W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halfmann, A., M. Kovacs, R. Hakenbeck, and R. Bruckner. 2007. Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: five out of 15 promoters drive expression of small non-coding RNAs. Mol. Microbiol. 66:110-126. [DOI] [PubMed] [Google Scholar]
- 26.Hickman, A. B., M. Chandler, and F. Dyda. 2010. Integrating prokaryotes and eukaryotes: DNA transposases in light of structure. Crit. Rev. Biochem. Mol. Biol. 45:50-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higuchi, M., Y. Yamamoto, and Y. Kamio. 2000. Molecular biology of oxygen tolerance in lactic acid bacteria: functions of NADH oxidases and Dpr in oxidative stress. J. Biosci. Bioeng. 90:484-493. [PubMed] [Google Scholar]
- 28.Hooper, S. D., K. Mavromatis, and N. C. Kyrpides. 2009. Microbial co-habitation and lateral gene transfer: what transposases can tell us. Genome Biol. 10:R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horn, C., S. Jenewein, L. Sohn-Bosser, E. Bremer, and L. Schmitt. 2005. Biochemical and structural analysis of the Bacillus subtilis ABC transporter OpuA and its isolated subunits. J. Mol. Microbiol. Biotechnol. 10:76-91. [DOI] [PubMed] [Google Scholar]
- 30.Jones, A. L., R. H. Needham, and C. E. Rubens. 2003. The delta subunit of RNA polymerase is required for virulence of Streptococcus agalactiae. Infect. Immun. 71:4011-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Juang, Y. L., and J. D. Helmann. 1994. The delta subunit of Bacillus subtilis RNA polymerase. An allosteric effector of the initiation and core-recycling phases of transcription. J. Mol. Biol. 239:1-14. [DOI] [PubMed] [Google Scholar]
- 32.Korithoski, B., C. M. Levesque, and D. G. Cvitkovitch. 2008. The involvement of the pyruvate dehydrogenase E1alpha subunit, in Streptococcus mutans acid tolerance. FEMS Microbiol. Lett. 289:13-19. [DOI] [PubMed] [Google Scholar]
- 33.Kreikemeyer, B., M. D. Boyle, B. A. Buttaro, M. Heinemann, and A. Podbielski. 2001. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol. Microbiol. 39:392-406. [DOI] [PubMed] [Google Scholar]
- 34.Kreth, J., L. Zhu, J. Merritt, W. Shi, and F. Qi. 2008. Role of sucrose in the fitness of Streptococcus mutans. Oral Microbiol. Immunol. 23:213-219. [DOI] [PubMed] [Google Scholar]
- 35.Lau, P. C., C. K. Sung, J. H. Lee, D. A. Morrison, and D. G. Cvitkovitch. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193-205. [DOI] [PubMed] [Google Scholar]
- 36.Lemme, A., H. Sztajer, and I. Wagner-Dobler. 2010. Characterization of mleR, a positive regulator of malolactic fermentation and part of the acid tolerance response in Streptococcus mutans. BMC Microbiol. 10:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemos, J. A., and R. A. Burne. 2008. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology 154:3247-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, J. M., and A. Camilli. 2010. A broadening world of bacterial small RNAs. Curr. Opin. Microbiol. 13:18-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopez de Saro, F. J., A. Y. Woody, and J. D. Helmann. 1995. Structural analysis of the Bacillus subtilis delta factor: a protein polyanion which displaces RNA from RNA polymerase. J. Mol. Biol. 252:189-202. [DOI] [PubMed] [Google Scholar]
- 41.Lopez de Saro, F. J., N. Yoshikawa, and J. D. Helmann. 1999. Expression, abundance, and RNA polymerase binding properties of the delta factor of Bacillus subtilis. J. Biol. Chem. 274:15953-15958. [DOI] [PubMed] [Google Scholar]
- 42.Lynch, D. J., T. L. Fountain, J. E. Mazurkiewicz, and J. A. Banas. 2007. Glucan-binding proteins are essential for shaping Streptococcus mutans biofilm architecture. FEMS Microbiol. Lett. 268:158-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mangold, M., M. Siller, B. Roppenser, B. J. Vlaminckx, T. A. Penfound, R. Klein, R. Novak, R. P. Novick, and E. Charpentier. 2004. Synthesis of group A streptococcal virulence factors is controlled by a regulatory RNA molecule. Mol. Microbiol. 53:1515-1527. [DOI] [PubMed] [Google Scholar]
