Abstract
Transcription of spa, encoding the virulence factor protein A in Staphylococcus aureus, is tightly controlled by a complex regulatory network, ensuring its temporal expression over growth and at appropriate stages of the infection process. Transcriptomic profiling of XdrA, a DNA-binding protein that is conserved in all S. aureus genomes and shares similarity with the XRE family of helix-turn-helix, antitoxin-like proteins, revealed it to be a previously unidentified activator of spa transcription. To assess how XdrA fits into the complex web of spa regulation, a series of regulatory mutants were constructed; consisting of single, double, triple, and quadruple mutants lacking XdrA and/or the three key regulators previously shown to influence spa transcription directly (SarS, SarA, and RNAIII). A series of lacZ reporter gene fusions containing nested deletions of the spa promoter identified regions influenced by XdrA and the other three regulators. XdrA had almost as strong an activating effect on spa as SarS and acted on the same spa operator regions as SarS, or closely overlapping regions. All data from microarrays, Northern and Western blot analyses, and reporter gene fusion experiments indicated that XdrA is a major activator of spa expression that appears to act directly on the spa promoter and not through previously characterized regulators.
Global gene regulation is essential for the pathogenic success of Staphylococcus aureus. The tightly regulated expression of virulence factors enables it to adapt to hostile environments and to cause a wide range of infections. The temporal modulation of virulence factors occurs throughout infection progression in vivo and has also been widely documented in vitro (1, 11). Virulence gene expression is coordinated by a highly complex, interconnected regulatory network that responds to both endogenous and external stimuli (8). This network consists of well-characterized loci, such as the agr two-component quorum sensor system (1, 54); the various members of the SarA regulatory protein family, including SarS, SarT, SarU, SarV, SarX, SarZ, Rot, MgrA, and TcaR (17); the main stress response alternative sigma factor, SigB (7); and two-component sensor-transducer systems saeRS (44), arlRS (42), and srrAB (61, 76). Other loci with different attributed primary functions, or whose main functions are unknown, e.g., ccpA (70, 71), spoVG (69), codY (60), msrR (33), and msa (65), have also been shown to contribute to virulence and/or to modulate virulence factor expression, creating links between virulence and other cellular traits such as metabolism and antibiotic resistance phenotypes.
Protein A, encoded by spa, is one of the most widely studied and characterized staphylococcal cell wall-anchored surface components (21, 51). It is best characterized for its ability to bind the Fc region of IgG, attenuating antibody-mediated opsonization (24, 59). Protein A has also been shown to bind to von Willebrand factor, to activate tumor necrosis factor (29, 31), and to induce inflammation by triggering B-cell proliferation (4). The contribution of protein A to virulence has been debated, but studies have shown that protein A-deficient S. aureus strains are phagocytosed more efficiently in vitro and show decreased virulence in murine models of septic arthritis and pneumonia (58).
Numerous regulatory elements modulate spa gene expression either directly or indirectly (30). The SarS DNA-binding transcriptional regulator, encoded upstream of spa, is the main activator and has been shown to bind directly to the spa promoter (56). Activation is counteracted by SarA (20, 72) and by the effector molecule of the agr system, RNAIII (20, 55), both of which bind to the spa promoter and repress transcription. RNAIII-dependent spa repression occurs not only at the transcriptional level but also at the posttranscriptional level via an antisense mechanism (34). Most other loci shown to influence spa transcription belong to intricate regulatory cascades that eventually control spa indirectly, through modulation of SarS, SarA, or RNAIII. Some regulators, such as MgrA and Rot, have been proposed to act both directly and indirectly, although their exact roles in direct regulation have not been well characterized (56). Comparative levels of spa transcription during growth, and the influence of different regulators on expression levels, differ enormously among different strain backgrounds (12, 36). Most spa regulation studies have been performed on strains derived from NCTC8325, which have mutations reducing SigB activity (41) and inactivating TcaR (52), both of which indirectly affect spa transcription. SarT and SarU, which are part of the cascade leading to spa regulation, make up part of the variable S. aureus core genome and are also absent from some S. aureus clonal complexes (12, 43).
We recently identified a new DNA-binding transcriptional regulator encoded by open reading frame (ORF) SA1665 in the S. aureus N315 genome (23). We have named this protein XdrA (XRE-like DNA-binding regulator, A) because it shares similarity with XRE (xenobiotic response element) family helix-turn-helix, antitoxin-like proteins and is not homologous to Sar family proteins. Deletion of xdrA increased β-lactam resistance in all clinical methicillin-resistant S. aureus (MRSA) isolates tested (23). Although the essential prerequisite for methicillin resistance is the expression of PBP2a, encoded by the mecA gene, more than 40 additional chromosomal loci, including both regulatory and structural genes, are known to modulate resistance phenotypes (5, 6). Some of these factors directly or indirectly influence processes impacting resistance, such as cell wall biosynthesis and autolysis, but the functions of several others and/or their roles in resistance are unknown. XdrA was found to bind to the mecA promoter/operator region but not to alter mecA transcription or PBP2a production (23), leaving the mechanism by which xdrA deletion increased resistance levels unclear.
Here we further characterized this protein by performing microarray analysis to identify the XdrA transcriptome in the clinical isolate CHE482, a rapidly growing MRSA strain which belongs to clonal complex 45 (CC45) and which is highly prevalent among MRSA strains circulating in Zurich, Switzerland (62). The gene with the greatest fold difference in transcription levels was spa. Here we confirmed the influence of xdrA deletion on spa transcription in different strain backgrounds and analyzed the interplay of XdrA with other key spa regulators. Our results indicated that XdrA acts as a direct activator of spa transcription.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Bacteria were grown on sheep blood or Luria-Bertani (LB) (Difco Laboratories, Detroit, MI) agar plates, and liquid cultures were grown in LB medium with shaking at 180 rpm at a broth/flask volume ratio of 1:4. Media were supplemented with the following antibiotics when appropriate: 50 μg/ml kanamycin, 10 μg/ml erythromycin, 10 μg/ml tetracycline, or 100 μg/ml ampicillin. Cell growth was measured by absorbance (optical density at 600 nm [OD600]).
TABLE 1.
Strains and plasmids
| Strain or plasmida | Relevant genotypeb | Reference or source |
|---|---|---|
| Strains | ||
| S. aureus | ||
| RN4220 | Restriction-negative derivative of NCTC8325-4 | 40 |
| CHE482 (1) | Clinical MRSA isolate; CC45, ST45, SCCmec type N1; blaZ SarT− SarU− | 23 |
| CHE482ΔxdrA (2) | CHE482 containing a markerless deletion of xdrA | 23 |
| ZH44 | Clinical MRSA isolate; CC8, ST8, SCCmec type II; kanamycin resistant | 23 |
| ZH44ΔxdrA | ZH44 containing a markerless deletion of xdrA | This study |
| COLn | Early clinical MRSA isolate COL cured of plasmid pT181; CC8, ST250, SCCmec type I | 39 |
| COLnΔxdrA | COLn containing a markerless deletion of xdrA | This study |
| Newman | Clinical methicillin-susceptible S. aureus strain (ATCC 25904); CC8, ST8 | 2, 22 |
| NewmanΔxdrA | Newman containing a markerless deletion of xdrA | This study |
| RN6911 | RN4220 Δagr::tetM | 55 |
| LR12 | RN4220 ΔsarA::tetL | This study |
| NM520 | CHE482 ΔsarA::ermB | This study |
| NM521 | CHE482 Δagr::ermB | This study |
| sarA strain (3) | CHE482 ΔsarA::tetL | This study |
| sarS strain (4) | CHE482 sarS::psarS | This study |
| agr strain (5) | CHE482 Δagr::tetM | This study |
| xdrA sarA strain (6) | CHE482 ΔxdrA ΔsarA::tetL | This study |
| xdrA sarS strain (7) | CHE482 ΔxdrA sarS::psarS | This study |
| sarS sarA strain (8) | CHE482 sarS::psarS ΔsarA::tetL | This study |
| xdrA agr strain (9) | CHE482 ΔxdrA Δagr::tetM | This study |
| sarS agr strain (10) | CHE482 sarS::psarS Δagr::tetM | This study |
| sarA agr strain (11) | CHE482 ΔsarA::ermB Δagr::tetM | This study |
| xdrA sarS sarA strain (12) | CHE482 ΔxdrA sarS::psarS ΔsarA::tetL | This study |
| xdrA sarS agr strain (13) | CHE482 ΔxdrA sarS::psarS Δagr::tetM | This study |
| xdrA sarA agr strain (14) | CHE482 ΔxdrA ΔsarA::ermB Δagr::tetM | This study |
| sarS sarA agr strain (15) | CHE482 sarS::psarS ΔsarA::ermB Δagr::tetM | This study |
| xdrA sarS sarA agr strain (16) | CHE482 ΔxdrA sarS::psarS ΔsarA::ermB Δagr::tetM | This study |
| E. coli DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 λ− | Invitrogen |
| Plasmids | ||
| pBUS1 | S. aureus-E. coli shuttle vector; tetL | 63 |
| pAW17 | S. aureus-E. coli shuttle vector; aac-aph | 63 |
| pAZ106 | Suicide vector containing promoterless lacZ reporter gene; ermB | 14 |
| pBUS-lacZ | pBUS1 containing a 4.5-kb EcoRI/Asp718 fragment from pAZ106, carrying the promoterless lacZ reporter gene; tetL | This study |
| pEC1 | E. coli plasmid; ori ColE1 bla ermB | 9 |
| pAD21 | S. aureus suicide vector derived from pAW17 by removal of pAMα1-ori | 52 |
| pAD21Δagr | pAD21 with a 3.3-kb fragment comprising ermB flanked by the upstream and downstream sequences of the agr operon; aac-aph | This study |
| pAD21ΔsarA | pAD21 with a 3.3-kb fragment comprising ermB flanked by the upstream and downstream sequences of sarA; aac-aph | This study |
| psarS | pAD21 with a 390-bp insert containing an internal portion of the COL sarS gene; aac-aph | 52 |
| pME26 | pAW17 containing the xdrA gene; aac-aph | 23 |
| pME27 | pBUS1 containing the xdrA gene; tetL | 23 |
Numbers in parentheses are alternate strain designations.
CC, clonal complex; ST, sequence type; SCCmec, staphylococcal cassette chromosome mec.
Microarrays.
Overnight cultures of S. aureus were diluted 100-fold into fresh prewarmed LB medium and were grown to an OD of 0.5. Cells were then mixed (1:2) with RNAprotect bacterial reagent (Qiagen), incubated at room temperature for 5 min, harvested by centrifugation at 4°C, and snap-frozen. RNA isolation was performed as described by Cheung et al. (16), and the RNeasy kit, with on-column DNase I digestion (Qiagen), was used for RNA purification. RNA integrity was checked on an Agilent BioAnalyser, model 2100. cDNA was synthesized using SuperScript II reverse transcriptase and random hexamer primers (Invitrogen) and was labeled with either Cy3- or Cy5-dCTP (GE Healthcare). Microarray analysis was performed using the Bacterial Microarray Group at St. George's University of London (BμG@S) SAv1.1.0 microarray, and hybridization was carried out as described by Witney et al. (75). The array design is available in BμG@Sbase (http://bugs.sgul.ac.uk/A-BUGS-17) and ArrayExpress under accession no. A-BUGS-17. Slides were scanned using an Affymetrix 428 scanner, and data were extracted using BlueFuse, version 3.0 (BlueGnome, Cambridge, United Kingdom). Three biological replicates of both wild-type and mutant strains were hybridized against each other in dye-swap experiments, and data from all six arrays were normalized and analyzed using GeneSpring, version 7.2 (Silicon Genetics). The list of differentially expressed genes (see Table 3) includes those with a 3-fold or greater change in the expression level and P values of ≤0.05 by the t test, with the Benjamini and Hochberg false-discovery-rate correction applied.
Detection of the sarT-sarU genomic region.
Primers sarT-U.F and sarT-U.R (Table 2) were used to detect the core variable RD5 genomic region by PCR. These primers were also used to amplify a single digoxigenin (DIG)-labeled probe for the simultaneous detection of sarT and sarU transcripts by Northern blot analysis.
TABLE 2.
Primers used in this study
| Primer function and name | Nucleotide sequence (5′-3′)a | Source or reference |
|---|---|---|
| Construction of plasmids for allelic replacement | ||
| agr.upF | ATTTA GAATTC ATATGAATGCTGAAGTAGAT | This study |
| agr.upR | ATTAA GGATCC GTTATCTTCGTATAGTACTA | This study |
| agr.downF | ATTTA CTGCAG TTCAATTGTAAATCTTGTTG | This study |
| agr.downR | AATAT AAGCTT TTGATACATCTAGTATAGAA | This study |
| sarA.upF | ATTTA GAATTC GAGACTTTATTCATATGCTT | This study |
| sarA.upR | ATTTA GGATCC CTTAAGAATGAGTTGACTAT | This study |
| sarA.downF | ATTTA CTGCAG TGATAGATGATACATTCTAT | This study |
| sarA.downR | ATTAT AAGCTT TTATCTTCACGTACAAGATT | This study |
| Primer extension | ||
| spa.PEbio1 | BIO-TGTTACGCCACCAGATATAA | This study |
| spa.PEbio3 | BIO-ACCTAGTTTACGAATTGAATA | This study |
| Amplification of DIG-labeled probes | ||
| SA1665F | TTCGTATAGAGGCTGGTTAG | 23 |
| SA1665R | AATTGGTTGGTTATCTGGAT | 23 |
| spaF | TGTAGGTATTGCATCTGTAA | 52 |
| spaR | AAGTTAGGCATATTCAAGAT | 52 |
| sarSF | TTGATGAGCGTAATACTTAC | 52 |
| sarSR | GAGCTAATAATTGTTCAGCA | 52 |
| RNAIII.F | TATAACTAGATCACAGAGATG | This study |
| RNAIII.R | TTAGTACTATACGAAGATAAC | This study |
| sarA.F3 | TATTGACATACATCAGCGAA | This study |
| sarA.R1 | GTTTGCTTCAGTGATTCGTT | This study |
| fadXF | TAGCGAATGATATTCATAGT | This study |
| fadXR | TAGTTGATAGTCGTCAACTA | This study |
| SAR0995F | ATCATTGAACGCTCTTGCAA | This study |
| SAR0995R | ATAGCGTGTCAATGCTTCAA | This study |
| SAR0996F | CTAATGTTAAACATACAACT | This study |
| SAR0996R | TATTCTGCCGTATATTGATT | This study |
| SAR1741F | GTATGAATTAATGCCGATTA | This study |
| SAR1741R | CGTCACCATAACCAATATAT | This study |
| SAR2413F | CAACGCTTACAAGGCTATAA | This study |
| SAR2413R | GCTACACAGTTCACTCTAAT | This study |
| SAR2589F | GGTTATGCATTAATGCAAGT | This study |
| SAR2589R | TAACGTGGAATACTTCTGTT | This study |
| mtlAF | CCAATGTTACTTGGTGCAAT | This study |
| mtlAR | TAAGTTGCAACACCAGTCAT | This study |
| sarT-U.F | GATTGGTCTATACCGATATA | This study |
| sarT-U.R | GTTGTATTGGAGTAACTGAA | This study |
| Amplification of spa promoter fragments for lacZ reporter gene constructs | ||
| spa.lacZF1 | AATTT GGATCC GAATCAATTATTAGCAGATAA | This study |
| spa.lacZF2 | AATTT GGATCC TCAGCACATAATGAACAACTT | This study |
| spa.lacZF3 | AATTT GGATCC TGTATTAAACCGCTTTCATTA | This study |
| spa.lacZF4 | AATTT GGATCC ACTTCCTGAATAAATCTTTCA | This study |
| spa.lacZF5 | AATTT GGATCC AAATATCTCTATATTTTATC | This study |
| spa.lacZF6 | AATTT GGATCC TATTAATCGAAATAGCGTGA | This study |
| spa.lacZF7 | AATTT GGATCC TTATAAGTTGTAAAACTTAC | This study |
| spa.lacZF8 | AATTT GGATCC AATATAGATTTTAGTATTGC | This study |
| spa.lacZF9 | AATTT GGATCC GTATTGCAATACATAATTCG | This study |
| spa.lacZR | AATTT GAATTC ACCTAGTTTACGAATTGAATA | This study |
Restriction sites are underlined; BIO denotes 5′ biotinylation.
Deletion mutants.
The original sarA and agr mutations in strains LR12 and RN6911, respectively, both carried a tetracycline resistance marker. In order to create a complete set of regulatory mutants, both loci were also deleted by allelic replacement, using the erythromycin resistance gene (ermB) from pEC1. To create the new agr operon and sarA deletion mutants, regions from both upstream and downstream of these loci were amplified from S. aureus COL and cloned into pEC1 on either side of the ermB gene. To delete the agr operon, a 1.256-kb region upstream of RNAIII was amplified using primers agr.upF and agr.UpR, and a 1.455-bp fragment downstream of agrA was amplified using primers agr.downF and agr.downR. To delete the sarA gene, a 1.103-kb fragment upstream of sarA was amplified using primers sarA.upF and sarA.upR, and a 1.204-kb region downstream of sarA was amplified using primers sarA.downF and sarA.downR. The primers are listed in Table 2.
pEC1 constructs isolated from E. coli DH5α were then digested with HindIII and EcoRI, and the fragments containing the upstream and downstream regions flanking ermB were subcloned into the S. aureus suicide plasmid pAD21 to create pAD21Δagr and pAD21ΔsarA. These plasmids were electroporated into S. aureus strain RN4220, and erythromycin-resistant transformants were screened for loss of kanamycin resistance.
All regulatory mutations (interruption of sarS with pAD21, replacement of sarA by insertion of either ermB or tetL, or replacement of the agr operon by insertion of either ermB or tetM) were transduced into either CHE482 or CHE482ΔxdrA using phage 80α, in order to create the full set of single, double, triple, and quadruple regulatory mutants. The genotypes of all mutants were confirmed by Southern blotting (data not shown).
Northern blotting.
Overnight cultures were diluted to an OD of 0.05 in fresh prewarmed LB medium. To monitor the temporal patterns of gene transcription, cultures were sampled at five different growth stages. Total RNA was extracted as described by Cheung et al. (16). RNA samples (8 μg) were separated in a 1.5% agarose-20 mM guanidine thiocyanate gel. DIG-labeled probes were amplified using the PCR DIG probe synthesis kit (Roche). Table 2 lists the primer pairs used for the amplification of DIG-labeled probes. All Northern blot experiments were repeated at least twice using at least two independently isolated RNA samples.
Western blotting.