- 44.Martin, M. E., R. C. Strachan, H. Aranha, S. L. Evans, M. L. Salin, B. Welch, J. E. Arceneaux, and B. R. Byers. 1984. Oxygen toxicity in Streptococcus mutans: manganese, iron, and superoxide dismutase. J. Bacteriol. 159:745-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maruyama, F., M. Kobata, K. Kurokawa, K. Nishida, A. Sakurai, K. Nakano, R. Nomura, S. Kawabata, T. Ooshima, K. Nakai, M. Hattori, S. Hamada, and I. Nakagawa. 2009. Comparative genomic analyses of Streptococcus mutans provide insights into chromosomal shuffling and species-specific content. BMC Genomics 10:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nan, J., E. Brostromer, X. Y. Liu, O. Kristensen, and X. D. Su. 2009. Bioinformatics and structural characterization of a hypothetical protein from Streptococcus mutans: implication of antibiotic resistance. PLoS One 4:e7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nelson, D. L., and M. M. Cox. 2004. Lehninger principles of biochemistry, 4th ed., p. 80-84. W. H. Freeman & Company, New York, NY.
- 48.Nelson, D. L., and M. M. Cox. 2004. Lehninger principles of biochemistry, 4th ed., p. 851-852. W. H. Freeman & Company, New York, NY.
- 49.Nishimura, Y., H. Adachi, M. Kyo, S. Murakami, S. Hattori, and K. Ajito. 2005. A proof of the specificity of kanamycin-ribosomal RNA interaction with designed synthetic analogs and the antibacterial activity. Bioorg. Med. Chem. Lett. 15:2159-2162. [DOI] [PubMed] [Google Scholar]
- 50.Paik, S., A. Brown, C. L. Munro, C. N. Cornelissen, and T. Kitten. 2003. The sloABCR operon of Streptococcus mutans encodes an Mn and Fe transport system required for endocarditis virulence and its Mn-dependent repressor. J. Bacteriol. 185:5967-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts, S. A., and J. R. Scott. 2007. RivR and the small RNA RivX: the missing links between the CovR regulatory cascade and the Mga regulon. Mol. Microbiol. 66:1506-1522. [DOI] [PubMed] [Google Scholar]
- 53.Rolerson, E., A. Swick, L. Newlon, C. Palmer, Y. Pan, B. Keeshan, and G. Spatafora. 2006. The SloR/Dlg metalloregulator modulates Streptococcus mutans virulence gene expression. J. Bacteriol. 188:5033-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Romby, P., and E. Charpentier. 2010. An overview of RNAs with regulatory functions in gram-positive bacteria. Cell Mol. Life Sci. 67:217-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seepersaud, R., R. H. Needham, C. S. Kim, and A. L. Jones. 2006. Abundance of the delta subunit of RNA polymerase is linked to the virulence of Streptococcus agalactiae. J. Bacteriol. 188:2096-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Senadheera, D., K. Krastel, R. Mair, A. Persadmehr, J. Abranches, R. A. Burne, and D. G. Cvitkovitch. 2009. Inactivation of VicK affects acid production and acid survival of Streptococcus mutans. J. Bacteriol. 191:6415-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheng, J., and R. E. Marquis. 2007. Malolactic fermentation by Streptococcus mutans. FEMS Microbiol. Lett. 272:196-201. [DOI] [PubMed] [Google Scholar]
- 58.Sturn, A., J. Quackenbush, and Z. Trajanoski. 2002. Genesis: cluster analysis of microarray data. Bioinformatics 18:207-208. [DOI] [PubMed] [Google Scholar]
- 59.Svensater, G., B. Sjogreen, and I. R. Hamilton. 2000. Multiple stress responses in Streptococcus mutans and the induction of general and stress-specific proteins. Microbiology 146:107-117. [DOI] [PubMed] [Google Scholar]