Cell envelope proteins were prepared as previously described (68). Bacteria were harvested at an OD of 4.0 and were resuspended in SMM buffer (0.5 M sucrose, 0.02 M maleate, 0.02 M MgCl2 [pH 6.5]). Lysostaphin (100 mg/liter) was added, and cells were lysed at 37°C. Protoplasts were centrifuged, and the supernatant containing cell envelope proteins was precipitated with 10% trichloroacetic acid. Proteins were resuspended in phosphate-buffered saline (PBS). Proteins (500 ng per sample) were separated in 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and were electroblotted onto polyvinylidene difluoride (PVDF) membranes (Millipore, Zug, Switzerland). Protein A was detected using a goat anti-mouse IgG antibody conjugated to horseradish peroxidase (dilution, 1:5,000). Protein detection was performed using SuperSignal West Pico chemiluminescent substrate (Thermo Scientific, Lausanne, Switzerland).
Hemolysis.
Hemolytic activities were compared on sheep blood agar plates. Strains were grown overnight in LB medium in a 96-well microtiter dish at 37°C. Cultures were then stamped onto a sheep blood agar plate, which was incubated first at 37°C for 24 h and then at 4°C for 48 h before being scanned.
Primer extension.
RNA was extracted from CHE482 cultures that were grown to an OD of 1.0, as described above. Primer extension reactions were performed with 20 μg of total RNA and 3 pmol of the 5′-biotin-labeled primers spa.PEbio1 and spa.PEbio3 (Table 2), using Superscript II reverse transcriptase (Invitrogen), according to the manufacturer's instructions. Sequencing reactions were performed using the Thermo Sequenase cycle sequencing kit (U.S. Biochemicals). The Biotin Chromogenic Detection kit (Fermentas) was used for detection.
lacZ reporter gene fusions.
Nine different-length fragments of the spa promoter region were amplified, all using the reverse primer spa.lacZR and one of the forward primers spa.lacZF1 to -F9. Fragments were ligated upstream of the promoterless lacZ gene in the E. coli-S. aureus shuttle vector pBUS-lacZ, and ligations were transformed into E. coli DH5α. The resulting fusion constructs, pspap-lacZ1 to pspap-lacZ9 (p1 to p9), were then transformed into S. aureus RN4220 before being transduced into CHE482 and its corresponding xdrA, sarS, sarA, and agr regulatory mutants.
β-Galactosidase activity.
β-Galactosidase activity was compared qualitatively by patching strains containing spa promoter-lacZ fusions onto LB medium containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (Xgal; 40 μg/ml) (Fermentas) and incubating the plates overnight at 37°C. β-Galactosidase activity was also measured quantitatively by performing o-nitrophenyl-β-d-galactopyranoside (ONPG) (Sigma) cleavage assays according to the standard protocol (28).
Microarray data accession numbers.
Fully annotated microarray data from this study have been deposited in BμG@Sbase (http://bugs.sgul.ac.uk/E-BUGS-105) and ArrayExpress under accession no. E-BUGS-105.
RESULTS
Differential gene expression resulting from xdrA deletion.
Transcriptional profiling had previously shown that xdrA was strongly expressed during early growth stages, with transcription beginning to decrease between ODs of 1.0 and 2.0 (23). Therefore, microarrays to compare the whole-genome transcriptional profiles of wild-type S. aureus CHE482 versus CHE482ΔxdrA were performed on RNA harvested at an OD of 0.5, at maximal xdrA expression. Statistical analysis of the results indicated that potentially 24 ORFs were upregulated and 26 downregulated by a factor of 3-fold or more in the xdrA deletion mutant (Table 3).
TABLE 3.
ORFs differentially regulated by xdrA deletion in S. aureus CHE482
| Gene IDa | Name | Gene product | Fold change in expressione |
|---|---|---|---|
| Upregulated genes | |||
| SAR0153 | capC | Capsular polysaccharide synthesis enzyme | 4.2 |
| SAR0154 | capD | Capsular polysaccharide synthesis enzyme | 3.4 |
| SAR0179 | Putative transporter protein | 3.8 | |
| SAR0227* | fadX | Putative acetyl-CoA transferase | 9 |
| SAR0228 | Putative glutamine amidotransferase class I | 3 | |
| SAR0420 | Putative membrane protein | 3.5 | |
| SAR0995* | Putative regulatory protein | 13.4 | |
| SAR1740 | DNA repair protein (partial) | 13.5 | |
| SAR1741* | Type III leader peptidase family protein | 14.7 | |
| SAR1812 | acuA | Acetoin utilization protein | 5.5 |
| SAR1813 | Histone deacetylase family protein | 4.1 | |
| SAR2396 | DeoR family regulatory protein | 4.1 | |
| SAR2413* | Putative short chain dehydrogenase | 8.7 | |
| SAR2587 | Hypothetical protein | 4.3 | |
| SAR2610 | Putative l-serine dehydratase, alpha chain | 4.7 | |
| SAR2611 | Putative l-serine dehydratase, beta chain | 4.9 | |
| SAR2612 | Putative membrane protein | 5.3 | |
| SAR2643 | crtM | Squalene desaturase (pseudogene) | 4 |
| SAR2645 | Putative glycosyl transferase | 5 | |
| SAR2646 | Putative phytoene dehydrogenase related protein | 4.3 | |
| SAR2647 | Putative membrane protein | 3.6 | |
| SAR2762 | Hypothetical protein | 3.7 | |
| MW0377b | Conserved hypothetical protein | 12 | |
| MW0378b | Conserved hypothetical protein | 14.3 | |
| Downregulated genes | |||
| SAR0114* | spa | Immunoglobulin G binding protein A precursor | 32.5 |
| SAR0178 | Putative d-isomer specific 2-hydroxyacid dehydrogenase | 3.2 | |
| SAR0435 | Exotoxin | 7.8 | |
| SAR0696 | Putative exported protein | 3.1 | |
| SAR0787 | sstA | FecCD transport family protein | 3.1 |
| SAR0788 | sstB | FecCD transport family protein | 3.0 |
| SAR0789 | sstC | ABC transporter ATP-binding protein | 3.2 |
| SAR0790 | sstD | Lipoprotein | 3.3 |
| SAR0806 | Putative S30EA family ribosomal protein | 5.3 | |
| SAR0866 | Hypothetical protein | 3.2 | |
| SAR0996* | Conserved hypothetical protein | 7.7 | |
| SAR1041 | Conserved hypothetical protein | 3.4 | |
| SAR1042 | purQ | Putative phosphoribosylformylglycinamidine synthase I | 4.3 |
| SAR1043 | purL | Putative phosphoribosylformylglycinamidine synthase II | 3.2 |
| SAR1450 | tdcB | Putative threonine dehydratase | 3.5 |
| SAR1451 | ald2 | Alanine dehydrogenase 2 | 4.8 |
| SAR1849 | Proline dehydrogenase | 3.2 | |
| SAR1938 (SA1665; xdrA) | Putative DNA-binding protein | 71.4 | |
| SAR2244* | mtlA | PTS system, mannitol-specific IIBC component | 15.2 |
| SAR2245 | Putative transcriptional antiterminator | 5.5 | |
| SAR2247 | mtlD | Putative mannitol-1-phosphate 5-dehydrogenase | 3.5 |
| SAR2589* | Putative transporter protein | 6.4 | |
| SAR2593 | Putative transcriptional regulator | 3.5 | |
| SAR2605 | ddh | d-Specific d-2-hydroxyacid dehydrogenase | 3.3 |
| SACOL0478c | Exotoxin 3 | 8.6 | |
| SA0393d | set15 | Exotoxin 15 | 8.1 |
Gene identifications (IDs) refer to S. aureus strain MRSA252 unless otherwise indicated. Genes selected for Northern blot analysis are marked with asterisks.
Gene probes on microarrays were obtained from S. aureus MW2.
Gene probes on microarrays were obtained from S. aureus COL.
Gene probes on microarrays were obtained from S. aureus N315.
P < 0.05.
Confirmation of microarray results by Northern blot analysis.
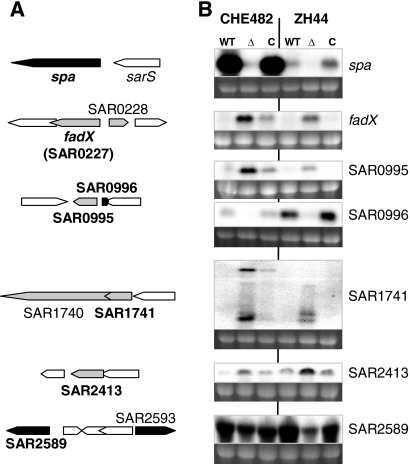
Northern blotting was used to confirm the transcriptional profiles of regulated ORFs. Four upregulated and four downregulated ORFs with the highest differential expression were selected (Table 3, asterisks). Transcription was compared in two different strain backgrounds: CHE482 (CHE482, CHE482ΔxdrA, and the trans-complemented mutant CHE482ΔxdrA pME26) and ZH44 (ZH44, ZH44ΔxdrA, and the trans-complemented mutant ZH44ΔxdrA pME27). The selected ORFs and their genomic contexts, along with the corresponding Northern blots, are shown in Fig. 1.
FIG. 1.
Northern blotting of ORFs up- or downregulated from microarrays of CHE482 against CHE482ΔxdrA. (A) Locus maps of selected ORFs either upregulated (gray) or downregulated (black) in CHE482ΔxdrA. The designations of ORFs used as probes for Northern blotting are shown in boldface. (B) Northern blots of RNA extracted from strains CHE482 and ZH44: WT, wild type; Δ, ΔxdrA deletion mutant; C, mutant complemented with xdrA in trans. ORFs/genes used as probes are given on the right. Ethidium bromide-stained 16S rRNA bands are shown beneath the transcripts as an indication of RNA loading.