- 60.Sztajer, H., A. Lemme, R. Vilchez, S. Schulz, R. Geffers, C. Y. Yip, C. M. Levesque, D. G. Cvitkovitch, and I. Wagner-Dobler. 2008. Autoinducer-2-regulated genes in Streptococcus mutans UA159 and global metabolic effect of the luxS mutation. J. Bacteriol. 190:401-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tao, L., I. C. Sutcliffe, R. R. Russell, and J. J. Ferretti. 1993. Transport of sugars, including sucrose, by the msm transport system of Streptococcus mutans. J. Dent. Res. 72:1386-1390. [DOI] [PubMed] [Google Scholar]
- 62.Thaker, M., P. Spanogiannopoulos, and G. D. Wright. 2010. The tetracycline resistome. Cell Mol. Life Sci. 67:419-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tipper, D. J. 1985. Mode of action of beta-lactam antibiotics. Pharmacol. Ther. 27:1-35. [DOI] [PubMed] [Google Scholar]
- 64.Tjaden, B. 2008. TargetRNA: a tool for predicting targets of small RNA action in bacteria. Nucleic Acids Res. 36:W109-W113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tremblay, Y. D., H. Lo, Y. H. Li, S. A. Halperin, and S. F. Lee. 2009. Expression of the Streptococcus mutans essential two-component regulatory system VicRK is pH and growth-phase dependent and controlled by the LiaFSR three-component regulatory system. Microbiology 155:2856-2865. [DOI] [PubMed] [Google Scholar]
- 66.Tsui, H. C., D. Mukherjee, V. A. Ray, L. T. Sham, A. L. Feig, and M. E. Winkler. 2010. Identification and characterization of noncoding small RNAs in Streptococcus pneumoniae serotype 2 strain D39. J. Bacteriol. 192:264-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Waters, L. S., and G. Storz. 2009. Regulatory RNAs in bacteria. Cell 136:615-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wen, Z. T., C. Browngardt, and R. A. Burne. 2001. Characterization of two operons that encode components of fructose-specific enzyme II of the sugar:phosphotransferase system of Streptococcus mutans. FEMS Microbiol. Lett. 205:337-342. [DOI] [PubMed] [Google Scholar]
- 69.Wen, Z. T., H. V. Baker, and R. A. Burne. 2006. Influence of BrpA on critical virulence attributes of Streptococcus mutans. J. Bacteriol. 188:2983-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wettenhall, J. M., and G. K. Smyth. 2004. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics 20:3705-3706. [DOI] [PubMed] [Google Scholar]
- 71.Yamada, T., and J. Carlsson. 1975. Regulation of lactate dehydrogenase and change of fermentation products in streptococci. J. Bacteriol. 124:55-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zeng, L., Z. T. Wen, and R. A. Burne. 2006. A novel signal transduction system and feedback loop regulate fructan hydrolase gene expression in Streptococcus mutans. Mol. Microbiol. 62:187-200. [DOI] [PubMed] [Google Scholar]
- 73.Zeng, L., and R. A. Burne. 2008. Multiple sugar:phosphotransferase system permeases participate in catabolite modification of gene expression in Streptococcus mutans. Mol. Microbiol. 70:197-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zeng, L., and R. A. Burne. 2010. Seryl-phosphorylated HPr regulates CcpA-independent carbon catabolite repression in conjunction with PTS permeases in Streptococcus mutans. Mol. Microbiol. 75:1145-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao, Y., H. Li, Y. Hou, L. Cha, Y. Cao, L. Wang, X. Ying, and W. Li. 2008. Construction of two mathematical models for prediction of bacterial sRNA targets. Biochem. Biophys. Res. Commun. 372:346-350. [DOI] [PubMed] [Google Scholar]
- 76.Zheng, M., B. Doan, T. D. Schneider, and G. Storz. 1999. OxyR and SoxRS regulation of fur. J. Bacteriol. 181:4639-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu, M., D. Ajdic, Y. Liu, D. Lynch, J. Merritt, and J. A. Banas. 2009. Role of the Streptococcus mutans irvA gene in GbpC-independent, dextran-dependent aggregation and biofilm formation. Appl. Environ. Microbiol. 75:7037-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.