All upregulated ORFs tested, including fadX (encoding a putative acetyl coenzyme A [acetyl-CoA] transferase), SAR0995 (encoding a conserved hypothetical protein), SAR1741 (encoding a type III leader peptidase family protein), and SAR2413 (encoding a putative short chain dehydrogenase), gave very weak signals in the parent strains but clear upregulation upon XdrA inactivation, with at least partial restoration to wild-type levels in the trans-complemented mutants, confirming the microarray data. The microarrays indicated that the largest transcriptional alteration was the downregulation of spa in CHE482ΔxdrA, by a factor of 32.2 fold. Northern blot analysis confirmed strong attenuation of spa transcription in both strain backgrounds, with wild-type levels of transcription restored by trans-complementation. Other downregulated ORFs that were confirmed included SAR0996, encoding a putative regulatory protein, which is transcribed divergently from the upregulated ORF SAR0995, and SAR2589, encoding a protein of unknown function with similarity to small-molecule transporters. SAR2589 was strongly expressed in both strain backgrounds, and transcription decreased by significant amounts in both xdrA mutants, increasing again upon complementation with xdrA in trans. Decreased transcription of SAR0996 by XdrA inactivation, and complementation in trans, was also confirmed in both strains. The inverse regulation of SAR0996 and the divergently transcribed SAR0995 suggested that there may be promoter exclusion controlled by the presence/absence of XdrA, although this requires further investigation. Northern blotting could not, however, confirm the 15-fold downregulation of mtlA, which forms part of a mannitol-specific phosphotransferase (PTS) system (data not shown); the reasons for this discrepancy also need further exploration.
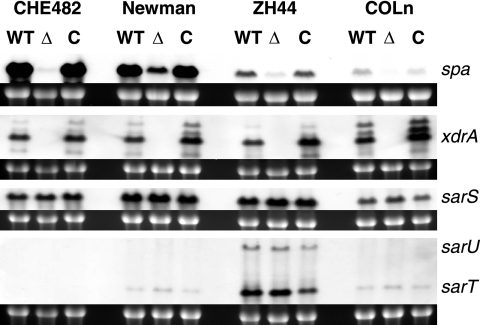
Effect of xdrA deletion on the transcription of spa, sarS, sarU, and sarT.
The effect of xdrA deletion on the transcription of spa was monitored and compared in different strain backgrounds, including strain CHE482, which belongs to CC45, and three strains belonging to CC8: strain Newman, the clinical isolate ZH44, and the highly methicillin resistant strain COLn. Although levels of spa expression differed greatly between these strains, transcription was always significantly decreased by xdrA deletion and fully restored by trans-complementation (Fig. 2). Levels of spa transcription were also compared to levels of xdrA, sarS, sarT, and sarU within these different strain backgrounds. These Northern blot analyses showed that levels of xdrA were very consistent in all wild-type strains tested. A PCR screen revealed that the core variable region containing sarT and sarU was absent from CHE482 but present in the other three strains. Northern blot analyses confirmed that sarT and sarU were not present in CHE482 and showed variability in the levels of sarS, sarT, and sarU among the different strain backgrounds; however, no specific relationships between the levels of these regulators and the overall levels of spa transcription could be ascertained (Fig. 2).
FIG. 2.
Northern blot comparison of spa transcription in different strain backgrounds. RNA was extracted from strains CHE482, Newman, ZH44, and COLn. WT, wild type; Δ, ΔxdrA deletion mutant; C, mutant complemented with xdrA in trans. Levels of spa transcription differed greatly among the four different strain backgrounds; however, spa levels were significantly decreased in all xdrA deletion mutants and were restored to wild-type levels by trans-complementation. Northern blotting was also performed to compare the levels of spa with levels of xdrA, sarS, sarU, and sarT present in each of the strains. Probes used for hybridization are given on the right. Ethidium bromide-stained 16S rRNA bands are shown beneath the transcripts as an indication of RNA loading.
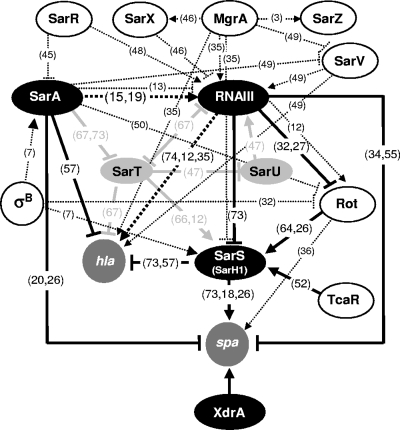
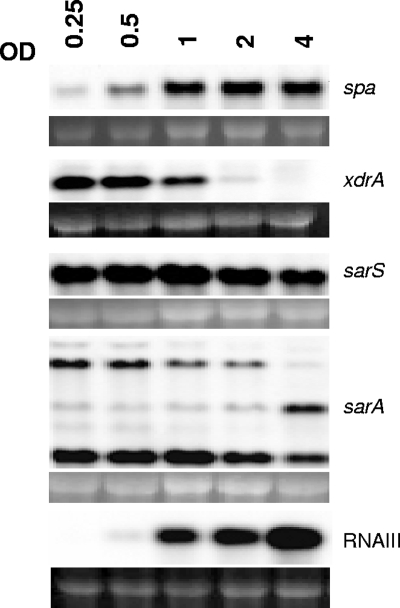
Transcriptional profiling of spa and selected regulators in CHE482 throughout growth.
The regulatory network controlling spa transcription is extensive (25, 26, 30, 34, 56, 61, 72). Currently at least 15 different regulatory elements have been shown to influence levels of spa transcription; some of these are shown in Fig. 3. So far, SarS, SarA, and RNAIII have been found to be the main regulators shown to act directly on the spa promoter; most other regulators influence transcription indirectly, through modulation of one or more of these regulators. The microarray results indicated that XdrA did not influence spa indirectly via one of its known regulators, because the transcriptome data set for XdrA did not overlap significantly with those previously published for any other regulator in the SAMMD database (53). Like that of many MSCRAMM proteins, protein A expression is growth stage dependent. RNA of CHE482 was sampled throughout growth, at ODs of 0.25, 0.5, 1, 2, and 4, and was used in Northern blot analyses to compare the temporal transcription of spa to those of its direct regulators sarS, sarA, and RNAIII, and to that of xdrA (Fig. 4). spa was transcribed weakly during early growth, with transcription increasing during exponential growth and decreasing only slightly by the final sampling point at an OD of 4.0. The major spa transcriptional activator SarS was transcribed at a consistently strong level throughout all growth stages, decreasing slightly at an OD of 4.0. SarA is proposed to displace SarS during the post-exponential growth phase, consequently decreasing spa transcript activation (26). Northern blotting of sarA showed transcription over all growth stages measured; however, sarA transcription profiles are difficult to interpret and correlate with SarA activity because of growth-phase dependent modulation in transcript initiation from the three different sarA promoters (15). RNAIII transcription was only barely detectable at an OD of 0.5 but then increased steadily throughout exponential growth and into post-exponential growth. This correlates with the published mechanism of spa repression by RNAIII, whereby RNAIII accumulation during post-exponential growth represses spa transcription. Although the microarray results and Northern blot analyses all indicated that XdrA had a positive regulatory effect on spa transcription, Fig. 4 shows an inverse relationship between spa and xdrA transcription, with xdrA transcripts steadily decreasing over growth, as spa transcripts increase.
FIG. 3.
Regulatory network controlling spa and hla expression. Regulatory loci directly or indirectly influencing spa transcription, either positively (arrows) or negatively (blocked arrows), are indicated. Regulators analyzed here are represented by filled ovals. SarT and SarU, shown as light shaded ovals, form part of the S. aureus core variable region RD5, which is absent from strain CHE482. Other regulatory connections reported in the literature are shown by broken lines. These regulatory connections are documented in references 3, 7, 12, 13, 15, 18-20, 26, 27, 32, 34-36, 45-50, 52, 55, 57, 64, 66, 67, 73, and 74. Reference numbers are given in parentheses on the figure.
FIG. 4.
Transcriptional profiles of spa and the regulators xdrA, sarS, sarA, and RNAIII over exponential growth and into early stationary phase. Northern blotting was performed on RNA extracted from strain CHE482 at the OD values indicated. Ethidium bromide-stained 16S rRNA bands are shown beneath the transcripts as an indication of RNA loading.
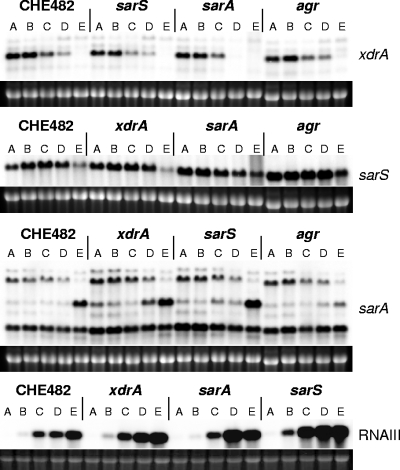
Impact of regulator mutations on spa transcription.
A series of mutants were constructed to monitor spa transcription in the presence and absence of the four regulators: SarA, SarS, RNAIII, and XdrA. Fifteen mutants were constructed, including single mutants of all regulators, all combinations of double and triple mutants, and a quadruple mutant devoid of all four regulators (Table 1). Because of differences in the growth rates of some mutants, RNA was sampled at several time points over growth at standardized ODs, in order to produce comprehensive transcriptional profiles for each of the strains (Fig. 5A). Northern blot profiling of all mutants required the loading of samples onto four different gels. Wild-type CHE482 samples were loaded onto each gel as a control, to enable transcription comparisons between different gels (Fig. 5B). Profiling of the single regulatory mutants showed that xdrA deletion led to a massive decrease in spa transcription over all growth stages tested, and an even larger decrease, with no distinct spa transcripts detected, was observed in sarS mutants. Deletion of sarA or the agr operon interfered with the temporal expression of spa. In the sarA mutant, transcription was much stronger than in the wild type at the first two sampling stages, although it was very similar to that in the wild type over the last three samples, indicating that SarA represses spa transcription during early-exponential growth. In the agr mutant, transcription levels also appeared higher in both the first and the last samples, indicating derepression of spa during both early and late growth stages.
FIG. 5.
Northern blot analysis of spa transcription in CHE482 regulatory mutants. (A) Growth curves of regulatory mutants, showing the five OD sampling points for RNA extraction. (B) Profiles of spa transcription over growth in wild-type CHE482 and strains containing single, double, triple, or quadruple mutations in the regulatory loci xdrA, sarS, sarA, and agr. RNA was extracted from cultures harvested at OD values of 0.25, 0.5, 1, 2, and 4 (lanes A to E, respectively). Ethidium bromide-stained 16S rRNA bands are shown beneath the transcripts as an indication of RNA loading.
Transcription patterns became much more complex in the double, triple and quadruple mutants. Analysis of the double mutants showed some expected results, in that when both activators were absent (SarS and XdrA), there was no detectable transcription, whereas when both repressors (SarA and RNAIII) were deleted, there was derepression in both the earliest and the latest samples. Deletion of both repressors did not appear to have an additive effect: transcript levels in the double mutant did not appear significantly higher than those in either of the single mutants. Complicated patterns emerged, however, when combinations of activators and repressors were inactivated together. For instance, analysis of single mutants indicated that SarS was essential for initiating spa transcription; however, when one or more of the repressors were also deleted, transcription could once again be detected. Transcription in the xdrA mutant also increased significantly when sarA, agr, or both were deleted. Transcription in both sarS and xdrA mutants was higher throughout growth when sarA was deleted than when agr was missing, suggesting that SarA is a stronger repressor of spa than RNAIII, and was much greater when both repressors were absent. Transcription was also higher when repressors were inactivated in the xdrA mutant background than in the sarS mutant background, suggesting again that SarS is the stronger spa activator. In double and triple mutants, the effects of sarA and agr mutation on the growth phase-dependent expression of spa were even more exaggerated, with sarA mutation enhancing spa transcription during early growth and agr mutation increasing expression levels in the later samples. Residual levels of transcription were low but clearly detectable in the quadruple mutant.
Western blot analysis of protein A production.
Cell wall-associated proteins were isolated from the wild type and all 15 regulatory mutants at an OD of 4.0 (Fig. 6). Western blot analysis showed that the levels of protein A produced closely mirrored the levels of spa transcription in each of the strains tested. The xdrA deletion greatly decreased amounts of cell wall-associated protein A in all combinations of regulatory mutants tested, albeit with the weakest effect in the agr mutant. Proteolytic cleavage of protein A was observed in all sarA mutants, suggesting an upregulation of proteases, as is generally observed in sarA mutants (37, 38).
FIG. 6.
Protein A expression in spa regulatory mutants. Cell envelope proteins were harvested at an OD of 4.0 and were probed with a goat anti-mouse IgG antibody conjugated to horseradish peroxidase.
Effect of xdrA deletion on transcription of sarA, sarS, and RNAIII.
The microarray results indicated that XdrA did not influence any previously characterized S. aureus regulatory loci. Northern blotting confirmed that deletion of xdrA did not significantly influence the expression of sarS, sarA, or RNAIII, nor did mutation of any of these three regulators influence xdrA transcription (Fig. 7). Because of differences in growth kinetics between mutants, sampling points were standardized at defined ODs. It seems that agr deletion enhanced sarS transcription, as reported previously by Tegmark et al. (73), and that sarS inactivation, conversely, resulted in a slight derepression of RNAIII. sarS upregulation in the agr mutant may also contribute to the higher levels of protein A in agr and agr xdrA mutants than in the wild type (Fig. 6).
FIG. 7.
Transcriptional profiles of xdrA, sarS, sarA, and RNAIII in wild-type CHE482 and in the single xdrA, sarS, sarA, and agr regulatory mutants. RNA was extracted from cultures harvested at OD values of 0.25, 0.5, 1, 2, and 4 (lanes A to E, respectively). Ethidium bromide-stained 16S rRNA bands are shown beneath the transcripts as an indication of RNA loading.
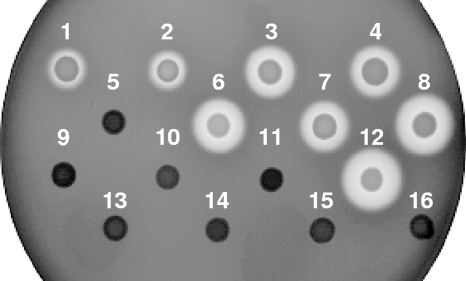
Hemolysis.
Hemolysis of CHE482 and the 15 regulatory mutants on sheep blood agar plates (Fig. 8) confirmed previously published findings, in that deletion of agr severely decreased hemolysis while deletion of sarS or sarA increased hemolysis (57, 74). Deletion of xdrA had no effect on hemolysis in CHE482, further indicating that XdrA influences spa directly and not through known regulators that also control hla expression.
FIG. 8.
Hemolytic activities of CHE482 regulatory mutants. Hemolysis zones surrounding colonies of regulatory mutants were compared on sheep blood agar. Numbers correspond to alternate strain designations in Table 1. Deletion of xdrA had no visible effect on autolysis. Disruption of sarS or deletion of sarA increased hemolysis, with an additive effect seen when both were missing (strains 8 and 12). Deletion of the agr operon had an overriding effect, abolishing hemolysis in all mutants.
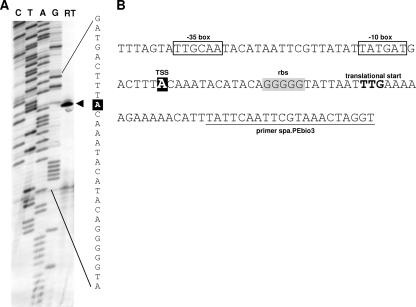
spa promoter analysis.
Primer extension was performed to confirm the transcriptional start site (TSS) for spa. The results from two different primers both showed transcription initiating at an adenine nucleotide (nt), 25 nt upstream of the spa TTG translational start codon. This TSS was 7 nt upstream of the previously published spa TSS (26) but corresponded to a faint band described in those investigators’ primer extension experiments (Fig. 9). DNase I footprinting experiments and electrophoretic mobility shift assays (EMSA) were performed on several biotin-labeled DNA fragments, covering different overlapping regions of the spa promoter. Unfortunately, the data from these experiments gave inconclusive results, which did not allow us to identify a localized XdrA binding site within the spa promoter (data not shown).
FIG. 9.
Primer extension determination of the spa TSS. (A) Lanes C, T, A, and G show the dideoxy-terminator sequencing ladder obtained using complementary dideoxy terminators; lane RT contains the reverse transcription product obtained using primer spa.PEbio3. The TSS is indicated by an arrowhead, and the corresponding nucleotide is highlighted in white on a black background. (B) Sequence of the spa promoter region. The TSS is shown in white on a black background; predicted −10 and −35 regions are boxed; the predicted ribosome binding site (rbs) is highlighted in gray; and the translational start site (TTG) of spa is shown in boldface. The sequence of primer spa.PEbio3 is underlined.
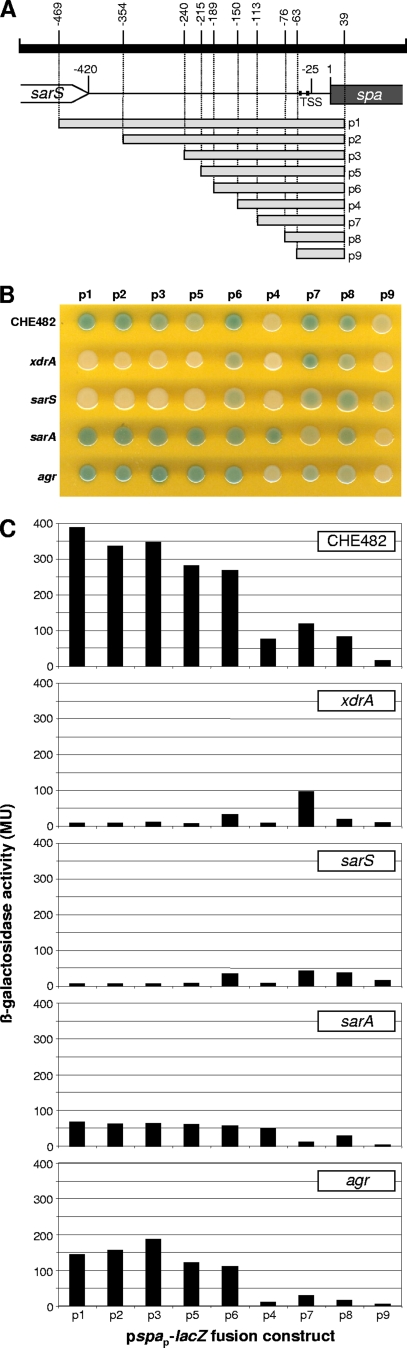
Therefore, a series of nine different lacZ fusion plasmids were constructed, containing successively smaller fragments of the spa promoter (Fig. 10A). These reporter gene plasmids, pspap-lacZ1 to pspap-lacZ9 (p1 to p9), were then transformed into RN4220, from which they were transduced into CHE482 and the single xdrA, sarS, sarA, and agr regulatory mutants. The phenotypes of all fusion-containing strains were then compared by growth on LB agar containing Xgal (Fig. 10B) and by ONPG cleavage assays (Fig. 10C).
FIG. 10.
Identification of regulatory regions within the spa promoter of CHE482. (A) Map showing regions of the spa promoter that were fused to lacZ to create fusion constructs p1 to p9. Fusion constructs were introduced into wild-type CHE482 and the single regulatory mutants lacking xdrA, sarS, sarA, or the agr operon. (B) Phenotypes of fusion-containing strains grown on LB agar containing Xgal. (C) β-Galactosidase activity detected in 8-h cultures of fusion-containing strains.
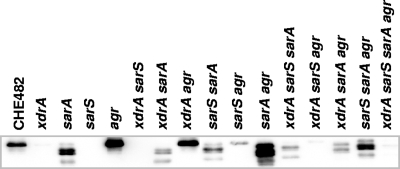
The results from fusion p1, containing the full-length spa promoter, closely mirrored those from Northern blotting; the wild type and the agr and sarA mutants all appeared dark blue on LB-Xgal plates, and the xdrA and sarS mutants appeared white (Fig. 10B). The results from the remaining fusion constructs, representing a series of nested spa promoter deletions, identified the regions of the promoter influenced by inactivation of the four regulatory loci. Presumably, increased β-galactosidase activity represented either increased activator binding, reduced repressor binding, or a combination of both.
All strains containing fusion p9 produced very low levels of β-galactosidase, ranging from 3.6 to 16.9 Miller units (MU). These values, however, were higher than the negligible background levels of β-galactosidase detected from cultures of CHE482 containing the empty fusion vector pBUS-lacZ (<0.4 MU). The p9 construct contained a promoter fragment of 63 nt, extending just 3 nt upstream of the predicted −35 box. Consistently low level expression of this fusion in all strains indicated that this truncated promoter contained no cis-acting regulatory elements, and the low levels of basal spa transcription corresponded well with transcript levels seen in Northern blots when all four regulators were absent (Fig. 5B).
The expression of the fusion constructs in CHE482 showed how different promoter lengths influenced spa expression in the wild type. On LB-Xgal plates, expression levels appeared to be consistently high in plasmids p1 to p6, through truncation of the promoter from 469 to 189 nt, indicating that this region did not play a significant role in regulation. Expression then decreased significantly in fusion p4, once the region between nt −150 and −189 had been deleted, suggesting that this region is important for activator binding. Expression then increased again once the promoter had been truncated to 113 nt in fusion p7, decreasing again to a basal level in fusion p9. A similar pattern of expression was reflected in the ONPG hydrolysis results, with expression decreasing sharply in fusion p4 and increasing again in p7 and p8, although only to levels approximately one-half to one-third of those seen in the full-length promoter (Fig. 10C).
xdrA and sarS mutants gave very similar reporter gene expression profiles, indicating that they act in similar ways on the same or closely overlapping regions of the promoter. As in Northern blot analyses, there was minimal spa expression from full-length promoters in both sarS and xdrA mutants. When the promoters became truncated to 189 nt, expression increased significantly in both mutants. Transcription in these two backgrounds also increased in Northern blots when one or both of the repressors were missing, thereby indicating that when the promoter is truncated to 189 nt, one or more of the repressors can no longer bind. Hence, the promoter region between 189 to 215 nt appears important for repressor binding. The strongest levels of transcription in both sarS and xdrA mutants were seen when the promoter was truncated to between 76 and 150 nt, once again suggesting limited repressor binding when promoters were truncated to this length. The levels of ONPG hydrolysis from fusions in xdrA and sarS mutants agreed well with the Northern blot analysis results, with expression levels from p1 to p5 being lower in the sarS mutant than in the xdrA mutant (Fig. 10C).
The patterns of reporter gene expression in the agr and sarA mutants indicated that these two repressors acted on similar promoter regions. In both mutants, expression was consistently high in fusions p1 to p6. Expression of p4 decreased in the agr mutant, as it did in the other four strains; however, it remained high in the sarA mutant, indicating that the region between −113 and −150 was not as essential for transcript activation when sarA was missing. This could correspond with the higher levels of expression in Northern blots for the sarA xdrA sarS triple mutant than for the agr xdrA sarS triple mutant (Fig. 5B). When strains containing fusions p1 to p6 were grown on LB-Xgal plates, levels of β-galactosidase in sarA and agr mutants appeared as strong as in the wild type, if not stronger, which would be consistent with the levels of spa transcription observed in Northern blot analyses (Fig. 10B). However, ONPG cleavage assays, while showing expression profiles consistent with those seen on LB-Xgal plates, showed lower levels of β-galactosidase activity in 8-h cultures of the sarA and agr mutants than in the wild type (Fig. 10C). These relatively low levels compared to those in the wild type were also observed in ONPG assays performed on 16-h and 24-h cultures (data not shown). The reasons for the low levels of β-galactosidase activity in these cultures, especially for the sarA mutant, are unknown but are probably linked to changes in the growth phenotype and global regulation of strains lacking SarA.
DISCUSSION
The regulon of XdrA was found to comprise potentially more than 50 ORFs in S. aureus CHE482. None, however, had previously been shown to influence β-lactam resistance levels. Therefore, more in-depth characterization of the XdrA regulon, to identify which member(s) contributes to enhanced β-lactam resistance in the absence of xdrA, is ongoing.
The gene with the largest fold change in transcription was spa, suggesting that the spa promoter is one of the major targets of XdrA. Decreased spa transcription and successful trans-complementation to wild-type levels was confirmed in four different strains that differed greatly in their levels of spa transcription and belonged to two unrelated clonal complexes; suggesting that the function of XdrA as a positive regulator of spa is widely conserved in S. aureus.
Strain CHE482 was chosen for extended analysis of spa regulation because of its high level of spa transcription and the subsequent dramatic decrease upon xdrA deletion. Regulation was also likely to be somewhat simplified in CHE482, since it lacked the RD5 core variable genomic region containing the regulators SarT and SarU, which play a central role in feedback regulatory circuits between sarA, sarS, and RNAIII (12, 66, 67). Northern blot analyses showed that the regulatory loci SarS, SarA, RNAIII, and XdrA had little transcriptional influence on one another in strain CHE482.
To assess how XdrA fits into the complex web of spa regulation, spa transcription levels throughout growth were compared in a series of regulatory mutants. The temporal expression patterns of spa have been shown to be highly strain variable, especially during post-exponential growth phases, with transcription decreasing significantly in some backgrounds while remaining much more stable in others (10). In CHE482, spa transcription increased steadily throughout exponential growth and decreased only slightly thereafter. The transcriptional profiles of single sarS, sarA, and agr mutants correlated well with previously published findings (73); no discernible spa transcripts were present in the sarS mutant, in contrast with increased transcription in the sarA and agr mutants. Transcription was markedly decreased in the xdrA mutant at all sampling points.
Levels of spa transcription increased again in both sarS and xdrA mutant backgrounds when either sarA or agr, or both, was also deleted. Mutants containing different combinations of activators and repressors gave rise to different levels and temporal patterns of spa transcription, showing the relative contributions of each of the regulatory loci at the different growth stages tested. Overall, spa transcript levels were lowest when SarS was inactivated, followed by the levels observed when xdrA was deleted; levels were increased, especially at later sampling points, when agr was absent and were highest when sarA was deleted. The effect of agr deletion on spa transcription in CHE482 was much less pronounced than that reported for NCTC8325-derived strains, substantiating previous reports that the effect of agr on the temporal expression of spa is strongly strain dependent (12).
High levels of spa transcription in sarS sarA double mutants previously led Gao and Stewart (26) to hypothesize that the role of SarS was not to activate the transcription of spa but to occlude repressor binding. Our results, from double, triple, and quadruple regulatory mutants, partially agree, in that they show that neither SarS nor XdrA is essential for the initiation of spa transcription and that relatively high levels of transcription are reached in corresponding sarA or agr double or triple mutants. However, our results also showed that transcription levels in double, triple, and quadruple mutants lacking one or more of the activators (SarS or XdrA) are never as high as in the wild type. The low levels of transcription in the xdrA sarS sarA and xdrA sarS agr triple mutants and in the quadruple mutant, which contains neither repressor, suggest that SarS and XdrA do stimulate transcription and that their function is not only to obstruct repressor binding. Otherwise, wild-type transcription should be restored by deleting both repressors. The same logic argues that XdrA can activate spa transcription in the absence of the other three regulators, since transcription in the sarS sarA agr triple mutant is significantly higher than in the quadruple mutant, where xdrA is also deleted.
Levels of β-galactosidase activity from reporter gene fusions, containing nested deletions of the spa promoter, identified potential cis-acting regulatory regions. Consistently low levels of p9 expression in all strains indicated that this minimal promoter, extending only 3 nt upstream of the predicted −35 promoter element, initiated a basal level of expression but lacked any cis-acting regulatory regions required for induction. The relative expression levels of other fusion plasmids in the wild-type strain CHE482 and in mutants lacking xdrA, sarS, sarA, or agr indicated that regions between p5 and p6 (nt −215 to −189) and p4 and p7 (nt −150 to −113) were involved in repressor binding; the second region corresponds to a previously identified SarA-responsive promoter element, which would lie between nt −121 and −115 (26). The fusions also indicated that the region between p6 and p4 (nt −189 to −150) was required for activator binding. Expression from fusions p7 and p8 was higher than that from p9 in all strains, indicating that this region enhanced transcription, although various levels of p7 and p8 expression in the five strains made its exact role in regulation difficult to interpret. Fusion results from CHE482, showing lower overall expression in fusions p4, p7, and p8, correlated with previous observations that this operator region, extending from immediately upstream of the −35 box to approximately nt −137, was required for SarS binding and hence for full activation of spa transcription (26, 73).
For the scope of this study, however, the most important results are those showing that xdrA deletion and sarS inactivation had very similar effects on spa promoter fragments, indicating that they act on the same or closely overlapping regulatory sequences. This potential interaction of XdrA and SarS at the same cis elements is likely to account for the apparent redundancy between these two activators when one or more of the repressors are absent. Such a redundancy would, once again, account for the lower levels of spa transcription in the triple and quadruple mutants when both XdrA and SarS are missing. No such redundancy can be seen, however, when both repressors SarA and RNAIII are present, since no spa transcription was detected in the single sarS mutant, and only very low levels were detected in the single xdrA mutant.
Therefore, the results presented here indicate that XdrA is a major activator of spa that acts on the same cis-regulatory elements as SarS, or closely overlapping elements, within the spa promoter. All the current evidence suggests that XdrA regulates spa directly and does not join the interconnected regulatory network linking other well-characterized regulators of spa.
Acknowledgments
This study has been carried out with financial support from the Commission of the European Communities, specifically the Infectious Diseases research domain of the Health theme of the 7th Framework Programme, contract 241446, “The effects of antibiotic administration on the emergence and persistence of antibiotic-resistant bacteria in humans and on the composition of the indigenous microbiotas at various body sites” to N.M., and from the Swiss National Science Foundation (grant 31-117707 to B.B.-B.). We are grateful to the Wellcome Trust for funding of BμG@S (Bacterial Microarray Group at St George's, University of London) to provide microarray facilities and support.
Footnotes
Published ahead of print on 30 July 2010.
REFERENCES
- 1.Arvidson, S., and K. Tegmark. 2001. Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291:159-170. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., T. Bae, O. Schneewind, F. Takeuchi, and K. Hiramatsu. 2008. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 190:300-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballal, A., B. Ray, and A. C. Manna. 2009. sarZ, a sarA family gene, is transcriptionally activated by MgrA and is involved in the regulation of genes encoding exoproteins in Staphylococcus aureus. J. Bacteriol. 191:1656-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bekeredjian-Ding, I., S. Inamura, T. Giese, H. Moll, S. Endres, A. Sing, U. Zahringer, and G. Hartmann. 2007. Staphylococcus aureus protein A triggers T cell-independent B cell proliferation by sensitizing B cells for TLR2 ligands. J. Immunol. 178:2803-2812. [DOI] [PubMed] [Google Scholar]
- 5.Berger-Bächi, B., M. M. Senn, M. Ender, K. Seidl, J. Hübscher, B. Schulthess, R. Heusser, P. Stutzmann Meier, and N. McCallum. 2009. Resistance to beta-lactam antibiotics, p. 170-192. In K. B. Crossley, K. K. Jefferson, G. L. Archer, and V. G. Fowler (ed.), Staphylococci in human disease, 2nd ed. Wiley-Blackwell, West Sussen, United Kingdom.
- 6.Berger-Bächi, B., and M. Tschierske. 1998. Role of Fem factors in methicillin resistance. Drug Resist. Updat. 1:325-335. [DOI] [PubMed] [Google Scholar]
- 7.Bischoff, M., P. M. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bächi, and S. J. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus σB regulon. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bronner, S., H. Monteil, and G. Prévost. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28:183-200. [DOI] [PubMed] [Google Scholar]
- 9.Brückner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 10.Burian, M., C. Wolz, and C. Goerke. 2010. Regulatory adaptation of Staphylococcus aureus during nasal colonization of humans. PLoS One 5:e10040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burlak, C., C. H. Hammer, M. Robinson, A. R. Whitney, M. J. McGavin, B. N. Kreiswirth, and F. R. DeLeo. 2007. Global analysis of community-associated methicillin-resistant Staphylococcus aureus exoproteins reveals molecules produced in vitro and during infection. Cell. Microbiol. 9:1172-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cassat, J., P. M. Dunman, E. Murphy, S. J. Projan, K. E. Beenken, K. J. Palm, S. Yang, K. C. Rice, K. W. Bayles, and M. S. Smeltzer. 2006. Transcriptional profiling of a Staphylococcus aureus clinical isolate and its isogenic agr and sarA mutants reveals global differences in comparison to the laboratory strain RN6390. Microbiology 152:3075-3090. [DOI] [PubMed] [Google Scholar]
- 13.Chakrabarti, S. K., and T. K. Misra. 2000. SarA represses agr operon expression in a purified in vitro Staphylococcus aureus transcription system. J. Bacteriol. 182:5893-5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan, P. F., S. J. Foster, E. Ingham, and M. O. Clements. 1998. The Staphylococcus aureus alternative sigma factor σB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J. Bacteriol. 180:6082-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheung, A. L., M. G. Bayer, and J. H. Heinrichs. 1997. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J. Bacteriol. 179:3963-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung, A. L., K. J. Eberhardt, and V. A. Fischetti. 1994. A method to isolate RNA from Gram-positive bacteria and mycobacteria. Anal. Biochem. 222:511-514. [DOI] [PubMed] [Google Scholar]
- 17.Cheung, A. L., K. A. Nishina, M. P. Trotonda, and S. Tamber. 2008. The SarA protein family of Staphylococcus aureus. Int. J. Biochem. Cell Biol. 40:355-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung, A. L., K. Schmidt, B. Bateman, and A. C. Manna. 2001. SarS, a SarA homolog repressible by agr, is an activator of protein A synthesis in Staphylococcus aureus. Infect. Immun. 69:2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chien, Y., A. C. Manna, and A. L. Cheung. 1998. SarA level is a determinant of agr activation in Staphylococcus aureus. Mol. Microbiol. 30:991-1001. [DOI] [PubMed] [Google Scholar]
- 20.Chien, Y., A. C. Manna, S. J. Projan, and A. L. Cheung. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 274:37169-37176. [DOI] [PubMed] [Google Scholar]
- 21.DeDent, A. C., M. McAdow, and O. Schneewind. 2007. Distribution of protein A on the surface of Staphylococcus aureus. J. Bacteriol. 189:4473-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duthie, E. S., and L. L. Lorenz. 1952. Staphylococcal coagulase: mode of action and antigenicity. J. Gen. Microbiol. 6:95-107. [DOI] [PubMed] [Google Scholar]
- 23.Ender, M., B. Berger-Bächi, and N. McCallum. 2009. A novel DNA-binding protein modulating methicillin resistance in Staphylococcus aureus. BMC Microbiol. 9:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forsgren, A., and J. Sjoquist. 1966. “Protein A” from S. aureus: I. Pseudo-immune reaction with human γ-globulin. J. Immunol. 97:822-827. [PubMed] [Google Scholar]
- 25.Fournier, B., and A. Klier. 2004. Protein A gene expression is regulated by DNA supercoiling which is modified by the ArlS-ArlR two-component system of Staphylococcus aureus. Microbiology 150:3807-3819. [DOI] [PubMed] [Google Scholar]
- 26.Gao, J., and G. C. Stewart. 2004. Regulatory elements of the Staphylococcus aureus protein A (Spa) promoter. J. Bacteriol. 186:3738-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geisinger, E., R. P. Adhikari, R. Jin, H. F. Ross, and R. P. Novick. 2006. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol. Microbiol. 61:1038-1048. [DOI] [PubMed] [Google Scholar]
- 28.Giacomini, A., V. Corich, F. J. Ollero, A. Squartini, and M. P. Nuti. 1992. Experimental conditions may affect reproducibility of the beta-galactosidase assay. FEMS Microbiol. Lett. 79:87-90. [DOI] [PubMed] [Google Scholar]
- 29.Gómez, M. I., M. O'Seaghdha, M. Magargee, T. J. Foster, and A. S. Prince. 2006. Staphylococcus aureus protein A activates TNFR1 signaling through conserved IgG binding domains. J. Biol. Chem. 281:20190-20196. [DOI] [PubMed] [Google Scholar]
- 30.Gustafsson, E., S. Karlsson, J. Oscarsson, P. Sögård, P. Nilsson, and S. Arvidson. 2009. Mathematical modelling of the regulation of spa (protein A) transcription in Staphylococcus aureus. Int. J. Med. Microbiol. 299:65-74. [DOI] [PubMed] [Google Scholar]
- 31.Hartleib, J., N. Kohler, R. B. Dickinson, G. S. Chhatwal, J. J. Sixma, O. M. Hartford, T. J. Foster, G. Peters, B. E. Kehrel, and M. Herrmann. 2000. Protein A is the von Willebrand factor binding protein on Staphylococcus aureus. Blood 96:2149-2156. [PubMed] [Google Scholar]
- 32.Hsieh, H., C. Tseng, and G. C. Stewart. 2008. Regulation of Rot expression in Staphylococcus aureus. J. Bacteriol. 190:546-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hübscher, J., N. McCallum, C. D. Sifri, P. A. Majcherczyk, J. M. Entenza, R. Heusser, B. Berger-Bächi, and P. Stutzmann Meier. 2009. MsrR contributes to cell surface characteristics and virulence in Staphylococcus aureus. FEMS Microbiol. Lett. 295:251-260. [DOI] [PubMed] [Google Scholar]
- 34.Huntzinger, E., S. Boisset, C. Saveanu, Y. Benito, T. Geissmann, A. Namane, G. Lina, J. Etienne, B. Ehresmann, C. Ehresmann, A. Jacquier, F. Vandenesch, and P. Romby. 2005. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 24:824-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ingavale, S., W. van Wamel, T. T. Luong, C. Y. Lee, and A. L. Cheung. 2005. Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect. Immun. 73:1423-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jelsbak, L., L. Hemmingsen, S. Donat, K. Ohlsen, K. Boye, H. Westh, H. Ingmer, and D. Frees. 2010. Growth phase-dependent regulation of the global virulence regulator Rot in clinical isolates of Staphylococcus aureus. Int. J. Med. Microbiol. 300:229-236. [DOI] [PubMed] [Google Scholar]
- 37.Jones, R. C., J. Deck, R. D. Edmondson, and M. E. Hart. 2008. Relative quantitative comparisons of the extracellular protein profiles of Staphylococcus aureus UAMS-1 and its sarA, agr, and sarA agr regulatory mutants using one-dimensional polyacrylamide gel electrophoresis and nanocapillary liquid chromatography coupled with tandem mass spectrometry. J. Bacteriol. 190:5265-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karlsson, A., P. Saravia-Otten, K. Tegmark, E. Morfeldt, and S. Arvidson. 2001. Decreased amounts of cell wall-associated protein A and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to up-regulation of extracellular proteases. Infect. Immun. 69:4742-4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katayama, Y., H. Zhang, and H. F. Chambers. 2004. PBP2a mutations producing very-high-level resistance to beta-lactams. Antimicrob. Agents Chemother. 48:453-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kreiswirth, B. N., S. Löfdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 41.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang, X., L. Zheng, C. Landwehr, D. Lunsford, D. Holmes, and Y. Ji. 2005. Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J. Bacteriol. 187:5486-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindsay, J. A., C. E. Moore, N. P. Day, S. J. Peacock, A. A. Witney, R. A. Stabler, S. E. Husain, P. D. Butcher, and J. Hinds. 2006. Microarrays reveal that each of the ten dominant lineages of Staphylococcus aureus has a unique combination of surface-associated and regulatory genes. J. Bacteriol. 188:669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mainiero, M., C. Goerke, T. Geiger, C. Gonser, S. Herbert, and C. Wolz. 2010. Differential target gene activation by the Staphylococcus aureus two-component system saeRS. J. Bacteriol. 192:613-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manna, A., and A. L. Cheung. 2001. Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect. Immun. 69:885-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manna, A. C., and A. L. Cheung. 2006. Expression of SarX, a negative regulator of agr and exoprotein synthesis, is activated by MgrA in Staphylococcus aureus. J. Bacteriol. 188:4288-4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manna, A. C., and A. L. Cheung. 2003. sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect. Immun. 71:343-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manna, A. C., and A. L. Cheung. 2006. Transcriptional regulation of the agr locus and the identification of DNA binding residues of the global regulatory protein SarR in Staphylococcus aureus. Mol. Microbiol. 60:1289-1301. [DOI] [PubMed] [Google Scholar]
- 49.Manna, A. C., S. S. Ingavale, M. Maloney, W. van Wamel, and A. L. Cheung. 2004. Identification of sarV (SA2062), a new transcriptional regulator, is repressed by SarA and MgrA (SA0641) and involved in the regulation of autolysis in Staphylococcus aureus. J. Bacteriol. 186:5267-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manna, A. C., and B. Ray. 2007. Regulation and characterization of rot transcription in Staphylococcus aureus. Microbiology 153:1538-1545. [DOI] [PubMed] [Google Scholar]
- 51.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40:1049-1057. [DOI] [PubMed] [Google Scholar]
- 52.McCallum, N., M. Bischoff, H. Maki, A. Wada, and B. Berger-Bächi. 2004. TcaR, a putative MarR-like regulator of sarS expression. J. Bacteriol. 186:2966-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagarajan, V., and M. Elasri. 2007. SAMMD: Staphylococcus aureus microarray meta-database. BMC Genomics 8:351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Novick, R. P., and E. Geisinger. 2008. Quorum sensing in staphylococci. Annu. Rev. Genet. 42:541-564. [DOI] [PubMed] [Google Scholar]
- 55.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. N. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oscarsson, J., C. Harlos, and S. Arvidson. 2005. Regulatory role of proteins binding to the spa (protein A) and sarS (staphylococcal accessory regulator) promoter regions in Staphylococcus aureus NTCC8325-4. Int. J. Med. Microbiol. 295:253-266. [DOI] [PubMed] [Google Scholar]
- 57.Oscarsson, J., A. Kanth, K. Tegmark-Wisell, and S. Arvidson. 2006. SarA is a repressor of hla (alpha-hemolysin) transcription in Staphylococcus aureus: its apparent role as an activator of hla in the prototype strain NCTC 8325 depends on reduced expression of sarS. J. Bacteriol. 188:8526-8533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palmqvist, N., F. Foster, A. Tarkowski, and E. Josefsson. 2002. Protein A is a virulence factor in Staphylococcus aureus arthritis and septic death. Microb. Pathog. 33:239-249. [DOI] [PubMed] [Google Scholar]
- 59.Peterson, P. K., J. Verhoef, L. D. Sabath, and P. G. Quie. 1977. Effect of protein A on staphylococcal opsonization. Infect. Immun. 15:760-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pohl, K., P. Francois, L. Stenz, F. Schlink, T. Geiger, S. Herbert, C. Goerke, J. Schrenzel, and C. Wolz. 2009. CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J. Bacteriol. 191:2953-2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pragman, A. A., J. M. Yarwood, T. J. Tripp, and P. M. Schlievert. 2004. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J. Bacteriol. 186:2430-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qi, W., M. Ender, F. O'Brien, A. Imhof, C. Ruef, N. McCallum, and B. Berger-Bachi. 2005. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Zurich, Switzerland (2003): prevalence of type IV SCCmec and a new SCCmec element associated with isolates from intravenous drug users. J. Clin. Microbiol. 43:5164-5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rossi, J., M. Bischoff, A. Wada, and B. Berger-Bächi. 2003. MsrR, a putative cell envelope-associated element involved in Staphylococcus aureus sarA attenuation. Antimicrob. Agents Chemother. 47:2558-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saïd-Salim, B., P. M. Dunman, F. M. McAleese, D. Macapagal, E. Murphy, P. J. McNamara, S. Arvidson, T. J. Foster, S. J. Projan, and B. N. Kreiswirth. 2003. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185:610-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sambanthamoorthy, K., M. S. Smeltzer, and M. O. Elasri. 2006. Identification and characterization of msa (SA1233), a gene involved in expression of SarA and several virulence factors in Staphylococcus aureus. Microbiology 152:2559-2572. [DOI] [PubMed] [Google Scholar]
- 66.Schmidt, K. A., A. C. Manna, and A. L. Cheung. 2003. SarT influences sarS expression in Staphylococcus aureus. Infect. Immun. 71:5139-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmidt, K. A., A. C. Manna, S. Gill, and A. L. Cheung. 2001. SarT, a repressor of alpha-hemolysin in Staphylococcus aureus. Infect. Immun. 69:4749-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schneewind, O., D. Mihaylova-Petkov, and P. Model. 1993. Cell wall sorting signals in surface proteins of gram-positive bacteria. EMBO J. 12:4803-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schulthess, B., S. Meier, D. Homerova, C. Goerke, C. Wolz, J. Kormanec, B. Berger-Bächi, and M. Bischoff. 2009. Functional characterization of the σB-dependent yabJ-spoVG operon in Staphylococcus aureus: role in methicillin and glycopeptide resistance. Antimicrob. Agents Chemother. 53:1832-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seidl, K., S. Muller, P. Francois, C. Kriebitzsch, J. Schrenzel, S. Engelmann, M. Bischoff, and B. Berger-Bächi. 2009. Effect of a glucose impulse on the CcpA regulon in Staphylococcus aureus. BMC Microbiol. 9:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seidl, K., M. Stucki, M. Ruegg, C. Goerke, C. Wolz, L. Harris, B. Berger-Bächi, and M. Bischoff. 2006. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 50:1183-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sterba, K. M., S. G. Mackintosh, J. S. Blevins, B. K. Hurlburt, and M. S. Smeltzer. 2003. Characterization of Staphylococcus aureus SarA binding sites. J. Bacteriol. 185:4410-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tegmark, K., A. Karlsson, and S. Arvidson. 2000. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 37:398-409. [DOI] [PubMed] [Google Scholar]
- 74.Vandenesch, F., J. Kornblum, and R. P. Novick. 1991. A temporal signal, independent of agr, is required for hla but not spa transcription in Staphylococcus aureus. J. Bacteriol. 173:6313-6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Witney, A. A., G. L. Marsden, M. T. Holden, R. A. Stabler, S. E. Husain, J. K. Vass, P. D. Butcher, J. Hinds, and J. A. Lindsay. 2005. Design, validation, and application of a seven-strain Staphylococcus aureus PCR product microarray for comparative genomics. Appl. Environ. Microbiol. 71:7504-7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yarwood, J. M., J. K. McCormick, and P. M. Schlievert. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183:1113-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]